Abstract
DA (D-blood group of Palm and Agouti, also known as Dark Agouti) and F344 (Fischer) are two inbred rat strains with differences in several phenotypes, including susceptibility to autoimmune disease models and inflammatory responses. While these strains have been extensively studied, little information is available about the DA and F344 genomes, as only the Brown Norway (BN) and spontaneously hypertensive rat strains have been sequenced to date. Here we report the sequencing of the DA and F344 genomes using next-generation Illumina paired-end read technology and the first de novo assembly of a rat genome. DA and F344 were sequenced with an average depth of 32-fold, covered 98.9% of the BN reference genome, and included 97.97% of known rat ESTs. New sequences could be assigned to 59 million positions with previously unknown data in the BN reference genome. Differences between DA, F344, and BN included 19 million positions in novel scaffolds, 4.09 million single nucleotide polymorphisms (SNPs) (including 1.37 million new SNPs), 458,224 short insertions and deletions, and 58,174 structural variants. Genetic differences between DA, F344, and BN, including high-impact SNPs and short insertions and deletions affecting >2500 genes, are likely to account for most of the phenotypic variation between these strains. The new DA and F344 genome sequencing data should facilitate gene discovery efforts in rat models of human disease.
Keywords: BN, DA, F344, Rattus norvegicus, whole-genome sequencing, next-generation whole-genome sequencing (NGS)
THE laboratory rat (Rattus norvegicus) has been a model organism for the study of human biology and diseases for nearly 200 years (Jacob 1999). Rats differing in susceptibility to disease models and other traits have been extensively studied to better understand human physiology, pharmacology, toxicology, nutrition, behavior, immunology, and diseases such as diabetes, autoimmunity, arthritis, and cancer. These traits have a strong genetic component, making rat models of human disease highly useful for the identification and validation of causative genes and pathways, as well as for testing new therapeutic approaches.
The sequencing of the Brown Norway (BN/SsNHsdMcwi) rat genome was a milestone for the identification, positional cloning, and study of disease model and trait regulatory genes. The BN rat genome was first drafted using a strategy that combined bacterial artificial chromosome (BAC) end sequencing, whole-genome shot gun sequencing, and BAC fingerprinting mapping (Gibbs et al. 2004). The BN rat genome was later expanded and reassembled, leading to the draft assembly RGSC v3.4 (Worley et al. 2008). The BN strain was chosen because it has been commonly used in many different fields and studies and was also a founder strain for panels of consomic and recombinant inbred rat strains (Worthey et al. 2010).
The DA (D-blood group of Palm and Agouti, also known as Dark Agouti) and the F344 (Fischer) strains have been extensively studied due to their phenotypic differences in complex traits as diverse as nociception and behavior (Brodkin et al. 1999; Terner et al. 2006), resistance to infections and parasites (Ishih 1994; Suzuki et al. 2006; Zhang et al. 2011), severity of autoimmune and inflammatory diseases such as arthritis (Dahlman et al. 1998; Sun et al. 1999; Wilder et al. 1999), oxygen-induced retinopathy (van Wijngaarden et al. 2007), muscular strength (Biesiadecki et al. 1998), bone mineral density (Turner et al. 2001), taste preference (Tordoff et al. 2008), cellular phenotypes (Brenner et al. 2007; Laragione et al. 2007, 2008; Zhang et al. 2011), metabolic traits (van Den Brandt et al. 2000), and costicosterone levels (Potenza et al. 2004). These and other complex traits have been mapped in linkage studies, and the Rat Genome Database (http://rgd.mcw.edu) presently curates 257 quantitative trait loci (QTL) in crosses involving DA and 362 QTL in crosses involving F344 rats, including congenics. Yet detailed genomic information for DA and F344 is lacking and would be instrumental for the identification of the genes accounting for each QTL and for the understanding of the genetic regulation of several complex traits.
Next-generation whole-genome sequencing (NGS) technology enables ultrahigh depth and high-resolution sequencing projects at a cost significantly lower than the traditional dideoxynucleotide-based capillary method. NGS has been successfully used to resequence the human (Bentley et al. 2008; Wang et al. 2008; Wheeler et al. 2008; Ahn et al. 2009; Kim et al. 2009; G. Li et al. 2009; Fujimoto et al. 2010; Tong et al. 2010), mouse (Keane et al. 2011; Yalcin et al. 2011), and the spontaneously hypertensive rat (SHR) (Atanur et al. 2010) genomes. Here we report the high-depth sequencing of the DA and F344 strains using NGS to generate the first two de novo assemblies of the rat genome and the identification of >2 million new variants likely to account for many of the phenotypic differences between DA, F344, and BN.
Materials and Methods
Rats and DNA
DA (DA/BklArbNsi) rats were originally purchased from Bantin and Kingman, transferred to the Arthritis and Rheumatism Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, and maintained since 2002 at the Laboratory of Experimental Rheumatology at the Feinstein Institute for Medical Research (formerly North Shore-Long Island Jewish Research Institute) via brother–sister mating. F344 (F344/NHsd) rats were purchased from Harlan Laboratories. Genomic DNA was extracted from the liver of one male DA and one male F344 rat using the phenol–chloroform–isoamyl alcohol method (Strauss 2001). The quantity of DNA was determined using a NanoDrop spectrophotometer (Thermo Scientific), and the integrity was evaluated using electrophoresis.
Construction and sequencing of DNA libraries
Illumina pair-end index libraries were constructed according to the manufacturer’s protocol. Briefly, ∼3 μg of DNA was randomly fragmented by nebulization with compressed nitrogen gas. Overhangs (5′ or 3′) of double-stranded DNA fragments were converted to blunt ends using T4 DNA polymerase and Klenow polymerase. An “A” base was added to the end of double-stranded DNA fragments using exo- Klenow polymerase, followed by ligation to adaptors with a “T” base overhang. After electrophoresis, DNA fragments of 500 bp on average were gel-purified. To minimize bias in library preparation, two DNA libraries were built for each sample. The adaptor-modified DNA fragments were loaded on an Illumina Cluster Station and underwent 10 cycles of bridge amplification PCR to generate sequencing template clusters on flow cells. Samples were processed on the HiSequation 2000 platform (Illumina) according to the manufacturer’s instructions for template hybridization, isothermal amplification, linearization, blocking and denaturing, and hybridization of the sequencing primers. Base-calling was done using Illumina’s pipeline, HiSeq Control (HCS) + OLB + GAPipeline-1.6 (Illumina), and the sequences of each lane were generated as 90-bp reads. Data were processed and analyzed according to a pipeline summarized in Supporting Information, Figure S9 and described in detail below.
