Abstract
Fusarium is one of the important phytopathogenic genera of microfungi causing serious losses on cucurbit plants in Kermanshah province, the largest area of cucurbits plantation in Iran. Therefore, the objectives in this study were to isolate and identify disease-causing Fusarium spp. from infected cucurbit plants, to ascertain their pathogenicity, and to determine their phylogenetic relationships. A total of 100 Fusarium isolates were obtained from diseased cucurbit plants collected from fields in different geographic regions in Kermanshah province, Iran. According to morphological characters, all isolates were identified as Fusarium oxysporum, Fusarium proliferatum, Fusarium equiseti, Fusarium semitectum and Fusarium solani. All isolates of the five Fusarium spp. were evaluated for their pathogenicity on healthy cucumber (Cucumis sativus) and honeydew melon (Cucumis melo) seedlings in the glasshouse. F. oxysporum caused damping-off in 20–35 days on both cucurbit seedlings tested. Typical stem rot symptoms were observed within 15 days after inoculation with F. solani on both seedlings. Based on the internal transcribed spacer (ITS) regions of ribosomal DNA (rDNA) restriction fragment length polymorphism (RFLP) analysis, the five Fusarium species were divided into two major groups. In particular, isolates belonging to the F. solani species complex (FSSC) were separated into two RFLP types. Grouping among Fusarium strains derived from restriction analysis was in agreement with criteria used in morphological classification. Therefore, the PCR-ITS-RFLP method provides a simple and rapid procedure for the differentiation of Fusarium strains at species level. This is the first report on identification and pathogenicity of major plant pathogenic Fusarium spp. causing root and stem rot on cucurbits in Iran.
Keywords: Fusarium, Pathogenicity, PCR, ITS-RFLP, Cucurbits, Iran
1. Introduction
Cucurbit plants (Cucurbitaceae) are the main agricultural crops, particularly in the Kermanshah province in Iran. Annually it is estimated that over 3000 ha of agricultural land in the province are under cucurbits. Root and stem rots of cucurbits have significantly increased in incidence and severity in the past 20 years and they are a yield-limiting factor in many intensive cucurbit production, especially in cucumber (Cucumis sativus), watermelon (Citrullus lunatus) and honeydew melon (Cucumis melo), resulting sudden death and complete destruction of these economic plants (Alymanesh et al., 2009). Diseased plants were characterized by yellowing of the leaves, stem necrotic lesions, phloem discolorations, and collapse. There are many pathogens capable of producing these vine decline symptoms in cucurbits (Boughalleb et al., 2005).
The most important pathogens that cause root and stem rots in cucurbit plants are Fusarium spp., which are responsible for vascular wilts, such as those on melons (cantaloupe and muskmelon) caused by Fusarium oxysporum f. sp. melonis. Fusarium proliferatum and Fusarium solani f. sp. cucurbitae cause crown and foot rots of summer squash, melon, pumpkin, and a fruit rot of pumpkin (Pivonia et al., 1997; Namiki et al., 1994). There are two “races” of F. solani f. sp. cucurbitae causing fruit, crown and foot rots in cucurbit plants. F. solani f. sp. cucurbitae race 1 (Fsc1) causes crown, fruit, and root rots of cucurbits whereas F. solani f. sp. cucurbitae race 2 (Fsc2) causes only a fruit rot (Hawthorne et al., 1992). Fsc1 and Fsc2 are not easily distinguished morphologically, but identification requires mating tests, pathogenicity tests and molecular assays (Mehl and Epstein, 2007; Hawthorne et al., 1994; Boughalleb et al., 2005). F. solani f. sp. cucurbitae race 1 has been reported as the causal agent of melon root and foot rot from Khorasan province in the eastern part of Iran (Alymanesh et al., 2009).
Characterization of the population structure of fungal pathogens is important for understanding the biology of the organism and for development of disease-control strategies (Malvick and Percich, 1998), and for molecular studies among individuals, which is one of the components of population structure (Leung et al., 1993). Universally, F. solani species complex (FSSC) has an extensive host range and very high levels of diversity in pathogenicity and morphology (Brasileiro et al., 2004). However, the classification system based only on morphology has not provided an accurate tool for the identification of FSSC, neither has morphological classification system resolved the relationship of isolates within FSCC. So, a molecular approach is promising in establishing the objective (O’Donnell and Gray, 1995; Zhang et al., 2006; O’Donnell et al., 2008). Among the methods which researchers have used to analyze the phylogenetics of F. solani species are rDNA-IGS, rDNA-ITS regions, large submit RNA gene and translation elongation factor-alpha (tef) (Zhang et al., 2006). Internal transcribed spacer (ITS) region is probably the most widely sequenced region of DNA in fungi. rDNA-ITS and rDNA-IGS (intergenic spacer) regions show a higher degree of diversity than other ribosomal regions such as small subunits (SSU) and large subunits (LSU) (O’Donnell and Gray, 1995; Depriest and Been, 1992; O’Donnell, 2000; Brasileiro et al., 2004). Therefore, the objectives of this study were: (i) to isolate and identify disease causing Fusarium spp. from infected cucurbit plants in Kermanshah province; (ii) to determine their pathogenicity; and (iii) to determine phylogenetic relationships and usefulness of the PCR-ITS-RFLP as a genetic marker within the Fusarium spp.
2. Materials and methods
2.1. Sample collection
Infected cucurbit plants were collected from different regions of Kermanshah province, Iran (Table 1). Each sample were stored in a paper envelope and kept in a cool box with dry ice. In the laboratory, roots and stems of diseased samples were washed in running tap water and cut into small blocks (1.5 cm) for isolation.
Table 1.
