Abstract
The propagation of all organisms depends on the accurate and orderly segregation of chromosomes in mitosis and meiosis. Budding yeast has long served as an outstanding model organism to identify the components and underlying mechanisms that regulate chromosome segregation. This review focuses on the kinetochore, the macromolecular protein complex that assembles on centromeric chromatin and maintains persistent load-bearing attachments to the dynamic tips of spindle microtubules. The kinetochore also serves as a regulatory hub for the spindle checkpoint, ensuring that cell cycle progression is coupled to the achievement of proper microtubule–kinetochore attachments. Progress in understanding the composition and overall architecture of the kinetochore, as well as its properties in making and regulating microtubule attachments and the spindle checkpoint, is discussed.
Keywords: budding yeast, kinetochore, microtubules, spindle checkpoint, biorientation
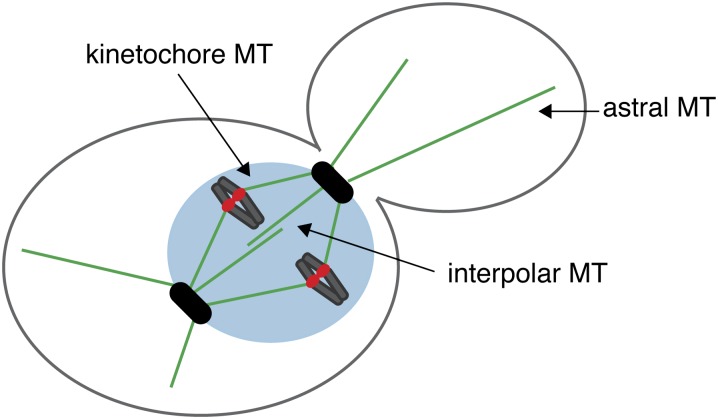
CHROMOSOME segregation is mediated by the interaction between spindle microtubules and kinetochores, the macromolecular structures that assemble at a unique chromosomal locus called the centromere (Westermann et al. 2007). Microtubules are dynamic polymers that grow and shrink by the addition and removal of tubulin dimers from their tips (Mitchison and Kirschner 1984). They switch stochastically between phases of assembly and disassembly, a behavior called dynamic instability (Mitchison and Kirschner 1984). Microtubules are nucleated by the centrosome, which is called the spindle pole body (SPB) in yeast (Winey and Bloom 2012). Microtubules have an inherent polarity with the minus end embedded in the SPB and the dynamic plus end distal. In yeast, microtubule growth and shrinkage appears to occur exclusively at the plus end (Maddox et al. 2000). Because the yeast nuclear envelope does not break down, the SPB is embedded in the nuclear envelope throughout the cell cycle. The SPB nucleates three populations of yeast microtubules that facilitate proper chromosome segregation (Figure 1). In the cytoplasm, astral microtubules position the nucleus throughout the cell cycle. Within the nucleus, kinetochore microtubules attach to the kinetochore at their plus ends, and interpolar microtubules interdigitate to connect the poles and stabilize the spindle during mitosis. The zone of overlap between interpolar microtubules is called the spindle midzone; a number of proteins specifically localize to the midzone to facilitate spindle assembly and disassembly.
Figure 1.
Key structures that mediate chromosome segregation. A cartoon of a budding yeast cell shows three populations of microtubules in green (astral, kinetochore, and interpolar) that emanate from the spindle pole bodies (SPBs). The nucleus is shown in blue with SPBs embedded in its nuclear envelope (black) and the kinetochores on the chromosomes are shown in red.
Stages of chromosome alignment and segregation
Yeast kinetochores are assembled and bind to microtubules for almost the entire cell cycle, with the exception of a brief window during S phase when they disassemble and rapidly reassemble (Kitamura et al. 2007). This may be the time when the replication fork travels through the centromere, although this has not yet been directly tested. Yeast kinetochores thus cluster near the spindle pole for most of the cell cycle (Heath 1980; Jin et al. 2000; Kitamura et al. 2007). This proximity led to the initial identification of many kinetochore components through SPB purifications (Wigge et al. 1998). Each budding yeast kinetochore binds to a single microtubule (Winey et al. 1995), which greatly simplifies studies because a kinetochore is either attached or unattached to a microtubule at any given time. In contrast, most eukaryotic kinetochores have from 3 to 30 microtubule binding sites, which can be partially occupied (Walczak et al. 2010). Replication creates sister chromatids, which become physically linked together by protein complexes called cohesin (Oliveira and Nasmyth 2010). Proper segregation requires sister kinetochores to biorient and attach to microtubules from opposite poles (Tanaka 2010). Once every pair of chromosomes biorients, the linkage between the sister chromatids is destroyed and the spindle physically pulls sister chromatids to opposite poles.
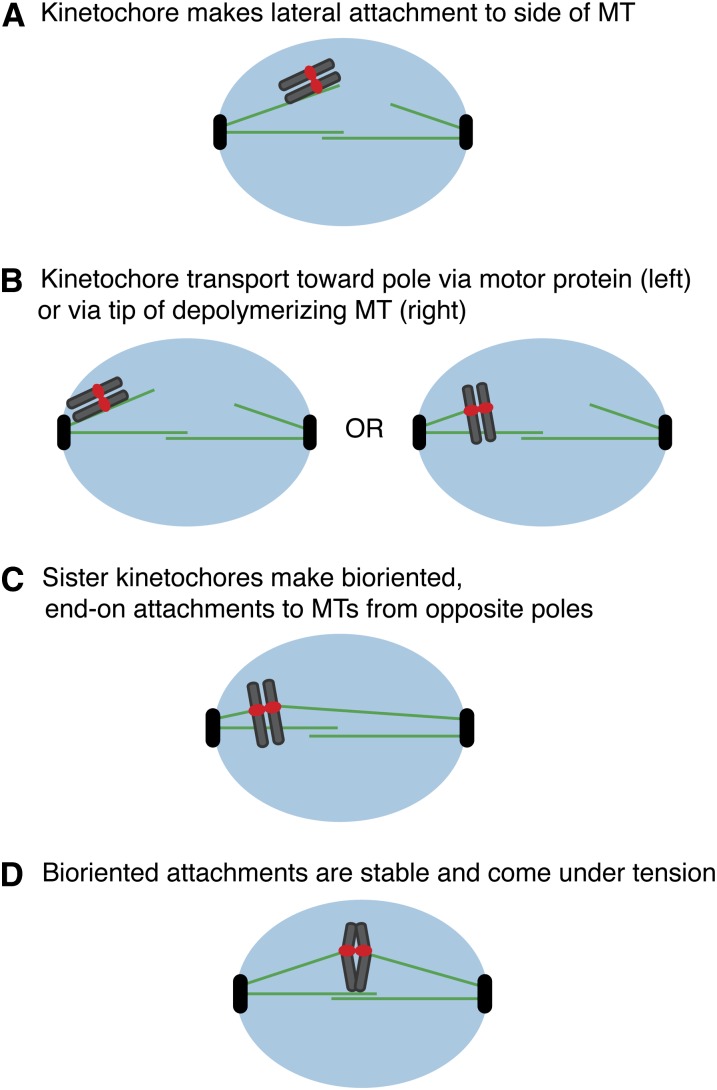
The small size of the yeast nucleus and difficulty in visualizing yeast chromosomes by microscopy makes it difficult to directly examine the steps of chromosome segregation. The assays used to examine the process therefore require cells to be arrested in conditions that may or may not reflect the normal course of events (Tanaka et al. 2005). Regardless, these studies revealed that budding yeast appear to initially make lateral attachments to the sides of microtubules like other eukaryotes (Figure 2A) (Hayden et al. 1990; Merdes and De Mey 1990; Rieder and Alexander 1990; Tanaka et al. 2005). Kinetochores appear to also directly nucleate microtubules, which may facilitate the capture of microtubules emanating from poles in yeast (Kitamura et al. 2010). Laterally attached yeast kinetochores are subsequently transported poleward by motor proteins and regulators where they become attached to the end of microtubules (Figure 2B) (Tanaka et al. 2005). Although motor-driven transport toward the pole is often slower than microtubule disassembly, the kinetochores do not detach from the microtubules. Instead, the kinetochore either establishes an end-on attachment when it meets the microtubule (Figure 2B) or else it promotes rescue of the shrinking microtubule. In this way, the kinetochore ensures that it stays bound until a proper end-on attachment can be achieved. Rescue is mediated by the Stu2 protein (XMAP215/Dis), which binds to tubulin dimers via TOG domains and facilitates microtubule growth (Wang and Huffaker 1997; Al-Bassam et al. 2006; Brouhard et al. 2008). Stu2 also helps kinetochores nucleate microtubules, a feature that appears to help establish lateral attachments through microtubule–microtubule interactions that are eventually converted to plus end attachments at the kinetochores (Kitamura et al. 2010; Tanaka 2010). Once the kinetochores travel back to the pole, the sister kinetochores make bioriented attachments to the tips of microtubules and come under tension due to pulling forces that are opposed by the linkage between the sisters (Figure 2, C and D). The kinetochores then maintain persistent load-bearing attachments to the continually growing and shrinking tips of the microtubules.
Figure 2.
Steps leading to bioriented kinetochore attachments. (A) The kinetochore initially makes a lateral attachment to a microtubule. (B) The kinetochore is transported toward the pole. The transport can either be mediated by motor and regulatory proteins (left), or the microtubule can depolymerize until the kinetochore is attached to the end of the microtubule (right). Note that sometimes the microtubule polymerizes to prevent the kinetochore from detaching if a proper end-on attachment is not made. (C) Once the chromosomes are near the pole, the sister kinetochores attach to microtubules from opposite poles. (D) The sister kinetochores make stable, bioriented attachments that are under tension until anaphase is initiated.
Establishing kinetochore biorientation
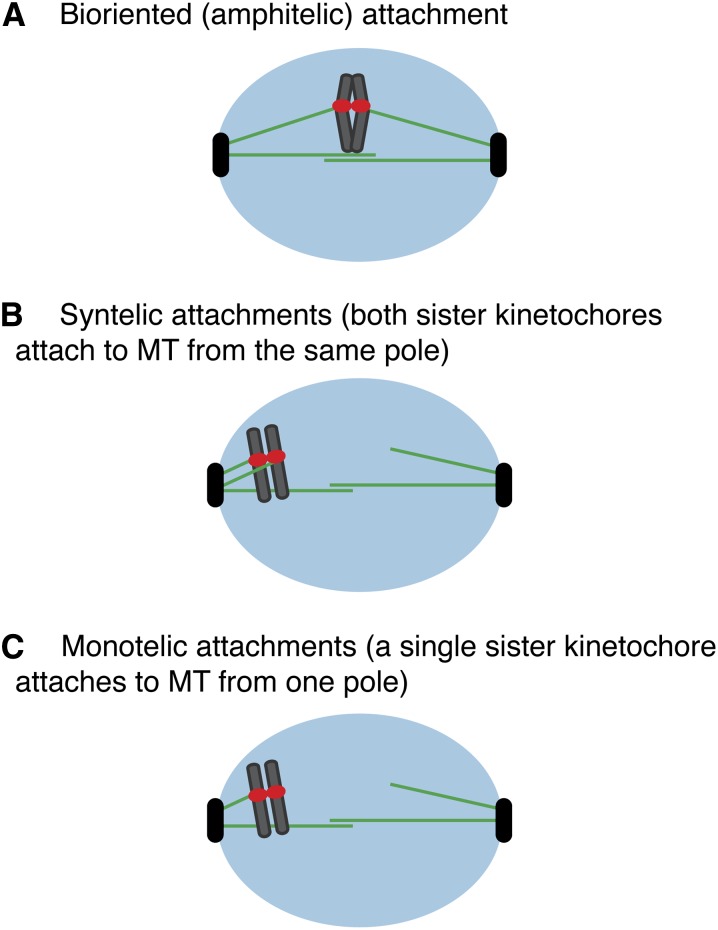
The process of making bioriented kinetochore–microtubule attachments is inherently error prone (Nicklas 1997). Kinetochores can make syntelic attachments where both sisters attach to microtubules from the same pole or monotelic attachments in which one of the two sister kinetochores attaches to a microtubule from one pole (Figure 3). Most eukaryotic kinetochores can also make merotelic attachments where a single kinetochore binds to microtubules from both poles (Cimini 2008), but this is not possible in budding yeast where there is only one microtubule-binding site on each kinetochore (Winey et al. 1995). Because syntelic or monooriented attachments will lead to errors in segregation, the cell has mechanisms to detect and correct inappropriate microtubule attachments. A variety of evidence suggests that the cell monitors the tension generated when sister kinetochores achieve biorientation (Nicklas and Koch 1969; Nicklas 1997). When kinetochores lack tension, the conserved Aurora B protein kinase phosphorylates kinetochore proteins (discussed below, kinetochore biorientation), leading to their release from microtubules so the cell can attempt biorientation again (Biggins et al. 1999; Cheeseman et al. 2002; Tanaka et al. 2002). In addition, tension prolongs the lifetime of kinetochore–microtubule interactions in vitro, suggesting that tension directly stabilizes microtubule attachments (Franck et al. 2007; Akiyoshi et al. 2010). Elegant computer modeling supports the role of tension in stabilizing attachments (Gardner et al. 2005).
Figure 3.
Types of kinetochore–microtubule attachments. (A) Bioriented (amphitelic) attachments occur when sister kinetochores bind to microtubules from opposite poles. (B) Syntelic attachments occur when both sister kinetochores attach to microtubules from the same pole. (C) Monotelic attachments occur when a single sister kinetochore binds to a microtubule from one pole.
Once all kinetochores biorient, the cohesin between sisters is cleaved, allowing the chromosomes to be separated and moved to the poles at anaphase. If even a single pair of chromosomes lacks tension or attachment, a signal transduction system called the spindle checkpoint prevents anaphase (Zich and Hardwick 2010; Murray 2011; Musacchio 2011). To date, it is still controversial whether there is a single upstream signal that triggers the checkpoint or whether tension and attachment are separately monitored (discussed below, The Spindle Checkpoint).
