Abstract
In this study, the gfp fragment as a reporter gene had integrated into the form plasmid vector pBC-hygro which contains an expressive promoter of the fungus to facilitate the transformation of Fusarium oxysporum. The resultant plasmid pBC-hygro-GFP was identified by digestion with enzymes. Binary plasmids pBC-hygro-GFP were transformed into F. oxysporum by using the PEG–CaCl2 mediated transformation technique. Results show that the recombinant plasmid pBC-hygro-GFP was constructed correctly. The gfp gene was stably maintained and did not convey any significant loss of phenotype which would affect the survival and behaviour of the tagged strains. Introduction of the gfp gene into F. oxysporum provides a simple, specific and cost-effective method of strain identification for ecological studies. Transcriptional reporter vectors were constructed for using the green fluorescent protein (GFP) reporter.
Keywords: GFP, Fusarium oxysporum, PBC-hygro, PEG–CaCl2
1. Introduction
Fusarium wilt is a worldwide soil-borne disease, which is caused by Fusarium oxysporum Schl. It is also the most serious disease on watermelon, muskmelon, cucumber, wax gourd and other melons which has caused huge economic losses by threatening crop production. However, exceptions to the concept of formae specials have been reported in these formae specials (Chalfie et al., 1994). On the basis of the cross-infectivity of these formae specials, the genetic relationships within and among these formae specials have been questioned.
Green fluorescent protein (GFP) is a small protein found in the jellyfish Aequorea victoria. It has the property of fluorescing when excited by UV light (Chalfie et al., 1994). GFP-marked strains of Pseudomonas Green fluorescent protein (GFP) expression is now a widely used tool in the molecular analysis of filamentous fungi, as reviewed by Lorang (McMillan, 1986). This inert reporter is excited by UV light (395 nm) and emits green light (509 nm) making it more easily visualized using epifluorescence microscopy employing commonly available filter sets. GFP has the additional advantage in that it does not require a substrate, eliminating associated solubility, toxicity, or permeability problems. The GFP marker has been introduced into Rhizobium meliloti and Rhizobium leguminosarum bv. trifolii to visualize the tagged bacteria in the rhizosphere and on root surfaces (Gage et al., 1996; Prayitno et al., 1999). Most of these fungi have been ascomycetes, with new reports of expression being commonplace: recent species to express GFP include F. graminearum (Lorang et al., 2001), Colletotrichum acutatum (Skadsen and Hohn, 2004), and Verticillium fungicola (Horowitz et al., 2002), Trichoderma reesei (Richard et al., 2002).
The aim of this work was the construction of a GFP-based vector for the analysis of transcriptional fusions in F. oxysporum, and lay a foundation for further studying the pathogenic mechanisms of F. oxysporum, find out the reason for the existence of pathogenic differences in formae specials.
2. Materials and methods
2.1. Strains, plasmids and culture conditions
F. oxysporum strain F-H.SY0975 isolated from cucumber. Escherichia coli strain DH5a (procured from Takara program) was used in the construction and transformation of recombinant plasmids. All clonings were done using E. coli strain DH5a grown at 37 °C in Luria broth (Sambrook et al., 1989) liquid media supplemented with 100 mg/ml ampicillin or chloramphenicol, and on plates solidified with 1.5% agar. Plasmid PBC-hygro stored in E. coli (procured from Biovector Science Lab), plasmid pEGFP(containing gfp gene) stored in E. coli. pEGFP and PBC-hygro were extracted using plasmid Mini kit(OMEGA, D6943–01). E. coli strain Trans 109 was used for F. oxysporum transformations (procured from Takara program).
2.2. Details of vector construction
2.2.1. Primer design and gfp fragments amplified
Primers were designed according to the gfp gene search from NCBI and the multiple cloning site of PBC-hygro using Software Primer 5.0. Restriction enzyme sites were added to primer GFPup1 and Down1. The gfp gene was amplified from pEGFP by PCR using the primer GFPup1: GGGAAGCTTAGTGCTGAAACCTCCGTAT (contain HindIII enzyme site) and GFPdown1: GGCGAATTCCACCTTGATGCCG (containing ECOlI enzyme site).
PCR was performed in 50 μl volume of 10 × buffer 2 μl, dNTP 0.6 μl (10 mM), Taq 0.5 μl (5 U/μl), primers 1.5 μl (10 μmol), DNA 0.5 μl, and ddH2O 45.4 μl. The cycling parameters were as follows: DNA denaturation at 94 °C 3 min; at 94 °C for 30 s, at 52 °C for 60 s, at 72 °C for 60 s, run for 35 cycles; and the final extension step was performed at 72 °C for 10 min.
2.2.1.1. E. coli transformation
The resulting PCR fragment was digested with HindIII and ECOlI, and the digested products were purified by using Universal DNA Purification Kit (TIAN GEN, DP214–02), cloned into the Trans 109 (destroyed by blunting with T4 DNA polymerase).
2.2.1.2. Protoplast preparation and transformation
Protoplast were prepared from F. oxysporum and co-transformed with 25 μl of plasmid pBC-hygro-GFP, which harbours the homologous gfp gene, using the PEG-CaCl2 mediated transfer of DNA as described previously (Bae and Knudsen, 2000).
2.2.2. Screening for GFP expression
Putative resistant transformants were transferred to PDA containing 300 mg/ml hygromycin B and screened for GFP expression. GFP expression was initially detected in F. oxysporum by microscopic screening using NIKON TS-100 with excitation filters at 450–490 nm, using 488 nm excitation line.
3. Results
3.1. The construction of pBC-hygro-GFP
3.1.1. Amplified gfp fragment
pEGFP was used as the template to augment PCR, whose products went through agarose gel electrophoresis to produce a 900 bp strip. As the fragment corresponded with what was required, it was indicated that the primer design and PCR reaction procedure conformed to the requirements of augmentation. Through enzyme digestion of PCR products with HindIII and ECOlI, cohesive terminus matched with the fragment was acquired (enzyme digestion products map of electrophoresis).
3.1.2. Extraction and enzyme digestion of PBC-hygro plasmid
PBC-hygro plasmid with hygromycin resistant gene was extracted, and after detected by agarose gel electrophoresis, a plasmid fragment of about 6.8 kb was obtained, the size of which corresponding to that showed in the plasmid profile. Through the enzyme digestion of plasmid PBC-hygro with HindIII and ECOlI, two fragment with 2 kb and 4.8 kb were obtained, and the 4.8 kb fragment purified.
3.1.3. Transformation of E. coli
After linking with gfp fragment by T4 ligase, the recycled 4.8 kb fragment was transformed to E. coli. The bacterial colony growing on the chloramphenicol medium was showed in Fig. 6. Extracted transformant plasmid was used to recombine the plasmid through enzyme digestion with HindIII and ECOlI, which was linked with the two fragments to produce two new fragments with the same size of 6.8 kb (detected by electrophoresis). The size of the plasmids indicated the right insertion.
Figure 6.
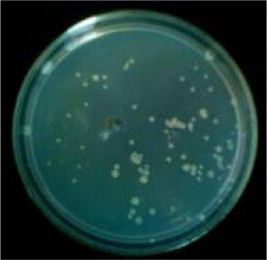
DH5a transformants.
3.1.4. GFP transformation of F. oxysporum strain F-H.SY0975
3.1.4.1. Preparation of protoplast
Microscopy was used to observe the production of protoplast after going through one hour enzymolysis with Driselase buffer. The morphology of the protoplast was shown in Fig. 8a, whose product mode was: top-end release (Fig. 8b), and intermediate release (Fig. 8c).
Figure 8.