Reference genome
The rat (R. norvegicus) reference genome (RGSC v3.4) was downloaded from the University of California at Santa Cruz database (http://genome.ucsc.edu/) along with data on gene annotation, ESTs, gaps, repeats, and position of the centromeres. Single nucleotide polymorphisms (SNPs) were downloaded from dbSNP build 136.
Read filtering and mapping
The raw data were refined using two filtering steps: (1) Contaminant filtering: adapter sequences may be introduced into raw reads during the library construction process. Therefore reads containing sequences similar to the adapter (mismatch ≤3) were considered contaminated and discarded, as were reads <30 bp in length. (2) Quality value filtering: to obtain high-quality data, reads with 40% or more low-confidence bases (quality value = 2) were discarded. All cleaned reads were mapped onto the BN rat reference genome using SOAP2.21 (Li et al. 2009b), allowing a maximum of five mismatches for each read. The alignment parameters were the following: -a –b –D –o -2 –u –m –x (-g) –l 32 –s 30 –v 3. Duplicated reads caused by PCR were removed using an in-house C++ script.
Detection of SNPs
To identify SNPs against the reference genome, the genotype probability of each site in DA and F344 was calculated using SOAPsnp (Li et al. 2009a), which is based on the Bayesian statistical model. A consensus sequence (CNS) was generated to contain the genotype with the highest probability for each position. SNPs between the reference sequence and the CNS were considered high-quality SNPs when they fulfilled all of the following criteria: (1) quality value >20 (indicating an inferred base call accuracy >99%); (2) estimated copy number of flanking sequences <2; (3) minimum distance between adjacent SNPs of 5 bp; (4) at least six uniquely mapped reads supporting homozygous SNPs or three for each allele of heterozygous SNPs; and (5) a maximum depth of each site of 75 (depth value was limited to twice the mean depth to avoid incorrect SNP calls supported by reads in repeats). DA and F344 genomic DNA were extracted from male rats, so we considered all SNP sites in chromosome X to be hemizygous and required them to be covered by only two reads.
Detection of short insertions/deletions
The clean reads were realigned to the BN genome with SOAP2 set to tolerate gaps of up to 10 bp. Then we clustered mapped read pairs containing gaps in only one end to detect insertions/deletions (indels) of up to 5 bp. Candidate indels overlapping SNP sites were filtered out. The remaining candidate events were considered high-quality indels when supported by 15–55 reads.
Experimental validation of SNPs and indels
Primers were designed to cover 1045 variants (SNPs and indels) on the chromosome 4 locus Cia3d (Brenner et al. 2011) and on the chromosome 10 loci Cia5a and Cia5d (Brenner et al. 2005). PCR products were generated using AmpliTaq Gold (Life Technologies) and 10 ng of genomic DNA. Excess primers and dNTPs were removed from the PCR reaction by treatment with Exosap-IT (USB) according to the manufacturer’s instructions. Samples were then diluted to 20–40 ng/μl and sequenced at Genewiz, Inc. (South Plainfield, NJ) using BigDye Terminator v.3.1 on a 3730xl capillary analyzer (Life Technologies). Base calls were manually determined using LaserGene v.8 (Dnastar, Madison, WI).
Detection of structural variation and copy-number variation candidates
We identified structural variation using the paired-end method (Wang et al. 2008). The accuracy of this method depends on the distribution of the insert size of the DNA library. A Perl script was written to compile the mean and the standard deviation of the insert sizes used for the paired-end mapping. Paired-end reads that could both be aligned but did not meet the insert size and/or orientation inferred from the reference genome were classified as abnormal paired-end reads. Regions supported by at least three abnormal paired-end reads and differing from the inferred insert size by at least 3 standard deviations were considered to contain structural variation. Abnormal paired-end reads were analyzed by clustering, and structural variants were categorized as insertions, tandem or dispersed duplications, deletions, and combinations of inversions and deletions.
Segmental duplication or deletion events are also evident as regions of increased or decreased copy number (Yoon et al. 2009). To locate copy-number variation (CNV) candidates using the alignment results, we first obtained the depth of each base along the reference genome using SOAPcoverage (http://soap.genomics.org.cn/). We then used CNVDetector (Chen et al. 2008), a program developed by BGI, to calculate the mean depth of 100-bp sliding windows along each chromosome and to select the candidate regions of CNV based on the difference of depth between each consecutive window and the overall mean. Events with a high absolute difference in depth (i.e., outside the 0.75- to 1.25-fold range) and >10 kb were considered an effective CNV candidate. Some candidate regions had to be subdivided because of gaps (N-region) in the BN genome.
Simulation
To evaluate our optimal sequencing depth and the accuracy of our methods, we simulated short reads of different lengths using the BN genome. We also simulated mismatch sequencing errors using a sampling of the quality scores from the DA and F344 sequencing data, as well as SNPs, indels, and structural variants (separately and with occurrence rates of 1 × 10−3, 1 × 10−4, and 1 × 10−5, respectively). The length of indels ranged from 1 to 10 bp and the length of structural variants ranged from 100 bp to 100 kb. The simulated reads were then realigned back to the whole BN genome. We used the rate of misplacement to calculate the sensitivity and specificity to detect SNPs, indels, and structural variation and how their detection rates were affected by coverage and quality scores.
GapCloser Tool
The GapCloser tool (http://soap.genomics.org.cn/) (Li et al. 2010) adopts a greedy algorithm to fill gaps. It extends contig ends iteratively by using reads overlapping with the contig end. Contig-end extension terminates when (1) the extended sequence overlaps with the other contig end at the other side of a gap, (2) an extended sequence with no overlap with the contig end at the other side of gap is 1000 bp longer than the size of the gap in the reference genome, or (3) no reads can be found to make a new round of extension. If extension of one strand fails to close a given gap, GapCloser will perform another extension on the complementary strand.
Construction of the DA and F344 genome drafts
The DA and F344 genomes were assembled using the reference-aided assembly method (RAM), a novel strategy for genome assembly based on resequencing data. RAM contains three main steps: (1) construction of semifinished genome, (2) independent de novo assembly to generate contigs and scaffolds, and (3) generation of the genome draft by anchoring scaffolds onto the semifinished genome.
Cleaned sequencing reads were aligned onto the BN reference genome using SOAPaligner (Li et al. 2009b) to construct DA and F344 CNS equal in length to the BN genome but tolerating SNPs at a rate of 10−3. Then gaps in each chromosome’s CNS were closed with each line’s own clean sequencing reads using GapCloser (Li et al. 2010). Each gap-closed CNS constituted a coordinated, semifinished genome.
To obtain the de novo genome assembly of each line, SOAPdenovo (Li et al. 2010) was used to reassemble clean reads and to generate contigs and scaffolds for DA and F344. Gaps between scaffolds were closed using GapCloser.