Fusarium species isolated from different hosts in different sampling locations in Kermanshah province.
| Place of sample collection | Host | Source | Fusarium species |
|---|---|---|---|
| Dorood Faraman-Maoqufeh | Watermelon | Root | F. ox, F. eq, F. so |
| Road Kamyaran-Varmele | Honeydew melon | Crown and Stem | F. ox, F. eq, F. so |
| Miandarband-JaFar Abad | Watermelon | Root | F. ox, F. pr, F. so |
| Qazanchi-Ahmad Abad | Melon | Crown and Stem | F. ox, F. eq, F. se |
| Qazanchi-Ahmad Abad | Honeydew melon | Root | F. ox, F. eq, F. so |
| Road Allah-yari | Honeydew melon | Root | F. ox, F. so |
| Road Ravansar-Kamyaran | Honeydew melon | Root | F. ox, F. eq, F. se |
| Kangavar-Pol Shekasteh | Honeydew melon | Root | F. ox, F. eq, F. so |
| Kangavar-Rahmat Abad | Honeydew melon | Root | F. ox, F. pr, F. so |
| Kangavar-Gaodin | Cucumber | Crown | F. ox, F. eq, F. so |
| Gaodin | Honeydew melon | Root and Crown | F. ox, F. eq, F. pr |
| Sonqor | Honeydew melon | Root | F. ox, F. se, F. so |
| Sonqor | Honeydew melon | Root | F. ox, F. eq, F. so |
| Road Sonqor- Asad Abad | Honeydew melon | Root | F. ox, F. eq, F. so |
| Harsin | Honeydew melon | Root | F. ox, F. se, F. pr |
| Dinavar-Shirkhan | Honeydew melon | Root and Crown | F. ox, F. eq, F. so |
| Bisotun-Barnaj | Honeydew melon | Root | F. ox, F. pr, F. so |
| Bisotun-Hosein Abad | Honeydew melon | Root | F. ox, F. eq, F. so |
| Bisotun-Chehr | Pumpkin | Root | F. ox, F. so |
| Bisotun | Pumpkin | Root | F. ox, F. eq, F. so |
| Road Kermanshah Sarab NilooFar | Pumpkin | Root and Crown | F. ox, F. eq, F. so, F. se |
| Road Kermanshah- Sarab NilooFar | Honeydew melon | Root | F. ox, F. pr, F. so |
| Sarab NilooFar | Honeydew melon | Stem | F. eq, F. so, F. se |
| Road Koozaran-Boor Boor | Honeydew melon | Crown and Stem | F. se, F. eq, F. so |
| Road Koozaran-Chehar Zabar | Honeydew melon | Stem | F. pr, F. eq, F. so |
| Gilan Garb | Cucumber | Root and Crown | F. ox, F. eq, F. so |
| Qasr Shirin | Cucumber | Stem | F. so, F. se |
| Road Paveh-Ravansar’ | Honeydew melon | Stem | F. so |
| Paveh- Shemshir | Cucumber | Crown and Stem | F. pr, F. eq, F. so |
| Sarpol Zohab | Watermelon | Stem | F. so |
F. ox = F. oxysporum, F. eq = F. equiseti, F. so = F. solani, F. pr = F. proliferatum, F. se = F. semitectum.
2.2. Isolation and identification
For isolation of Fusarium spp., the blocks were surface-sterilized with 1% sodium hypochlorite for 3 min and rinsed with several changes of sterile water. The sterilized samples were placed onto a general medium (water agar) (Burgess et al., 1994) and a semi-selective medium for Fusarium, i.e., peptone-pentachloronitrobenzene agar (PPA) plates (Nash and Snyder, 1962), and incubated under a standard growth condition (Salleh and Sulaiman, 1984). The resulting Fusarium colonies were single-spored and transferred onto potato dextrose agar (PDA), carnation leaf agar (CLA) (Fisher et al., 1982), spezieller nahrstoffarmer agar (SNA) (Nirenberg, 1976), and potassium chloride agar (KClA) plates (Fisher et al., 1983) for morphological identification (Leslie and Summerell, 2006).
2.3. Pathogenicity test
All isolates of the Fusarium species were tested for their pathogenicity on apparently healthy and uniform 20 days-old seedlings of cucumber (C. sativus) and honeydew melon (C. melo) in the glasshouse. Roots and stems of the cucumber and honeydew melon seedlings were washed in running tap water before inoculation. Conidial suspension of each individual isolate was prepared by pouring sterile distilled water and gently scraping the conidia of 7 days-old cultures on PDA plates grown under the standard growth condition (Salleh and Sulaiman, 1984). The concentration of the pooled suspension was adjusted to 2 × 106 conidia/ml by using a haemocytometer. The roots of the seedlings were soaked in 20 ml conidial suspension for 20 min for root inoculation technique. For stem inoculation technique, 20 ml of the conidial suspension of each Fusarium species was sprayed on the stems. The control plants were inoculated by booth techniques with 20 ml of sterile distilled water. Three replicates were performed for each isolate and the experiment was repeated twice. All the inoculated and controls seedlings were placed in the glasshouse with day and night temperatures of 30–35 °C and 23–30 °C, respectively. Development of symptoms on inoculated and control seedlings were observed every 2 days for 45 days. The inoculated fungi were re-isolated from the infected plants to prove the Koch’s postulates.
2.4. Growth condition and DNA extraction
All Fusarium isolates were grown in 250 ml of potato dextrose broth (PDB) (Difco) in a rotary shaker at 180 rpm for 48 h at 28 °C. After vacuum filtration, the mycelium of each isolate was dried, ground with sterile sea sand in a mortal and pestle, and stored at −20 °C for further studies. Genomic DNA was extracted using a modified method of Kim et al. (1992). Approximately 0.5 g of the ground mycelium was suspended in CTAB extraction buffer (0.7 M NaCl, 50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% 2-mercaptoethanol, 1% CTAB), and extracted with phenol/chloroform/isoamylalcohol (25:24:1) and chloroform/isoamylalcohol (24:1). RNA was degraded by treatment with RNase (Qiagen) (50 mg/ml) for 30 min at 37 °C. DNA was then precipitated by adding 2.5 volumes of absolute ethanol and pelleted by centrifugation for 15 min at 12,000 rpm. The pellet was washed with 70% ethanol, air-dried and re-suspended in 1 mM TE buffer {10 mM Tris–HCl, 1 mM EDTA (pH 8.0)}. DNA concentration and purity was measured using a spectrophotometer (Shimadzu UV-120) at 260 and 280 nm.
