Abstract
Internal transcribed spacer (ITS) region of nuclear ribosomal DNA from 16 populations of Persicaria barbata (L.) H. Hara (Polygonaceae) belonging to five geographical locations of India (Arunachal Pradesh, Himachal Pradesh, Bihar, Karnataka and Andaman Island) was sequenced. Analysis of nucleotide sequences reveals polymorphism among the populations. UPGMA analysis conducted on the ITS datasets shows that the sampled populations of P. barbata are grouped according to their geographic locations and are supposed to be evolved under reproductive isolation which most probably are due to the long distance distribution and population fragmentation.
Keywords: Persicaria barbata, Genetic diversity, ITS, nrDNA
1. Introduction
Persicaria Mill. (Polygonaceae) is comprised of about 100 species of annual or perennial herbs or vines distributed around the northern temperate and into tropical regions of the world (Brandbyge, 1993). The genus is represented in all continents, but most of them are of restricted distribution, and several are localized endemics. In India, they are mainly distributed in the Himalayas. Earlier, they were circumscribed under the broad genus Polygonum L. s.l., but the anatomical and molecular studies (Haraldson, 1978; Ronse Decraene et al., 2000) proved them to be separate entities. They are basically characterized by the presence of spicate or capitate inflorescence and tepals with three main nervatures departing from base (Haraldson, 1978; Decraene and Akeroyd, 1988; Kim and Donoghue, 2008). The data about the number of species and their distribution in the Eastern and Western Himalayas are gradually increasing since the time of Hooker (1886). It is apparent that though the distribution is mainly throughout the Himalayas but concentration varies in East, West and Central Himalayas (Nepal). It has the greater concentration in Western and Eastern Himalayas. However, they are poorly represented in the rest of India including the Andaman and Nicobar islands. Till date, there is no any updated record on the genus Persicaria in India. Tentatively, it is represented by c. 30 species many of which are used for medicinal purposes in different regions. The seeds of Persicaria barbata (L.) H. Hara are used for griping colic pain in Kalahandi area of Orissa state (Singh and Jain, 2003) whereas, the aerial parts possess antinociceptive, anti-inflammatory and diuretic properties (Abdul et al., 2009). Roots are used as an astringent and cooling remedy (Chehregani et al., 2009). Decoction of leaves and shoots is used as a stimulating wash for ulcers, acting as a good healer of the scarred tissue (Kirtikar and Basu, 1935; Gorsi and Sharma, 2002). A paste of the root is used externally in the treatment of scabies (Manandhar, 2002).
Phylogeny estimation has been a central theme of macro and micro evolutionary studies ever since cladistic and nucleotide sequencing techniques became available (Castelloe and Templeton, 1994). During the last decade, most researches have focused on phylogeny above the species level using various molecular markers of nuclear as well as organelle genomes (Avise, 1994). Evolutionary biologists have turned their attention to the subspecies level (Kooistra et al., 1992; Vogler and DeSalle, 1994). Gene trees provide powerful data to investigate population-level phenomena, such as gene flow, founder events and the history of lineages (Castelloe and Templeton, 1994). Internal transcribed spacers of nuclear ribosomal DNA (nrDNA) have been widely used for resolving phylogenetic relationships among closely related species of angiosperms. They have frequent insertions/deletions which could be phylogenetically informative (Baldwin et al., 1995).
An important prerequisite for the development of an effective conservation strategy is the proper evaluation of the distribution and study at the level of genetic variation (Milligan et al., 1994). ITS sequence of nrDNA data of P. barbata was earlier worked out for molecular systematic studies (Kim and Donoghue, 2008) but this did not include the genetic variation of the species within different populations. Hitherto, information on population structure and genetic variation of P. barbata is lacking. Hence, the main objectives of the present study were to utilize the nucleotide data of the combined ITS1-5.8S-ITS2 sequences to evaluate the degree of differentiation among P. barbata population from India and its relationship with geographical distribution.