Assays to study yeast chromosome segregation
Cytological assays
Historically, one of the greatest difficulties in studying yeast chromosome segregation has been that the 16 budding yeast chromosomes cannot be distinguished by classical cytological techniques. Instead, they appear as a single amorphous nuclear mass that splits into two at anaphase when stained with dyes. This makes it impossible to monitor the fate of sister chromatids at anaphase and to determine whether individual chromosomes are attached or unattached to microtubules. One of the biggest technical advances was the development of a system to fluorescently mark individual chromosomes with a GFP tag in vivo (Straight et al. 1996; Michaelis et al. 1997). These systems exploit the ability to integrate tandem arrays of lactose or tetracycline operators from bacteria into the yeast genome into strains-containing GFP fusions to the lactose or tetracycline repressors, respectively. The GFP fusions bind to the operators and fluorescently mark the chromosomal locus, and the operators can be easily moved to any genomic position using homologous recombination. This technique revealed that yeast chromosome arms are held in close proximity until anaphase (Straight et al. 1996; Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000; Pearson et al. 2001). However, centromeres transiently separate and reassociate prior to anaphase, and this splitting can be detected with probes integrated up to 38 kb from the centromere (Goshima and Yanagida 2000; He et al. 2000; Pearson et al. 2001). Because the splitting depends on microtubules and its frequency increases as the probe is moved toward the centromere, it is presumably generated by microtubule pulling forces on bioriented kinetochores. Fluorescently marking a single centromere is therefore a powerful technique to monitor the kinetics of biorientation and separation to the poles.
GFP fusions to kinetochore proteins have been another major advance in assaying kinetochore function. Although this technique means that all kinetochores are marked rather than individual sisters, it is informative because yeast kinetochores cluster. A tagged kinetochore protein exhibits a single fluorescent focus prior to biorientation that splits into two foci upon biorientation (Goshima and Yanagida 2000; He et al. 2000; Pearson et al. 2001). When anaphase ensues, the foci move to opposite poles as the spindle elongates. However, if kinetochore function is disrupted, the GFP foci often have a reduced intensity due to a decreased association with the centromere, and the kinetochores often decluster because they detach from microtubules (Pinsky et al. 2006). In this case, it is obvious that the kinetochores no longer colocalize with microtubules. The disadvantage to this assay is that it is currently impossible to know whether a GFP focus represents one or more kinetochores, so the fate of a pair of sister chromatids cannot be monitored.
Genetic and genomic assays
There are also a number of genetic and genomic assays for kinetochore function. Chromatin immunoprecipitation (ChIP) assays and ChIP-sequencing techniques clearly determine if a protein is associated with the centromere, an issue that was difficult to confirm in the past (Meluh and Koshland 1995; Lefrancois et al. 2009; Krassovsky et al. 2012). A fruitful genetic assay exploits the ability to monitor the segregation of a nonessential ectopic chromosome containing a centromere by colony color (Koshland and Hieter 1987; Shero et al. 1991). This sectoring assay has been used in numerous screens to identify segregation genes and to quantify chromosome loss rates in mutant strains (Spencer et al. 1990; Doheny et al. 1993; Warren et al. 2002). Another useful assay is a conditional dicentric assay where a second centromere is integrated into the chromosome (Hill and Bloom 1987). Although dicentric chromosomes are normally unstable and lost during cell division, the galactose promoter controls the second centromere in this assay and transcription through the centromere abolishes its function. When the cells are shifted into glucose, transcription through the centromere is halted allowing a second kinetochore to form, which can subsequently be assayed for the de novo assembly of kinetochore proteins and other kinetochore functions (Tanaka et al. 1999; Mythreye and Bloom 2003; Collins et al. 2005). One-hybrid assays can also identify kinetochore proteins (Ortiz et al. 1999).
Biochemical, structural, and biophysical assays
While studies in vivo have been essential for the identification of kinetochore components and functions, dissecting the underlying mechanism of chromosome movement depends on experiments in vitro that allow individual events to be monitored and manipulated (Akiyoshi and Biggins 2012; Umbreit and Davis 2012). A number of biochemical and biophysical assays for kinetochore function have therefore been developed. Gel shift assays using centromeric DNA originally identified the inner centromere binding proteins (Lechner and Carbon 1991). “Minimal” kinetochores containing centromeric DNA and some inner kinetochore proteins have helped to dissect functions (Kingsbury and Koshland 1991; Sorger et al. 1994; Biggins et al. 1999; Sandall et al. 2006), and large kinetochore particles were recently isolated (Akiyoshi et al. 2010). In the past decade, the development of biophysical assays to analyze the functions of both individual subcomplexes and larger kinetochore assemblies has led to major mechanistic insights (Gestaut et al. 2010). The use of total internal reflection microscopy (TIRF) allows complexes to be visualized at the single particle level in the presence or absence of microtubules. Optical trapping is powerful because tension can be applied to linkages between complexes and microtubules, mimicking the forces that kinetochores sustain in vivo (Asbury et al. 2006; Grishchuk et al. 2008a; Franck et al. 2010). Finally, structural biology has played a key role in elucidating the organization and architecture of many kinetochore assemblies, including the two major microtubule binding complexes in the yeast kinetochore (Miranda et al. 2005; Wei et al. 2005, 2006, 2007; Westermann et al. 2005, 2006; Wang et al. 2007, 2008; Maskell et al. 2010; Hornung et al. 2011).
The Centromere
Centromere structure
The budding yeast centromere was first identified by its ability to confer mitotic and meiotic stability to a plasmid (Clarke and Carbon 1980). In contrast to most eukaryotic centromeres that span megabases of DNA (Burrack and Berman 2012), the functional yeast centromere is defined by a ∼200-bp nuclease resistant region containing a ∼125-bp “point” centromere, with regularly spaced nucleosomes positioned on either side (Bloom and Carbon 1982; Fitzgerald-Hayes et al. 1982; Clarke and Carbon 1985). There are three conserved centromere-determining elements (CDE): an 8-bp palindrome called CDEI, a 78- to 86-bp stretch of AT-rich (>90%) DNA called CDEII, and a conserved 26-bp element called CDEIII (Figure 4) (Clarke 1998). Although most eukaryotic centromeres are maintained epigenetically (Black et al. 2010; Henikoff and Furuyama 2010), yeast centromeres are genetically specified by DNA sequence. The CDEI consensus sequence (PuTCACPuTG) binds to the helix-loop-helix protein Cbf1 (Cai and Davis 1989; Baker and Masison 1990; Cai and Davis 1990), a transcription factor that also binds to other elements throughout the genome. CDE1 and Cbf1 contribute to kinetochore function but are not essential. The CDEIII consensus (TGTTT(T/A)TGNTTTCCGAAANNNAAAAA) binds to the CBF3 complex via a conserved CCG motif that is essential for centromere function (Jehn et al. 1991; Lechner and Carbon 1991). The small size and sequence specificity of the budding yeast centromere has made yeast a powerful organism for its study because the sequences can be easily mutated to identify the important functional regions. It also facilitates techniques such as ChIP, which cannot be easily performed on the highly repetitive centromeres in other organisms. In addition, the centromere can be moved to other genomic regions, allowing the construction of artificial chromosomes and plasmids as well as tools such as conditional centromeres (Murray and Szostak 1983; Hill and Bloom 1989).
Figure 4.
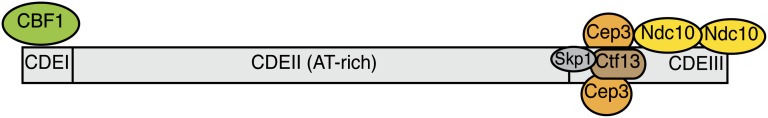
Schematic of the yeast centromere. The conserved structure is ∼120 bp and contains three elements, CDEI, CDEII, and CDEIII. CDE1 is 8–10 bp and binds to the Cbf1 protein. CDEIII is 26 bp and binds to the CBF3 complex that consists of Ndc10, Cep3, Ctf13, and Skp1. The CDEII element is AT rich and wraps around the centromeric nucleosome.
Like other eukaryotes, the budding yeast centromere replicates early in S phase (McCarroll and Fangman 1988). The early replication is due to the presence of the centromere, but it is not yet known what aspect of the centromere or kinetochore dictates early origin activity (Pohl et al. 2012). While it is not yet clear whether early centromere replication is important for subsequent kinetochore function, one possibility is that it ensures that the kinetochore has enough time to assemble prior to mitosis. This might be especially important in budding yeast where there is no clear G2 phase of the cell cycle, resulting in little time for kinetochore assembly prior to mitosis (Kitamura et al. 2007).
Most eukaryotic centromeres contain arrays of canonical and specialized centromereic nucleosomes that are embedded in pericentric heterochromatin (Choo 2001; Kniola et al. 2001). Budding yeast lack many of the characteristic hallmarks of pericentric heterochromatin, including histone H3–K9 methylation and the associated transcriptional silencing of genes. However, similar to other eukaryotes, cohesin is enriched within a 20- to 50-kb domain around centromeres (Blat and Kleckner 1999; Glynn et al. 2004; Weber et al. 2004). Strikingly, the pericentric cohesins in budding yeast appear to be arranged as a cyclindrical array around the spindle (Yeh et al. 2008), which may be due to the formation of an intramolecular C loop on each sister chromatid that extends ∼25 kb (Yeh et al. 2008). Cohesin would therefore encircle a single chromatid rather than sisters in this region, resolving the apparent “cohesin” paradox where the highest levels of cohesin reside in the areas that are physically split at metaphase. At least one function of pericentric cohesion is to facilitate kinetochore biorientation by resisting the pulling forces of microtubules and/or by promoting the architecture of sister kinetochores (Eckert et al. 2007; Fernius and Marston 2009; Ng et al. 2009; Bloom and Joglekar 2010). Consistent with this, the geometry and elasticity of the pericentromere and inner kinetochore can change in response to alterations in microtubule dynamics (Haase et al. 2012; Stephens et al. 2013). These properties are regulated by the Bub1 and Sgo1 proteins as well as various chromatin-remodeling complexes (Haase et al. 2012; Verdaasdonk et al. 2012). While heterochromatin recruits pericentric cohesin in some organisms (Bernard et al. 2001; Fukagawa et al. 2004), components of the kinetochore itself direct cohesion enrichment in budding yeast (Megee et al. 1999; Tanaka et al. 1999; Weber et al. 2004; Eckert et al. 2007; Fernius and Marston 2009; Ng et al. 2009; Fernius et al. 2013).
The pericentromere also contributes to segregation by localizing key regulators of kinetochore biorientation and the checkpoint. The Bub1 kinase, originally identified as a spindle checkpoint protein (see below), phosphorylates H2A in the pericentromeres (Hoyt et al. 1991; Kawashima et al. 2010; Yamagishi et al. 2010). This phosphorylation recruits the Sgo1 protein that facilitates kinetochore biorientation and the spindle checkpoint when kinetochores lack tension (Indjeian et al. 2005; Kitajima et al. 2005; Fernius and Hardwick 2007; Indjeian and Murray 2007). In most organisms, the Haspin kinase phosphorylates H3 to recruit the chromosome passenger complex (CPC), which contains the Aurora B protein kinase that regulates biorientation and the checkpoint (Dai et al. 2005; Kelly et al. 2010). However, the budding yeast Haspin kinases, Alk1 and Alk2, are not known to have a role in chromosome segregation. The CPC may act in a distinct pathway from Bub1 and Sgo1 in budding yeast (Storchova et al. 2011), and it is still unclear how it is recruited to budding yeast pericentromeres.
Budding yeast centromeres have a defined centromeric DNA sequence, leading to the assumption that epigenetic mechanisms do not contribute to their propagation. However, at least two findings using the conditional centromere suggest there is an epigenetic component. First, cohesin enrichment around centromeres exhibits a greater dependence on kinetochore function in newly activated conditional centromeres than previously established endogenous centromeres (Tanaka et al. 1999). This observation suggests that cohesin levels are maintained at least in part by an epigenetic mechanism. Second, the Chl4 kinetochore protein is required for the function of a newly established kinetochore but not a previously formed kinetochore (Mythreye and Bloom 2003), suggesting that epigenetic signals allow cells to bypass the need for Chl4 at established kinetochores. The underlying mechanisms for these observations are not yet known.
Centromeric chromatin
A hallmark of all eukaryotic centromeres is a specialized chromatin structure (Carroll and Straight 2006). Classical chromatin mapping experiments showed that the budding yeast centromere contains a 160- to 220-bp nuclease resistant core flanked by positioned nucleosomes (Bloom and Carbon 1982; Bloom et al. 1984). While most of the chromosome contains nucleosomes made of histone octamers composed of two copies of H2A, H2B, H3, and H4 wrapped by two turns of DNA, centromeres contain a specialized nucleosome where H3 is replaced by a histone H3 variant originally named CENP-A (Earnshaw and Rothfield 1985; Palmer et al. 1987). The budding yeast centromeric histone H3 variant is Cse4 and was initially shown to localize to the centromere by ChIP experiments (Stoler et al. 1995; Meluh et al. 1998). Higher resolution techniques later determined that there is a single, well-positioned nucleosome containing Cse4 that resides over CDEII (Furuyama and Biggins 2007; Lefrancois et al. 2009; Cole et al. 2011; Krassovsky et al. 2012). There are also additional Cse4 molecules around centromeres (Coffman et al. 2011; Lawrimore et al. 2011; Lefrancois et al. 2013), and a challenge for the field is to determine the properties and number of Cse4 nucleosomes that contribute to kinetochore assembly and function. Cse4 can also incorporate into euchromatin, especially at sites of high histone turnover (Collins et al. 2004; Lefrancois et al. 2009; Krassovsky et al. 2012). Cse4 does not stably incorporate into euchromatin because its protein levels are tightly controlled by proteolysis via the Psh1 E3 ubiquitin ligase and additional mechanisms (Collins et al. 2004; Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013). In the absence of proteolysis, Cse4 levels increase and its overexpression in these cells leads to mislocalization throughout euchromatin and subsequent lethality.
Like all histones, Cse4 is recognized and deposited into chromatin by a histone chaperone called Scm3 in budding yeast (HJURP in human cells) (Stoler et al. 2007; Dunleavy et al. 2009; Foltz et al. 2009). Scm3 recognizes Cse4 through the centromere-targeting domain (CATD) in the histone fold and mediates its incorporation into chromatin in vivo and in vitro (Camahort et al. 2007; Shivaraju et al. 2011). Scm3 also interacts with the Ndc10 component of the CBF3 complex, which can explain the specific deposition of Cse4 at centromeres (Camahort et al. 2007). It is not yet known what chaperone incorporates Cse4 into euchromatin. In most organisms, CENP-A is deposited in G1 when Cdk1 activity is low (Jansen et al. 2007; Silva et al. 2012), and the timing of Cse4 deposition is probably similar. Fluorescence recovery after photobleaching (FRAP) experiments showed it is deposited during late G1 or early S phase (Pearson et al. 2004). Although it was reported that Cse4 is also deposited during anaphase (Shivaraju et al. 2012), the marker used for anaphase may not distinguish between late anaphase and G1. Consistent with this, other groups have not observed anaphase incorporation (Pearson et al. 2004; Coffman et al. 2011; Lawrimore et al. 2011).