2.5. PCR condition
The ITS region of Fusarium spp. was amplified with primers ITS1 (5′-TCCGTTGGTGAACCAGCG G-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). Amplification was performed in 100 μl of reaction mixture containing 50 pmol of primers, 2.5 units of Taq DNA polymerase (Takara), 200 mM of each dNTP, 10 ml of 10× PCR buffer and 0.2 mg of template DNA. The mixture was subjected to PCR in a Thermo cycler. The PCR cycles began with an initial denaturation for 3 min at 95 °C, followed by 30 cycles of annealing for 40 s at 58 °C, extension for 40 s at 72 °C and denaturation for 40 s at 94 °C before a final extension for 5 min at 72 °C. The PCR product obtained was run on 1.4% agarose gels, stained with ethidium bromide (EtBr) and visualized under a UV transilluminator (Brand and company).
2.6. Enzyme digestion
Aliquots of 12 μl of PCR products were digested with 10 units of restriction enzymes EcoRI, SphI, PstI, HaeIII, HinfI, MspI and SmaI according to the manufacturer’s instructions (Fermentas). The restriction fragments were separated on 2.5% agarose gel, run for 140 min at 80 V, 400 mA and stained with EtBr. The restriction fragments were visualized under the UV visualizer and 100 bp DNA ladder (GeneRulers™, Fermentas) was used to estimate the size of the restriction fragments. The restriction analysis was repeated twice.
2.7. Data analysis
The molecular size of each fragment was estimated using a standard curve of migration versus the log of the molecular size of 100 bp ladder. Each fragment was scored on the basis of the presence (1) or absence (0) of particular fragments. A data matrix was constructed based on the presence or absence of the fragments and converted to a similarity matrix. The similarity matrix was then subjected to the unweighted pair group method with arithmetical mean (UPGMA) cluster analysis based on simple matching coefficient (SMC) (Romesburg, 1994). The data analysis was performed by using the Numerical Taxonomy and Multivariate Analysis System (NTSYS-pc, version 2.1) (Rohlf, 2000) to analyze the relationship among all isolates of Fusarium species.
3. Results
3.1. Occurrence and pathogenicity
In this study, a total of 100 Fusarium isolates were obtained from diseased cucurbit plants in 30 different locations in the Kermanshah province (Table 1), and based on their morphological characteristics, these isolates were identified as F. oxysporum, F. proliferatum, Fusarium equiseti, Fusarium semitectum, and F. solani species complex (FSSC) (Table 2, Figs. 1 and 2). On the basis of mean lesion sizes on stems, the pathogenicity of each Fusarium isolates was classified into three groups: virulent (2.5–1.80 cm2), moderately virulent (0.8–0.5 cm2) and nonvirulent (0.0–0.0 cm2) (Table 3).
Table 2.
Morphological characters of Fusarium spp. isolated from cucurbit plants.
| Fusarium Species | Chla. shape | Pigmentation on PDA | Number of septa in macroconidia | Microconidia shape | Types of conidiogenous cells |
General morphology |
Macroconidia size (μm) | ||
|---|---|---|---|---|---|---|---|---|---|
| Poly | Mono | Apical cell | Basal cell | ||||||
| F. equiseti | ro | Brown | 5–7 | − | − | + | Tapered, elongated | Fs | 44–78 × 3.3–5.6 |
| F. oxysporum | sm | Violet | 3 | ov to el | − | + | Curved | Fs | 32–56 × 3.1–5.7 |
| F. proliferatum | − | Violet | 3–5 | cl, na | + | + | Curved | Pdfs | 25–58 × 3.0–5.0 |
| F. semitectum | ro | Brown | 3–5 | − | + | + | Curved and tapered | Fs | 37–58 × 3.0–5.0 |
| F. solani (morphotype II) | sm | Red | 5 | el to tr, cl | − | + | Tapered | Fs | 32–68 × 3.6–6.0 |
| F. solani (morphotype I) | ro | White | 3 | re | − | + | Rounded and curved | Nfs | 30–50 × 3.5–5.7 |
+ = presence, − = absence, Poly = polyphialidic, Mono = monophialidic, Pdfs = poorly developed foot shape, Nfs = Notch or foot shape, Fs = foot shape, Lfs = long foot shape, Chla = chlamydospore, ro = rough, sm = smooth, el = ellipsoid, tr = truncate, cl = clavate, re = reniform, ov = oval.
Figure 1.
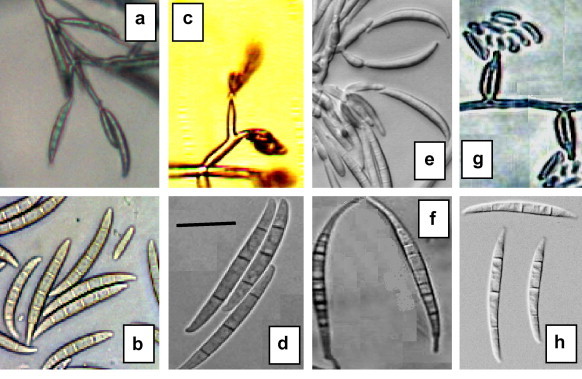
Conidiophores and macroconidial characters of Fusarium spp. a and b = F. semitectum, c and d = F. proliferatum, e and f = F. equiseti, g and h = F. oxysporum (scale bar = 25 μm).
Figure 2.