The final step to obtain the genome drafts of DA and F344 was anchoring the scaffolds onto the semifinished genome. To avoid scaffold contamination, only qualified scaffolds—>200 bp and containing <50% of Ns—were selected for anchoring. Tag sequences with a length of 100 bp and containing no Ns were extracted from each end of the qualified scaffolds, with additional tags extracted for >5000 bp. Tag sequences were mapped to the semifinished genomes using BLAST (Altschul et al. 1990), and the aligned tag sequences were filtered according to the following criteria: (a) e-value <1 × 10−40, (b) identity value >95, (c) alignment length >95, and (d) number of mismatches fewer than five. We used these qualified tag sequences to anchor the high-confidence scaffolds onto the semifinished genome and thus obtain the genome assembly for each strain.
To evaluate the accuracy of the DA and F344 genome drafts, we retrieved all 194,363 ESTs available in the rat genome and aligned them to the assembled drafts using BLAST, set to cover at least 95% of each EST. We also estimated the single-base error for each genome draft by comparing their sequence to the corresponding positions containing homozygous SNPs at the same strain’s semifinished genome.
Data access
All reads have been deposited in the European Bioinformatics Institute (EBI)/NCBI Short Read Archive (accession no. SRA046343). All DA and F344 data have been released for public use and can be freely accessed at NCBI’s Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra or at http://dx.doi.org/10.5524/100042). The data set includes all reads, semifinished genome sequences, genome drafts, annotation of variants including SNPs, short indels (1–5 bp), structural variations, and the bioinformatics tools used.
Results
Sequencing
Genomic DNA was extracted from the liver of male DA (DA/BklArbNsi, The Feinstein Institute for Medical Research) and F344 (F344/NHsd, Harlan Laboratories) rats using the phenol–chloroform–isoamyl alcohol method (Strauss 2001). Massively parallel whole-genome sequencing was performed using the Illumina HiSeq2000 sequencing platform. To minimize systematic bias in library preparation, for each genome we prepared two paired-end DNA libraries with a read length of 90 bp and insert sizes of 475–504 bp (Table S1). A total of 1.02 billion reads from the DA and 1.07 billion reads from the F344 genomes were generated, corresponding to 92.03 and 96.80 Gb of sequencing data, respectively. The proportion of high-quality data (Q-score ≥20) obtained for DA was 96.67% and for F344 was 97.63%.
The current assembly of the BN reference genome has an effective size of 2.57 Gb. Using the Short Oligonucleotide Alignment Program (SOAP) (Li et al. 2008), 83.87 Gb of DA and 88.18 Gb of F344 sequence—91.3% of each strain’s reads—aligned with the BN genome. These reads covered 98.9% of the BN reference genome with at least one read and 98.0% with a sequencing depth of three or more reads and resulted in genome-wide average sequencing depths of 32.68-fold for DA and 34.36-fold for F344 reads (Table S2). The sequencing depth did not vary significantly between autosomes, indicating euploidy (Figure S1). Sequencing depth followed a Poisson distribution, and regions of lower depth correlated with extremes of GC content (Figure S2).
Regions of sequence ambiguity and breaks between contigs in the BN genome form 876,652 gaps that limit alignment with DA and F344 reads. These gaps contain 267.83 million positions of undetermined sequence (Ns). Using the GapCloser tool (Li et al. 2010) to bridge gaps with aligned reads, we were able to assign sequences to 59.31 million positions in DA and to 59.70 million positions in F344 and to effectively close 359,392 gaps in DA and 361,412 in F344 (Figure 1, Table S2).
Figure 1.
Genetic variation in the DA and F344 genomes. Distribution and frequency of (A) SNPs, (B) short insertion/deletions (InDel), (C) structural variants (SV), (D) copy-number variant (CNV) candidates, and (E) filled gaps along the rat genome (numbers outside the circle represent each chromosome), using the BN genome as reference, are shown. The F344 genome is in dark blue, and DA is in light blue.
SNPs
We used SOAPsnp (Li et al. 2010) to identify the SNP sets for each inbred line based on the alignment results of all sequencing data with the BN genome sequence. Unreliable sites were excluded from the analysis by filtering the SNPs for quality, copy number, distance between SNPs, number of supporting reads for each allele, and total depth. After filtering, we identified 2,964,158 high-quality nuclear DNA SNPs in DA and 2,973,513 in F344, compared with the BN genome (Figure 2A). We also identified 156 mitochondrial SNPs in DA and 163 in F344. Mitochondrial SNPs had a frequency of 9.8 × 10−3.
Figure 2.
Variation between DA, F344, and BN. (A) DA (blue, light blue) and F344 (red, pink) each had 1.03 million unique homozygous SNPs and shared alleles for 1,786,600 homozygous SNPs (purple, light purple). Forty-eight percent of the strain-specific SNPs and 20% of the shared SNPs were novel and were not present in dbSNP v.136, including 502,994 SNPs with alleles unique to DA (blue), 496,368 SNPs with alleles unique to F344 (red), and 370,879 SNPs for which DA and F344 had the same alleles (purple). (B) DA and F344 had the same alleles for 146,502 homozygous indels; 143,058 were unique to DA, and 149,621 were unique to F344. Most homozygous indels were 1 bp long, and distribution of short insertions and deletions according to size was similar in DA and F344. (C) DA and F344 had the same alleles covering 80% of 30,978 structural variants; 15,151 were unique to DA, and 17,575 were unique to F344. The most frequent structural variants were deletions and insertions, followed by tandem and dispersed duplications. (D) There were 2594 CNV candidates unique to DA, 3611 unique to F344, and 994 identical between DA and F344. Most CNV candidates were in the 10- to 20-kb range (blue: DA; red: F344; purple or gray: shared).
We detected a total of 5,632,694 homozygous SNPs: 2,816,017 in the DA set and 2,816,677 in the F344 set. A total of 2,059,492 homozygous SNPs were polymorphic between DA and F344, and 1,786,600 homozygous SNPs were identical (Table 1, Figure 2A). The frequency of homozygous SNPs was 1.1 × 10−3 for both strains. More than 1.37 million homozygous SNPs were new and not represented in the dbSNP database (build 136). These novel SNPs included 502,994 SNPs with alleles unique to DA (F344 and BN carried the same allele), 496,368 SNPs with alleles unique to F344 (DA and BN carried the same allele), and 370,879 SNPs with alleles unique to BN (DA and F344 carried the same allele). A percentage of 39.38 of DA and F344 homozygous SNPs mapped to repeat regions, in agreement with the 40% interspersed repetitive DNA described in the rat genome (Gibbs et al. 2004). To estimate the accuracy of our SNP set, we sequenced three gene regions of chromosomes 4 and 10 using the Sanger method and confirmed 99.68% of 933 homozygous SNPs (Table S3).
