Abstract
Detoxification of Cr(VI) under alkaline pH requires attention due to the alkaline nature of many effluents. An alkaliphilic gram-positive Bacillus subtilis isolated from tannery effluent contaminated soil was found to grow and reduce Cr(VI) up to 100% at an alkaline pH 9. Decrease in pH to acidic range with growth of the bacterium signified the role played by metabolites (organic acids) in chromium resistance and reduction mechanism. The XPS and FT-IR spectra confirmed the reduction of Cr(VI) by bacteria into +3 oxidation state. Chromate reductase assay indicated that the reduction was mediated by constitutive membrane bound enzymes. The kinetics of Cr(VI) reduction activity derived using the monod equation proved (Ks = 0.00032) high affinity of the organism to the metal. This study thus helped to localize the reduction activity at subcellular level in a chromium resistant alkaliphilic Bacillus sp.
Keywords: Cr(VI) reduction, Bacillus sp., Chromate reductase, Membrane bound proteins, 16S rRNA identification
1. Introduction
Hexavalent chromium is a known pollutant originating from industrial effluents, such as paints and pigments, leather, metal plating, wood preservation, etc. (Baldi et al., 1990). Although chromium is an essential micronutrient soluble Cr(VI) is a carcinogen and toxic to all forms of life since the toxicity of chromium is dependent on its oxidation state (James, 1996). Detoxification of hexavalent chromium was known to be carried out by variety of bacteria under both aerobic and anaerobic conditions e.g. Pseudomonas fluorescens LB 300 (Bopp et al., 1983), Enterobacter cloacae HO1 (Wang et al., 1989), Bacillus sp. (Wang and Xiao, 1995). It was carried out intracellularly by the bacterial enzymes, either constitutive or induced. The location of these enzymes could be either in particulate fraction (probably in the cytoplasmic membranes) and/or in soluble fraction (Laxman and More, 2002).
The speciation of chromium is dependent on the pH, with chromate as the dominant species in an aqueous environment at pH 6.5–9 (McLean and Beveridge, 2001) and generally mobile in soil–water systems (Losi et al., 1994). Effluents released containing toxic metals are under alkaline or acidic pH. Earlier Cr(VI) detoxification studies mediated by bacteria were reported at neutral/near-neutral pH and very few studies were reported under alkaline condition (Ye et al., 2004, Stewart et al., 2007). Cr(VI) reduction at high pH conditions is important for certain bioremediation efforts because Cr(VI) contamination has been reported in high pH soils. (Kamaludeen et al., 2003, Van Engelen et al., 2008). Also the efficiency of gram-positive bacteria in Cr(VI) detoxification was less patronized compared to gram negative bacteria. Bacteria that can survive under highly alkaline conditions and can detoxify metals need to be identified. This study for the first time strongly focuses on the Cr(VI) detoxification efficiency by the gram-positive Bacillus subtilis under alkaline pH in conjunction with its subcellular localization.
2. Materials and methods
2.1. Characterization of Bacillus sp.
The isolation of the organism and identification through biochemical tests were described in an earlier study (Mary et al., 2008). Further characterization of the isolate was done through 16S rRNA sequencing. The genomic DNA was isolated using QIAamp kit and the 16S rRNA gene fragment was amplified using RW01 and dg74 primers and sequenced. Sequence was initially analyzed at NCBI server (http://www.ncbi.nlm.nih.gov) using BLAST(n) tool and corresponding neighbour sequences were downloaded from NCBI database. All sequences were aligned using CLUSTALW program (http://www.ebi.ac.uk/clustalw). The phylogenetic tree was constructed using the aligned sequences by the neighbour joining (NJ) method using Jukes–Cantor evolutionary distances and evaluated by performing bootstrap analyses of 1000 replicates in Molecular Evolutionary Genetics Analysis (MEGA version 4.0) software.
2.2. Chromium uptake studies by Bacillus sp.
2.2.1. Medium for chromium uptake studies
Chromium uptake studies were carried out in CA-M9 Minimal Media with the following composition: Na2HPO4 – 0.65 g/L, KH2PO4 – 1.5 g/L, NaCl – 0.25 g/L, NH4Cl – 0.5 g/L, MgSO4 – 0.12 g/L, Casamino Acid – 10 g/L, Glucose – 5 g/L. A 1000 mg/L stock solution of potassium dichromate was used as a source of Cr(VI) in the experiment.
2.2.2. Methodology for Cr(VI) analysis
The decrease in Cr(VI) concentration with time was estimated spectrophotometrically using 1,5-Diphenyl carbazide at 540 nm according to the method adopted by Urvashi and Datta (2005). The measure of residual chromium concentration in the supernatant indicates the chromium reducing activity.
2.2.3. Influence of initial pH and Cr(VI) concentration on the reduction efficiency
Seed culture (5%, v/v) inoculated into the CA-M9 media containing 50 mg/L Cr(VI) and adjusted to pH 6, 7, 8 and 9 was incubated at 30 °C under agitation (100 rpm). Aliquots of sample were withdrawn at intervals, centrifuged at 6000 rpm and the supernatant analyzed for residual Cr(VI). The initial Cr(VI) concentration was varied at constant pH of 9 to monitor the effect on growth and reduction efficiency. Simultaneously the change in pH of the media with the reduction of Cr(VI) was observed at regular intervals. Uninoculated media containing Cr(VI) served as control. All the experiments were done in triplicates.
2.3. Characterization of Bacillus sp. cells after chromium uptake studies
2.3.1. SEM/EDX analysis of Bacillus sp. cells
The cells grown in the presence of Cr(VI) were washed with ultrapure water and smeared onto glass slides and dried. Then it was fixed in 2.5% glutaraldehyde for 12 h at 4 °C followed by rinsing in distilled water three times to remove traces of glutaraldehyde. Later it was dehydrated in a series of ethanol concentrations (30%, 50%, 75%, 85%, 95% and 100%), dried and kept in a desiccator until use. The samples were subsequently mounted on aluminium stubs and sputter coated with gold. Specimens were examined using a XL 30 ESEM Scanning Electron Microscope equipped with an Energy Dispersive X-ray spectrophotometer (EDX).