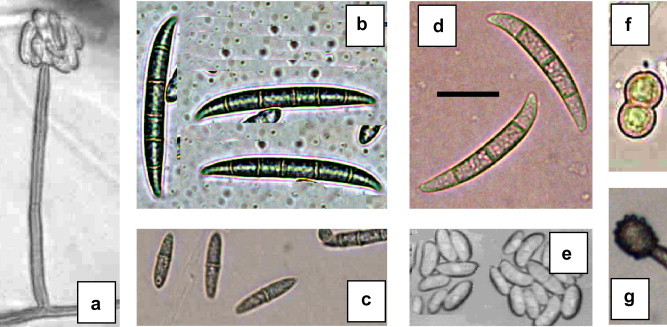
Morphological characters of Fusarium solani. a = Conidiophores; b, c and f = macroconidia, microconidia and chlamydospores of FSQA isolate (morphotype II); d, e and g = macroconidia, microconidia and chlamydospores of FSMV isolate (morphotype I) (scale bar = 25 μm).
Table 3.
Relative virulence of Fusarium spp. based on the pathogenicity test on cucurbit seedlings.
| Name of species | Number isolate | Pathogenicity test |
||
|---|---|---|---|---|
| Virulent | Moderately virulent | Non virulent | ||
| F. equiseti | FEQ101 | + | ||
| F. equiseti | FEQ21 | + | ||
| F. equiseti | FEQ51 | + | ||
| F. equiseti | FEQUI | + | ||
| F. equiseti | FEQ22 | + | ||
| F. equiseti | FEQUII | + | ||
| F. equiseti | FEQ10 | + | ||
| F. equiseti | FEQ2 | + | ||
| F. equiseti | FEQ5 | + | ||
| F. equiseti | FEQ104 | + | ||
| F. equiseti | FEQ27 | + | ||
| F. oxysporum | FOX142 | + | ||
| F. oxysporum | FOAY | + | ||
| F. oxysporum | FOX247 | + | ||
| F. oxysporum | FOX142 | + | ||
| F. oxysporum | FOX148 | + | ||
| F. oxysporum | FOX27 | + | ||
| F. oxysporum | FOX112 | + | ||
| F. oxysporum | FOX148 | + | ||
| F. oxysporum | FOX247 | + | ||
| F. oxysporum | FOX112 | + | ||
| F. oxysporum | FOX118 | + | ||
| F. oxysporum | FOX271 | + | ||
| F. oxysporum | FOX121 | + | ||
| F. oxysporum | FOX188 | + | ||
| F. oxysporum | FOX278 | + | ||
| F. oxysporum | FORK | + | ||
| F. oxysporum | FOX158 | + | ||
| F. oxysporum | FOX257 | + | ||
| F. oxysporum | FOX182 | + | ||
| F. oxysporum | FOX127 | + | ||
| F. oxysporum | FOX186 | + | ||
| F. oxysporum | FOX273 | + | ||
| F. oxysporum | FOX122 | + | ||
| F. oxysporum | FOX158 | + | ||
| F. oxysporum | FOX287 | + | ||
| F. oxysporum | FOX152 | + | ||
| F. oxysporum | FOX1228 | + | ||
| F. oxysporum | FOX1258 | + | ||
| F. oxysporum | FOX277 | + | ||
| F. oxysporum | FOX127 | + | ||
| F. oxysporum | FOX187 | + | ||
| F. oxysporum | FOX278 | + | ||
| F. oxysporum | FOX127 | + | ||
| F. oxysporum | FOX188 | + | ||
| F. oxysporum | FOX278 | + | ||
| F. oxysporum | FOX189 | + | ||
| F. oxysporum | FOX278 | + | ||
| F. proliferatum | FPRFI | + | ||
| F. proliferatum | FPR78 | |||
| F. proliferatum | FPRFII | + | ||
| F. proliferatum | FPR78 | |||
| F. proliferatum | FPR811 | |||
| F. proliferatum | FPR785 | + | ||
| F. proliferatum | FPR88 | |||
| F. proliferatum | FPR711 | |||
| F. proliferatum | FPR885 | + | ||
| F. proliferatum | FPR88 | + | ||
| F. proliferatum | FPR811 | + | ||
| F. semitectum | FSEM | + | ||
| F. semitectum | FSE186 | + | ||
| F. semitectum | FSE287 | + | ||
| F. semitectum | FSE168 | + | ||
| F. semitectum | FSE16 | + | ||
| F. semitectum | FSE76 | + | ||
| F. semitectum | FSE287 | + | ||
| F. semitectum | FSE16 | + | ||
| F. solani | FSQA | + | ||
| F. solani | FSO89 | + | ||
| F. solani | FSO22 | + | ||
| F. solani | FSO694 | + | ||
| F. solani | FSO639 | |||
| F. solani | FSO193 | + | ||
| F. solani | FSO1 | + | ||
| F. solani | FSO298 | + | ||
| F. solani | FSO23 | + | + | |
| F. solani | FSO559 | + | ||
| F. solani | FSO55 | + | ||
| F. solani | FSO18 | |||
| F. solani | FSO288 | |||
| F. solani | FSO273 | + | ||
| F. solani | FSO585 | + | ||
| F. solani | FSO17 | + | ||
| F. solani | FSMV | + | ||
| F. solani | FSM555 | + | ||
| F. solani | FSO57 | + | ||
| F. solani | FSO288 | + | ||
| F. solani | FSO237 | + | ||
| F. solani | FSM555 | + | ||
| F. solani | FSO58 | |||
| F. solani | FSO253 | |||
| F. solani | FSRS | + | ||
| F. solani | FSPS | + | ||
| F. solani | FSO237 | + | ||
| F. solani | FSO555 | + | ||
| F. solani | FSO55 | + | ||
| F. solani | FSO287 | |||
| F. solani | FSO239 | + | ||
| F. solani | FSO558 | + | ||
| F. solani | FSO98 | + | ||
+ = virulent (mean of lesion size = 2.5–1.80 cm2), moderately virulent (mean of lesion size = 0.8–0.5 cm2), non virulent (mean of lesion size = 0.0–0.00 cm2).