Although Cse4 is an essential component of centromeric chromatin, the precise composition of the Cse4 nucleosome is controversial (Henikoff and Furuyama 2012). Cse4 is released from minichromosomes in 0.3 M NaCl, conditions that do not affect the binding of canonical H3 to DNA (Akiyoshi et al. 2009b). In addition, Cse4 protects a smaller region of DNA at the centromere than a traditional H3 octamer when treated with the enzyme micrococcal nuclease (MNase), suggesting that the centromeric nucleosome is atypical (Cole et al. 2011; Krassovsky et al. 2012). Consistent with this, it has been proposed that the centromeric nucleosome might exist as a hemisome (containing a single copy of H2A, H2B, CENP-A, and H4) for at least a portion of the cell cycle (Dalal et al. 2007; Dimitriadis et al. 2010; Shivaraju et al. 2012). This was further supported by the observation that centromeric nucleosomes induce positive supercoiling at centromeres in vivo (Furuyama and Henikoff 2009), which has been observed in archaeal tetrameric nucleosomes and is not compatible with the presence of negatively supercoiled histone octamers (Musgrave et al. 1991). However, alternative structures have also been proposed that could explain the smaller protected region of centromeric DNA. One posited that centromeric nucleosomes completely lack H2A and H2B and instead contain two copies of Scm3 (Mizuguchi et al. 2007). The demonstration that Scm3 is a chaperone for Cse4/H4 and that Cse4/H4 cannot simultaneously bind to DNA and Scm3 eliminated this model (Cho and Harrison 2011; Dechassa et al. 2011; Shivaraju et al. 2011; Xiao et al. 2011; Zhou et al. 2011). In a revised model, the centromeric nucleosome was proposed to be a tetramer containing two copies of Cse4 and H4 and completely lacking H2A and H2B (Xiao et al. 2011). However, H2A and H2B have been detected at centromeres making this model less likely (Krassovsky et al. 2012; Lochmann and Ivanov 2012; Shivaraju et al. 2012). Although H3 was also reported to localize to centromeres in budding yeast (Lochmann and Ivanov 2012), the region of DNA analyzed was large enough to contain two nucleosomes so the H3 detected may be in the neighboring nucleosome rather than the centromeric nucleosome. Consistent with this, depletion of H3 in budding yeast has little effect on kinetochore function compared to H4 depletion or H2A mutations that lead to defects in kinetochore–microtubule attachments (Pinto and Winston 2000; Bouck and Bloom 2007; Verdaasdonk et al. 2012). Together, these data suggest that H3 does not reside at the point centromere, although it is important for accurate segregation through its role in recruiting Sgo1 to the pericentromere and facilitating inner kinetochore function (Luo et al. 2010; Verdaasdonk et al. 2012). Finally, it was argued that Cse4 is part of an octameric nucleosome at the centromere based on sequential immunoprecipitation experiments, but the starting material for these experiments was not pure mononucleosomes (Camahort et al. 2009). Therefore, the ability to detect octamers could be due to Cse4 incorporation into neighboring euchromatin. Recently, it was reported that Cse4 exists as a hemisome for most of the cell cycle and then transitions into an octamer at anaphase (Shivaraju et al. 2012). Although this model is attractive because it would reconcile different findings, none of the experiments in this manuscript directly measure Cse4 incorporation into nucleosomes. Instead, these conclusions are based on fluorescence correlation microscopy measurements that may reflect changes in the positioning of kinetochores at anaphase, as well as sequential immunoprecipitations that were not internally consistent because doubling of the H2A histone was not observed when Cse4 doubled (Shivaraju et al. 2012). In sum, the composition of the centromeric nucleosome is still unclear although many of its properties are clearly different from canonical nucleosomes. Because kinetochores may alter the accessibility of the centromeric nucleosome to MNase, affect crosslinking accessibility, or change the wrap of DNA, settling the debate requires that assays be performed on the centromeric nucleosome in vivo in the absence of the kinetochore. Cse4 octameric nucleosomes and hemisomes can both be assembled in vitro (Mizuguchi et al. 2007; Camahort et al. 2009; Dechassa et al. 2011; Kingston et al. 2011; Furuyama et al. 2013). Resolving the structure therefore requires studies on centromeric nucleosomes isolated from cells, but there is no current way to isolate them in the absence of the kinetochore. In the future, it will be critical to apply higher resolution techniques to assay the material at the centromere or to develop a method to isolate the centromeric nucleosomes specifically from the kinetochore, to fully understand their composition. Ultimately, the key issue is to understand how the structure of the centromeric nucleosome specifies and contributes to the assembly and functions of the kinetochore.
Composition of the Budding Yeast Kinetochore
All kinetochores are composed of a number of distinct subcomplexes that can be reconstituted from recombinant proteins or purified from cells as individual complexes (Table 1). While the overall sequence similarity of kinetochore proteins is highly divergent, there is often strong conservation of three-dimensional structure and function of the complexes. In budding yeast, stable subcomplexes that make up the “core” kinetochore include the conserved Ndc80, KNL1/Spc105, Mtw1/MIND/Mis12, COMA/Ctf19, CENP-T, CENP-W, and Cse4 complexes, as well as the yeast-specific Dam1/DASH/DDD and CBF3 complexes. In addition, there are many more conserved proteins, such as motors and spindle checkpoint proteins, which associate with kinetochores depending on the purification conditions and cell cycle stage. Because distinct subcomplexes can be individually purified, it has been suggested that the kinetochore is assembled in a hierarchal manner on centromeric DNA (De Wulf et al. 2003). However, it is still unclear how and where the various subcomplexes assemble into larger complexes to form a kinetochore. Because artificial kinetochores can be formed by tethering the Dam1 or CENP-T complexes to ectopic sites (in the absence of a centromeric nucleosome) (Kiermaier et al. 2009; Lacefield et al. 2009; Schleiffer et al. 2012), the minimal requirements for kinetochore assembly are unclear. While the yeast kinetochore is often suggested to contain three domains (inner, middle, and outer), I refer to proteins as either “inner” to reflect those close to the chromatin or “outer” to reflect roles in mediating microtubule attachment.
Table 1. Kinetochore proteins in budding yeast.
| Complex | Components | Human names |
|---|---|---|
| CBF3 | Ndc10 (Lechner and Carbon 1991; Goh and Kilmartin 1993) | |
| Cep3 (Lechner and Carbon 1991) | ||
| Ctf13 (Lechner and Carbon 1991) | ||
| Skp1 (Connelly and Hieter 1996; Stemmann and Lechner 1996) | ||
| CCAN | Mif2 (Meluh and Koshland 1995) | CENP-C |
| Cse4 (Meluh et al. 1998) | CENP-A | |
| Ctf19 (Ortiz et al. 1999) | CENP-P | |
| Okp1 (Ortiz et al. 1999) | CENP-Q | |
| Mcm21 (Ortiz et al. 1999) | CENP-O | |
| Ame1 (De Wulf et al. 2003) | CENP-U | |
| Chl4 (Mythreye and Bloom 2003; Pot et al. 2003) | CENP-N | |
| Cnn1 (De Wulf et al. 2003) | CENP-T | |
| Wip1 (Schleiffer et al. 2012) | CENP-W | |
| Mhf1 (Schleiffer et al. 2012) | CEN-S | |
| Mhf2 (Schleiffer et al. 2012) | CENP-X | |
| Mcm16 (Measday et al. 2002) | CENP-H | |
| Ctf3 (Measday et al. 2002) | CENP-I | |
| Mcm22 (Measday et al. 2002) | CENP-K | |
| Iml3/Mcm19 (Pot et al. 2003) | CENP-L | |
| Nkp1 (Cheeseman et al. 2002) | ||
| Nkp2 (Cheeseman et al. 2002) | ||
| Ybp2 (Ohkuni et al. 2008) | ||
| Cbf1 | ||
| CPC | Ipl1(Biggins and Murray 2001) | Aurora B |
| Sli15 (Widlund et al. 2006) | INCENP | |
| Nbl1 (Nakajima et al. 2009) | Borealin | |
| Bir1 (Widlund et al. 2006) | Survivin | |
| Mis12 | Mtw1 (Goshima and Yanagida 2000) | Mis12 |
| Dsn1 (De Wulf et al. 2003; Nekrasov et al. 2003; Pinsky et al. 2003) | Dsn1 | |
| Nnf1 (De Wulf et al. 2003; Nekrasov et al. 2003) | Nnf1 | |
| Nsl1 (De Wulf et al. 2003; Nekrasov et al. 2003) | Nsl1 | |
| Ndc80 | Ndc80 (Janke et al. 2001; Wigge and Kilmartin 2001) | Ndc80 |
| Nuf2 (Janke et al. 2001; Wigge and Kilmartin 2001) | Nuf2 | |
| Spc24 (Janke et al. 2001; Wigge and Kilmartin 2001) | Spc24 | |
| Spc25 (Janke et al. 2001; Wigge and Kilmartin 2001) | Spc25 | |
| Spc105 | Spc105 (Nekrasov et al. 2003) | KNL-1 |
| Ydr532 (Nekrasov et al. 2003) | Zwint | |
| Dam1 | Ask1 (Cheeseman et al. 2001a; Janke et al. 2002) | |
| Dad1 (Enquist-Newman et al. 2001) | ||
| Dad2 (Cheeseman et al. 2001a; Janke et al. 2002) | ||
| Dad3 (Cheeseman et al. 2002) | ||
| Dad4 (Cheeseman et al. 2002) | ||
| Dam1 (Enquist-Newman et al. 2001) | ||
| Duo1 (Enquist-Newman et al. 2001) | ||
| Spc19 (Cheeseman et al. 2001a; Janke et al. 2002) | ||
| Spc34 (Cheeseman et al. 2001a; Janke et al. 2002) | ||
| Hsk1 (Cheeseman et al. 2001a; Cheeseman et al. 2002; Li et al. 2002) | ||
| Spindle Checkpoint | Mad1 (Gillett et al. 2004) | Mad1 |
| Mad2 (Gillett et al. 2004) | Mad2 | |
| Bub1 (Gillett et al. 2004) | Bub1 | |
| Bub3 (Gillett et al. 2004) | Bub3 | |
| Mps1 (Jones et al. 2001) | Mps1 | |
| Motor proteins | Kip1 (Tytell and Sorger 2006) | BimC family |
| Kip3 (Tytell and Sorger 2006) | Kinesin-8 | |
| Cin8 (He et al. 2001) | Kinesin-5 | |
| Kar3 (Tanaka et al. 2005) | Kinesin-14 | |
| MAPS | Slk19 (Zeng et al. 1999) | |
| Bik1 (He et al. 2001) | CLIP-170 | |
| Stu1 (Ortiz et al. 2009) | CLASP | |
| Stu2 (He et al. 2001) | XMAP215 |
References are for the initial localization of the component to the kinetochore.
Inner centromere binding proteins
The “inner centromere” proteins are those that are most closely associated with centromeric chromatin. Purification of CENP-A and other inner centromere proteins in vertebrates identified a network of associated components that were collectively termed the constitutive centromere associated network (CCAN) (Obuse et al. 2004; Foltz et al. 2006; Izuta et al. 2006; Okada et al. 2006; Hori et al. 2008). The CCAN consists of various subcomplexes that include the following proteins: CENP-C, CENP-H/I/K, CENP-L/M/N, CENP-O/P/Q/R/U, and the histone fold complexes CENP-T/W and CENP-S/X (McAinsh and Meraldi 2011; Perpelescu and Fukagawa 2011; Takeuchi and Fukagawa 2012). As discussed below, budding yeast inner centromeres contain orthologs of most of these CCAN proteins as well as a yeast-specific complex called CBF3. The composition and deposition of the Cse4 centromeric nucleosome are discussed above.
CBF3:
The CBF3 complex was the first yeast kinetochore subcomplex identified due to its sequence-specific binding activity for centromeric DNA sequences containing CDEIII (Ng and Carbon 1987; Lechner and Carbon 1991; Sorger et al. 1995). The complex contains four essential proteins that are most commonly referred to as Ndc10 (Cbf3a/Cbf2/Ctf14/p110) (Doheny et al. 1993; Goh and Kilmartin 1993; Jiang et al. 1993), Cep3 (Cbf3b/p64) (Lechner 1994; Strunnikov et al. 1995), Ctf13 (Cbf3c/p58) (Doheny et al. 1993), and Skp1 (Cbf3/p19) (Connelly and Hieter 1996; Stemmann and Lechner 1996). Cep3 has a Zinc-cluster motif found in transcription factors (Dhawale and Lane 1993; Strunnikov et al. 1995; Schjerling and Holmberg 1996) and Ndc10 was recently shown to have structural similarity to tyrosine DNA recombinases (Cho and Harrison 2012; Perriches and Singleton 2012), although it does not exhibit catalytic activity or DNA base sequence specificity. Consistent with this, Ndc10 (in the absence of CBF3) can also bind to the CDEII element in vitro as well as other genomic regions that are AT rich, although these activities are not known to be relevant to CBF3 assembly in vivo (Espelin et al. 2003). The stoichiometry of the CBF3 complex bound to centromeres appears to consist of a Cep3 homodimer, a Skp1-Ctf13 heterodimer, and an Ndc10 homodimer (Espelin et al. 1997; Pietrasanta et al. 1999; Russell et al. 1999; Cho and Harrison 2012) (Figure 4). The minimal CBF3 binding region in vitro is a 57-bp core that covers CDEIII and additional base pairs on the right side of the element (Ng and Carbon 1987; Lechner and Carbon 1991; Sorger et al. 1995; Cho and Harrison 2012). Cep3 appears to contact the essential CCG motif in CDEIII, consistent with its similarity to transcription factors containing Zn2Cys6 clusters (Espelin et al. 1997; Purvis and Singleton 2008). Recent structural studies on Ndc10 reveal that the dimer binds to independent DNA fragments, leading to the model that it might stabilize a loop at the centromere (Cho and Harrison 2012 and see below). This is consistent with the observation of bending of the DNA upon CBF3 binding by atomic force microscopy (Pietrasanta et al. 1999).