2.3.2. XPS analysis of Bacillus sp. cells
For XPS analysis, the cells exposed to Cr(VI) were smeared onto Titanium substrate and dried. The X-ray Photoelectron Spectrometer (SPECS make) used to record the spectra has a monochromatised X-ray source using Al Kα line and a hemispherical energy analyzer coupled with a detector. High resolution spectra were recorded with pass energy of 12 eV and an energy sweep step size of 0.02 V.
2.3.3. FT-IR analysis of Bacillus sp. cells
Fourier Transform InfraRed (FT-IR) spectra of pristine and chromium absorbed cells exposed to different concentrations of Cr(VI) were recorded on a Perkin Elmer FT-IR (Spectrum One) spectrometer in the region of 500–4000 cm−1. The cells grown overnight in the absence and presence of chromium were harvested by centrifugation. Then they were dried in the hot air oven at 60 °C to complete dryness. The dried biomass is ground to a fine powder using a mortar and pestle. The powdered sample was pressed into spectroscopic quality KBr pellet with a sample/KBr ratio of 1/100.
2.4. Characterization of chromium reducing activity of Bacillus sp.
2.4.1. Chromium reducing activity by resting and permeabilized cells
Bacterial cells grown for 24 h in minimal media (pH 9, OD24h = 2.46) were harvested by centrifuging at 6000 rpm and at 4 °C for 15 min. The cells were washed twice with 10 mM Tris–HCl buffer (pH 7) and resuspended in the same buffer. Cr(VI) was added to a final concentration of 10 mg/L and incubated at 30 °C under agitation for 24 h. To study the effect of permeabilization, 1% Toluene and 2% Triton X were added separately to a known volume of resuspended cells and vortexed. Heat killed cells served as control. Aliquots of sample were withdrawn at regular intervals, centrifuged and the supernatant analyzed for Cr(VI). Similar experiment was carried out using 10 mM Tris–HCl adjusted to pH 6 and 9. All experiments were done in triplicates.
2.4.2. Chromium reducing activity with cell free extract
To prepare the crude cell free extract, bacterial cells grown overnight in minimal media (pH 9) without and with Cr(VI) were harvested separately by centrifugation, washed twice and resuspended in 40 mL of 10 mM Tris–HCl buffer (pH 7) each. The cells were placed on ice bath and disrupted by sonication for 10 min (20 × 30 s) (Sonic Vibra Cell). The resultant homogenate was centrifuged (8000 rpm, 30 min, 4 °C) and the supernatant was filter sterilized and used for the chromate reductase assay. Total protein content of the cytosolic fraction was analyzed by a modified method of Lowry et al. (Hartee, 1972). The initial concentration of Cr(VI) used was 10 mg/L. The pellet consisting of particulate fraction was resuspended in 40 mL of 10 mM Tris–HCl (pH 7) and assayed for Cr(VI) reduction at an initial concentration of 10 mg/L. Crude extract and pellet heated at 100 °C served as control. All the flasks were incubated at 30 °C for 24 h. Aliquots of sample were withdrawn at intervals and analyzed for Cr(VI). The assay was carried out in triplicates at varied pH of 6 and 9.
2.5. Evaluation of kinetic parameters of Cr(VI) reduction by Bacillus sp.
The kinetic parameters of Cr(VI) reduction by the Bacillus sp. were evaluated. The Monod equation (1) was used to describe the reduction experiment where μ and μmax are the measured and maximum specific reduction rates (mg metal/(mg protein day)), respectively, M is the metal concentration (mg metal), and Ks is the half saturation constant (mg metal):
| (1) |
To solve μmax and Ks, the Lineweaver–Burk method was used. Time course data at different Cr(VI) concentrations were subjected to an exponential decay equation [y = a * exp(−bx)] to estimate the rate constant (b) of Cr reduction.
3. Results
3.1. Characterization of the microorganism
The microorganism was isolated from the tannery effluent exposed soil with an aim to screen and isolate Cr(VI) resistant organism. This microorganism which grew well in CA-M9 minimal media, amended with Cr(VI), adjusted to pH 9 was found to have a Minimum Inhibitory Concentration (MIC) of 800 mg/L of Cr(VI). Identification based on biochemical tests performed earlier (Mary et al., 2008) and 16S rRNA sequencing confirmed the test organism as B. subtilis. Using the 750 bp sequence of amplified 16S rRNA gene fragment and the sequences retrieved from NCBI database, the phylogenetic tree was constructed through the neighbour joining program in MEGA 4.0 software (Fig. 1). The tree provided evidence of 100% sequence homology of the test organism to B. subtilis. The sequence was deposited in NCBI GenBank database with accession number FJ178872.
Figure 1.

Phylogenetic tree based on 16S rRNA gene sequence: shows the relationship between members of family Bacillaceae and G7 (test organism). The bar represents distance values calculated in MEGA and values at nod represent percentage of 1000 bootstrap replicates. Numbers in bracket represent Genbank accession numbers.
3.2. Optimisation of pH for Cr(VI) reduction
Metal ion sorption on both non-specific and specific sorbents is pH dependent, as the pH affects the availability of metal ions in solution (speciation), as well as the metal binding sites onto cell surface (Zouboulis et al., 2004). The optimum condition for complete reduction of Cr(VI) by the Bacillus sp. was observed at pH 9. These results show that with a gradual increase in pH from 6 to 9 the amount of Cr(VI) reduced was calculated to be 32%, 63%, 82% and 96% (Fig. 2). The rate of reduction of Cr(VI) increased with the increasing pH confirming that the alkaline condition favours the reduction of Cr(VI) compared to neutral or acidic conditions.
Figure 2.

Reduction of Cr(VI) at different pH.
3.3. Cr(VI) reduction with varied initial concentration
The growth of the Bacillus sp. and its Cr(VI) reduction efficiency monitored at varied concentrations showed that 50 mg/L of Cr(VI) was reduced to near zero in 65 h (Fig. 3A), whereas 100, 150 and 200 mg/L were reduced by 71%, 62%, 27% in 144 h (Fig. 3B–D), respectively. Further increase in contact time showed no significant difference in the rate of reduction. Higher concentration of Cr(VI) increased the time for total reduction but had a significant effect on initial reduction rate. The rate of reduction in the presence of 50 mg/L Cr(VI) during the first 10 h was found to be 1.73 mg Cr(VI) L−1 h−1. But the rate during the initial 10 h increased to 2.69 mg Cr(VI) L−1 h−1, 2.07 mg Cr(VI) L−1 h−1 with an increase in concentration to 100 and 150 mg/L Cr(VI). And with even higher concentration of 200 mg/L Cr(VI) the rate again decreased to 1.46 mg Cr(VI) L−1 h−1. The rate of Cr(VI) reduction was slower during the lag (0–4 h) and stationary phase (>15 h) compared to the log phase (4–15 h). The growth of the bacterium monitored simultaneously with the reduction of Cr(VI) showed a faster rate of growth in control cultures than when exposed to high concentrations. The growth in the presence of 50 mg/L Cr(VI) (Fig. 3A) was higher than in control but in higher concentrations the culture reached a stationary phase early compared to control (Fig. 3B–D).