2. Materials and methods
2.1. DNA extraction, amplification, and sequencing
The leaf material of P. barbata was collected from natural population during floristic exploration trips to different geographical regions of India. Leaves were dried in Silica gel prior to DNA extraction. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN Inc., Crawley, West Sussex, UK). ITS sequences of nrDNA were amplified using primers of White et al. (1990) using AccuPower HF PCR PreMix (Bioneer, Daejeon, South Korea) in 20 μL volumes containing 2 μL of 10× buffer, 300 μM dNTPs, 1 μL of a 10 pM solution of each primer, 1 unit of HF DNA polymerase. One round of amplification consisting of denaturation at 94 °C for 5 min followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 49 °C for 1 min and extension at 72 °C for 1 min with a final extension step of 72 °C for 5 min. The PCR products were purified with the SolGent PCR Purification Kit-Ultra (SolGent, Daejeon, South Korea) prior to sequencing. The purified fragments were directly sequenced using dye terminator chemistry following the manufacturer’s protocol. The sequencing reaction was performed in a 10 μL final volume with the BigDye Terminator cycle sequencing kit (Perkin–Elmer, Applied Biosystems). Cycle sequencing was conducted using same primers used in amplification and BigDye version 3 reagents and an ABI PRISM 3100 DNA Analyzer (Perkin–Elmer, Applied Biosystems). Cycling conditions included an initial denaturing set at 94 °C for 5 min, followed by 30 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. Sequenced product was precipitated with 17 μL of deionized sterile water, 3 μL of 3 M NaOAc, and 70 μL of 95% EtOH. Polyacrylamide gel electrophoresis was conducted with long ranger single packs (FMC BioProducts) and an ABI 3100 automated DNA sequencer (Perkin–Elmer, Applied Biosystems). Each sample was sequenced in the sense and antisense direction and analyzed with ABI Sequence Navigator software (Perkin–Elmer/Applied Biosystems). Nucleotide sequences of both DNA strands were obtained and compared to ensure accuracy.
2.2. Alignments and phylogenetic analyses
Sequence alignments were performed using ClustalX version 1.81 (Thompson et al., 1997). Alignments were subsequently adjusted manually using BioEdit (Hall, 1999). Insertion-deletions (Indels) were scored as single characters when we had confidence in positional homology. The boundaries between the ITS1, 5.8S, and ITS2 were determined by comparisons with earlier published sequences available at NCBI (www.ncbi.nlm.nih.gov). Gaps were treated as missing data in phylogenetic analyses. All sequences generated in the present study were deposited in GenBank (HQ709145 to HQ709160). Nucleotide polymorphism, as measured by θw (Watterson, 1975) and diversity, as measured by π (Nei, 1978) were calculated using DnaSP v4.5 (Rozas and Rozas, 1999). Analysis of molecular variance (AMOVA) was performed using GenAlEx 6.1 (Peakall and Smouse, 2006) to assess genotypic variations across all the populations studied. This analysis, apart from partitioning of total genetic variation into within-group and among-group variation components, also provided a measure of intergroup genetic distance as the proportion of the total variation residing between populations. The significance of the analysis was tested using 999 random permutations. The cladistic analysis of aligned sequences was performed by UPGMA method using MEGA5 (Tamura et al., 2007).
3. Results and discussion
ITS sequences of nrDNA regions from 16 individuals of P. barbata were sampled from five geographical locations of India, i.e., Arunachal Pradesh, Himachal Pradesh, Bihar, Karnataka and Andaman Island. The absence of other variable regions in the nuclear DNA of plants that could provide useful markers at both intra-family (Baldwin and Markos, 1998) and intra-genomic level differentiation (Feliner et al., 2004), makes ITS ostensibly the best marker for phylogenetic studies. The amplified region of ITS1-5.8S-ITS2 in P. barbata was found to have 646 bases (GC content 59–60%) in all the accession used for the present analysis. Data matrix has a total number of 646 characters, of which invariable (monomorphic) sites are 616, variable (polymorphic) sites are 30 (total number of mutations are 31), singleton variable sites are 07 and parsimony informative sites are 23. The substitution probabilities are given in Table 1. The overall transition/transversion bias (R) was found to be 0.527.
Table 1.
Maximum composite likelihood estimate of the nucleotide substitution. Each entry shows the probability of substitution from one base (row) to another base (column) (Tamura et al., 2004). Rates of different transitional substitutions are shown in bold and those of transversional substitutions are shown in italics. The nucleotide frequencies are 0.212 (A), 0.193 (T/U), 0.316 (C), and 0.28 (G). The transition/transversion rate ratios are k1 = 1.021 (purines) and k2 = 0.796 (pyrimidines).
| A | T | C | G | |
|---|---|---|---|---|
| A | – | 6.64 | 10.86 | 9.82 |
| T | 7.28 | – | 8.65 | 9.62 |
| C | 7.28 | 5.29 | – | 9.62 |
| G | 7.43 | 6.64 | 10.86 | – |
Polymorphism was observed among the populations [π = 0.01312, θw = 0.01400 (0.00256) and total variance 3.202]. π and θw refer to nucleotide diversity according to Nei and Li (1979) and Watterson’s parameter (Watterson, 1975), respectively. Figures in parenthesis denote standard deviation. Variability within the nuclear ribosomal transcription unit (NRTU) usually depends upon the number of gene copies, rates of mutation, concerted evolution, number and chromosomal location of NRTU clusters, and the proportion of sexual and asexual reproduction (Dover et al., 1993). Polymorphism may arise when concerted evolution is not fast enough to homogenize repeats in the face of high rates of mutation (Appels and Honeycutt, 1986) or by the loss of sexual recombination (Campbell et al., 1997).