Table 1. SNPs and indels in the DA and F344 consensus assemblies.
| SNPs |
Short indels |
|||||
|---|---|---|---|---|---|---|
| Sample | Homozygous | Heterozygous | Knownab | Novelb | Homozygous | Heterozygous |
| DAc | 1,029,417 | 91,848 | 526,423 | 502,994 | 143,058 | 9,461 |
| F344d | 1,030,075 | 100,545 | 533,707 | 496,368 | 149,621 | 9,071 |
| Sharede | 1,786,600 | 56,293 | 1,415,721 | 370,879 | 146,502 | 511 |
| Total | 3,846,092 | 248,686 | 2,475,851 | 1,370,241 | 439,181 | 19,043 |
Homozygous SNPs mapping to SNP positions in dbSNP 136.
Include only homozygous SNPs.
Allele is unique to DA (F344 allele = BN allele)
Allele is unique to F344 (DA allele = BN allele)
DA and F344 have the same allele, which is different from BN.
A percentage of 5.14 of the SNPs were detected in the heterozygous state, with a genomic distribution rate of 5.9 × 10−5. Heterozygous SNPs were predominantly detected in regions with high alignment rates (median sequencing depth; homozygous SNPs = 28-fold for DA and 27-fold for F344, heterozygous SNPs = 40-fold for both strains; Figure S3) and mapped to repeat regions at a rate significantly higher than homozygous SNPs (60.02% vs. 39.38%, respectively; P < 0.001, chi-square test), suggesting that improperly aligned reads might account for some of heterozygous SNP calls. To confirm heterozygous SNPs in these highly inbred strains, 45 heterozygous SNPs were sequenced with Sanger methodology, and 6 (13.33%) were in fact homozygous SNPs, while 39 (86.87%) were false SNP calls (Table S3). Therefore, heterozygous SNPs were not included in subsequent analyses.
Many of the homozygous SNPs detected had the potential to impact gene function, including 422 SNPs predicted to cause loss or gain of start codons, 231 SNPs impacting splicing sites, and 140 SNPs causing loss or gain of stop codons. Additionally, 15,477 SNPs were nonsynonymous, mapping to 3174 Refseq genes and 4724 Ensembl genes in the DA genome and to 3074 Refseq genes and 4632 Ensembl genes in the F344 genome (Table S4 and Table S5). A total of 4.3 million SNPs (88.4% of all homozygous SNPs) were intergenic, intronic, or synonymous.
Indels
We detected indels using SOAP2 (Li et al. 2009b). Indels were defined as alignment gaps of up to 5 bp, supported by three or more nonredundant pairs of reads and present in at least one-third of reads for autosomic indels or in all of the reads for X-chromosome indels. In total, we identified 299,532 indels in DA and 305,705 in F344 (Figure 2B, Table 1). Of the indels, 96.8% were homozygous and had a genomic distribution rate of 1.1 × 10−4, and 3.2% were heterozygous and had a genomic distribution rate of 3.8 × 10−6. Insertions or deletions of 1 bp accounted for 67.76% of the indels (Figure S4). Sanger sequencing confirmed 100% of 77 homozygous indels tested (Table S6).
While DA and F344 shared 146,502 homozygous indels, 292,679 were polymorphic between these two strains with 143,058 indels unique to DA and 149,621 unique to F344 (Figure 2B). Most indels were intronic or intergenic (Figure S5), but 605 homozygous indels were predicted to cause codon insertions/deletions or frameshift in coding genes. Of these, 204 transcript-affecting indels were unique to DA, 155 were unique to F344, and 246 were found in both DA and F344 (Table 2).
Table 2. Genetic variation annotation of DA and F344.
| SNPsa |
Indels |
Structural variant |
||||||
|---|---|---|---|---|---|---|---|---|
| DA | F344 | Shared | DA | F344 | Shared | DA | F344 | |
| Intergenic region | 658,646 | 660,019 | 1,150,467 | 85,753 | 91,326 | 97,035 | — | — |
| Intron | 485,087 | 489,738 | 837,376 | 70,958 | 73,231 | 75,716 | 17516 | 19607 |
| Downstream (up to 5 kb) | 71,596 | 71,646 | 123,030 | 10,565 | 10,620 | 10,722 | — | — |
| Upstream (up to 5 kb) | 70,831 | 71,107 | 119,219 | 10,109 | 10,413 | 9,853 | — | — |
| 3′ UTR | 4,094 | 3,766 | 6,955 | 685 | 723 | 811 | 291 | 305 |
| 5′ UTR | 752 | 647 | 1,061 | 48 | 37 | 47 | 226 | 239 |
| Start gained in 5′ UTR | 128 | 122 | 157 | — | — | — | — | — |
| Coding sequences and splice sites | 2572 | 2829 | ||||||
| Synonymous coding | 7,146 | 6,981 | 12,104 | — | — | — | — | — |
| Nonsynonymous coding | 4,230 | 4,060 | 7,187 | — | — | — | — | — |
| Frameshift | — | — | — | 156 | 135 | 217 | — | — |
| Start lost | 1 | 6 | 8 | — | — | — | — | — |
| Stop gained | 35 | 35 | 59 | — | — | — | — | — |
| Stop lost | 4 | 1 | 6 | — | — | — | — | — |
| Splice-site acceptor | 30 | 31 | 59 | 35 | 21 | 58 | — | — |
| Splice-site donor | 26 | 27 | 58 | 32 | 16 | 62 | — | — |
| Synonymous stop | 3 | 5 | 10 | — | — | — | — | — |
| Nonsynonymous start | 1 | 0 | 2 | — | — | — | — | — |
| Codon deletion | — | — | — | 19 | 10 | 14 | — | — |
| Codon change + codon deletion | — | — | — | 10 | 2 | 6 | — | — |
| Codon insertion | — | — | — | 12 | 4 | 5 | — | — |
| Codon change plus codon insertion | — | — | — | 7 | 4 | 4 | — | — |
| Within noncoding gene | ||||||||
| Nonsynonymous coding | 434 | 428 | 866 | — | — | — | — | — |
| Synonymous coding | 232 | 228 | 395 | — | — | — | — | — |
| Stop gained | 24 | 14 | 27 | — | — | — | — | — |
| Stop lost | 6 | 15 | 18 | — | — | — | — | — |
| Synonymous stop | 3 | 0 | 6 | — | — | — | — | — |
| Start lost | 3 | 1 | 1 | — | — | — | — | — |
| Synonymous start | 1 | 0 | 0 | — | — | — | — | — |
| Codon change + codon deletion | — | — | — | 3 | 0 | 2 | — | — |
| Codon deletion | — | — | — | 2 | 1 | 2 | — | — |
| Codon insertion | — | — | — | 1 | 0 | 2 | — | — |
| Codon change + codon insertion | — | — | — | 0 | 1 | 0 | — | — |
Calculated using SNPEff v.1.9.5 (Cingolani et al. 2012) and Ensembl’s R. norvegicus build 3.4.64.