The assembly of the CBF3 complex is highly regulated in vivo and there has been more work on its assembly than any other yeast kinetochore subcomplex. Data suggest that the complex assembles prior to binding to DNA (Lechner and Carbon 1991; Russell et al. 1999). Ctf13 must be “activated” to form a functional CBF3 complex. The activation process requires binding to Skp1, a protein that is also a component of the SCF ubiquitin ligase complex (Bai et al. 1996; Connelly and Hieter 1996; Kaplan et al. 1997). Although the activation process was initially thought to require Ctf13 phosphorylation by a Skp1-interacting kinase (Kaplan et al. 1997), later work showed that phosphorylation is not required for CBF3 assembly on centromeres (Stemmann et al. 2002). Instead, an Hsp90-Sgt1 co-chaperone complex binds to Skp1, which enhances Skp1 binding to Ctf13 (Stemmann et al. 2002; Bansal et al. 2004; Rodrigo-Brenni et al. 2004). Hsp90 and Sgt1 are not core kinetochore components and only transiently associate with Ctf13. Although a variety of complexes containing these components exist, the relevant intermediate complexes that form in vivo to generate activated Ctf13 are not known. Once Ctf13 is activated by Skp1, a complex containing Cep3, Ctf13, and Skp1 assembles rapidly in vivo, and the rate-limiting step in CBF3 formation is the addition of Ndc10 (Russell et al. 1999; Rodrigo-Brenni et al. 2004). At this time, it is still not known what precise changes occur to activate Ctf13 to allow it to form in CBF3. Because cells can form active CBF3 complexes throughout the cell cycle (Rodrigo-Brenni et al. 2004), there is careful control over the total levels of the complex via Ctf13 proteolysis (Kaplan et al. 1997). When Ctf13 does not form a complex with Cep3, it is degraded in a Skp1-dependent manner (Kaplan et al. 1997; Russell et al. 1999). Ctf13 has an F-box that binds to Skp1, consistent with its role as both an SCF scaffold and a substrate (Zhou and Howley 1998; Galan and Peter 1999). This likely prevents accumulation of misassembled complexes, a behavior associated with many Hsp90 clients.
Once CBF3 associates with the centromere, it is stably bound (Espelin et al. 1997). In fact, even when soluble CBF3 complexes cannot form due to defects in the assembly pathway, previously associated centromere-bound CBF3 is stable (Rodrigo-Brenni et al. 2004). Consistent with this, the ndc10-1 mutation that is commonly used to prevent kinetochore assembly requires that cells go through S phase to remove the mutant CBF3 complexes from the centromere (Poddar et al. 2004). At this time, there is no additional structural data on larger assemblies of the CBF3 complex. A major challenge for the future is to understand precisely how the components of the CBF3 complex interact with each other and how the entire complex binds to DNA to nucleate kinetochore assembly.
CCAN components:
Mif2
Additional budding yeast inner centromere proteins include many orthologs of the vertebrate CCAN (see Table 1). Because the sequence identity is very low, many of these proteins were not identified as CCAN components until very recently (Schleiffer et al. 2012). One of the major conserved components is Mif2, the budding yeast ortholog of CENP-C, an essential inner kinetochore protein (Earnshaw and Rothfield 1985; Meeks-Wagner et al. 1986; Brown 1995; Meluh and Koshland 1995). Mif2 dimerizes and fluorescence measurements in vivo suggest that a single Mif2 dimer binds to each centromere at CDEIII (Meluh and Koshland 1995, 1997; Ortiz et al. 1999; Joglekar et al. 2006; Cohen et al. 2008). Mif2 can bind to CDEIII directly in vitro in a manner that requires a stretch of A:T bases instead of the CCG motif required for CBF3 binding (Cohen et al. 2008). While vertebrate CENP-C binds to CENP-A nucleosomes in vitro (Carroll et al. 2010), less is known about the precise manner in which Mif2 binds to the yeast centromere. The Cse4 nucleosome co-purifies with Mif2 (Westermann et al. 2003) and the centromere localization of Mif2 requires both Cse4 and CBF3 (Meluh and Koshland 1997; Westermann et al. 2003), consistent with the possibility that Mif2 recognizes an aspect of yeast centromeric nucleosome structure. Mif2 also requires a functional Mis12 complex for centromere localization (Westermann et al. 2003), similar to vertebrate requirements for CENP-C localization (Fukagawa et al. 2001).
COMA and interacting proteins
Additional components of the yeast inner kinetochore include the COMA subcomplex (Ctf19, Okp1, Mcm21 and Ame1), as well as many additional interacting proteins (see Table 1) (Kroll et al. 1996; Sanyal et al. 1998; Hyland et al. 1999; Ortiz et al. 1999; Poddar et al. 1999; Cheeseman et al. 2002; Measday et al. 2002; De Wulf et al. 2003; Pot et al. 2003; Ohkuni et al. 2008; Schleiffer et al. 2012). With the exception of Okp1 and Ame1, most of these proteins are nonessential and may have redundant functions. A Ctf19/Mcm21 crystal structure of recombinant Kluyveromyces lactis proteins has been solved and shows that each protein contains double “RWD” domains that are interaction motifs in a variety of proteins (Nameki et al. 2004; Schmitzberger and Harrison 2012). Four of the CCAN components contain histone fold domains (HFD) that form two subcomplexes: Cnn1/Wip1 (orthologs of CENP-T/W) and Mhf1/Mhf2 (orthologs of CENP-S/X) (Bock et al. 2012; Schleiffer et al. 2012). In vertebrates, these two complexes form a heterotetramer that contacts DNA, suggesting it may be a novel nucleosome-like structure at the centromere (Hori et al. 2008; Nishino et al. 2012). However, it is not yet known whether these complexes form nucleosome-like structures in budding yeast, nor how they might be positioned relative to the centromeric nucleosome. In contrast to other organisms and yeast kinetochore proteins, the copy number of these proteins at the kinetochore appears to increase at anaphase (Bock et al. 2012; Schleiffer et al. 2012). Cnn1 interacts with the outer kinetochore complex Ndc80, and recent evidence suggests that it may be a receptor for Ndc80 in anaphase (Bock et al. 2012; Schleiffer et al. 2012; Malvezzi et al. 2013). Fluorescence microscopy measurements suggest there are approximately three COMA complexes that constitutively associate with the centromere (Joglekar et al. 2006), but the relative stoichiometry of most of the other inner kinetochore CCAN components has not been analyzed.
Model for the inner kinetochore:
Combined data from many studies has led to a potential model for inner kinetochore structure in yeast (Figure 5) (Yeh et al. 2008; Cho and Harrison 2012). A key aspect of the model is based on the observation that an Ndc10 dimer binds to independent DNA segments as well as to multiple kinetochore proteins through other domains (Cho and Harrison 2012). Because Ndc10 binds to CDEIII as well the CDEI binding protein Cbf1, an attractive idea is that Ndc10 can loop the centromeric DNA so that CDEI and CDEIII are in proximity (Cho and Harrison 2012). Cbf1 is nonessential, so Ndc10 may maintain this structure even in its absence. Ndc10 binds to the Cse4 chaperone, Scm3, through a different domain to localize the centromeric nucleosome (Camahort et al. 2007; Stoler et al. 2007; Cho and Harrison 2012), and Ndc10 looping may help to position the centromeric DNA around the nucleosome. Cbf3 also recruits Mif2 and the other CCAN components, but their precise locations relative to CBF3 and the centromeric nucleosome core are not yet known. Together, these data explain why CBF3 is a key nucleating factor for the yeast kinetochore. Although other organisms do not have CBF3, the overall conservation of inner kinetochore proteins suggests similar functions. One possibility is that CCAN components have acquired CBF3 activities in other organisms. Consistent with this, the requirement for CBF3 function to stabilize minichromosomes can be bypassed in yeast by artificially tethering the Cnn1 (CENP-T) kinetochore protein to the minichromosome (Schleiffer et al. 2012).
Figure 5.
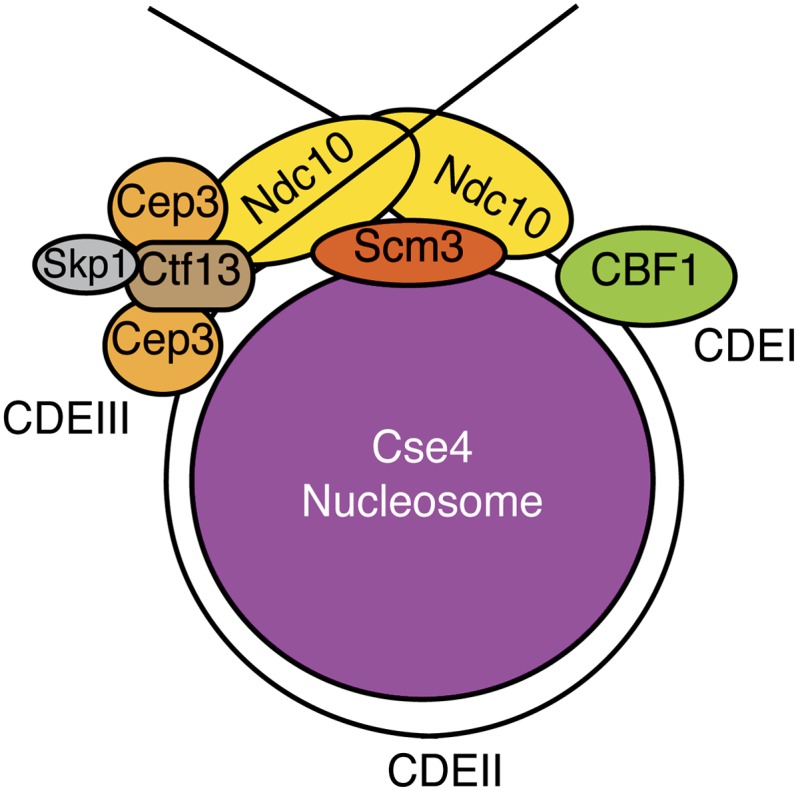
Model for the inner kinetochore. One possible model, based on Cho and Harrison (2012), suggests that the Ndc10 homodimer within the CBF3 complex interacts with CDEI and CDEIII to loop the centromeric DNA. Ndc10 also recruits the Scm3 chaperone that deposits Cse4, leading to the specialized inner centromere structure.
The CPC complex is also part of the inner kinetochore, although it is not a core kinetochore complex. Composed of the Ipl1 protein kinase (Aurora B), Sli15 (INCENP), Bir1 (Survivin), and Nbl1 (Borealin) proteins, this complex associates with kinetochores from G1 until anaphase (Widlund et al. 2006; Carmena et al. 2012). The CPC association with the inner kinetochore is mediated via its interaction with CBF3 through Bir1 (Yoon and Carbon 1999; Sandall et al. 2006), and a separate pool may be localized through binding COMA via Sli15 (Knockleby and Vogel 2009). At anaphase, the CPC dissociates from the kinetochore and localizes to the spindle and spindle midzone. The dynamic localization of the CPC reflects its numerous functions in chromosome segregation, including kinetochore biorientation and spindle function (Carmena et al. 2012 and below). While CPC association with the spindle requires dephosphorylation of the Sli15 microtubule-binding domain (Pereira and Schiebel 2003), the mechanisms that control the timing of its localization to kinetochores and pericentromeric chromatin in budding yeast have not been elucidated.
Outer kinetochore proteins
The outer kinetochore contains the microtubule-binding activity and consists of the essential subcomplexes Mtw1/Mis12/MIND, Spc105/Knl-1/Blinkin, Ndc80, Dam1/DASH/DDD, as well as nonessential proteins such as motors and checkpoint components (Table 1). For simplicity, I use the most common yeast complex names, Mis12, Spc105, and Dam1.
KMN:
The Mis12 (composed of Mtw1, Dsn1, Nnf1, and Nsl1 at a 1:1:1:1 stoichiometry (Euskirchen 2002; De Wulf et al. 2003; Nekrasov et al. 2003; Pinsky et al. 2003; Westermann et al. 2003; Maskell et al. 2010; Hornung et al. 2011), Spc105 (composed of Spc105 and Ydr532/Kre28 at a 1:2 Spc105:Kre28 ratio (Nekrasov et al. 2003; Pagliuca et al. 2009) and Ndc80 (composed of a 1:1:1:1 ratio of Ndc80, Nuf2, Spc24, and Spc25 (Janke et al. 2001; Wigge and Kilmartin 2001; Ciferri et al. 2005; Wei et al. 2005) subcomplexes form a larger, highly conserved network called KMN that contains the core microtubule binding activity of the kinetochore (Cheeseman et al. 2006). Consistent with this, yeast mutants in KMN fail to make kinetochore–microtubule attachments (Wigge et al. 1998; Nekrasov et al. 2003; Pinsky et al. 2006; Pagliuca et al. 2009). The entire KMN complex is likely a 1:1:1 stoichiometry of Mis12, Ndc80, and Spc105 subcomplexes (Cheeseman et al. 2006; Joglekar et al. 2006), although this has not been precisely determined in any organism. The Mis12 complex is composed of heterodimers of Mtw1/Nnf1 and Dsn1/Nsl1 (Maskell et al. 2010; Hornung et al. 2011) and does not exhibit microtubule binding activity on its own (Cheeseman et al. 2006; Hornung et al. 2011). The complex is a 21–25 nm long elongated bilobed complex (Maskell et al. 2010; Hornung et al. 2011). All four components appear to contribute to a larger globular domain at the head that is connected to an extended rod most likely composed of the Nnf1 and Mtw1 subunits (Maskell et al. 2010). The Dsn1/Nsl1 heterodimer interacts directly with the globular C-terminal domains of the Ndc80 complex Spc24/25 heterodimer (Maskell et al. 2010). The Spc105 complex has not been reconstituted so structural work on recombinant proteins has not been performed. Spc105 purified from yeast exhibits weak microtubule binding activity (Pagliuca et al. 2009). The microtubule binding activity within the ortholog KNL1 appears to be mediated by its N terminus (Cheeseman et al. 2006; Kiyomitsu et al. 2007; Pagliuca et al. 2009; Welburn et al. 2010). The C terminus of Spc105 interacts with the Mis12 complex, likely through multiple Mis12 components (Maskell et al. 2010). In addition to contributing to KMN function, Spc105 also appears to be a scaffold for other outer kinetochore proteins. It recruits the Bub1 and Bub3 proteins to the kinetochore, and it may be a regulatory subunit for PP1 at the kinetochore (Kiyomitsu et al. 2007, 2011; Liu et al. 2010; Rosenberg et al. 2011) (discussed below, The Spindle Checkpoint). The Ndc80 complex has two globular head domains that are connected by a long rod (Wei et al. 2005; Ciferri et al. 2008). One head contains Nuf2 and Ndc80, which each contain positively charged calponin-homology domains (CH) that facilitate binding to the negative microtubule surface (Wei et al. 2005; Cheeseman et al. 2006; Wei et al. 2007; Ciferri et al. 2008). CH domains have diverse functions and have been identified in other microtubule binding proteins (Hayashi and Ikura 2003; Dougherty et al. 2005). An unstructured N-terminal tail on Ndc80 enhances the microtubule binding activity of the complex (Wei et al. 2005; DeLuca et al. 2006; Wei et al. 2007; Ciferri et al. 2008; Miller et al. 2008; Alushin et al. 2010), although it is not essential for yeast viability due to redundancy with Dam1 (Kemmler et al. 2009; Demirel et al. 2012; Lampert et al. 2013). The interaction between Ndc80 and microtubules is largely electrostatic and requires the C-terminal tails of tubulin (Ciferri et al. 2008). Spc24 and Spc25 fold into a single globular domain that links the Ndc80 complex to the kinetochore through the Mis12 complex (Wei et al. 2006). The Ndc80 coiled-coil rod is interrupted by a stretch of residues that are not predicted to form a coiled coil and appear to loop out, possibly facilitating a geometry needed for microtubule binding, tension sensing, and/or serving as a protein interaction motif (Wang et al. 2008). The loop is required to recruit the Dam1 complex to kinetochores in vivo (Maure et al. 2011), but it is not necessarily a direct binding site and the requirement may be due to a structural change that occurs when the loop is deleted. In other organisms, the loop has been implicated in interacting with the Ska1 complex, the Dis1/TOG/Stu2 protein, and the Cdt1 replication factor (Hsu and Toda 2011; Varma et al. 2012; Zhang et al. 2012), so its precise role is unclear.