Regenerstion of protoplasts (a: a single protoplast morphology; b: top-end release; c: intermediate release).
3.1.4.2. Stability verification of F. oxysporum F-H.SY0975 green fluorescent protein marking transformants
In order to detect the genetic stability of green fluorescent protein gene in transformant genome, the third culture of F. oxysporum GFP transformants was inoculated onto the PDA medium without hygromycin. After continuous culturing for five generations, it was later inoculated onto the hygromycin medium. The lines witnessing normal growth were a stable transformant. GFP expression was maintained in mycelia (results not presented).
3.1.4.3. Fluorescence detection of green fluorescent protein marking transformants
Fluorescence expression of stable transformants was observed under fluorescence microscope, with results being that the three transformants had a stronger fluorescence expression and some had a weaker expression. For transformants with hygromycin resistance, a small amount of hyphae and spore were selected to be observed under fluorescence microscope. It was discovered that GFP had stronger expression in young hyphae and weaker expression in aging hypha, while the expression was weak in conidium.
3.1.4.4. GFP transformants PCR confirmation
Four transformants having fluorescence properties strongly and two wild strains being genomous, were abstracted to prove that hygromycin-resistant transformants contain the inserted gfp gene. As a template, the application of amplified gfp fragment primers. GFPup1 and GFPdown1 of the PCR amplification results showed that the four transformants contain gfp gene, but the other two wild strains do not contain gfp gene.
4. Discussion
In this study, binary plasmids pBC-hygro-GFP were transformed into F. oxysporum by the PEG-mediated transformation technique and E. coli-mediated methods succeeded in F. oxysporum B-resistant transformants.
In the F. oxysporum transformation, the size of prepared protoplast is very important for protoplast transformation, too big or too small cannot reach the transformation requirements.1.2 × 108 spore/ml is the optimal concentration. The fluorescence expression of transformants, the GFP expression was not detectable in any of the hygromycin resistant pBC-hygro-GFP transformants, despite GFP DNA has been amplified by PCR. Fluorescence expression in different transformants is inconsistent. With is introducing the exogenous gene GFP, this difference may be due to the GFP insert, and insert the copy number.
In this experiment, it was observed that GFP marked transformants of F. oxysporum line had a very weak fluorescence expression. The conidiums without significant expression was outotheca isolated to detect the fluorescence expression of colony growing out of a single conidium. It was discovered that the hyphae of colony grown out of cultured conidium without significant fluorescence expression were able to express fluorescence normally, which was deduced that the weak fluorescence expression of conidium was caused by the small number of protoplasm, as little GFP protein was expressed. What could be known from the literature was that after marking fungus with GFP, there was a difference in GFP for conidium of different fungi. Some might have significant green fluorescence expression such as the conidium of PyrenoPhora tritici-repentis and Cochliobolus Sativus (Prayitno et al., 1999). The difference might be due to the different protoplasm and protein contents in the conidium of different fungi, for only the protoplast of higher content and water could enable the stimulated blue light to pass through the conidium easily to get GFP protein of inner expression stimulated. We have also used successful GFP expression in F. oxysporum to extend the range of available promoters for use in fungus transformation, and in doing so we have shown that the gfp gene can express stably in the F. oxysporum strain. The vector we have developed will facilitate further analysis of transformation in Fusaruim and other fungi (see Figs. 1–5, 7 and 9–12).
Figure 1.