Figure 3.
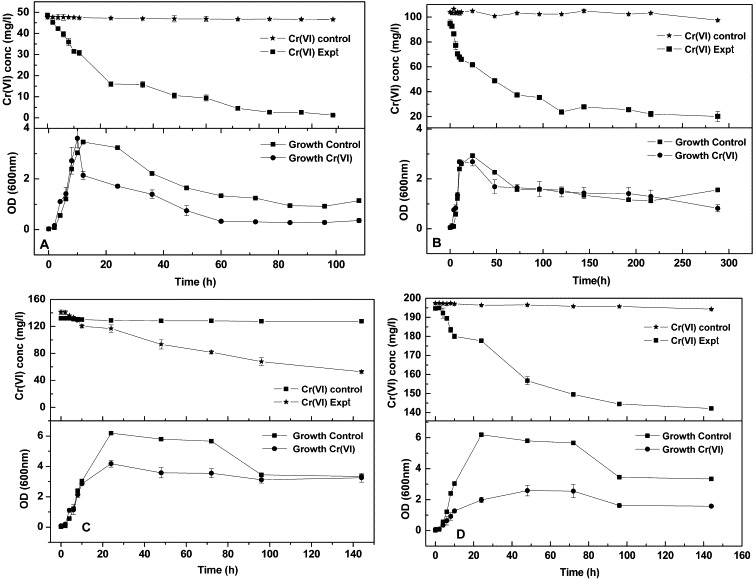
Cr(VI) reduction vs. growth over a period of time (in mg/L) (A) 50; (B) 100; (C) 150; (D) 200.
3.4. SEM/EDX and XPS analysis of Bacillus sp. cells
A peak was exhibited in the EDX spectrum that corresponds to chromium region (Fig. 4). The SEM image (Fig. 4A, Inset) shows the presence of extracellular substances adhered to the surface of the rod shaped cells. To verify the oxidation state of the chromium bound to the cells studied, XPS was employed in this study (Fig. 5). The spectrum of the bacterial sample treated with chromium has shown a convoluted peak at the chromium region. Deconvolution of the chromium peak contained components at binding energies 578.3, 577.8 and 576 eV which can be attributed to Cr2O3 (major part) and Cr(OH)3 (minor part). These correspond to +6 oxidation state and +3 oxidation state of chromium as per the PHI Handbook of X-ray photoelectron spectroscopy (Puzon et al., 2002).
Figure 4.

(A) EDX spectrum of the Bacillus cells shows presence of chromium on the cell surface. Inset: SEM images of Bacillus cells.
Figure 5.

XPS spectrum of cells shows presence of chromium in +3 oxidation state.
3.5. FT-IR analysis of Bacillus sp. cells
The FT-IR spectra (Fig. 6a) of the Bacillus sp. native and metal loaded bacteria were taken to obtain information on the nature of the possible cell–metal ions interactions. In the present investigation, the hexavalent chromium was expected to undergo reduction via complexation with carboxyl or amide or hydroxyl moieties of Bacillus sp. To evident the possible interaction, the FT-IR spectra from 400 to 4000 cm−1 wavenumber ranges were recorded. Table 1 depicts the assignments of various IR frequencies from the native and metal loaded bacteria. The FT-IR spectrum of the pure Bacillus sp. showed the presence of characteristic bacterial signatures as expected (Fig. 6a). Fig. 6a displayed a broad stretching peak around 2932 cm−1 characteristic of weak C–H stretching band from alkyl groups. An asymmetrical stretching peak that was noticed around 1700 cm−1 suggests the presence of ester C O groups. Further, the spectrum showed the presence of prominent carboxyl (around 1400 cm−1) and amide groups (1234, 1548 and 1648 cm−1), which are preferentially expected for bacterial cultures. As far as the FT-IR spectra of metal loaded Bacillus sp. are concerned, they showed some subtle changes. The FT-IR spectra of metal loaded bacteria showed a highly significant shift in frequency to lower range from 1403 to 1380 cm−1 echoing the strong interaction of the –O–C O group in the chromium binding by the Bacillus sp. The peak at 1292 cm−1 frequency becomes more prominent and significant on exposure to Cr(VI) thus suggesting the involvement of either the phosphate moiety or the C O group in the interaction with chromium. Obvious shift of the phosphate linkage frequency (1234 cm−1) to lower frequency (1229 cm−1) was observed with 50 mg/L of Cr(VI), whereas the peak disappeared on exposure to 100 mg/L of Cr(VI). This strongly implicated the involvement of phosphate linkage in chromium binding. The peak observed at 1057 cm−1 corresponding to the C O stretching remained the same in the biomass whether unexposed or exposed to chromium. Strengthening of the peak at 1728 cm−1 on exposure to 50 and 100 mg/L Cr(VI) implied the involvement of protonated carboxylic groups in chromium adsorption on the biomass. While the slight shift from 1648 cm−1 to higher frequency 1653 cm−1 indicated the intervention of C O group of the amide I bond (CO–NH), the peak position at 1547 cm−1 remained unaltered thus indicating the non-involvement of amide II bond in the chromium adsorption process. In order to quantify the effective reduction of the biomass while interacting with the Cr(VI) moiety, the area values of various functional groups in the deconvoluted spectra and protein/lipid ratios of Bacillus sp. in the presence and absence of hexavalent chromium were compared (Fig. 6b and Table 2). Both the area values and protein/lipid ratios showed marked difference in the biomass with the increase in chromium(VI) concentration. Though FT-IR spectra were able to give information on the biomass, it did not trace out the chromium compounds. Fig. 6b is the deconvoluted FT-IR spectra which clearly showed the differences between the chromium loaded bacterial samples and the pure bacterial culture. As can be seen from the spectra amides I and II bands remained unaltered, whereas noticeable reduction in the absorbance and area value of hydroxyl group was observed. Probably hydroxyl groups must have undergone oxidation, while they played a role in the reduction of chromium. It indicates the tendency of the hydroxyl groups to get involved in the chromium reduction for the subsequent conversion of hydroxyl groups into acids. This is supported by the enhancement of COO− and C O peaks in the metal loaded spectra. Also, the absorbance value for the alkyl stretch around 2964 cm−1 showed slight reduction in the metal loaded samples. Further, the spectra evidenced the sufficient participation of phosphate groups in the chromium reduction. This is attributed by the increase of phosphate frequencies in the spectra. The table depicts that the areas of the carboxyl and ester C O increased with the metal addition, whereas the areas of hydroxyl and alkyl groups decreased with the addition of chromium. The band areas of the amide groups did not change much irrespective of the chromium additions. The FT-IR spectra of the 50 and 100 mg/L metal loaded samples were found to be similar. The protein/lipid ratio was also calculated by taking the ratio of the areas of the bands arising from lipids and proteins. In the present case, we considered the ratio of the area of the amides I and II bands (1657 and 1541 cm−1) to the area of the ester C O stretching (1730 cm−1). We observed marked reduction in the protein/lipid ratio when hexavalent chromium was added to the system. It was observed that the ratio was comparatively lesser, indicating lesser protein content in the metal loaded bacterial samples. Although, amide I and amide II bands were unaltered and their areas were almost the same in all the cases, the reduction in protein/lipid ratio gave indirect evidence for the participation of proteins in the chromium reduction.