The major microtubule binding activity within KMN is via the Ndc80 globular N-terminal domain and its extension (Cheeseman et al. 2006; DeLuca et al. 2006; Wei et al. 2007; Ciferri et al. 2008; Powers et al. 2009; Alushin et al. 2010; Hornung et al. 2011; Sundin et al. 2011). Although the Caenorhabditis elegans KMN enhances microtubule binding of the individual components in a cooperative manner, this has not been directly tested with yeast proteins due to the inability to purify recombinant Spc105 and reconstitute yeast KMN (Cheeseman et al. 2006). The microtubule binding activity within the nematode KNL1 appears to be important for spindle checkpoint silencing in vivo rather than kinetochore–microtubule coupling activity (Espeut et al. 2012). A goal for the future is therefore to determine how the Mis12 and Spc105 subcomplexes contribute to enhancing microtubule-binding activity.
Dam1 complex:
The Dam1 complex is an essential 10 component yeast-specific complex (Ask1, Dad1, Dad2, Dad3, Dad4, Dam1, Duo1, Hsk3, Spc19, and Spc34 (Hofmann et al. 1998; Jones et al. 1999; Cheeseman et al. 2001a,b; Enquist-Newman et al. 2001; Janke et al. 2002; Li et al. 2002; De Wulf et al. 2003; Li et al. 2005; Miranda et al. 2005; Westermann et al. 2005) that requires the function of KMN and microtubules for kinetochore localization (Janke et al. 2002; Li et al. 2002; Tanaka et al. 2005; Maure et al. 2011). Consistent with this, Ndc80 has been implicated in its localization and microscopy studies show that Dam1 is the outermost kinetochore complex (Shang et al. 2003; Joglekar et al. 2006; Maure et al. 2011; Gonen et al. 2012; Lampert et al. 2013). The Dam1 complex can be reconstituted by coexpression of all components in bacteria (Miranda et al. 2005). Each protein is present at a single copy per complex and 16 complexes can assemble into a ring around microtubules in vitro in either orientation relative to the plus end (Miranda et al. 2005; Westermann et al. 2005; Wang et al. 2007; Ramey et al. 2011). However, small oligomers and other larger Dam1 structures can also attach to microtubules, making it unclear which structures are relevant to activity in vivo (Gestaut et al. 2008; Grishchuk et al. 2008b). At low concentrations, the Dam1 complex prefers to interact with microtubules through the C-terminal E-hook regions of tubulin (Westermann et al. 2005; Ramey et al. 2011). The diameter of the ring is ∼50 nm and appears to interact with microtubules through electrostatic interactions via “arms” that extend from the Dam1 complex (Miranda et al. 2005; Westermann et al. 2005). These interactions are at least partly mediated through the N terminus of Dam1 and possibly the Duo1 subunit, which also exhibits microtubule-binding activity (Hofmann et al. 1998; Cheeseman et al. 2001b; Wang et al. 2007; Ramey et al. 2011). Although there are no atomic structures for any Dam1 components, cryo-EM analyses indicate that the complex does not appear to undergo major rearrangements upon forming a ring around the microtubule (Ramey et al. 2011).
Other outer kinetochore proteins:
Additional proteins that localize to the outer kinetochore include the Stu1 and Stu2 proteins (orthologs of the vertebrate CLASP and XMAP215/Dis1 proteins) (He et al. 2001; Ortiz et al. 2009; Kitamura et al. 2010), the Slk19 protein (Zeng et al. 1999), the Bik1 protein (He et al. 2001), and the four nuclear motor proteins, Kar3, Cin8, Kip1, and Kip3 (Tanaka et al. 2005; Tytell and Sorger 2006; Pagliuca et al. 2009). The localization of these proteins to kinetochores has been assayed by ChIP and/or microscopy, so it is difficult to determine how closely associated each protein is with the core kinetochore. In addition, these proteins are not core proteins that are part of the constitutive structure, but are instead regulatory proteins that associate transiently. Some of these proteins may reside at microtubule plus ends rather than bind directly to the kinetochore (Shimogawa et al. 2006, 2010), but resolution limits make it difficult to directly test this in budding yeast. Because many of these proteins may affect microtubule dynamics and/or kinetochore–microtubule interactions, it will be important to understand their roles at the kinetochore in the future. Additional regulatory proteins, such as the checkpoint proteins Mps1, Mad1, Mad2, Bub1, and Bub3, also associate with the outer kinetochore (see below, The Spindle Checkpoint).
Architecture of the kinetochore
While there has been significant progress identifying the components and structural details of kinetochore proteins and subcomplexes, the overall structural organization of the entire macromolecular complex is just beginning to be understood (Welburn and Cheeseman 2008; Alushin and Nogales 2011). One unresolved issue is the precise copy number of each subcomplex within the kinetochore. The best estimates have been made using high-resolution fluorescence microscopy measurements, which clearly show that there are more outer than inner kinetochore subcomplexes (Joglekar et al. 2006). However, the precise numbers are not clear because the initial estimates were based on the assumption that there is a single centromeric nucleosome with two copies of Cse4 (Joglekar et al. 2006). In this case, the inner kinetochore complexes range from 1–2 copies (Mif2) to up to 16 copies of the outer kinetochore complexes (Dam1) (Joglekar et al. 2006). KMN is estimated to be at 5–8 subcomplexes/kinetochore, consistent with EM data on isolated kinetochores showing 5–7 globular domains that may represent KMN (Joglekar et al. 2009; Gonen et al. 2012). However, if a different fluorescence standard is used, all kinetochore components are present at two- to threefold higher numbers, which greatly changes the overall size of the kinetochore (Lawrimore et al. 2011).
A model of overall kinetochore organization was proposed, which was based on the wealth of existing biochemical and genetic interaction data, combined with elegant microscopy experiments that measured the average distances between kinetochore subcomplexes (Figure 6A) (Joglekar et al. 2009). First, the kinetochore is built upon a centromeric chromatin base that contains CBF3, Mif2, and other CCAN components. CENP-C/Mif2 interacts with the Mis12 subcomplex in other eukaryotes (Petrovic et al. 2010; Przewloka et al. 2011), and this appears to be true in yeast (S. Westermann, personal communication). The COMA inner kinetochore complex also binds to Mis12, providing an additional bridge between centromeric chromatin and the outer kinetochore (Hornung et al. 2011). The existence of multiple inner kinetochore receptors for the Mis12 complex may explain how the copy number of the outer complexes increases relative to the inner kinetochore components. There are also multiple receptors for the Ndc80 complex, because it binds to both the Mis12 and Cnn1 complexes (Bock et al. 2012; Schleiffer et al. 2012; Malvezzi et al. 2013). The Spc24/25 proteins within the Ndc80 complex interact with similar motifs in Cnn1 and the Mis12 component Dsn1 (Malvezzi et al. 2013). The interaction with Dsn1 is essential and Dsn1 is the major receptor throughout the bulk of the cell cycle. Cnn1 may inhibit this interaction at anaphase, suggesting a potential change in KMN receptors for unknown reasons (Bock et al. 2012; Schleiffer et al. 2012; Malvezzi et al. 2013). Spc105 also binds to Mis12 (Maskell et al. 2010), but its localization in vivo does not depend on subcomplexes other than CBF3 (Pagliuca et al. 2009). The connections between Spc105 and the kinetochore are still not completely understood. Ndc80 orients and localizes the Dam1 complex, which is the outermost complex and may form a ring in vivo. In meiosis I, the kinetochore must change its behavior to coorient sister kinetochores rather than biorient to ensure that sister chromatids travel to the same pole. The Csm1/Lrs4 monopolin complex forms a clamp-like structure that binds to the Dsn1 protein (Corbett et al. 2010), leading to the idea that sister kinetochores can be crosslinked to behave as a single unit.
Figure 6.
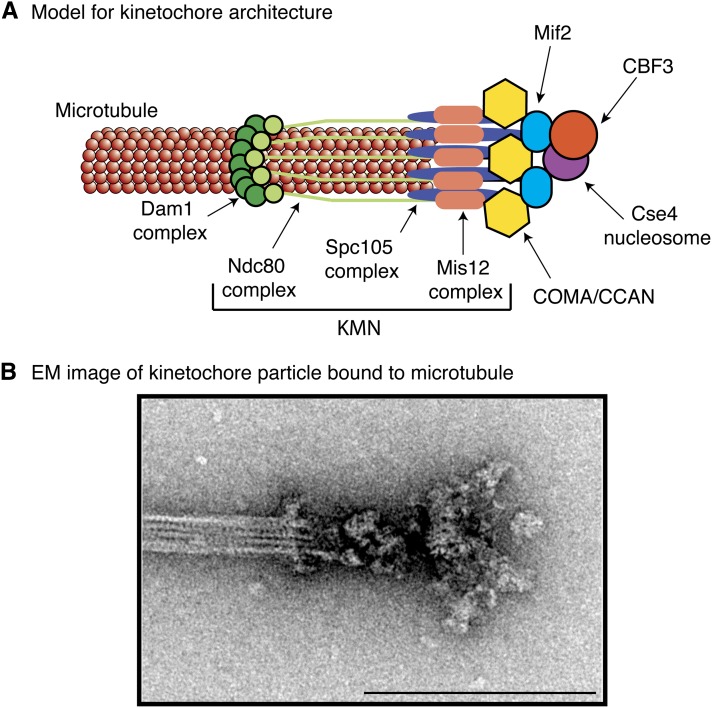
Model for the budding yeast kinetochore. (A) Schematic indicating the rough position and stoichiometry of the budding yeast kinetochore subcomplexes. (B) Electron microscope image of a purified yeast kinetochore particle bound to a microtubule, originally published in Gonen et al. (2012). There is a ring that encircles the microtubule and globular domains that could represent KMN that touch the microtubule. Bar, 200 μm.
While the precise number and arrangement of subcomplexes within the kinetochore are still unknown, isolated kinetochores were recently visualized by EM (Figure 6B) (Gonen et al. 2012). They appear to have a central hub surrounded by 5–7 globular domains that appear to contact the microtubule, consistent with their identity as KMN or a part of KMN. In support of this possibility, an extension that contains a kink and is the length of the Ndc80 complex extends from the globular domains. In some kinetochore particles, the extension is connected to a ring that encircles the microtubule and that depends on the presence of functional Dam1 complex (Gonen et al. 2012). Partial rings around microtubules were also recently visualized within cells by EM (McIntosh et al. 2013). Together, these data are consistent with the idea that the central hub represents the inner kinetochore and the surrounding globular domains represent KMN connected to a Dam1 ring. Although it has been difficult to visualize yeast kinetochores within cells by EM, puck-like structures at the end of microtubules that may correspond to kinetochores were recently described (McIntosh et al. 2013). In the future, higher resolution structural information will be critical to determining precisely where each kinetochore component exists in the kinetochore.
Kinetochore–microtubule attachments and coupling activity
One of the most outstanding questions in the field is the mechanistic basis for the persistent load-bearing attachment of kinetochores to the tips of dynamic microtubules. A variety of evidence suggests that an end-on attachment is different from a lateral attachment close to the tip of the microtubule (Asbury et al. 2006; Powers et al. 2009; Akiyoshi et al. 2010; Tanaka 2010). While the discovery of motor proteins at the kinetochore immediately suggested a mechanism to couple kinetochores to microtubules, we now know that motor proteins do not provide the major coupling activity (McIntosh 2012). Some motor proteins localize to yeast kinetochores and facilitate segregation, but the genes are all nonessential and the phenotypes of deletions do not lead to defects in kinetochore–microtubule attachments (Tanaka et al. 2005; Tytell and Sorger 2006; Pagliuca et al. 2009; Wargacki et al. 2010).
In the budding yeast kinetochore, Ndc80 and Dam1 are the major subcomplexes currently known to bind to microtubules (Cheeseman et al. 2001a; Janke et al. 2002; Miranda et al. 2005; Tanaka et al. 2005; Wei et al. 2005; Westermann et al. 2005; Asbury et al. 2006; Westermann et al. 2006; Gestaut et al. 2008; Powers et al. 2009). Unlike Ndc80, the Dam1 complex requires microtubules for kinetochore association (Li et al. 2002). Consistent with this, the Ndc80 complex is required for both lateral attachments and end-on attachments, while Dam1 is only required for proper end-on attachments (Tanaka et al. 2005; Shimogawa et al. 2006; Akiyoshi et al. 2010). Elegant experiments in vitro showed that Dam1 enhances the microtubule tip tracking activity of the Ndc80 complex under load, suggesting that it is a processivity factor for Ndc80 (Lampert et al. 2010; Tien et al. 2010). These data are consistent with the requirement for Dam1 to couple bioriented, end-on attached sister kinetochores, which experience the highest forces. It is currently unclear how the other budding yeast subcomplexes contribute to microtubule binding. Spc105 purified from yeast exhibits weak binding activity but the lack of recombinant complex has made it difficult to study its precise contributions (Pagliuca et al. 2009). Although kinetochore particles purified from spc105 mutant cells are defective in microtubule attachment, the particles also have substantially reduced Ndc80 levels (Akiyoshi et al. 2010). Similarly, although many kinetochore components are required for kinetochore–microtubule attachments in vivo (Tanaka et al. 2005), this may be a secondary effect of altered kinetochore composition. For example, mutants in COMA components (Okp1 and Ame1) exhibit segregation defects in vivo (Ortiz et al. 1999; Tanaka et al. 2005; Knockleby and Vogel 2009), but this may reflect their role in recruiting outer kinetochore proteins.