Homozygous SNPs.
The frequencies of homozygous SNPs along the DA and F344 genomes varied from 0 to 3 ± 10−3 and strongly correlated with that of homozygous indels (Figure 1, Figure S6), suggesting a progressive increase in variation density from shared haplotypes.
Structural variation
We used paired-end alignment to identify structural variation. Regions containing structural variants were detected when read pairs aligned to the reference genome abnormally—differing in orientation and/or inferred insert size with the support of at least three read pairs. We identified a total of 58,174 structural variants: 12,151 unique to DA, 17,575 unique to F344, and 30,978 present in both DA and F344 (Figure 2C, Figure S7, and Figure S8). Deletions and insertions >5 bp were the most frequently detected class of structural variants, followed by tandem duplication, dispersed duplication, and combined insertion–deletion. Structural variants overlapping coding sequences have a high potential to disrupt the function of those genes. In total, 2572 structural variants in the DA and 2829 in the F344 genomes overlapped coding sequences of Ensembl genes (Table 2). And 1398 structural variants in the DA and 1502 in the F344 genomes overlapped coding sequences of RefSeq genes (Table S7).
Based on the mean depth of 100-bp sliding windows along each chromosome, we detected 7199 candidate regions of copy-number variation: 2594 unique to DA, 3691 unique to F344, and 994 in both DA and F344 (Figure 1D, Table S8). Seventy-seven percent of copy-number variant candidates were in the 10- to 20-kb range (Figure 2D).
Sensitivity and specificity
To evaluate the accuracy of read mapping, we generated a variation of the BN genome containing SNPs, indel, and structural variants with frequencies similar to those observed in the DA and F344 sequencing data. We also simulated short reads of different lengths containing mismatch sequencing errors and quality scores similar to those in the DA and F344 sequences. We then aligned the simulated reads back to the BN reference assembly to quantify the precision of alignment for the detection of variants.
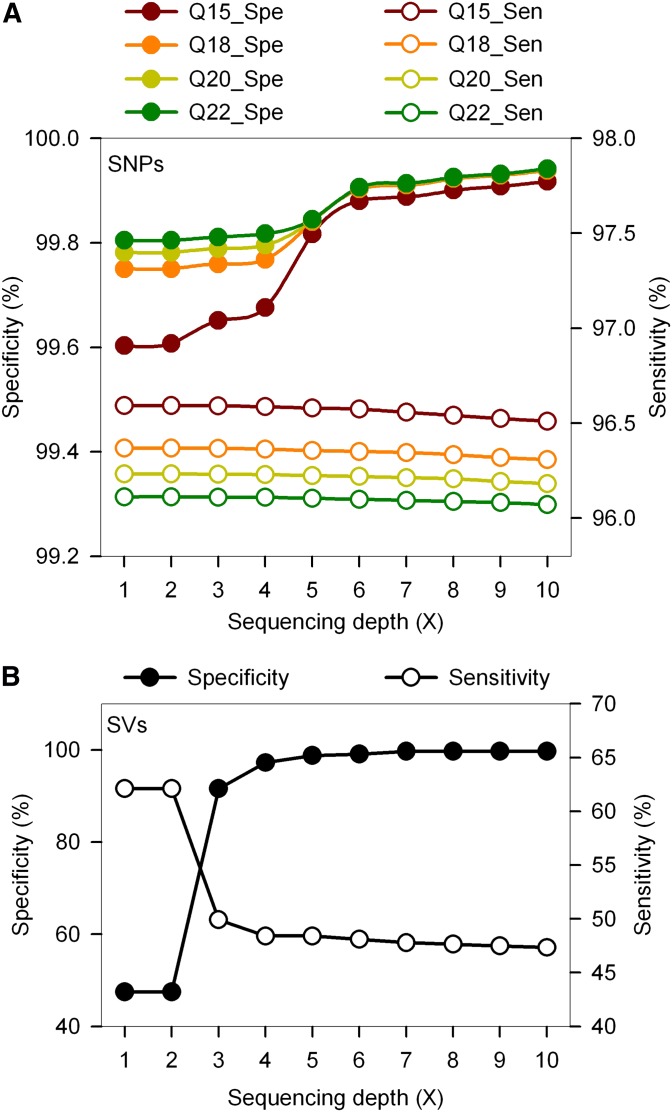
For an average 35-fold coverage with simulated reads, sensitivity for SNP detection was inversely proportional to read-quality threshold and varied from slightly over 96% for reads with Q = 22, to 96.6% and for reads with Q = 15. Specificity for SNP detection was more dependent on sequencing coverage, and it increased from 99.78% with a depth of 1-fold to 99.82% with a depth of 5-fold to 99.94% with a depth of 10-fold (Figure 3A). Sensitivity and specificity for indel detection were similar to those of the simulated SNPs (data not shown).
Figure 3.
Accuracy of variant detection. We produced a copy of the BN rat genome with a read coverage of 35-fold and aligned the simulated reads back onto the RGSC3.4 genome scaffold to measure the rate of misplacement. Simulated reads contained simulated mismatch sequencing errors, SNPs, indels, and structural variants to the RGSC3.4 reference at rates similar to those detected in the DA and F344 genomes. (A) SNP detection sensitivity (open circles) was inversely proportional to the read quality threshold. The SNP detection specificity (solid circles) was more dependent on the number of supporting reads. (B) The detection sensitivity for structural variants (open circles) was inversely proportional to the number of supporting reads. The detection specificity for structural variants (solid circles) increased with the number of supporting reads and remained >99% with six or more reads.
Specificity for the detection of structural variants increased sharply with the number of supporting reads from 47% (1–2 reads) to 91% (3 reads) and continued increasing at a lower rate to plateau at 99.68% with seven or more reads (Figure 3B). Sensitivity for the detection of structural variants was inversely correlated with the number of supporting reads, sharply declining from 62.1% (1 read) to 49.92% (3 reads) and then to 47.34% (10 reads).
Construction of the DA and F344 genome drafts
To generate the DA and F344 genome drafts, we created a new strategy for de novo genome assembly using NGS data: the reference-aided assembly method (Figure 4). Briefly, semifinished genomes were generated for each strain by aligning their reads to the BN genome using SOAPaligner (Li et al. 2009a) to form a consensus sequence, followed by assembly of reads to bridge gaps in the BN genome using GapCloser (Li et al. 2010). In parallel, contigs and scaffolds were independently assembled for each strain using SOAPdenovo (Li et al. 2010), followed by closure of gaps between scaffolds using GapCloser. Finally, sequences from both ends of each scaffold were mapped onto each coordinated semifinished genome using BLAST to anchor the scaffolds and obtain the DA and F344 genome drafts.
Figure 4.