Figure 6.

(a) FT-IR spectra of the cells unexposed and exposed to Cr(VI). (b) Spectra showing the peak–functional group assignment and comparison of peak area.
Table 1.
The observed FT-IR band assignments for Bacillus sp.
| Wave numbers (cm−1) | Assignments | Probable site for functional group |
|---|---|---|
| 3293, 3410 | N–H and O–H stretching vibrations from polysaccharides and proteins | Cell wall – direct interaction of OH with Cr (Doshi et al., 2007) |
| 2932 | CH3 asymmetric stretching from lipids, proteins, polysaccharides and nucleic acids | Proteins and carbohydrates in the cell wall (Das and Guha, 2007) |
| 1728, 1726 | C O stretching from lipids and triglycerides | Carbohydrates – protonated carboxylic groups (or) ester groups (Das and Guha, 2007) |
| 1648, 1653 | Amide I (protein C O stretching) | Peptides – amino acids/amides (Doshi et al., 2007) |
| 1547, 1548 | Amide II (protein N–H bending and C–N stretching) | Peptides – cell wall (Das and Guha, 2007) |
| 1380, 1383, 1402 | COO− symmetric stretching from aminoacid side chains and fatty acids | Amino acid – cell wall of the bacterial cells (Das and Guha, 2007) |
| 1291, 1292 | or C O bending or asymmetric stretching | Cell wall (Doshi et al., 2007) |
| 1229 | asymmetric stretching mainly from nucleic acids with a little contribution from phospholipids | Teichoic acids – cell wall (Doshi et al., 2007) |
| 1057 | C–O (or) SO and PO stretching vibrations | Peptidoglycan – cell wall (Aravindhan et al., 2004), cell wall (Loukidou et al., 2004) |
| Around 622 | Glycogen units, polysaccharides | Cell wall (Nakamoto, 1963) |
Table 2.
The FT-IR band area values of some functional groups and the protein/lipid ratios of Bacillus sp. in absence and presence of hexavalent chromium.
| Functional groups | Bacillus sp. | Bacillus sp. with 50 ppm Cr(VI) | Bacillus sp. with 100 ppm Cr(VI) |
|---|---|---|---|
| Band area values | |||
| Ester (C O) stretching | 2.38 | 3.6 | 3.6 |
| Amide I | 22.96 | 23.19 | 23.19 |
| Amide II | 8.47 | 8.23 | 8.23 |
| COO− symmetric stretching | 16.03 | 22.17 | 22.17 |
| Hydroxyl | 63.60 | 49.7 | 49.40 |
| Alkyl stretching | 11.45 | 8.95 | 8.95 |
| Protein/lipid ratio | |||
| Amides I + amides II/ester C O stretching | 11.94 | 8.7 | 8.7 |
3.6. Alteration of pH with reduction of Cr(VI)
The speciation of chromium being pH dependent, the alterations in pH during the reduction was investigated. During the log phase of bacterial growth (in control) the pH of the medium diminished from alkaline to acidic range (pH decrease: 9–6.01 for 50 mg/L; 9–5.96 for 100 mg/L; 9–6.23 for 150 mg/L; 9–6.24 for 200 mg/L) Cr(VI) and increased during the stationary phase (pH 6.01–9.02). But in the presence of Cr(VI) (Fig. 7A) the medium tend to remain in acidic condition (pH 6.0) until the concentration of Cr(VI) was completely reduced after which it gradually increased. The drop in pH was in concordance with the Cr(VI) reduction even with higher concentrations (Fig. 7B, C and D).
Figure 7.

Reduction of pH and chromium at different initial Cr(VI) concentrations (in mg/L) (A) 50; (B) 100; (C) 150; (D) 200.
3.7. Characterization of chromate reductase activity in Bacillus sp.
Assays carried out using resting and permeabilized cells of the Bacillus sp. at different pH under a constant Cr(VI) concentration proved the involvement of enzymes in reduction. The data on Cr(VI) reduction showed that complete reduction was observed at pH 7 in 24 h, compared to pH 6 and 9 with the resting cells, whereas the permeabilization do not favour Cr(VI) reduction even at pH 7 (Table 3). Permeabilization with Toluene (1%) showed 53% and 41% reduction at pH 7 and 9, whereas permeabilization by Triton X (2%) gave only 23% and 2% reduction even after 24 h incubation. Heat killed cells used as control do not show any reduction in Cr(VI) concentration. Statistical analysis of the data using Tukey–Kramer multiple comparison test confirmed that the reduction at pH 7 was highly significant with p-value <0.001. Even though statistical analysis showed that difference was extremely significant (p < 0.001) at pH 7 after 24 h when compared to the rest of the pH, it was only significant (p < 0.05) when comparison was made between Cr(VI) reduction values of resting and permeabilized assay at pH 7. Assays conducted with cytosolic and particulate fraction of the Bacillus sp. at different pH confirmed the role of membrane bound proteins in Cr(VI) reduction (Table 4). The cytosolic fraction showed a maximum reduction of 45% after 24 h at pH 7 in comparison to pH 6 and 9, whereas the particulate fraction gives a reduction of 95% in 24 h (Protein content 1.8 mg/mL). The cytosolic fraction of cells grown in the presence and absence of Cr(VI) showed very minimal difference in Cr(VI) reduction activity (5%), whereas the particulate fraction of cells grown in the presence of Cr(VI) showed less reduction (60%) compared to cells grown without Cr(VI) (96%) (Table 5). The statistical analysis of the data for Cr(VI) reduction by cytosolic and particulate fraction of cells grown in the presence and absence of Cr(VI) showed a p-value <0.001 thus stating that the difference is highly significant.