Because it is difficult to distinguish direct effects on microtubule binding from secondary effects on kinetochore composition in vivo, elucidating the mechanism of kinetochore–microtubule attachments requires studies in vitro (Akiyoshi and Biggins 2012). Considerable progress in reconstituting the kinetochore–microtubule interface in vitro has provided experimental support for two major coupling models (Asbury et al. 2011). The first proposes a biased diffusion mechanism in which the kinetochore contains multiple weak microtubule binding elements that together have enough total energy to maintain an attachment (Hill 1985). As long as the elements are able to quickly diffuse along the microtubule, they can maintain kinetochore attachment to the dynamic microtubule tip and harness the energy of microtubule dynamics to move the chromosome. Any motion that brings more of the binding elements into contact with the microtubule will favor the attachment and thus provide a biased direction for the diffusion. A variety of data support this model. First, both the Ndc80 and the Dam1 complexes are able to diffuse rapidly along the microtubule lattice in vitro (Westermann et al. 2006; Gestaut et al. 2008; Powers et al. 2009). Additionally, these complexes (alone or in combination) as well as purified kinetochore particles maintain load-bearing attachments to dynamic microtubule tips (Westermann et al. 2006; Franck et al. 2007; Grishchuk et al. 2008a,b; Powers et al. 2009; Akiyoshi et al. 2010; Lampert et al. 2010; Tien et al. 2010; Volkov et al. 2013). Second, kinetochores contain numerous copies of the Ndc80 and Dam1 complexes, consistent with a multivalent attachment mechanism (Joglekar et al. 2006; Gonen et al. 2012). Although it is still not known how many of these elements within a single kinetochore might contact a microtubule at one time, static EM images show that multiple domains within a single kinetochore particle can contact a microtubule (Gonen et al. 2012). Although isolated kinetochore particles do not diffuse on the lattice (Akiyoshi et al. 2010), the rate of diffusion for a multivalent coupler is slower on the lattice than on a disassembling tip. Depending on the number of binding elements, the lattice rate can be negligibly slow, but the tip rate will remain fast enough to support tip tracking (Hill 1985; Powers et al. 2009).
The other major mechanism that has been supported by both theoretical considerations and experimental evidence is referred to as the “conformational wave” model (Koshland et al. 1988; Molodtsov et al. 2005; McIntosh et al. 2008). This model and a variation called the “forced walk” theorize that a portion of the kinetochore forms a ring or fibrils that are pushed on by depolymerizing filaments in the microtubule to move the kinetochore. The conformational wave model proposes a ring structure with sufficient diameter that it could freely slide along the microtubule, while the forced walk model suggests that fibrils would harness protofilament peeling. In either case, the underlying mechanism is similar. Support came from the exciting discovery that Dam1 complexes can self-assemble into rings with a 16-fold symmetry around microtubules in vitro (Miranda et al. 2005; Westermann et al. 2006). In addition, isolated kinetochore particles bound to microtubules sometimes contain rings encircling the microtubule (Gonen et al. 2012). The Dam1 complex exhibits a preference for the GTP-bound tips of microtubules (Westermann et al. 2005; Gestaut et al. 2008) and moves along microtubules in a processive manner, consistent with the sliding of a ring (Westermann et al. 2005, 2006; Asbury et al. 2006; Grishchuk et al. 2008a). It can also maintain load-bearing attachments to dynamic microtubules (Franck et al. 2007; Grishchuk et al. 2008a; Volkov et al. 2013), and quantitative fluorescence data indicate that there are sufficient Dam1 complexes at kinetochores in vivo to form rings (Joglekar et al. 2006). When Dam1 is tethered to beads in a manner that might mimic fibrils, it can maintain much greater load in vitro (Volkov et al. 2013), and fibril-like connections have been observed by tomography on mammalian cells (McIntosh et al. 2008). While these data support the conformational wave model, a single Dam1 complex is sufficient to diffuse along a microtubule and to attach to disassembling tips in vitro (Gestaut et al. 2008; Grishchuk et al. 2008b). A potential unifying view is that rings likely do exist in vivo, but that they are involved in a biased diffusion mechanism. This is supported by data showing that the Dam1 complex exhibits electrostatic interactions with the C-terminal tails of tubulin that likely promote biased diffusion rather than a forced-walk model (Westermann et al. 2005; Ramey et al. 2011). In addition, the conformational wave model is based on curved, peeling protofilaments and therefore predicts that kinetochores would be more stably attached to disassembling tips than assembling tips. However, isolated kinetochores as well as the Dam1 and Ndc80 subcomplexes all detach from disassembling tips more readily than assembling tips (Asbury et al. 2006; Franck et al. 2007; Powers et al. 2009; Akiyoshi et al. 2010; Tien et al. 2010). In addition, the Dam1 complex exhibits autonomous tracking with assembling tips, a property that is consistent with its preference for the GTP-bound microtubule tip but not with a requirement for peeling protofilaments (Westermann et al. 2006). EM data within cells and with purified kinetochores bound to microtubules also support the possibility of multivalent attachments, a basis for biased diffusion (Dong et al. 2007; Akiyoshi et al. 2010; McCwen and Dong 2010). In sum, experiments suggest that elements of both biased diffusion and the conformational wave may contribute, and further defining the coupling mechanism will be a major focus of future research.
Regulation of kinetochore attachments
Kinetochore biorientation
Accurate chromosome segregation requires pairs of sister kinetochores to biorient so that they attach to microtubules from opposite poles. Biorientation is a complicated process that requires cells to both detect and correct kinetochore–microtubule attachment errors. Biorientation generates tension on kinetochores due to the microtubule pulling forces on sister chromatids linked by cohesin. Consistent with this, cohesin is highly enriched in a 50-kb domain around yeast centromeres, presumably to resist the pulling forces of microtubules (Megee et al. 1999; Tanaka et al. 1999; Glynn et al. 2004). The kinetochore is required to recruit pericentromeric cohesin, and the COMA subcomplex has been specifically implicated in this process (Tanaka et al. 1999; Weber et al. 2004; Eckert et al. 2007; Fernius and Marston 2009; Ng et al. 2009; Fernius et al. 2013). However, the details of how cohesin spreads from kinetochores to a large domain around the centromere are still unknown.
Cells appear to monitor biorientation via the level of tension generated on the kinetochore. Attachments lacking tension in vivo are highly unstable, while those that come under tension are stably maintained (Nicklas 1997). A pioneering experiment that directly tested the effects of tension was performed in grasshopper spermatocytes cells by applying tension to a monooriented chromosome (Nicklas and Koch 1969). Once the chromosome came under tension, it maintained a stable attachment to the pole. Similarly elegant in vivo experiments were performed in budding yeast and showed that minichromosomes lacking tension destabilized their microtubule attachments and continued to reorient between spindle pole bodies, while those under tension were stably attached (Tanaka et al. 2002; Dewar et al. 2004). Direct support in vitro for the stabilization of attachments by tension came from the finding that isolated budding yeast kinetochores maintain attachments to microtubules for longer periods of time at higher forces (Akiyoshi et al. 2010).
One can imagine a variety of mechanisms that could regulate kinetochore biorientation, and there is support for at least three. Two mechanisms involve the selective destabilization of kinetochore attachments lacking tension, thereby giving the cell another chance to make a proper attachment. First, tension directly stabilizes attachments in vitro by modulating microtubule tip dynamics (Franck et al. 2007; Akiyoshi et al. 2010). As the level of force on kinetochore–microtubule attachments increases in vitro, the rate of catastrophes decreases and microtubule rescue is promoted. Strikingly, kinetochores maintain attachments to assembling tips for longer periods of time than disassembling tips. Together, these data suggest that tension directly promotes attachments by modulating microtubule tip dynamics to favor the state where kinetochores have a higher probability of staying bound. A second mechanism involves the destabilization of kinetochore–microtubule attachments by Aurora B (Ipl1 in yeast) kinase-mediated phosphorylation. In Aurora B mutant cells, the majority of kinetochore attachments are monooriented, and Aurora B activity is required for kinetochores to detach from microtubules when they lack tension (Biggins et al. 1999; Tanaka et al. 2002; Dewar et al. 2004; Tanaka et al. 2005). An important challenge in the field has been to identify the key Aurora B kinetochore substrates that are involved in biorientation and to understand how their phosphorylation leads to destabilization of attachments. While Aurora B-mediated phosphorylation of many kinetochore proteins has been reported (Cheeseman et al. 2002; Westermann et al. 2003; Maskell et al. 2010), the only substrates implicated in biorientation in budding yeast to date are the major microtubule binding complexes, Dam1 and Ndc80. Mutants in the Dam1 phosphorylation sites lead to biorientation defects in vivo (Cheeseman et al. 2002) and Aurora B phosphorylation site mutants in Ndc80 exhibit biorientation defects when Aurora B function is further compromised in vivo (Akiyoshi et al. 2009a). These data suggest that Ndc80 phosphorylation is important but redundant with additional substrates in yeast (Akiyoshi et al. 2009a; Demirel et al. 2012).
Phosphorylation has multiple effects on kinetochore behavior. Aurora B-mediated phosphorylation directly weakens the interaction between kinetochores and microtubules, a behavior consistent with the overall negative charge of the microtubule. Aurora B phosphorylation of S20 on the Dam1 subcomplex directly reduces the affinity of the subcomplex for microtubules and causes it to detach more frequently in vitro (Gestaut et al. 2008). In addition, purified kinetochores containing phosphomimetic mutants in Dam1 or Ndc80 exhibit weaker attachments to microtubules that have additive effects (Sarangapani et al. 2013). Phosphorylation of the Dam1 complex on sites other than S20 has further effects that lead to the destabilization of attachments. First, although phosphorylation of the Aurora B sites in Dam1 does not alter the structure of the monomeric Dam1 complex, it reduces its ability to oligomerize and to assemble rings in vitro (Wang et al. 2007). Second, Dam1 phosphorylation decreases its ability to interact with the Ndc80 complex on microtubules and confer tip-tracking activity (Lampert et al. 2010; Tien et al. 2010). Finally, kinetochore particles containing the Dam1 phosphomimetic mutants increase microtubule catastrophe rates, which could indirectly weaken kinetochore attachments by promoting microtubule disassembly (Akiyoshi et al. 2010; Sarangapani et al. 2013). Because there are additional Aurora B sites in the Ndc80 and Dam1 complexes, as well as additional Aurora B kinetochore substrates, an important future goal will be to fully analyze the corresponding mutants in vivo and in vitro.
The other mechanism implicated in kinetochore biorientation is a steric consideration. If sister kinetochores assume a back-to-back geometry, it would ensure that once a kinetochore attaches to a microtubule, its sister would bind to a microtubule emanating from the opposite pole. Because there is not enough resolution to visualize sister kinetochores by EM in yeast to distinguish their geometry, it has been difficult to design a definitive experiment to address sister kinetochore geometry. An unreplicated minichromosome containing two centromeres efficiently biorients, suggesting that any linkage is sufficient to generate tension (Dewar et al. 2004). While this finding led to the conclusion that kinetochore geometry was not a factor in biorientation (Dewar et al. 2004), the experiment did not rule out the possibility that the two centromeres on the minichromosome assumed a specific geometry. A different observation suggests that geometry may indeed contribute to biorientation in yeast (Indjeian and Murray 2007). Sgo1 is required for biorientation when kinetochores attach to microtubules coming from unseparated spindle poles, but not from separated poles. This differential requirement suggests that yeast kinetochores are intrinsically biased to attach to opposite poles so active error correction by Sgo1 is only required in situations prone to form monoorientated attachments (such as unseparated poles). Although this experiment lends support to a specific geometry of sister kinetochores, it does not explain why there is an essential requirement for the Aurora B kinase in reorientation regardless of the timing of pole separation. One possibility is that Sgo1 becomes important for reorientation later in the cell cycle due to its role in regulating Aurora B, a function that may be carried out by other proteins earlier in the cell cycle (Kawashima et al. 2007; Yamagishi et al. 2010).
The Mps1 kinase is also essential for yeast kinetochore biorientation and its substrates have started to be identified (Shimogawa et al. 2006; Maure et al. 2007; Kemmler et al. 2009; London et al. 2012). Mps1 phosphorylates a number of sites in the Dam1 protein, and cells expressing a S218A S221A phosphomutant appear to biorient sister kinetochores and satisfy the checkpoint without making end-on microtubule attachments (Shimogawa et al. 2006, 2010). Although Mps1 and Aurora B have varied effects on the activity of each other in other organisms (Jelluma et al. 2008; Saurin et al. 2011; van der Waal et al. 2012), there is no data in yeast suggesting that they act in the same pathway (Maure et al. 2007; Storchova et al. 2011). The mechanism by which Mps1 regulates biorientation has therefore not yet been elucidated and it will be important to understand its contribution in the future.
Mps1 and Aurora B localize to kinetochores, so it is critical to understand how they are regulated by a lack of tension to promote biorientation. An elegant model for Aurora B activity was proposed that is based on its spatial distance from substrates (Tanaka et al. 2002). Because Aurora B localizes to the inner kinetochore and its key substrates appear to be on the outer kinetochore, it was proposed that it would only be able to phosphorylate its substrates when kinetochores lack tension. As kinetochores biorient, tension will pull the kinetochores apart and move Aurora B away from its substrates, thus stabilizing attachments. While this model is attractive and has been supported by some experiments (Keating et al. 2009; Liu et al. 2009; Welburn et al. 2010), it does not explain how initial attachments that lack tension would be made in the presence of high kinase activity. Protein phosphatase I activity opposes Aurora B (Francisco et al. 1994), so the kinase/phosphatase balance must be carefully regulated if this model is correct. In addition, it was recently shown that cells lacking the inner centromere localization of Aurora B can still biorient kinetochores (Campbell and Desai 2013). In the future, it will be critical to determine where the active enzyme is localized to understand whether there is spatial control over its activity. Mps1 appears to localize to the outer kinetochore (Kemmler et al. 2009), suggesting that its regulation will be different from Aurora B.