Construction of the DA and F344 genome drafts using the Reference-Aided Assembly Method. The strategy to construct the DA and F344 genome drafts from NGS data consisted of (1) generating a coordinated, semifinished genome, (2) producing a de novo assembly, and (3) anchoring the de novo assembly onto the semifinished genome. Each semifinished genome was created by alignment of reads onto the BN reference genome, inference of a consensus sequence, and closure of gaps (left arm). In parallel, reads were assembled independently into scaffolds, followed by closure of gaps and extraction of tag sequences (right arm). Tag sequences were then mapped onto the semifinished genome using BLAST, anchoring the affiliated scaffolds to finalize each genome draft (bottom of diagram).
The DA and F344 genome drafts include 2,616,053,766 and 2,615,410,193 effective bases and are 1.94% and 1.91% larger than the BN genome, respectively. The DA and F344 genome drafts also contain 49.76 and 49.11 million novel base pairs bridging 391,057 and 401,069 gaps of the BN genome. Of the novel base pairs, 20.47 million (41.13%) and 19.35 (39.41%) million base pairs are in novel scaffolds. And 2.55% and 2.42% more reads could be mapped to each coordinated draft compared with the consensus sequences (Table 3).
Table 3. Construction of the DA and F344 genome drafts.
| Genome size (bp) |
Novel scaffolds (bp) |
Reads mapped |
|||||
|---|---|---|---|---|---|---|---|
| Total | Effectivea | Gapsb | Total | Effectivea | DA (%) | F344 (%) | |
| BN | 2,834,127,293 | 2,566,294,765 | 876,652 | — | — | 91.49 | 91.84 |
| DA | 2,798,712,224 | 2,616,053,766 | 485,595 | 20,558,331 | 20,465,987 | 94.03 | — |
| F344 | 2,793,938,348 | 2,615,410,193 | 475,583 | 19,441,505 | 19,355,322 | — | 94.27 |
Genome length without Ns.
Number of gaps in each genome draft.
We evaluated the quality of DA and F344 genome drafts using two methods. First, we retrieved all 194,363 ESTs available in the rat genome and aligned them to the assembled drafts using BLAST to cover at least 95% of each EST. Of the ESTs, 97.97% aligned to each de novo assembly, and 836 (0.43%) and 1088 (0.56%) ESTs aligned exclusively to novel scaffolds in DA and F344, respectively (Table S9). Second, we estimated the single-base error rates for de novo assemblies by comparing the draft genome sequences to corresponding positions containing homozygous SNPs in the semifinished genome of each strain. The estimated single-base error for these two newly assembled drafts was 3.06 × 10−5 for DA and 2.99 × 10−5 for F344 (Table S10).
Discussion
DA and the F344 rats have unique dichotomous phenotypes that have been used to better understand development, human physiology, and disease. DA rats are highly susceptible to autoimmunity, including models of rheumatoid arthritis, multiple sclerosis, and uveitis (Dahlman et al. 1998; Sun et al. 1999; Wilder et al. 1999). DA rats are also susceptible to bladder and tongue carcinomas (Kitano et al. 1992), have reduced variation in circadian corticosteroid production (Brodkin et al. 1999), and are more easily addicted to morphine (Brodkin et al. 1999). F344 rats, on the other hand, are typically resistant to the above conditions, but are susceptible to chemically induced hepatocarcinoma and lymphoma (Lu et al. 1999; De Miglio et al. 2006) and have decreased bone mineral density (Turner et al. 2001). Genetic variation between these strains accounts for most of such strain-specific phenotypes. Therefore, sequencing the DA and F344 genomes constitutes a major step toward identifying the genetic causes and pathogenic processes underlying these traits and models of human diseases such as rheumatoid arthritis, multiple sclerosis, and cancer (Table S11) and should facilitate the development of novel disease treatments and biomarkers, as well as new pathways to be tested for disease prevention.
The sequencing of the DA and F344 genomes identified a large number of variants between each of these two strains and BN. The 5.6 million SNPs identified in the DA and F344 genomes increased the total number of known SNPs between these two strains and BN by 150-fold from 19,326 (Saar et al. 2008) to 2.2 million SNPs between DA and F344 and 2.9 million SNPs between BN and each of the other two strains. Furthermore, 1.37 million SNPs and 0.44 million indels were novel. The addition of these novel variants significantly expands known variation in the rat genome.
A large number of variants were predicted to significantly disrupt gene structure. High-impact variants included deletion of coding sequences, loss of start codons, premature stops, frameshifts, codon insertions/deletions, nonsynonymous SNPs, and changes at splicing sites. In addition to these effects on gene structure, other variants can potentially alter gene expression. Upstream and 5′-UTR variants can disrupt epigenetic regulation and transcription factor-binding sites, 3′-UTR variants can modify messenger RNA stability (Boffa et al. 2008), intronic SNPs can influence expression breadth (Park et al. 2012), and synonymous SNPs can affect translation efficiency (Plotkin and Kudla 2011). The DA and F344 genome sequencing provides a detailed framework for future studies aimed at characterizing how these variants alter gene function.
At 32- and 34-fold redundancy, the DA and F344 genomes were assembled at a sequencing depth almost five times that of the BN genome (Gibbs et al. 2004) and three times that of the SHR genome (Atanur et al. 2010). The DA and F344 genome assemblies also used stringent quality criteria to define variants. The combination of high-quality and high-sequencing depth resulted in increased accuracy to detect SNPs and indels, as was confirmed with Sanger sequencing and in silico simulations. The frequency of SNPs was 10-fold higher than that of indels, revealing a SNP/indel ratio similar to that of other resequencing projects (Ahn et al. 2009; Atanur et al. 2010). SNPs in mitochondrial DNA were 8.9-fold more frequent than in nuclear DNA in agreement with its 9–25 times higher mutation rate (Lynch et al. 2006).
A small percentage of the SNPs (5%) and indels (3%) were detected as heterozygous, and Sanger-based resequencing showed that a fraction were in fact homozygous SNPs, while the majority were false calls. Misalignment of reads mapping to repeats or highly homologous segmental duplications and sequencing errors may have contributed to false detection of heterozygous SNPs in the DA and F344 genomes. Eventual residual heterozygosity cannot be entirely excluded; it might result from selection against recessive alleles that are embryonically lethal or are associated with infertility or unproductive breeding behavior (Bailey 1977; Saar et al. 2008).