Table 3.
Reduction of Cr(VI) by permeabilized and resting cells at different pH 6, 7 and 9.
| Time (h) | Permeabilized cell assay (% Cr(VI) remaining) |
Resting cell assay (% Cr(VI) remaining) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Toluene 1% |
Triton X 2% |
pH 6 |
pH 7 |
pH 9 |
||||||||||||||
| pH 6 |
pH 7 |
pH 9 |
pH 6 |
pH 7 |
pH 9 |
Ctrl | Expt | Ctrl | Expt | Ctrl | Expt | |||||||
| Ctrl | Expt. | Ctrl | Expt. | Ctrl | Expt. | Ctrl | Expt. | Ctrl | Expt. | Ctrl | Expt. | |||||||
| 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 6 | 98.77 | 98.75 ± 0.18 | 95.34 | 66.70 ± 0.24 | 91.62 | 86.51 ± 0.13 | 99.25 | 98.75 ± 0.16 | 98.12 | 81.49 ± 0.31 | 99.94 | 99.70 ± 0.04 | 100 | 96.22 ± 0.88 | 97.71 | 59.77 | 99.75 | 82.17 ± 0.06 |
| 12 | 98.60 | 94.72 ± 0.21 | 95.67 | 57.14 ± 0.07 | 91.24 | 63.42 ± 0.57 | 99 | 96.12 ± 0.1 | 96.76 | 79.88 ± 0.06 | 99.93 | 92.96 ± 0.26 | 97.77 | 90.29 ± 1.08 | 96.94 | 26.07 ± 0.12 | 96.12 | 55.17 |
| 24 | 98.40 | 90.49 ± 0.34 | 93.56 | 46.80 ± 0.41 | 91.24 | 58.95 ± 0.12 | 99.3 | 95.37 ± 0.35 | 95.83 | 76.77 ± 0.13 | 99.93 | 98.54 ± 0.21 | 91.31 | 77.76 ± 1.5 | 96.07 | 0 | 95.92 | 14.36 ± 0.06 |
Table 4.
Reduction of Cr(VI) by cytosolic (extract) and particulate (pellet) fraction at different pH 6, 7 and 9.
| Time (h) | Cell free extract and particulate fraction (% Cr(VI) remaining) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 6 |
pH 7 |
pH 9 |
||||||||||
| Extract |
Pellet |
Extract |
Pellet |
Extract |
Pellet |
|||||||
| Control | Expt. | Control | Expt. | Control | Expt. | Control | Expt. | Control | Expt. | Control | Expt. | |
| 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 6 | 96.06 | 96.83 ± 0.05 | 96.91 | 93.49 ± 0.39 | 98.02 | 98.02 ± 0.13 | 96.23 | 74.46 ± 0.17 | 98.94 | 97.40 ± 0.05 | 93.10 | 81.88 ± 0.18 |
| 12 | 98.95 | 96.2 ± 0.05 | 93.82 | 84.67 ± 0.21 | 95.05 | 90.03 ± 0.1 | 90.56 | 18.44 ± 0.1 | 98.13 | 95.75 ± 0.18 | 97.69 | 62.14 ± 0.13 |
| 24 | 96.06 | 93.17 ± 0.11 | 87.62 | 41.48 ± 0.22 | 95.05 | 55.33 ± 1.78 | 89.62 | 5.31 ± 0.08 | 98.88 | 96.81 ± 0.15 | 95.41 | 42.38 ± 0.13 |
Table 5.
Reduction of Cr(VI) by cytosolic (extract) and particulate (pellet) fraction of cells grown in absence and presence of Cr(VI) at pH 7.
| Time (h) | Grown in absence of Cr(VI) (% Cr(VI) remaining) |
Grown in presence of Cr(VI) (% Cr(VI) remaining) |
||||||
|---|---|---|---|---|---|---|---|---|
| Extract |
Pellet |
Extract |
Pellet |
|||||
| Control | Expt. | Control | Expt. | Control | Expt. | Control | Expt. | |
| 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 6 | 97.55 | 99.62 ± 0.48 | 91.26 | 69.13 ± 0.16 | 95.25 | 89.34 ± 0.13 | 92.67 | 87.85 ± 0.17 |
| 12 | 96.70 | 92.91 ± 0.44 | 90.86 | 16.87 ± 0.33 | 95.25 | 83.27 ± 0.19 | 92.99 | 77.10 ± 0.05 |
| 24 | 95.11 | 64.80 ± 0.38 | 91.86 | 4.16 ± 0.14 | 95.36 | 59.79 ± 0.19 | 92.67 | 40.53 ± 0.41 |
3.8. Kinetic parameters
The μmax and Ks values for the metal reduction experiments using whole cells were calculated to be 0.862 mg Cr(VI) day−1 mg protein−1 and 0.00032 mg Cr(VI), respectively. The time course reduction rate constant (b) for different Cr(VI) concentrations showed a decrease with an increase in concentration of Cr(VI). The value showed a trend of 0.012 for 50 mg/L (r2 = 0.93) to 0.0005 for 200 mg/L (r2 = 0.94) Cr(VI).
4. Discussion
A chromium resistant microorganism identified through 16S rRNA as B. subtilis (Fig. 1) was isolated from tannery effluent contaminated soil since chromium contamination was known to exert a selective pressure on the indigenous microbial flora of that soil (Viti et al., 2003). Earlier studies report tolerance range of such Cr(VI) resistant microorganisms to be around 100–4000 mg/L Cr(VI) (Urvashi et al., 2007, Zhu et al., 2008). The Bacillus sp. isolated in this study falls within this range of tolerance. This variation in tolerance was due to the chemical composition of the medium used, which may have affected the actual Cr(VI) toxicity via a ‘masking’ effect (Desai et al., 2008, Caravelli et al., 2008). The toxicity effect was found to be more in liquid cultures (Shakoori et al., 2000) since metals are relatively more freely available in liquid than in solid medium.