AMOVA was used to partition the genetic diversity among the populations and tested whether there is any hierarchy of ITS sequence variation among individuals (Table 2; Figs. 1 and 2). The genetic differentiation between the populations is high (ΦST = 0.1994). (Nei (1978) classified GST > 0.15 as high, ΦST and GST both denote fixation index and are comparable).
Table 2.
Hierarchical analysis of molecular variance (AMOVA) within/among P. barbata populations. (d.f.: degrees of freedom; SSD: sum of squared deviations; ΦST: fixation index).
| Source of variation | d.f. | SSD | Estimated variance | Total variance (%) | ΦST |
|---|---|---|---|---|---|
| Among population | 4 | 3.813 | 0.206 | 41 | 0.411 |
| Within population | 11 | 3.250 | 0.295 | 59 | |
| Total | 15 | 7.063 | 0.502 |
Figure 1.
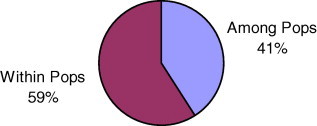
Percentage of molecular variance within and among population of P. barbata.
Figure 2.
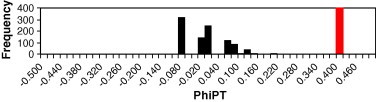
Frequency distribution of random PhiPT vs observed PhiPT among population of P. barbata.
The evolutionary history was inferred using the UPGMA method (Sneath and Sokal, 1973). The optimal tree with the sum of branch length = 0.04708052 is shown in Fig. 3. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) which are in the units of the number of base substitutions per site. Codon positions included were first + second + third + noncoding. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). The UPGMA tree reveals two major groups. In the first group, accessions collected from Bihar, Himachal Pradesh, Arunachal Pradesh and Karnataka were grouped together while in the second group accessions collected from Andaman island were grouped, which indicate that the sampled populations were grouped together according to their geographic locations (Bihar, bs 65%; Arunachal Pradesh, bs 69%; Himachal Pradesh, bs 93%; Karnataka, bs 84% and Andaman, bs 99%. It clearly appears that P. barbata evolved under reproductive isolation probably due to the long distance distribution and population fragmentation.
Figure 3.
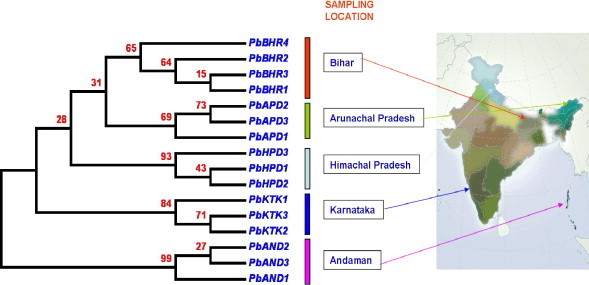
Evolutionary relationship among population of P. barbata inferred using UPGMA method. Number on the branches indicates bootstrap support under 1000 bootstrap replicates.