The results of the pathogenicity test revealed that 25 isolates of F. oxysporum were the major causal agent of cucumber and honeydew melon root rot. The symptoms were observed within 20–35 days after inoculation as necrosis and brown discoloration of the root phloem and yellowing of the canopy plants. Also, 14 isolates of F. solani, 3 isolates of F. proliferatum and 2 isolates of F. equiseti caused discoloration and necrosis and then brown rot of the stems. The plants were maintained in the glasshouse for 45 days for symptom development. Their initial symptoms were observed on the 15th day after inoculation as water-soaked lesions on the stems. The inoculated fungi were consistently isolated from the diseased plants but not from negative control plants and nonvirulent isolates (Table 3); thus fulfilled the Koch’s postulates.
3.2. Molecular analysis
A PCR product from each isolate of the three Fusarium species was amplified by using primer pairs ITS1 and ITS4. F. oxysporum, F. equiseti and F. semitectum produced approximately 550 bp band, F. solani species complex about 570 bp and F. proliferatum approximately 560 bp bands (Fig. 3). Table 4 and Fig. 3 shows estimated sizes of the restriction bands produced after digestion of the ITS+5.8S using EcoRI, SphI, PstI, HaeIII, HinfI, MspI and SmaI for F. oxysporum, F. equiseti, F. semitectum, F. solani species complex and F. proliferatum. Generally, the restriction patterns by the restriction enzymes could differentiate the five Fusarium species (Fig. 3).
Figure 3.
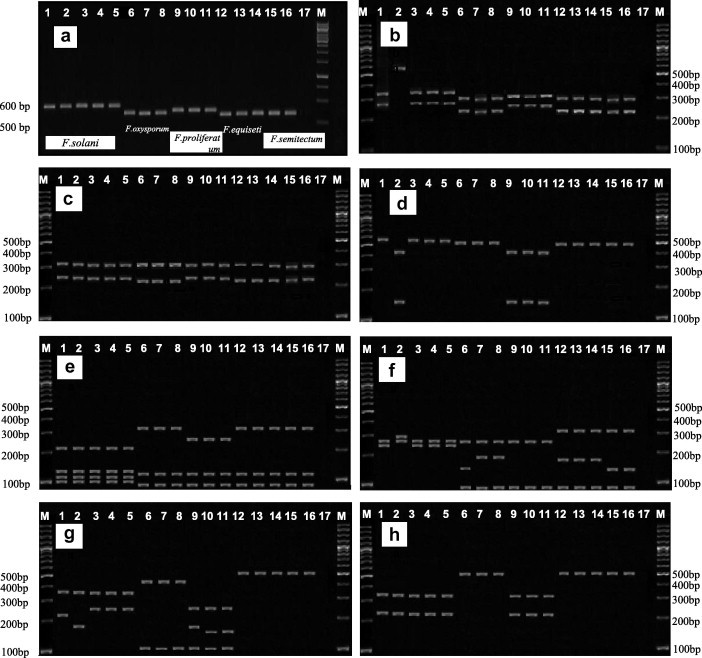
Agarose gels showing: a = amplification of the ITS region (ITS1, ITS2 and 5.8S) and restriction patterns of PCR-amplified rDNA digested with EcoRI (b), SphI (c), PstI (d), HaeIII (e), HinfI (f), MspI (g) and SmaI (h). M. DNA size marker of 100 bp ladder, (1) USM5708 (F. solani), (2) FSQA (F. solani), (3) FSMV (F. solani), (4) FSRS (F. solani), (5) FSPS (F. solani), (6) USM4484 (F. oxysporum), (7) FOAY (F. oxysporum), (8) FORK (F. oxysporum), (9) USM11558 (F. proliferatum), (10) FPRFI (F. proliferatum), (11) FPRFII (F. proliferatum), (12) USM14999 (F. equiseti), (13) FEQUI (F. equiseti), (14) FEQUII (F. equiseti), (15) USM11548 (F. semitectum), (16) FSEM (F. semitectum), (17) negative control.
Table 4.
Restriction fragment size (in bp) of Fusarium ITS region digested with EcoRI, SphI, PstI, HaeIII, HinfI, MspI and SmaI.
| Isolates | Isolates number | ITS total size | EcoRI | SphI | PstI | HaeIII | HinfI | MspI | SmaI |
|---|---|---|---|---|---|---|---|---|---|
| F. solani | USM5708 | 570 | 280, 320 | 250, 320 | 570 | 100, 110, 130, 230 | 270, 280 | 210, 360 | 230, 350 |
| F. solani | FSQA | 570 | 570 | 250, 320 | 150, 420 | 100, 110, 130, 230 | 280, 290 | 180, 360 | 230, 350 |
| F. solani | FSMV | 570 | 280, 320 | 250, 320 | 570 | 100, 110, 130, 230 | 270, 280 | 240, 360 | 230, 350 |
| F. solani | FSRS | 570 | 280, 320 | 250, 320 | 570 | 100, 110, 130, 230 | 270, 280 | 240, 360 | 230, 350 |
| F. solani | FSPS | 570 | 280, 320 | 250, 320 | 570 | 100, 110, 130, 230 | 270, 280 | 240, 360 | 230, 350 |
| F. oxysporum | USM4484 | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 150, 280 | 100, 450 | 550 |
| F. oxysporum | FOAY | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 200, 280 | 100, 450 | 550 |
| F. oxysporum | FORK | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 200, 280 | 100, 450 | 550 |
| F. proliferatum | USM11558 | 570 | 280, 310 | 250, 320 | 150, 420 | 90, 120, 280 | 100, 280 | 100, 180, 250 | 230, 350 |
| F. proliferatum | FPRFI | 570 | 280, 310 | 250, 320 | 150, 420 | 90, 120, 280 | 100, 280 | 100, 150, 250 | 230, 350 |
| F. proliferatum | FPRFII | 570 | 280, 310 | 250, 320 | 150, 420 | 90, 120, 280 | 100, 280 | 100, 150, 250 | 230, 350 |
| F. equiseti | USM14999 | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 200, 320 | 550 | 550 |
| F. equiseti | FEQUI | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 200, 320 | 550 | 550 |
| F. equiseti | FEQUII | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 200, 320 | 550 | 550 |
| F. semitectum | USM11548 | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 150, 320 | 550 | 550 |
| F. semitectum | FSEM | 550 | 250, 300 | 230, 320 | 550 | 90, 120, 340 | 100, 150, 320 | 550 | 550 |