The Spindle Checkpoint
It is essential that defects in kinetochore–microtubule attachments are monitored and corrected prior to cell division. The spindle checkpoint is the surveillance system that halts the cell cycle if there are defects in kinetochore–microtubule attachments, thus giving cells time to ensure that every chromosome makes a proper attachment (for recent reviews, see Zich and Hardwick 2010; Kim and Yu 2011; Musacchio 2011). Two budding yeast genetic screens identified the majority of checkpoint genes (Hoyt et al. 1991; Li and Murray 1991). The MPS1, MAD1, MAD2, MAD3 (BUBR1), BUB1, and BUB3 genes are highly conserved genes that are all required to arrest the cell cycle in response to defects in kinetochore–microtubule attachments (Hoyt et al. 1991; Li and Murray 1991; Weiss and Winey 1996). While the BUB2 gene was originally thought to be essential for the spindle checkpoint, later work showed that it monitors defects in spindle positioning (Wang and Burke 1995; Alexandru et al. 1999; Fesquet et al. 1999; Fraschini et al. 1999; Li 1999). The yeast checkpoint genes are nonessential with the exception of MPS1, which encodes a conserved protein kinase with roles in other essential cellular processes (Winey et al. 1991; Jones et al. 2001). Because the checkpoint genes are dispensable during a normal cell cycle, budding yeast cells do not normally need any delay to complete biorientation before anaphase. In contrast, these genes are essential in animal cells (Basu et al. 1999; Kitagawa and Rose 1999; Dobles et al. 2000; Kalitsis et al. 2000; Gillett et al. 2004). Despite being nonessential, the spindle checkpoint is active for at least a short duration because spindle checkpoint complexes assemble early during each S phase (Brady and Hardwick 2000). In addition, deletions of the spindle checkpoint genes result in chromosome segregation defects to varying degrees (Li and Murray 1991; Pangilinan and Spencer 1996; Warren et al. 2002). Mutations in the Bub1 and Bub3 proteins result in the strongest segregation defects (Warren et al. 2002), consistent with separate functions in kinetochore biorientation (Fernius and Hardwick 2007; Windecker et al. 2009; Kawashima et al. 2010; Storchova et al. 2011). Mad1 and mad2 mutants also exhibit segregation defects and genetic interactions that are stronger than a mad3 deletion, suggesting they also have separable segregation functions (Warren et al. 2002; Daniel et al. 2006). Mad1 was recently implicated in regulating nuclear transport, which may indirectly affect segregation because the import of the PP1/Glc7 phosphatase, discussed below, is affected (Cairo et al. 2013). However, this role is independent of Mad2 so there are likely additional functions for Mad1 and Mad2 in chromosome segregation.
The spindle checkpoint mediates cell cycle arrest by inhibiting the anaphase-promoting complex (APC) through inhibition of the Cdc20 activator (Li et al. 1997; Fang et al. 1998; Hwang et al. 1998; Kim et al. 1998). This prevents the APC from promoting the ubiquitylation of cyclin B and securin, two key substrates required for mitotic progression (Kim and Yu 2011). Thus, the checkpoint ensures that downstream mitotic events do not occur if kinetochore–microtubule attachments are defective via the inhibition of Cdc20-APC activity. While various subcomplexes between Cdc20 and checkpoint proteins exist, the most potent inhibitor of the APC is a soluble mitotic checkpoint complex (MCC) containing Mad2, Mad3, Bub3, and Cdc20 (Brady and Hardwick 2000; Hardwick et al. 2000; Fraschini et al. 2001b; Sudakin et al. 2001; Tang et al. 2001; Fang 2002; Chao et al. 2012). A variety of data suggest that the key inhibition comes from the binding of Mad2 and Mad3 to Cdc20 (Chao et al. 2012; Lau and Murray 2012).
There is strong evidence that the checkpoint signal is generated at the kinetochore. Classic laser ablation and micromanipulation experiments showed that kinetochores lacking tension or attachments produce an inhibitory signal that halts the cell cycle (Rieder et al. 1994; Li and Nicklas 1995). All of the yeast checkpoint proteins except Mad3 localize to the kinetochore (Gillett et al. 2004). While the Bub1 and Bub3 checkpoint proteins always localize to kinetochores at mitosis (presumably to regulate kinetochore biorientation), Mad1 and Mad2 are specifically recruited to unattached kinetochores (Chen et al. 1996; Gillett et al. 2004). FRAP and additional microscopy experiments in other organisms showed that Bub1 and Mad1 are stably bound to unattached kinetochores, while Mad2 exists two pools, one that is stable and one that rapidly exchanges (Howell et al. 2000, 2004; Shah et al. 2004). While most yeast mutants defective in centromere function and kinetochore–microtubule attachments trigger the spindle checkpoint (Spencer and Hieter 1992; Wang and Burke 1995; Pangilinan and Spencer 1996; Wells and Murray 1996; Gardner et al. 2001), the ndc10-1 mutant that completely blocks kinetochore assembly does not (Goh and Kilmartin 1993; Fraschini et al. 2001a; Poddar et al. 2004). The Ndc80 and COMA subcomplexes have also been implicated in the checkpoint, likely due to direct or indirect recruitment of checkpoint proteins (McCleland et al. 2003; Matson et al. 2012). The overexpression of the Mps1 checkpoint kinase can generate a sustained checkpoint signal in an ndc10-1 mutant that lacks kinetochores (Fraschini et al. 2001a; Poddar et al. 2004), but this may be due to constitutive downstream signaling events that have not yet been identified. Together, these data provide strong support that the kinetochore generates a checkpoint signal, which leads to the formation of a soluble cell cycle inhibitor.
One of the more controversial questions is the nature of the signal that triggers the checkpoint. While defects in many aspects of microtubule and kinetochore function lead to cell cycle arrest due to the checkpoint (Spencer and Hieter 1992; Wang and Burke 1995; Pangilinan and Spencer 1996; Hardwick et al. 1999; Gardner et al. 2001; Stern and Murray 2001), the underlying structural signal is difficult to precisely test. Unattached kinetochores clearly recruit checkpoint proteins and lead to a checkpoint-mediated arrest (Gillett et al. 2004). However, defects in tension can be generated in yeast by either completely preventing replication or preventing the cohesion between sister chromatids. In these cases, kinetochore–microtubule attachments appear to be made, but they lack tension and arrest the cell cycle in a checkpoint-dependent manner (Biggins and Murray 2001; Stern and Murray 2001). The Aurora B protein kinase and the Sgo1 protein are required to halt the cell cycle in response to these defects, but they are not required for the checkpoint in response to unattached kinetochores (Biggins and Murray 2001; Indjeian et al. 2005). A histone H3 mutant that mislocalizes Sgo1 from the centromeric region has a similar phenotype (Luo et al. 2010). While these data can be interpreted to mean that tension defects signal the spindle checkpoint independently from attachment defects, the lack of tension on kinetochores also destabilizes attachments (reviewed above). Therefore, it is difficult to determine whether tension directly triggers the checkpoint signal or indirectly creates unattached kinetochores that trigger the checkpoint signal. Consistent with this, Aurora B and Sgo1 destabilize inappropriate microtubule attachments (Biggins et al. 1999; Tanaka et al. 2002; Pinsky et al. 2006; Indjeian and Murray 2007), making it unclear whether they truly have a separate role in a tension-sensing checkpoint pathway (Pinsky and Biggins 2005). Further complicating the issue is recent data in mammalian cells that suggests that intrakinetochore tension, not interkinetochore tension, is coupled to the checkpoint response (Maresca and Salmon 2009; Uchida et al. 2009). Because tension and attachment are interdependent in vivo, dissecting this issue will require a complete reconstitution of the checkpoint in vitro to determine the upstream signal.
Another controversial question is whether Aurora B has a direct function in the checkpoint or whether its sole function is to create unattached kinetochores that trigger the checkpoint (Pinsky and Biggins 2005). In some organisms, Aurora B is required for the spindle checkpoint in response to defects in attachment as well as tension (Maresca 2011). It is not clear whether this reflects a difference in function between organisms or whether the yeast Aurora B alleles retain enough function to respond to defects in attachment. The Mad3 protein has Aurora B sites and mutants in these sites are defective in the response to tension defects but not attachment defects (Rancati et al. 2005; King et al. 2007). However, there could be other key Aurora B targets and/or the Mad3 mutant could be a hypomorph that can respond to strong but not weaker signals. A key Aurora B substrate that is defective in all aspects of spindle checkpoint signaling has not yet been identified in any organism.
Regardless of whether there is a single checkpoint signaling pathway or separate tension and attachment-dependent pathways, some of the key steps in the generation of the yeast checkpoint signal have been identified (Figure 7). First, the Mps1 kinase appears to be the most upstream signal and recruits downstream checkpoint components to kinetochores (Hardwick et al. 1996; Heinrich et al. 2012; London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). Mps1 copurifies with Ndc80 and interacts with its N-terminal domain in vitro (Kemmler et al. 2009), suggesting that Ndc80 may be the kinetochore receptor for Mps1. Phosphorylation of the Spc105 protein on conserved MELT motifs by Mps1 recruits the Bub1 and Bub3 proteins to the kinetochore (Kiyomitsu et al. 2007, 2011; Krenn et al. 2012; London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). Mps1, Bub1, and Bub3 are all required for the recruitment of the Mad1 and Mad2 proteins to kinetochores, although the Mad1 receptor has not yet been identified (Gillett et al. 2004; Heinrich et al. 2012). A prevailing model for checkpoint signaling is the “Mad2 template model” that requires its interactions with kinetochore-bound Mad1 (De Antoni et al. 2005). There are two forms of Mad2 that exist on the kinetochore: a pool stably bound to Mad1, and a pool that rapidly cycles on and off the kinetochore (Howell et al. 2000). When bound to Mad1, Mad2 adopts a “closed” form structure (Mad2-C or N2-Mad2) that remains stably bound to Mad1 (Luo et al. 2002; Sironi et al. 2002). This population of Mad2 serves as a receptor for soluble Mad2, which is in an “open” (Mad2-O or N1-Mad2) conformation (Luo et al. 2000, 2002; Sironi et al. 2002; De Antoni et al. 2005). Mad2-O dynamically cycles onto the kinetochore where it binds to Mad2-C, which promotes its conversation to Mad2-C. Mad1 and Cdc20 have similar Mad2 binding motifs, which means that Mad2-C will bind to Cdc20 (Luo et al. 2000, 2002; Mapelli et al. 2007). Therefore, the kinetochore Mad1-bound pool of Mad2 “templates” the formation of a soluble Mad2-Cdc20 structural copy, thus promoting APC inhibition.
Figure 7.
Spindle checkpoint pathway. Mps1 phosphorylates Spc105 to recruit the Bub1/3 proteins. Once Bub1/3 are bound to phosphorylated Spc105, the Mad1/Mad2 complex is recruited to the kinetochore. The open form of Mad2 is converted to a closed form, and the closed Mad2 binds to Cdc20 and eventually forms a mitotic checkpoint complex (containing Bub3, Mad2, Mad3, and Cdc20) that inhibits the progression into anaphase.
A key feature of the checkpoint is regulation by phosphorylation (Kang and Yu 2009). Mps1 phosphorylation of Spc105 is essential for the checkpoint (London et al. 2012; Shepperd et al. 2012; Yamagishi et al. 2012). Mps1 has also been implicated in phosphorylation of Mad1 and Ndc80 (Hardwick et al. 1996; Kemmler et al. 2009), but the functions of these phosphorylation events have not been identified. Mps1 autophosphorylation is required for its activation and kinetochore localization in some organisms (Kang et al. 2007; Mattison et al. 2007; Xu et al. 2009), although this has not been studied as much in budding yeast. It is unclear how many checkpoint substrates exist for Mps1, but its ability to halt the cell cycle in a kinetochore-independent manner suggests it likely has soluble targets downstream of the kinetochore that remain to be identified (Hardwick et al. 1996; Fraschini et al. 2001a; Poddar et al. 2004). In fission yeast, Mps1-mediated phosphorylation of Mad2 is important for its association with Mad3 and Cdc20, but Mad2 is not known to be phosphorylated in budding yeast (Zich et al. 2012). While Bub1 is also a kinase (Roberts et al. 1994), its catalytic activity is not required for the yeast spindle checkpoint (Sharp-Baker and Chen 2001; Warren et al. 2002). Although it was initially thought that the kinase domain is required for the checkpoint (Roberts et al. 1994), this conclusion was made using a catalytic site mutant that destabilizes the protein (Warren et al. 2002). A deletion of the kinase domain does not alter Bub1 levels and fully supports yeast checkpoint function (Warren et al. 2002; Fernius and Hardwick 2007). Consistent with these data, a Bub1 checkpoint substrate in yeast has not been identified. Although Bub1 phosphorylation of Cdc20 in mammalian cells has been implicated in the checkpoint (Tang et al. 2004; Kang et al. 2008), its kinase activity is more important for segregation than the checkpoint (Sharp-Baker and Chen 2001; Klebig et al. 2009). As mentioned above, Aurora B phosphorylation in yeast has also been implicated in the checkpoint because it phosphorylates Mad3 to generate a response to defects in tension (Rancati et al. 2005; King et al. 2007). However, an Aurora B substrate essential for all checkpoint responses has not been identified in any organism. A clear goal for the future is the identification of additional checkpoint phosphorylation events.
Just as phosphorylation is required to engage the checkpoint, dephosphorylation via protein phosphatase 1 (PP1) is required to silence the checkpoint (Pinsky et al. 2009; Vanoosthuyse and Hardwick 2009). PP1 reverses the Mps1-mediated phosphorylation on Spc105, and it likely has additional key substrates (Pinsky et al. 2009; London et al. 2012). Spc105 may be involved in a checkpoint feedback loop because it has been implicated as the kinetochore receptor for PP1 in budding and fission yeast (Meadows et al. 2011; Rosenberg et al. 2011). However, direct binding between the budding yeast Spc105 and PP1 proteins was not demonstrated in budding yeast (Rosenberg et al. 2011), and the Fin1 kinetochore protein also binds to PP1 (Akiyoshi et al. 2009b). It is therefore still unclear precisely how PP1 is recruited and regulated to silence the checkpoint in yeast. The Cdc14 phosphatase is also implicated in reversing the checkpoint through dephosphorylation of the Sli15 activator of Aurora B, which promotes Sli15/Aurora B relocalization to the spindle (Mirchenko and Uhlmann 2010). The destruction of the Cdc20, Mps1, and Bub1 proteins and the autoubiquitination of Cdc20 also ensure that the checkpoint is silenced (Pan and Chen 2004; Palframan et al. 2006; Goto et al. 2011; Foster and Morgan 2012). In other organisms, checkpoint silencing occurs via competition between the 31Pcomet protein and Mad3 for their interaction with Mad2 (Xia et al. 2004; Yang et al. 2007; Chao et al. 2012). This antagonizes MCC assembly, thus helping to silence the checkpoint. However, there is no obvious 31P ortholog in budding yeast. Dynein-mediated removal of Mad2 has also been implicated in other organisms (Howell et al. 2001), but budding yeast lack nuclear dynein.