Enzyme digestion of gfp fragment.
Figure 2.

Amplified gfp fragment.
Figure 3.

Plasmid PBC-hygro.
Figure 4.

Enzyme digestion PBC-hygro.
Figure 5.
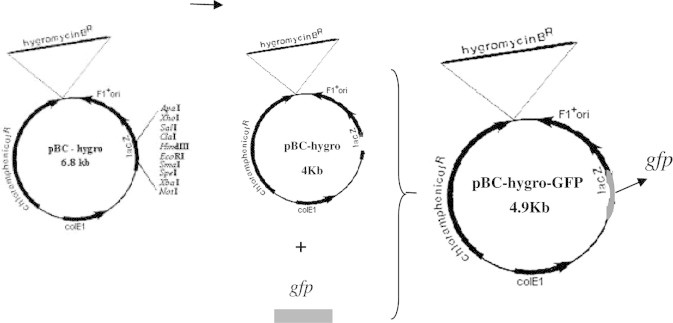
Schema of the vector pBC-hygro-GFP construction.
Figure 7.

Agarosegel analysis of three plasmid vectors by two restriction enzymes.
Figure 9.

Hypha regenerated from protoplast.
Figure 10.

Amplify gfp fragment from E. coli transformants.
Figure 11.
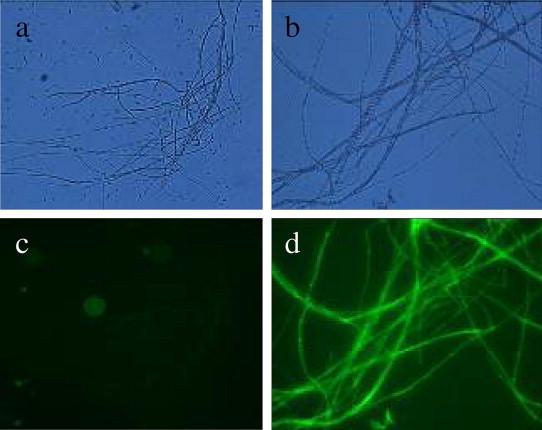
The mycelium of Fusarium oxysporum. (a) The mycelium of wild strain under UV (400×), (b) the morphology of mycelium under bright light (400×), (c) the mycelium of wild strain under bright light (400×) and (d) the morphology of mycelium under UV (400×).
Figure 12.

PCR analysis of transformants of Fusarium oxysporum with GFP gene.
Acknowledgements
This work was supported by the National 863 Project (2006CB101901), Research Projects in Liaoning Province (2011214002) and Shenyang Research Projects (F12-119-3-00).
Footnotes
Peer review under responsibility of King Saud University.

References
- Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prashar D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- McMillan R.T. Cross pathogenicity studies with isolates of Fusarium oxysporum from either cucumber or watermelon pathogenic to both crop species. Ann. Appl. Biol. 1986;109:101–105. [Google Scholar]
- Gage D.J., Bobo T., Long S.R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J. Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prayitno J., Stefaniak J., McIver J. Interaction of rice seedlings with bacteria isolated from rice roots. Aust. J. Plant. Physiol. 1999;26:521–535. [Google Scholar]
- Lorang J.M., Tuori R.P., Martinez J.P., Sawyer T.L. Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 2001;67:1987–1994. doi: 10.1128/AEM.67.5.1987-1994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skadsen R.W., Hohn T.M. Use of Fusarium graminearum transformed with gfp to follow infection patterns in barley and Arabidopsis. Physiol. Mol. Plant. Pathol. 2004;64:45–53. [Google Scholar]
- Horowitz D.S., Lee E.J., Mabon S.A., Misteli T. A cyclophilin functions in pre-mRNA splicing. EMBO J. 2002;21:470–480. doi: 10.1093/emboj/21.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C., Amey A., Claire B., Peter R., Andy B. PEG-mediated and Agrobacterium-mediated transformation in the mycopathogen Verticillium fungicola. Mycol. Res. 2002;106(1):4–11. [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Bae Y.S., Knudsen G.R. Cotransformation of Trichoderma harzianum with b-glucuronidase and green fluorescent protein genes provides a useful tool for monitoring fungal growth and activity in natural soils. Appl. Environ. Microbiol. 2000;66:810–815. doi: 10.1128/aem.66.2.810-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