Banding patterns of all Fusarium spp. after digestion analysis were presented in Table 4. Digestion of the PCR products with EcoRI generated two banding patterns in all Fusarium isolates except for isolate FSQA, which indicated that there is no restriction site for the restriction enzyme EcoRI within it. Digestion with EcoRI produced the same patterns for the F. oxysporum, F. semitectum and F. equiseti isolates. It also showed one common and one different band for F. proliferatum and F. solani isolates except for isolate FSQA. Digestion of the ITS+5.8S region with SphI showed two banding patterns in all isolates. The fragment of 320 bp was present in all isolates. The fragment of 250 bp was present in all F. proliferatum and F. solani isolates and the fragment of 230 bp was found in all F. oxysporum, F. semitectum and F. equiseti isolates. Digestion with enzyme PstI mostly indicated that there is no recognition site for the restriction except for F. solani isolate FSQA and F. proliferatum isolates with two fragments of 150 and 420 bp. Digestion with HaeIII produced identical patterns of four fragments for F. solani isolates, three identical fragments for the F. oxysporum, F. semitectum and F. equiseti isolates and three identical fragments for F. proliferatum isolates. Digestion with HinfI exhibited three banding patterns producing three fragments in the F. oxysporum, F. semitectum and F. equiseti isolates and two fragments in the F. solani and F. proliferatum isolates. The fragment of 270 was found in all F. solani isolates except for isolate FSQA that produced the fragment of 290 bp. Digestion with MspI indicated that there is no restriction site for F. semitectum and F. equiseti isolates whereas there are three banding patterns in the F. proliferatum isolates and two banding patterns in F. oxysporum and F. solani isolates. Digestion with SmaI exhibited two identical fragments (230 and 350 bp) in the F. solani and F. proliferatum isolates but there was no restriction site for F. oxysporum, F. semitectum and F. equiseti isolates.
The cluster analysis clearly discriminate the five Fusarium species into five separate clusters. F. solani isolates were clustered in subcluster A and F. proliferatum isolates in subcluster B in major cluster I and F. oxysporum isolates were clustered in subcluster C, F. equiseti isolates in subcluster D and F. semitectum isolates in subcluster E in major cluster II (Fig. 4).
Figure 4.
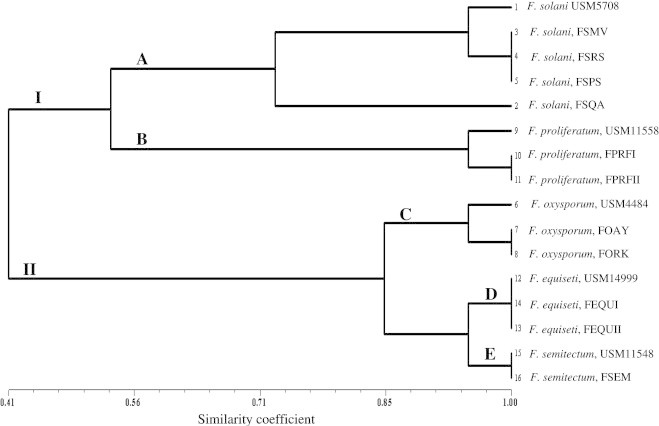
UPGMA dendrogram showing relationships among the 12 isolates of Fusarium based on restriction site data.
4. Discussion
Fusarium species are ubiquitous in roots and stalks of most plants, including cucurbits, and may exist as saprophytes in plant tissues or as opportunistic pathogens awaiting pre-disposed conditions such as stress in their hosts. Many species of Fusarium are viewed as opportunistic or weak pathogens that are capable of attacking only plants that were weakened previously by some other stress factors. Certainly, factors such as those induced by drought, wind and insects are known to affect the amount and disease incidence (Palmer and Kommedahl, 1960). Our observations in rainfed cucurbit fields in Kermanshah province showed that the severity of the disease may be increased under certain environmental conditions in the middle of the summer. There are also observations showing that in irrigated fields the severity of the disease may increased under flooding, and irregular furrow irrigation. Thus, the time and type of irrigation in the cucurbit fields in Kermanshah province may be considered as one of the feasible cultural practices for better disease control.
Fusarium species have been recorded from several parts of the world and they are known to be pathogenic to many plants, especially to cucurbits. The results of this study are in agreement with those of the previous literatures, e.g., Boughalleb et al. (2005), Mehl and Epstein (2007). The cluster analysis based on restriction bands formed two major groups, I and II. The group including F. solani and F. proliferatum isolates was supported at the similarity level of ca. 56%. The two subgroups of F. solani species complex (FSSC) were supported at the similarity level of 69%. The second group, that included F. oxysporum, F. semitectum and F. equiseti, was supported at the similarity level of ca. 80% similarity. Within this group, the group of F. semitectum and F. equiseti was supported at the similarity level of 90%. PCR–RFLP of ITS+5.8S have been used by Suga et al. (2000) in distinguishing formae speciales (f. spp.) of F. solani and Lee et al. (2000) for comparing genetic relationships between 12 Fusarium species from different sections. Digestion of F. solani PCRs product with EcoRI, PstI, HinfI and MspI produced variable restriction patterns. Digestion with HaeIII, SphI and SmaI generated the same restriction patterns for all F. solani isolates obtained from cucurbits showing root and stem rot symptoms in the field. According to the RFLP results, F. solani isolate FSQA was different from other isolates having a PstI, HinfI and MspI restriction site and the absence of the EcoRI restriction site. This result correlated with the report that F. solani f. sp. phaseoli is a distinct species within F. solani complex (O’Donnell and Gray, 1995). This result is in agreement with that of O’Donnell and Gray (1995), who reported that isolates F. solani f. sp. phaseoli from soybeans, could be easily distinguished from the FSSC shown by the presence of a unique PstI restriction site within the ITS2 region.