Moreover, the speciation chemistry of Cr(VI), in aqueous solution, favours negatively charged species, such as (pH between 1 and 6) and (pH above 7) (Stasicka and Kotas, 2000). Virtually all the earlier studies with different bacterial systems, including other strains of B. subtilis, on Cr(VI) reduction were conducted at near-neutral pH conditions. This study using a B. subtilis showed a unique quality to reduce Cr(VI) only under alkaline conditions (most favourable pH 9) (Fig. 2). The less growth observed in the presence of higher concentrations of Cr(VI) was attributed to the reduction of metabolism rate by Cr(VI) (Pal et al., 2005). The Cr(VI) reduction ability of the bacteria was found to be growth dependent (Fig. 3). The effects of different concentrations of Cr(VI) on the reduction trend showed that the mechanism of reduction was enzyme mediated since the overall rate of Cr(VI) reduction decreased with the increasing concentration of Cr(VI). However, the rate of Cr(VI) reduction was not inhibited by high levels of Cr(VI) during the early phase of reduction process. Similar trend was reported by Rahman et al. (2007) for the growth of Pseudomonas sp. C-171 at different concentrations of Cr(VI). Though the Arthrobacter sp. was observed to be more efficient in chromium reduction than in the Bacillus sp. according to Megharaj et al., 2003, the Arthrobacter oxydans was affected even at a very low concentration of 35 μg/mL of Cr(VI) (Asatiani et al., 2004). Thus the threshold concentration after which chromate displays a toxic effect on the bacterium varies depending on the species adaptability. The sporulating characteristic of this Bacillus sp. makes it possible to remain in stationary phase for a longer duration (Zouboulis et al., 2004) and carry out reduction though at a reduced rate. Results for control set of Cr(VI) reduction studies showed negligible reduction, thereby eliminating any probability of influence by external factors, such as temperature, pH and aeration, which were maintained constant during the study.
The first step in reduction was binding of chromium on to the surface of the cells, which was confirmed by the EDX spectrum and the extracellular substances on the surface possibly could have helped in the sequestration of chromium (Fig. 4). The oxidation state of the chromium sorbed to the biomass dictates to what extent the reduction has taken place. The XPS spectra confirm the presence of both the hexavalent and trivalent oxidation states of chromium, which suggest the mechanism of adsorption in conjunction with the reduction at work on the surface of the cells (Fig. 5). The trivalent form of chromium was known to readily precipitate as chromium hydroxide at pH 7 (Bopp et al., 1983). X-ray photoemission spectroscopy studies of batch cultures of Shewanella oneidensis with chromate were able to provide evidence for Cr(VI) intermediates at cell surfaces (Neal et al., 2002). The high density of electronegative sites within the wall gives B. subtilis its affinity for metals and its ability to form metal precipitates (Doyle et al., 1980).
The FT-IR spectra showed the presence of ionizable functional groups (i.e., carboxyl, hydroxyl, phosphate and amino groups) that are able to interact with metal ions (Table 1) (Naja et al., 2005). In many Gram-positive bacteria, teichoic acids provide highly charged anionic clusters due to the presence of repeating phosphodiester residues (Beveridge and Murray, 1976). The carboxyl groups of peptidoglycan were found to be primarily responsible for the interactions between cell walls and cations (Fig. 6). Marquis et al. (1976) have suggested that carboxyl groups are involved in the binding of metal ions to the cell wall of Streptococci. The phosphate moiety was the second main group responsible in chromium binding. Along with this broad hydroxyl group peaks and +3 oxidation state suggest the precipitation of Cr(OH)3 during reduction. These results confirmed that different functional groups of the biomass were involved in chemical reactions (Aravindhan et al., 2004), e.g., ionic interaction, hydrogen bonding or ion dipole interaction and complexation with chromate ions. The interaction of these groups with the metal ions was found to be favoured by the acidic pH (Fig. 7) due to the organic acids (confirmed by TLC) exuded by the Bacillus sp. during its growth. These acids thus contribute to the resistance and reduction of Cr(VI). Delhaize and Ryan (1995) also report the role of organic acids, such as citrate and malate in aluminium tolerance through extracellular chelation.
Characterization of chromate reductase activity (Table 3) in Bacillus sp. through permeabilization assay helped us to identify the activity with either soluble or membrane fraction. Toluene is known to permeabilise by dissolving the phospholipids and Triton X permeabilises by solubilising inner membrane proteins (Asenjo, 1990). Less reduction observed with Triton X was due to the solubilisation and inactivation of membrane bound proteins. The inability of the heat killed cells to reduce Cr(VI) showed that the reduction of Cr(VI) was mediated by cell membrane-bound or soluble proteins. Moreover in the absence of added electron donors, aerobic organisms may reduce Cr(VI) through the action of enzyme reductase using endogenous electron reserves or NADH as an electron donor. Suzuki et al. (1992) demonstrated that intracellular Cr(VI) accepts a single electron from an NADH molecule forming a Cr(V) intermediate which in turn accepts two electrons from two molecules of NADH to form stable Cr(III). Megharaj et al. (2003) reported similar results showing reduction to undetectable level by resting cells of Arthrobacter sp. and Bacillus sp. Thus it may be inferred that the reduction of Cr(VI) was associated more with the membrane bound proteins.
The results of the assay using cell free extract and particulate fraction (Table 4) concluded that the reduction activity was associated with membrane proteins to a higher degree and to a lesser degree associated with the soluble fraction. Moreover higher reduction rate by unexposed cells compared to exposed cells (Table 5) indicated that Cr(VI) reducing activity of the isolate was constitutive and maybe inducible to a lesser degree (Pal et al., 2005, McLean and Beveridge, 2001). Earlier reports also associate the chromate reduction activity with intracellular soluble fraction of the B. sphaericus and Providencia sp. cells (Pal et al., 2005, Urvashi et al., 2006). Our results though contrary to these reports were in accordance with the reports stating the association of chromate reduction activity with membrane fraction of E. cloacae HO1 (Wang et al., 1990) and P. fluorescens LB 300 (Bopp et al., 1983). Suzuki et al. (1992) has reported that some bacteria were known to use chromate as the terminal electron acceptor employing membrane bound enzymes, while others use soluble enzymes. Evaluation of the kinetic parameters shows that the low Ks value proved high affinity of the organism to Cr(VI). The effect of different concentrations of chromium signified that Cr(VI) reduction activity was essentially an enzyme mediated reaction, since the hyperbolic curve fit of the data showed clear evidence of dependence on Cr(VI).