References
- Abdul M.M., Datta B.K., Nahar L., Khairul Bashar S.A.M., Bachar S.C., Sarker S.D. Antinociceptive, anti-inflammatory and diuretic properties of Polygonum barbatum (L.) Hara var. barbata. Rev. Bras. Farmacogn. 2009;19(3):117–119. [Google Scholar]
- Appels R., Honeycutt R.L. RDNA: evolution over a billion years. In: Dutton S., editor. DNA Systematics. CRC Press; Boca Raton, FL, USA: 1986. pp. 81–135. [Google Scholar]
- Avise J.C. Chapman & Hall; New York: 1994. Molecular Markers, Natural History and Evolution. [Google Scholar]
- Baldwin B.G., Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) Mol. Phylogen. Evol. 1998;10:449–463. doi: 10.1006/mpev.1998.0545. [DOI] [PubMed] [Google Scholar]
- Baldwin B.G., Sanderson M.J., Porter J.M., Wojciechowski M.F., Campbell C.S., Donoghue M.J. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Miss. Bot. Gard. 1995;82:247–277. [Google Scholar]
- Brandbyge J. Polygonaceae. In: Kubitzki K., Rohwer J.G., Bittrich V., editors. vol. 2. Springer; Berlin, Heidelberg, New York, London, Paris, Tokio, Hong Kong, Barcelona, Budapest: 1993. pp. 531–544. (The Families and Genera of Vascular Plants. Flowering plants. Dicotyledons, Magnoliid, Hamamelid and Caryophyllid families). [Google Scholar]
- Campbell C.S., Wojciechowski M.F., Baldwin B.G., Alice L.A., Donoghue M.J. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae) Mol. Biol. Evol. 1997;14:81–90. doi: 10.1093/oxfordjournals.molbev.a025705. [DOI] [PubMed] [Google Scholar]
- Castelloe J., Templeton A.R. Root probability for intraspecific gene trees under neutral coalescent theory. Mol. Phylogenet. Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- Chehregani A., Noori M., Yazdi H.L. Phytoremediation of heavy-metal-polluted soils: screening for new accumulator plants in Angouran mine (Iran) and evaluation of removal ability. Ecotoxicol. Environ. Saf. 2009;72(5):1349–1353. doi: 10.1016/j.ecoenv.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Dover G.A., Linares A.R., Bowen T., Hancock J.M. Detection and quantification of concerted evolution and molecular drive. Methods Enzymol. 1993;224:525–541. doi: 10.1016/0076-6879(93)24039-w. [DOI] [PubMed] [Google Scholar]
- Feliner G.N., Larena B.G., Aguilar J.F. Fine scale geographic structure, intra-individual polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae) Ann. Bot. 2004;93:189–200. doi: 10.1093/aob/mch027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsi M.S., Sharma M. Ethnomedicinal survey of plants of khanabad village and its allied areas, district Gilgit. Asian J. Plant Sci. 2002;1(5):604–615. [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Haraldson K. Anatomy and taxonomy in Polygonaceae subfam. Polygonoideae Meisn. Emend. Jaretzky. Symb. Bot. Upsal. 1978;22(2):1–95. [Google Scholar]
- Hooker J.D. vol. 5. Gilbert & Livington Ltd; London: 1886. (Flora of British India). [Google Scholar]
- Kim S., Donoghue M.J. Incongruence between cpDNA and nrITS trees indicates extensive hybridization within genus Eupersicaria (Polygonaceae) Am. J. Bot. 2008;95(9):1122–1135. doi: 10.3732/ajb.0700008. [DOI] [PubMed] [Google Scholar]
- Kirtikar K.R., Basu B.D. vols. 1–4. Bishen Singh Mahendra Pal Singh; Dehra Dun, India: 1935. (Indian Medicinal Plants). [Google Scholar]
- Kooistra W.H.C.F., Stam W.T., Olson J.L., Van Den Hoek C. Biogeography of Cladophoropsis membranacea based on comparisons of nuclear rDNA ITS sequences. J. Phycol. 1992;28:660–668. [Google Scholar]
- Manandhar N.P. Timber Press; Oregon: 2002. Plants and People of Nepal. [Google Scholar]
- Milligan B.G., Leebens-Mack J., Strand A.E. Conservation genetics: beyond the maintenance of marker diversity. Mol. Ecol. 1994;3:423–435. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. GENALEX 6: genetic analysis in Excel, population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronse Decraene L.P., Akeroyd J.R. Generic limits in Polygonum and related genera (Polygonaceae) on the basis of floral characters. Bot. J. Linn. Soc., London. 1988;98(4):321–371. [Google Scholar]
- Ronse Decraene L.P., Hong S.P., Smets E. Systematic significance of fruit morphology and anatomy in tribes Persicarieae and Polygoneae (Polygonaceae) Bot. J. Linn. Soc., London. 2000;134(1–2):301–337. [Google Scholar]
- Rozas J., Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Singh V., Jain A.P. In: Singh V., Jain A.P., editors. vol. 2. Scientific Publishers; India: 2003. (Ethnobotany and Medicinal Plants of India and Nepal). [Google Scholar]
- Sneath P.H.A., Sokal R.R. Freeman; San Francisco: 1973. Numerical Taxonomy. [Google Scholar]
- Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 40. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins G.D. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acid Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler A.P., DeSalle R. Evolution and phylogenetic information content of the ITS-1 region in the tiger beetle Cicindela dorsalis. Mol. Biol. Evol. 1994;11:393–405. doi: 10.1093/oxfordjournals.molbev.a040121. [DOI] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Pop. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., Taylor J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M., Gelfand D., Sninsky J., White T., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]