Brasileiro et al. (2004) have used two restriction enzymes, EcoRI and HaeIII for studying the diversity of the ITS region of the F. solani. Also, four restriction enzymes have been used to compare formae speciales (f. spp.) of F. solani by Suga et al. (2000). In the present work, the PCR products of F. oxysporum isolates were undigested after digestion with PstI and SmaI and a band of 550 bp was observed which indicated that there was no restriction site for the restriction enzyme within the ITS+5.8S of the isolates. F. proliferatum isolates from cucurbits stem and root rot as well as from the stock culture showed similar restriction patterns when digested with EcoRI, SphI, PstI, HaeIII, HinfI and SmaI. Only MspI produced variable restriction patterns. There was no restriction site for the restriction enzymes MspI, PstI and SmaI within the ITS+5.8S of the F. semitectum and F. equiseti isolates. Also, F. semitectum and F. equiseti isolates showed similar restriction patterns when digested by using EcoRI, SphI and HaeIII. Only HinfI restriction patterns produced variable patterns within isolates of these two species. HinfI revealed the highest variation in both fragment sizes and restriction sites within all species. Intraspecies variations could be due to minor changes in nucleotide composition within the ITS+5.8S, which might lead to different restriction patterns. Similar results were obtained by Konstantinova and Yli-Mattila (2004), in their study using PCR–RFLP of ribosomal intergenic spacer region to analyse Fusarium species in section Sporotrichiella. Further studies, such as rDNA RFLP analysis with more restriction enzymes and different genes (e.g., IGS region) have demonstrated convincingly the morphological and molecular classifications of several Fusarium species including F. oxysporum, F. semitectum and F. proliferatum isolates (Paavanen-Huhtala et al., 1999; Hawa et al., 2010; Lee et al., 2000).
In this study, biological characterization, including culture morphology, pathogenicity, and molecular study were used to compare isolates of F. solani associated with cucurbit plants. These isolates caused typical disease symptoms on stems of the tested cucurbit seedlings. Molecular approach by PCR-ITS-RFLP analyses strongly supported the existence of two distinct clades among F. solani isolates in the present study. Based on morphological characters, the isolates USM5708, FSRS, FSMV, and FSPS are classifies as morphotype I while FSQA as morphotype II and this observation is in agreement with previous molecular studies, whereas based on morphological characters, there is no difference between isolates USM5708, FSRS, FSMV, FSPS. The results of the present taxonomical studies using molecular method and grouping among Fusarium strains derived from restriction analysis were in agreement with previous molecular and morphological classification criteria. Therefore, the PCR-ITS-RFLP method described in this paper provides a simple and rapid procedure for the differentiation of Fusarium strains at the species level. Further studies using ITS+5.8S sequence analysis and other sequence analyses, such as IGS sequences, would be necessary to compare the genetic variations observed in Fusarium isolates from root and stem rot of cucurbit plants.
5. Conclusion
Five Fusarium species were isolated from root and stem rot of naturally diseased cucurbit plants grown in Kermanshah province, Iran. Based on the morphological characteristics, the 100 isolates were identified as F. oxysporum, F. proliferatum, F. equiseti, F. semitectum and two morphotypes of Fusarium solani. From the pathogenicity test, F. oxysporum and other Fusarium species were the major causal agents of root and stem rot of cucurbit plants in the province. PCR–RFLP of ITS+5.8S analysis used in this study, offers a convenient tool for characterization and analyzing variations of Fusarium species associated with root and stem rot of cucurbit plants.
Acknowledgements
Khosrow Chehri acknowledges the Razi University, Iran and Universiti Sains Malaysia (USM), Penang, Malaysia for providing necessary facilities. We are grateful for the Research Grant 300/PBIOLOGI/811009 provided by USM.
References
- Alymanesh M.R., Falahatirastegar M., Jafarpour B., Mahdikhanimoghadam E. Genetic diversity in the fungus Fusarium solani f. sp. cucurbitae race 1, the casual agent of root and crown rot of cucurbits in Iran, using molecular markers. Pak. J. Biol. Sci. 2009;12:836–843. doi: 10.3923/pjbs.2009.836.843. [DOI] [PubMed] [Google Scholar]
- Boughalleb N., Armengol J., El Mahjoub M. Detection of races 1 and 2 of Fusarium solani f. sp. cucurbitae and their distribution in watermelon fields in Tunisia. J. Phytopathol. 2005;153:162–168. [Google Scholar]
- Brasileiro T.R.V.B., Coimbra M.R.M., Antonio de Morais M., Jr., Tinti de Oliveria N. Genetic variability within Fusarium solani species as revealed by PCR-fingerprinting based on PCR markers. Braz. J. Microbiol. 2004;35:205–210. [Google Scholar]
- Burgess, L.W., Summerell, B.A., Bullock, P., Backhouse, D., 1994. Laboratory Manual for Fusarium Research, third ed., Department of crop science, University of Sydney, Sydney, Australia, p. 133.