5. Conclusions
Investigating the efficiency of heavy metal reduction will help in understanding the mechanism adopted by different microorganisms and selecting a better strain. This study thus evaluated the reduction potential of an indigenous Bacillus sp. under alkaline condition and localized the chromium reducing activity at subcellular level. Resting cells were found to completely reduce Cr(VI), whereas permeabilized cells did not effectively reduce thus confirming the enzyme mediated mechanism. Further, enhanced reduction by particulate fraction compared to cell free extract indicated the role of membrane bound proteins in reduction mechanism. The derivation of kinetic parameter Ks (0.00032) suggested high affinity of the organism to the metal. Since Cr(VI) reduction products are the least soluble at pH 9 (Rai et al., 1987), alkaliphilic Cr(VI) reducing bacteria could potentially be useful in the remediation of these types of chromium contaminated sites.
Acknowledgements
The authors would like to thank Dr. C.S. Sundar, Head, MSD, Dr. R.K. Dayal, Head, CSTD and Dr. Rani George, CSTD for providing the facilities to carry out this work. We also sincerely thank Dr. Ramya, MDL in helping us to analyse the FT-IR spectra.
References
- Aravindhan R., Madhan B., Raghava Rao J., Unni Nair B., Ramasami T. Bioaccumulation of chromium from the tannery waste water: an approach for chrome recovery and reuse. Environ. Sci. Technol. 2004;38:300–306. doi: 10.1021/es034427s. [DOI] [PubMed] [Google Scholar]
- Asatiani N.V., Abuladze M.K., Kartvelishvili T.M., Bakradze N.G., Sapojnikova N.A., Tsibakhashvili N.Y., Tabatadze L.V., Lejava L.V., Asanishvili L.L., Holman H.Y. Effect of Cr(VI) action on Arthrobacter oxydans. Curr. Microbiol. 2004;49:321–326. doi: 10.1007/s00284-004-4351-2. [DOI] [PubMed] [Google Scholar]
- Asenjo J.A. vol. 9. CRC Press; New York: 1990. (Separation Processes in Biotechnology). [Google Scholar]
- Baldi F., Vaughan A.M., Olson G.J. Chromium(VI) resistant yeast isolated from a sewage treatment plant receiving tannery wastes. Appl. Environ. Microbiol. 1990;56:913–918. doi: 10.1128/aem.56.4.913-918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge T.J., Murray R.G.E. Uptake and retention of metals by cell walls of Bacillus subtilis. J. Bacteriol. 1976;127:1502–1518. doi: 10.1128/jb.127.3.1502-1518.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp L.H., Chakrabarty A.M., Ehrlich H.L. Chromate resistance plasmid in Psuedomonas fluorescens. J. Bacteriol. 1983;155:1105–1109. doi: 10.1128/jb.155.3.1105-1109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravelli A.H., Giannuzzi L., Zaritzky N.E. Reduction of Hexavalent chromium by Sphaerotilus natans a filamentous micro-organism present in activated sludges. J. Hazard. Mater. 2008;156:214–222. doi: 10.1016/j.jhazmat.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Das S.K., Guha A.K. Biosorption of chromium by Termitomyces clypeatus. Colloid. Surf. B: Biointerf. 2007;60:46–54. doi: 10.1016/j.colsurfb.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Delhaize E.P., Ryan R. Aluminum toxicity and tolerance in plants. Plant Physiol. 1995;107:315–321. doi: 10.1104/pp.107.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C., Jain K., Madamwar D. Evaluation of in vitro Cr(VI) reduction potential in cytosolic extracts of three indigenous Bacillus sp. isolated from Cr(VI) polluted industrial landfill. Bioresour. Technol. 2008;99:6059–6069. doi: 10.1016/j.biortech.2007.12.046. [DOI] [PubMed] [Google Scholar]
- Doshi H., Ray A., Kothari I.L. Biosorption of cadmium by live and dead spirulina: IR spectroscopic, kinetics, and SEM studies. Curr. Microbiol. 2007;54:213–218. doi: 10.1007/s00284-006-0340-y. [DOI] [PubMed] [Google Scholar]
- Doyle R.J., Matthews T.H., Streips U.N. Chemical basis for selectivity of metal ions by the Bacillus subtilis cell wall. J. Bacteriol. 1980;143:471–480. doi: 10.1128/jb.143.1.471-480.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartee E.E. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 1972;48:422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- James B.R. The challenge of remediating chromium contaminated soil. Environ. Sci. Technol. 1996;30:248–251. doi: 10.1021/es962269h. [DOI] [PubMed] [Google Scholar]
- Kamaludeen S.P.B., Megharaj M., Naidu R., Singleton I., Juhasz A.L., Hawke B.G., Sethunanthan N. Microbial activity and phospholipid fatty acid pattern in long-term tannery waste-contaminated soil. Ecotoxicol. Environ. Safety. 2003;56:302–310. doi: 10.1016/s0147-6513(02)00075-1. [DOI] [PubMed] [Google Scholar]
- Laxman R.S., More S. Reduction of hexavalent chromium by Streptomyces griseus. Miner. Eng. 2002;15:831–837. [Google Scholar]
- Losi M.E., Amrhein C., Frankenberger W.T. Environmental biochemistry of chromium. Rev. Environ. Contam. Toxicol. 1994;36:91–121. doi: 10.1007/978-1-4612-2656-7_3. [DOI] [PubMed] [Google Scholar]
- Loukidou M.X., Zouboulis A.I., Karapantsios T.D., Matis K.A. Equilibrium and kinetic modeling of chromium (VI) biosorption by Aeromonas caviae. Colloid. Surf. A. Physicochem. Eng. Aspects. 2004;242:93–104. [Google Scholar]
- Marquis R.E., Mayzel K., Carstensen E.l. Cation exchange in cell walls of gram positive bacteria. Can. J. Microbiol. 1976;22:975–982. doi: 10.1139/m76-142. [DOI] [PubMed] [Google Scholar]