The importance of copy-number and copy-neutral structural variants in the genome has only recently begun to be understood (Korbel et al. 2007). Structural variation accounts for an even higher proportion of the genetic diversity between individuals than SNPs (Li et al. 2011) and has been associated with disease in both rats and humans (Aitman et al. 2006). Copy number variants can also correlate with levels of gene expression in rats (Guryev et al. 2008; Charchar et al. 2010) and have been estimated to account for 20% of expression differences in humans (Stranger et al. 2007). We identified variants in the DA and F344 genomes that caused duplications, deletions, or potential disruptions of the structure of >2500 genes. This frequency of potentially gene-disrupting structural/copy-number variants has also been seen in other interstrain comparisons such as that described between DBA and B6 mice (Quinlan et al. 2010). Insert size of libraries can be a limiting factor for the identification of insertion events in NGS (Pang et al. 2010). And in fact, using the simulated reads we estimated that our method of detecting structural variants had a sensitivity of 45–50%. Therefore, DA and F344 structural variants are most likely underrepresented.
DA and F344 rats shared alleles for 60% of the SNPs, 50% of the indels, and 70% of the structural variants, an indication of the phylogenetic proximity between these two strains. The high levels of allele sharing between DA and F344 are in agreement with a previous observation that BN was the most divergent of 167 commonly used laboratory inbred strains, including DA and F344 (Saar et al. 2008).
We devised and employed a new strategy to generate the first de novo assembly of a rat genome using NGS technology data. As a result, the DA and F344 genome drafts are more extensive and more complete than the BN genome and should facilitate the study of discrepancies with genetic maps (Saar et al. 2008) and areas of sequencing collapse (Guryev et al. 2008). The new DA and F344 genome drafts contain 49 million base pairs of novel sequence each, nearly half the number of gaps present in the BN genome, and ∼1000 ESTs uniquely mapped to novel scaffolds of each strain.
The BN and SHR are the only rat nuclear genomes drafted to date. As additional rat genomes become available, investigators will be able to construct detailed haplotype maps, a key resource for both targeted and genome-wide studies in the rat. Sequencing additional genomes will help resolve regions of poor coverage in the BN and other rat genomes, as well as alignment and sequencing errors and undetected duplications.
Over 615 inbred rat strains and substrains are presently registered at the Rat Genome Database. These strains are an important resource for gene identification and studies of gene function and are currently being used by several laboratories worldwide. The SNPs, indels, and structural variants reported here compose a large collection of new informative markers that can be used to increase the precision of genetic mapping and genotype-guided breeding, as well as for studies in advanced intercross lines and for genome-wide association studies using heterogeneous stocks. Indeed, with an average density of one SNP per 0.86 kb, SNPs identified in this study will facilitate mapping at a resolution 100-fold higher than with previously available SNPs (Saar et al. 2008).
Supplementary Material
Acknowledgments
Funded by the National Institutes of Health grants R01-AR46213, R01-AR052439 (NIAMS) and R01-AI54348 (NIAID) to Dr. P. Gulko, and by The Shenzhen Municipal Government of China (grants JC201005260191A and CXB201108250096A) and from the National Gene Bank Project of China to Dr. Wang Jun.
Footnotes
Communicating editor: T. R. Magnuson
Literature Cited
- Ahn S. M., Kim T. H., Lee S., Kim D., Ghang H., et al. , 2009. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 19: 1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman T. J., Dong R., Vyse T. J., Norsworthy P. J., Johnson M. D., et al. , 2006. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851–855 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Atanur S. S., Birol I., Guryev V., Hirst M., Hummel O., et al. , 2010. The genome sequence of the spontaneously hypertensive rat: analysis and functional significance. Genome Res. 20: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D. W., 1977. Genetic drift: the problem and its possible solution by frozen-embryo storage. Ciba Found. Symp. 52: 291–303 [DOI] [PubMed] [Google Scholar]
- Bentley D. R., Balasubramanian S., Swerdlow H. P., Smith G. P., Milton J., et al. , 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiadecki B. J., Brand P. H., Metting P. J., Koch L. G., Britton S. L., 1998. Phenotypic variation in strength among eleven inbred strains of rats. Proc. Soc. Exp. Biol. Med. 219: 126–131 [DOI] [PubMed] [Google Scholar]
- Boffa M. B., Maret D., Hamill J. D., Bastajian N., Crainich P., et al. , 2008. Effect of single nucleotide polymorphisms on expression of the gene encoding thrombin-activatable fibrinolysis inhibitor: a functional analysis. Blood 111: 183–189 [DOI] [PubMed] [Google Scholar]
- Brenner M., Meng H. C., Yarlett N. C., Joe B., Griffiths M. M., et al. , 2005. The non-MHC quantitative trait locus Cia5 contains three major arthritis genes that differentially regulate disease severity, pannus formation, and joint damage in collagen- and pristane-induced arthritis. J. Immunol. 174: 7894–7903 [DOI] [PubMed] [Google Scholar]
- Brenner M., Laragione T., Yarlett N. C., Gulko P. S., 2007. Genetic regulation of T regulatory, CD4, and CD8 cell numbers by the arthritis severity loci Cia5a, Cia5d, and the MHC/Cia1 in the rat. Mol. Med. 13: 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M., Laragione T., Shah A., Mello A., Remmers E. F., et al. , 2011. Identification of two new arthritis severity loci that regulate levels of autoantibodies, IL-1beta and joint damage. Arthritis Rheum. 64: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodkin E. S., Kosten T. A., Haile C. N., Heninger G. R., Carlezon W. A., Jr, et al. , 1999. Dark Agouti and Fischer 344 rats: differential behavioral responses to morphine and biochemical differences in the ventral tegmental area. Neuroscience 88: 1307–1315 [DOI] [PubMed] [Google Scholar]
- Charchar F. J., Kaiser M., Bingham A. J., Fotinatos N., Ahmady F., et al. , 2010. Whole genome survey of copy number variation in the spontaneously hypertensive rat: relationship to quantitative trait loci, gene expression, and blood pressure. Hypertension 55: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. A., Liu H. F., Chao K. M., 2008. CNVDetector: locating copy number variations using array CGH data. Bioinformatics 24: 2773–2775 [DOI] [PubMed] [Google Scholar]
- Cingolani P., Patel V. M., Coon M., Nguyen T., Land S. J., et al. , 2012. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman I., Lorentzen J. C., de Graaf K. L., Stefferl A., Linington C., et al. , 1998. Quantitative trait loci disposing for both experimental arthritis and encephalomyelitis in the DA rat: impact on severity of myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis and antibody isotype pattern. Eur. J. Immunol. 28: 2188–2196 [DOI] [PubMed] [Google Scholar]
- De Miglio M. R., Virdis P., Calvisi D. F., Frau M., Muroni M. R., et al. , 2006. Mapping a sex hormone-sensitive gene determining female resistance to liver carcinogenesis in a congenic F344.BN-Hcs4 rat. Cancer Res. 66: 10384–10390 [DOI] [PubMed] [Google Scholar]