- Depriest P.T., Been M.D. Numerous group I introns with variable distributions in the ribosomal DNA of a lichen fungus. J. Mol. Biol. 1992;228:315–321. doi: 10.1016/0022-2836(92)90819-6. [DOI] [PubMed] [Google Scholar]
- Fisher N.L., Burgess L.W., Toussoun T.A., Nelson P.E. Carnation leaves as a substrate and for preserving Fusarium species. Phytopathology. 1982;72:151–153. [Google Scholar]
- Fisher N.L., Marasas W.F.O., Toussoun T.A. Taxonomic importance of microconidial chains in Fusarium section Liseola and effects of water potential on their formation. Mycologia. 1983;75:693–698. [Google Scholar]
- Hawa M.M., Salleh B., Latiffah Z. Characterization and intraspecific variation of Fusarium semitectum (Berkeley and Ravenel) associated with red-fleshed dragon fruit (Hylocereus polyrhizus [Weber] Britton and Rose) in Malaysia. Afr. J. Biotechnol. 2010;9:273–284. [Google Scholar]
- Hawthorne B.T., Rees-George J., Broadhurst P.G. Mating behavior and pathogenicity of New Zealand isolates of Nectria haematococca (Fusarium solani) NZ J. Crop Hortic. Sci. 1992;20:51–57. [Google Scholar]
- Hawthorne B.T., Ball R.D., Rees-George J. Genetic analysis of variation of pathogenicity in Nectria haematococca (Fusarium solani) on Cucurbita sp. Mycol. Res. 1994;98:1183–1191. [Google Scholar]
- Kim D.H., Martyn R.D., Magill C.W. Restriction fragment length polymorphism groups and physical map of mitochondrial DNA from Fusarium oxysporum f. sp. niveum. Phytopathology. 1992;82:346–353. [Google Scholar]
- Konstantinova P., Yli-Mattila T. IGS-RFLP analysis and development of molecular markers for identification of identification of Fusarium poae, Fusarium langsethiae, Fusarium sporotrichioides and Fusarium kyushuense. Int. J. Food Microbiol. 2004;95:321–331. doi: 10.1016/j.ijfoodmicro.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Lee Y.M., Choi L.Y., Min B.R. PCR–RFLP and sequence analysis of rDNA its region in the Fusarium spp. J. Microbiol. 2000;38:66–73. [Google Scholar]
- Leslie J.F., Summerell B.A. Blackwell Publish Ltd.; UK: 2006. The Fusarium Laboratory Manual. p. 388. [Google Scholar]
- Leung H., Nelson R.J., Leach J.E. Population structure of plant pathogenic fungi and bacteria. In: Andrews J.H., Tommerup I.C., editors. Advances in Plant Pathology. Academic Press; London: 1993. pp. 157–204. [Google Scholar]
- Malvick D.K., Percich J.A. Genotypic and pathogenic diversity among pea-infecting isolates of Aphanomyces euteiches from the central and western United States. Phytopathology. 1998;88:915–921. doi: 10.1094/PHYTO.1998.88.9.915. [DOI] [PubMed] [Google Scholar]
- Mehl H.L., Epstein L. Identification of Fusarium solani f. sp. cucurbitae race 1 and race 2 with PCR and production of disease-free pumpkin seeds. Plant Dis. 2007;91:1288–1292. doi: 10.1094/PDIS-91-10-1288. [DOI] [PubMed] [Google Scholar]
- Namiki F., Shiomi T., Kayamura T., Tsuge T. Characterization of the formae speciales of Fusarium oxysporum causing wilts of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 1994;60:2684–2691. doi: 10.1128/aem.60.8.2684-2691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash S.M., Snyder W.C. Quantitative and estimations by plat counts of propagules of the bean rot Fusarium in field soils. Phytopathology. 1962;73:458–462. [Google Scholar]
- Nirenberg H.L. Untersuchungen uber die morphologische und biologische differenzierung in der Fusarium section Liseola. Mitteilungen aus der biologischen bundesanstalt fur land-und forstwirtschaft (Berlin-Dahlem) 1976;169:1–117. [Google Scholar]
- O’Donnell K. Molecular phylogeny of the Nectria haematococca–Fusarium solani species complex. Mycologia. 2000;92:919–938. [Google Scholar]
- O’Donnell K., Gray L.E. Phylogenetic relationships of the soybean sudden death syndrome pathogen Fusarium solani f. sp. phaseoli inferred from rDNA sequence data and PCR primers for its identification. Mol. Plant Microbe Interact. 1995;8:709–716. doi: 10.1094/mpmi-8-0709. [DOI] [PubMed] [Google Scholar]
- O’Donnell K., Sutton D.A., Fothergill A., McCarthy D., Rinaldi M.G., Brandt M.E., Zhang N., Geiser D.M. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavanen-Huhtala S., Hyvönen J., Bulat S.A., Yli-Mattila T. RAPD–PCR, isozyme, rDNA RFLP and rDNA sequence analyses in identification of Finnish Fusarium oxysporum isolates. Mycol. Res. 1999;103:625–634. [Google Scholar]
- Palmer L.T., Kommedahl T. Root-infecting Fusarium species in relation to rootworm infestations in corn. Phytopathology. 1960;59:1613–1617. [Google Scholar]
- Pivonia S., Cohen R., Kafkafi U., Ben Ze’ev I.S., Katan J. Sudden wilt of melons in southern Israel: fungal agents and relationship with plant development. Plant Dis. 1997;81:1264–1268. doi: 10.1094/PDIS.1997.81.11.1264. [DOI] [PubMed] [Google Scholar]
- Rohlf F.J. Exeter Publishing Ltd.; Setauket, New York, USA: 2000. NTSYS-pc Numerical Taxonomy and Multivariate Analysis System, version 2.1. [Google Scholar]
- Romesburg H.C. Lifetime Learning Publications Belmont; California: 1994. Cluster Analysis for Researchers. [Google Scholar]
- Salleh B., Sulaiman B. Fusarium associated with naturally diseases plants in Penang. J. Plant Prot. Tropics. 1984;1:47–53. [Google Scholar]
- Suga H., Hasegawa T., Mitsui H., Kageyama K., Hyakumachi M. Phylogenetic analysis of the phytopathogenic fungus Fusarium solani based on the rDNA-ITS region. Mycol. Res. 2000;104:1175–1183. [Google Scholar]
- White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- Zhang N., O’Donnell K., Sutton D.A., Nalim F.A., Summerbell R.C., Padhye A.A., Geiser D.M. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006;44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