- Mary M.S., Gopal Judy, Tata B.V.R., Rao T.S., Vincent S. A confocal microscopic study on colony morphology and sporulation of Bacillus sp. World J. Microbiol. Biotechnol. 2008;24:2435–2442. [Google Scholar]
- McLean J., Beveridge T.J. Chromate reduction by a Psuedomonad isolated from a site contaminated with chromated copper arsenate. Appl. Environ. Microbiol. 2001;67:1076–1084. doi: 10.1128/AEM.67.3.1076-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megharaj M., Avudainayagam S., Naidu R. Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste. Curr. Microbiol. 2003;47:51–54. doi: 10.1007/s00284-002-3889-0. [DOI] [PubMed] [Google Scholar]
- Naja G., Mustin C., Berthelin B., Volesky B. Lead biosorption study with Rhizopus arrhizus using a metal-based titration technique. J. Colloid Interf. Sci. 2005;292:537–544. doi: 10.1016/j.jcis.2005.05.098. [DOI] [PubMed] [Google Scholar]
- Nakamoto K. John Wiley and Sons; New York: 1963. Infrared spectra of inorganic and co-ordination compounds. p. 107. [Google Scholar]
- Neal A.L., Lowe K., Daulton T.L., Jones-Meehan J., Little B.J. Oxidation state of chromium associated with cell surfaces of Shewanella oneidensis during chromate reduction. Appl. Surf. Sci. 2002;202:150–159. [Google Scholar]
- Pal A., Dutta S., Paul A.K. Reduction of hexavalent chromium by cell free extract of Bacillus sphaericus AND 303 isolated from serpentine soil. Curr. Microbiol. 2005;51:327–330. doi: 10.1007/s00284-005-0048-4. [DOI] [PubMed] [Google Scholar]
- Puzon G.J., Petersen J.N., Roberts A.G., Kramer M., Xun L. A bacterial flavin reductase system reduces chromate to a soluble chromium(III)-NAD+ complex. Biochem. Biophys. Res. Commun. 2002;294:76–81. doi: 10.1016/S0006-291X(02)00438-2. [DOI] [PubMed] [Google Scholar]
- Rahman M., Gul S., Ul Haq M. Reduction of chromium(VI) by locally isolated Pseudomonas sp. C-171. Turk. J. Biol. 2007;31:161–166. [Google Scholar]
- Rai D., Sass M., Moore D.A. Chromium(III) hydrolysis constants and solubility of chromium(III) hydroxide. Inorg. Chem. 1987;26:345–349. [Google Scholar]
- Shakoori A.R., Makhdoom M., Haq R.U. Hexavalent chromium reduction by a dichromate-resistant gram-positive bacterium isolated from effluents of tanneries. Appl. Microbiol. Biotechnol. 2000;53:48–351. doi: 10.1007/s002530050033. [DOI] [PubMed] [Google Scholar]
- Stasicka Z., Kotas J. Chromium occurrence in the environment and methods of its speciation. Environ. Pollut. 2000;107:1027–1042. doi: 10.1016/s0269-7491(99)00168-2. [DOI] [PubMed] [Google Scholar]
- Stewart D.I., Burke I.T., Mortimer R.J.G. Stimulation of microbially mediated chromate reduction in alkaline soil–water systems. Geomicrobiol. J. 2007;4:655–669. [Google Scholar]
- Suzuki T., Miyata N., Horitsu H., Kawai K. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate formed during the reduction of Cr(VI) to Cr(III) J. Bacteriol. 1992;174:5340–5345. doi: 10.1128/jb.174.16.5340-5345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urvashi T., Datta M. Reduction of toxic chromium and partial localization of chromium reductase activity in bacterial isolate DMI. World J. Microbiol. Biotechnol. 2005;21:891–899. [Google Scholar]
- Urvashi T., Parikh R., Shouche Y., Datta M. Hexavalent chromium reduction by Providencia sp. Process Biochem. 2006;41:1332–1337. [Google Scholar]
- Urvashi T., Parikh R., Shouche Y., Datta M. Reduction of chromate cell free extract of Brucella sp. isolated from Cr(VI) contaminated sites. Bioresour. Technol. 2007;98:1541–1547. doi: 10.1016/j.biortech.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Van Engelen M.R., Peyton B.M., Mormile M.R., Pinkart H.C. Fe(III), Cr(VI), and Fe(III) mediated Cr(VI) reduction in alkaline media using a Halomonas isolate from Soap Lake, Washington. Biodegradation. 2008;19:841–850. doi: 10.1007/s10532-008-9187-1. [DOI] [PubMed] [Google Scholar]
- Viti C., Pace A., Giovannetti L. Characterization of Cr(VI) resistant bacteria isolated from chromium contaminated soil by tannery activity. Curr. Microbiol. 2003;46:1–5. doi: 10.1007/s00284-002-3800-z. [DOI] [PubMed] [Google Scholar]
- Wang P.C., Mori T., Komori K., Sasatu M., et al. Isolation and characterization of Enterobacter cloacae strain that reduces hexavalent chromium under anaerobic conditions. Appl. Environ. Microbiol. 1989;55:665–1669. doi: 10.1128/aem.55.7.1665-1669.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.C., Mori T., Toda K., Ohtake H. Membrane associated chromate reductase activity from Enterobacter cloacae. J. Bacteriol. 1990;172:1670–1672. doi: 10.1128/jb.172.3.1670-1672.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiao C. Factors affecting hexavalent chromium reduction in pure cultures of bacteria. Water Res. 1995;24:2467–2474. [Google Scholar]
- Ye Q., Roh Y., Carroll S.L., Blair B., Zhou J., Zhang C.L., Fields M.W. Alkaline anaerobic respiration: isolation and characterization of a novel alkaliphilic and metal-reducing bacteria. Appl. Environ. Microbiol. 2004;70:5595–5602. doi: 10.1128/AEM.70.9.5595-5602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Yang Z., Ma Z., Chai L. Reduction of high concentrations of chromate by Leucobacter sp. CRB1 isolated from Changsha, China. World J. Microbiol. Biotechnol. 2008;24:991–996. [Google Scholar]
- Zouboulis A.I., Loukidou M.X., Matis K.A. Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process Biochem. 2004;39:909–916. [Google Scholar]