- Fujimoto A., Nakagawa H., Hosono N., Nakano K., Abe T., et al. , 2010. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat. Genet. 42: 931–936 [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Weinstock G. M., Metzker M. L., Muzny D. M., Sodergren E. J., et al. , 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521 [DOI] [PubMed] [Google Scholar]
- Guryev V., Saar K., Adamovic T., Verheul M., van Heesch S. A., et al. , 2008. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 40: 538–545 [DOI] [PubMed] [Google Scholar]
- Ishih A., 1994. Worm burden and mucosal mast cell response in DA and F344/N rat strains infected with Hymenolepis diminuta. Int. J. Parasitol. 24: 295–298 [DOI] [PubMed] [Google Scholar]
- Jacob H. J., 1999. Functional genomics and rat models. Genome Res. 9: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. I., Ju Y. S., Park H., Kim S., Lee S., et al. , 2009. A highly annotated whole-genome sequence of a Korean individual. Nature 460: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M., Hatano H., Shisa H., 1992. Strain difference of susceptibility to 4-nitroquinoline 1-oxide-induced tongue carcinoma in rats. Jpn. J. Cancer Res. 83: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O., Urban A. E., Affourtit J. P., Godwin B., Grubert F., et al. , 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laragione T., Yarlett N. C., Brenner M., Mello A., Sherry B., et al. , 2007. The arthritis severity quantitative trait loci Cia4 and Cia6 regulate neutrophil migration into inflammatory sites and levels of TNF-alpha and nitric oxide. J. Immunol. 178: 2344–2351 [DOI] [PubMed] [Google Scholar]
- Laragione T., Brenner M., Mello A., Symons M., Gulko P. S., 2008. The arthritis severity locus Cia5d is a novel genetic regulator of the invasive properties of synovial fibroblasts. Arthritis Rheum. 58: 2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ma L., Song C., Yang Z., Wang X., et al. , 2009. The YH database: the first Asian diploid genome database. Nucleic Acids Res. 37: D1025–D1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J., 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24: 713–714 [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Fang X., Yang H., Wang J., et al. , 2009a SNP detection for massively parallel whole-genome resequencing. Genome Res. 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T. W., Yiu S. M., et al. , 2009b SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li R., Zhu H., Ruan J., Qian W., Fang X., et al. , 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng H., Luo R., Wu H., Zhu H., et al. , 2011. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat. Biotechnol. 29: 723–730 [DOI] [PubMed] [Google Scholar]
- Lu L. M., Shisa H., Tanuma J., Hiai H., 1999. Propylnitrosourea-induced T-lymphomas in LEXF RI strains of rats: genetic analysis. Br. J. Cancer 80: 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Koskella B., Schaack S., 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311: 1727–1730 [DOI] [PubMed] [Google Scholar]
- Pang A. W., MacDonald J. R., Pinto D., Wei J., Rafiq M. A., et al. , 2010. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Xu K., Park T., Yi S. V., 2012. What are the determinants of gene expression levels and breadths in the human genome? Hum. Mol. Genet. 21: 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J. B., Kudla G., 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza M. N., Brodkin E. S., Joe B., Luo X., Remmers E. F., et al. , 2004. Genomic regions controlling corticosterone levels in rats. Biol. Psychiatry 55: 634–641 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Clark R. A., Sokolova S., Leibowitz M. L., Zhang Y., et al. , 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 20: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K., Beck A., Bihoreau M. T., Birney E., Brocklebank D., et al. , 2008. SNP and haplotype mapping for genetic analysis in the rat. Nat. Genet. 40: 560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E., Forrest M. S., Dunning M., Ingle C. E., Beazley C., et al. , 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, W. M., 2001 Preparation of genomic DNA from mammalian tissue. Curr. Protoc. Mol. Biol. Chapter 2: Unit 2.2. DOI: 10.1002/0471142727.mb0202s42. [DOI] [PubMed]
- Sun S. H., Silver P. B., Caspi R. R., Du Y., Chan C. C., et al. , 1999. Identification of genomic regions controlling experimental autoimmune uveoretinitis in rats. Int. Immunol. 11: 529–534 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Ishih A., Kino H., Muregi F. W., Takabayashi S., et al. , 2006. Chromosomal mapping of host resistance loci to Trichinella spiralis nematode infection in rats. Immunogenetics 58: 26–30 [DOI] [PubMed] [Google Scholar]
- Terner J. M., Barrett A. C., Lomas L. M., Negus S. S., Picker M. J., 2006. Influence of low doses of naltrexone on morphine antinociception and morphine tolerance in male and female rats of four strains. Pain 122: 90–101 [DOI] [PubMed] [Google Scholar]
- Tong P., Prendergast J. G., Lohan A. J., Farrington S. M., Cronin S., et al. , 2010. Sequencing and analysis of an Irish human genome. Genome Biol. 11: R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff M. G., Alarcon L. K., Lawler M. P., 2008. Preferences of 14 rat strains for 17 taste compounds. Physiol. Behav. 95: 308–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. H., Roeder R. K., Wieczorek A., Foroud T., Liu G., et al. , 2001. Variability in skeletal mass, structure, and biomechanical properties among inbred strains of rats. J. Bone Miner. Res. 16: 1532–1539 [DOI] [PubMed] [Google Scholar]
- van Den Brandt J., Kovacs P., Kloting I., 2000. Metabolic variability among disease-resistant inbred rat strains and in comparison with wild rats (Rattus norvegicus). Clin. Exp. Pharmacol. Physiol. 27: 793–795 [DOI] [PubMed] [Google Scholar]
- van Wijngaarden P., Brereton H. M., Coster D. J., Williams K. A., 2007. Genetic influences on susceptibility to oxygen-induced retinopathy. Invest. Ophthalmol. Vis. Sci. 48: 1761–1766 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Li R., Li Y., Tian G., et al. , 2008. The diploid genome sequence of an Asian individual. Nature 456: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. A., Srinivasan M., Egholm M., Shen Y., Chen L., et al. , 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Wilder R. L., Remmers E. F., Kawahito Y., Gulko P. S., Cannon G. W., et al. , 1999. Genetic factors regulating experimental arthritis in mice and rats. Curr. Dir. Autoimmun. 1: 121–165 [DOI] [PubMed] [Google Scholar]
- Worley K. C., Weinstock G. M., Gibbs R. A., 2008. Rats in the genomic era. Physiol. Genomics 32: 273–282 [DOI] [PubMed] [Google Scholar]
- Worthey E. A., Stoddard A. J., Jacob H. J., 2010. Sequencing of the rat genome and databases. Methods Mol. Biol. 597: 33–53 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Xuan Z., Makarov V., Ye K., Sebat J., 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 19: 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lin X., Koga K., Takahashi K., Linge H. M., et al. , 2011. Strain differences in alveolar neutrophil infiltration and macrophage phenotypes in an acute lung inflammation model. Mol. Med. 17: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.