Ultimately, the mechanism by which microtubules attach to kinetochores and come under tension must be integrated with the mechanism of spindle checkpoint signaling. The role of the KMN complex in both mediating microtubule attachment and recruiting checkpoint proteins makes it a prime candidate for the integration of these pathways. One possibility is that microtubule attachment itself disrupts the binding of one or more checkpoint proteins, making it impossible for the checkpoint to signal when microtubules are attached. This system would rely on error correction to ensure that attachments lacking tension are disrupted, thus signaling the checkpoint. Mad1 and Mad2 do not accumulate on kinetochores that are attached to microtubules (Gillett et al. 2004), suggesting they are the checkpoint proteins most sensitive to the state of kinetochore–microtubule attachments. In sum, there are significant questions about the amplification of the checkpoint signal and the underlying mechanisms that connect the state of kinetochore–microtubule attachment to the cell cycle that must be elucidated in the future.
Perspectives
The identification of a small, sequence-specific point centromere sequence in budding yeast led to the initial thought that kinetochores would be vastly different between organisms. Instead, we now know that the most important features of kinetochores are highly conserved. A unique underlying centromeric chromatin structure is the foundation for the assembly of many subcomplexes to be built into a dynamic kinetochore structure. The precise composition of the centromeric nucleosome is still debated and needs to be resolved, but future work needs to focus on understanding how it contributes to kinetochore assembly and function. It will also be key to understand how the newly identified histone fold proteins at the centromere contribute to kinetochore function. While the complexity of kinetochores is greater in most other eukaryotes, the yeast kinetochore is a simplified version that employs similar complexes and core mechanisms for its fundamental functions. Understanding how these complexes assemble into a large macromolecular structure at a single chromosomal locus is another area that needs to be tackled in the future. We still have relatively limited knowledge of the requirements and order of assembly of the kinetochore. These studies will be critical for the reconstitution of an entire kinetochore, a step necessary to allow high-resolution structural studies and additional biochemical and biophysical kinetochore assays to be performed. This work will be critical to ultimately understanding the mechanistic basis of kinetochore–microtubule attachments.
There has also been tremendous progress in understanding the regulation of kinetochore–microtubule attachments and the spindle checkpoint. We now know that multiple pathways lead to the destabilization of microtubule attachments lacking tension, but we are still just beginning to identify the underlying mechanisms and their regulation. The spindle checkpoint components have been identified and a major challenge for the future will be to understand the nature of the defects they detect, as well as to understand how signals from a single kinetochore are amplified into a soluble cell cycle inhibitor. The kinetochore must perform a different mode of segregation in meiosis I, so another challenge for the future is elucidating the mechanisms that alter kinetochore function depending on the cell division state. Given that we now have the component parts and a wealth of techniques to study kinetochore functions, these exciting questions can be addressed in the future.
Acknowledgments
I am very grateful to my colleagues in the field, Bungo Akiyoshi, Chip Asbury, Kerry Bloom, Trisha Davis, Kevin Hardwick, Ken Kaplan, and Stefan Westermann, who generously provided information prior to publication, and to Bungo Akiyoshi, Chip Asbury, Trisha Davis, Erica Hildebrand, Nitobe London, and Matt Miller for comments on the manuscript. I apologize to those colleagues whose work was not cited due to space limitations. The National Institutes of Health funded the work in my laboratory (R01 GM078079 and R01 GM064386).
Footnotes
Communicating editor: J. Boeke
Literature Cited
- Akiyoshi B., Biggins S., 2012. Reconstituting the kinetochore-microtubule interface: what, why, and how. Chromosoma 121: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C. R., Ranish J. A., Biggins S., 2009a Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics 183: 1591–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C. R., Ranish J. A., Biggins S., 2009b Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23: 2887–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Sarangapani K. K., Powers A. F., Nelson C. R., Reichow S. L., et al. , 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C., Hyman A., 2006. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172: 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G., Zachariae W., Schleiffer A., Nasmyth K., 1999. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 18: 2707–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G., Nogales E., 2011. Visualizing kinetochore architecture. Curr. Opin. Struct. Biol. 21: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G. M., Ramey V. H., Pasqualato S., Ball D. A., Grigorieff N., et al. , 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C. L., Gestaut D. R., Powers A. F., Franck A. D., Davis T. N., 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 103: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C. L., Tien J. F., Davis T. N., 2011. Kinetochores’ gripping feat: Conformational wave or biased diffusion? Trends Cell Biol. 21: 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., et al. , 2013. A novel role of the N terminus of budding yeast histone h3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Partha S., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Baker R. E., Masison D. C., 1990. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10: 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal P. K., Abdulle R., Kitagawa K., 2004. Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol. Cell. Biol. 24: 8069–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B. C., et al. , 1999. Mutations in the essential spindle checkpoint gene Bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146: 13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., et al. , 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A. W., 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., et al. , 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Jansen L. E., Foltz D. R., Cleveland D. W., 2010. Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb. Symp. Quant. Biol. 75: 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Kleckner N., 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms vs. the centric region. Cell 98: 249–259 [DOI] [PubMed] [Google Scholar]
- Bloom K., Joglekar A., 2010. Towards building a chromosome segregation machine. Nature 463: 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J., 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305–317 [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Amaya E., Carbon J., Clarke L., Hill A., et al. , 1984. Chromatin conformation of yeast centromeres. J. Cell Biol. 99: 1559–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock L. J., Pagliuca C., Kobayashi N., Grove R. A., Oku Y., et al. , 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14: 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K., 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17: 741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D. M., Hardwick K. G., 2000. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10: 675–678 [DOI] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., et al. , 2008. XMAP215 is a processive microtubule polymerase. Cell 132: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. T., 1995. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene 160: 111–116 [DOI] [PubMed] [Google Scholar]
- Burrack L. S., Berman J., 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28: 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W., 1989. Purification of a yeast centromere binding protein that is able to distinguish single base pair mutations in its recognition site. Mol. Cell. Biol. 9: 2544–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W., 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61: 437–446 [DOI] [PubMed] [Google Scholar]
- Cairo L. V., Ptak C., Wozniak R. W., 2013. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol. Cell 49: 109–120 [DOI] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S. K., Washburn M. P., et al. , 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., et al. , 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. S., Desai A., 2013. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature 497: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., Earnshaw W. C., 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13: 789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C. W., Straight A. F., 2006. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 16: 70–78 [DOI] [PubMed] [Google Scholar]
- Carroll C. W., Milks K. J., Straight A. F., 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W. C., Kulkarni K., Zhang Z., Kong E. H., Barford D., 2012. Structure of the mitotic checkpoint complex. Nature 484: 208–213 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Brew C., Wolyniak M., Desai A., Anderson S., et al. , 2001a Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Enquist-Newman M., Muller-Reichert T., Drubin D. G., Barnes G., 2001b Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152: 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., et al. , 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A., 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Waters J. C., Salmon E. D., Murray A. W., 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274: 242–246 [DOI] [PubMed] [Google Scholar]
- Cho U. S., Harrison S. C., 2011. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. USA 108: 9367–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U. S., Harrison S. C., 2012. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat. Struct. Mol. Biol. 19: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K. H., 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1: 165–177 [DOI] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K. J., Ristic D., et al. , 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280: 29088–29095 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., et al. , 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 1786: 32–40 [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J., 1980. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509 [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J., 1985. The structure and function of yeast centromeres. Annu. Rev. Genet. 19: 29–55. [DOI] [PubMed] [Google Scholar]
- Clarke L., 1998. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 8: 212–218 [DOI] [PubMed] [Google Scholar]
- Coffman V. C., Wu P., Parthun M. R., Wu J. Q., 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. L., Espelin C. W., De Wulf P., Sorger P. K., Harrison S. C., et al. , 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell 19: 4480–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole H. A., Howard B. H., Clark D. J., 2011. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl. Acad. Sci. USA 108: 12687–12692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Collins K. A., Castillo A. R., Tatsutani S. Y., Biggins S., 2005. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell 16: 5649–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C., Hieter P., 1996. Budding yeast SKP1 encodes a evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. D., Yip C. K., Ee L. S., Walz T., Amon A., et al. , 2010. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sultan S., Taylor S. S., Higgins J. M., 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19: 472–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y., Wang H., Lindsay S., Henikoff S., 2007. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. A., Keyes B. E., Ng Y. P., Freeman C. O., Burke D. J., 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 172: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C. G., Cimini D., Canman J. C., Sala V., et al. , 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15: 214–225 [DOI] [PubMed] [Google Scholar]
- Dechassa M. L., Wyns K., Li M., Hall M. A., Wang M. D., et al. , 2011. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., et al. , 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- Demirel P. B., Keyes B. E., Chaterjee M., Remington C. E., Burke D. J., 2012. A redundant function for the N-terminal tail of Ndc80 in kinetochore–microtubule interaction in Saccharomyces cerevisiae. Genetics 192: 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T. U., 2004. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428: 93–97 [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K., 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17: 2902–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale S. S., Lane A. C., 1993. Compilation of sequence-specific DNA-binding proteins implicated in transcriptional control in fungi. Nucleic Acids Res. 21: 5537–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis E. K., Weber C., Gill R. K., Diekmann S., Dalal Y., 2010. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 107: 20317–20322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M. L., Benezra R., Sorger P. K., 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635–645 [DOI] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., et al. , 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell 73: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K. J., Meng X., Khodjakov A., McEwen B. F., 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty G. W., Adler H. J., Rzadzinska A., Gimona M., Tomita Y., et al. , 2005. CLAMP, a novel microtubule-associated protein with EB-type calponin homology. Cell Motil. Cytoskeleton 62: 141–156 [DOI] [PubMed] [Google Scholar]
- Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., et al. , 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N., 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Eckert C. A., Gravdahl D. J., Megee P. C., 2007. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 21: 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman M., Cheeseman I. M., Van Goor D., Drubin D. G., Meluh P. B., et al. , 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin C. W., Kaplan K. B., Sorger P. K., 1997. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin C. W., Simons K. T., Harrison S. C., Sorger P. K., 2003. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell 14: 4557–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Cheerambathur D. K., Krenning L., Oegema K., Desai A., 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196: 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G. M., 2002. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot. Cell 1: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M. W., 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J., Hardwick K. G., 2007. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 3: e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J., Marston A. L., 2009. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 5: e1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J., Nerusheva O. O., Galander S., Alves Fde L., Rappsilber J., et al. , 2013. Cohesin-dependent association of Scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr. Biol. 23: 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick P. J., Johnson A. L., Kramer K. M., Toyn J. H., et al. , 1999. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18: 2424–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J., 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29: 235–244 [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, et al. , 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., 3rd, Bassett E. A., et al. , 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S. A., Morgan D. O., 2012. The APC/C Subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol. Cell 47: 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L., Wang W., Chan C. S., 1994. Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14: 4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A. D., Powers A. F., Gestaut D. R., Gonen T., Davis T. N., et al. , 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9: 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A. D., Powers A. F., Gestaut D. R., Davis T. N., Asbury C. L., 2010. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods 51: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti E., Lucchini G., Piatti S., 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145: 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Lucchini G., Piatti S., 2001a Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol. Genet. Genomics 266: 115–125 [DOI] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., et al. , 2001b Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20: 6648–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., et al. , 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20: 4603–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., et al. , 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6: 784–791 [DOI] [PubMed] [Google Scholar]
- Furuyama S., Biggins S., 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104: 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Henikoff S., 2009. Centromeric nucleosomes induce positive DNA supercoils. Cell 138: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Codomo C. A., Henikoff S., 2013. Reconstitution of hemisomes on budding yeast centromeric DNA. Nucleic Acids Res. 41: 5769–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Peter M., 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. D., Poddar A., Yellman C., Tavormina P. A., Monteagudo M. C., et al. , 2001. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics 157: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., Pearson C. G., Sprague B. L., Zarzar T. R., Bloom K., et al. , 2005. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell 16: 3764–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D. R., Graczyk B., Cooper J., Widlund P. O., Zelter A., et al. , 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D. R., Cooper J., Asbury C. L., Davis T. N., Wordeman L., 2010. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 95: 641–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett E. S., Espelin C. W., Sorger P. K., 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E. F., Megee P. C., Yu H. G., Mistrot C., Unal E., et al. , 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V., 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121: 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Akiyoshi B., Iadanza M. G., Shi D., Duggan N., et al. , 2012. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19: 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M., 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100: 619–633 [DOI] [PubMed] [Google Scholar]
- Goto G. H., Mishra A., Abdulle R., Slaughter C. A., Kitagawa K., 2011. Bub1-mediated adaptation of the spindle checkpoint. PLoS Genet. 7: e1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Efremov A. K., Volkov V. A., Spiridonov I. S., Gudimchuk N., et al. , 2008a The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA 105: 15423–15428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Spiridonov I. S., Volkov V. A., Efremov A., Westermann S., et al. , 2008b Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA 105: 6918–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J., Stephens A., Verdaasdonk J., Yeh E., Bloom K., 2012. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr. Biol. 22: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K. G., Weiss E., Luca F. C., Winey M., Murray A. W., 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273: 953–956 [DOI] [PubMed] [Google Scholar]
- Hardwick K. G., Li R., Mistrot C., Chen R. H., Dann P., et al. , 1999. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics 152: 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K. G., Johnston R. C., Smith D. L., Murray A. W., 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I., Ikura M., 2003. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278: 36430–36434 [DOI] [PubMed] [Google Scholar]
- Hayden J. H., Bowser S. S., Rieder C. L., 1990. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J. Cell Biol. 111: 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K., 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101: 763–775 [DOI] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K., 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195–206 [DOI] [PubMed] [Google Scholar]
- Heath I. B., 1980. Behavior of kinetochores during mitosis in the fungus Saprolegnia ferax. J. Cell Biol. 84: 531–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Windecker H., Hustedt N., Hauf S., 2012. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J. Cell Sci. : 4720–4727.125 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., 2010. Epigenetic inheritance of centromeres. Cold Spring Harb. Symp. Quant. Biol. 75: 51–60 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., 2012. The unconventional structure of centromeric nucleosomes. Chromosoma 121: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K., 1987. Genetic manipulation of centromere function. Mol. Cell. Biol. 7: 2397–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Bloom K., 1989. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 1368–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. L., 1985. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA 82: 4404–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C., Cheeseman I. M., Goode B. L., McDonald K. L., Barnes G., et al. , 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., et al. , 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Hornung P., Maier M., Alushin G. M., Lander G. C., Nogales E., et al. , 2011. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J. Mol. Biol. 405: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Hoffman D. B., Fang G., Murray A. W., Salmon E. D., 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150: 1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., et al. , 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., et al. , 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T., 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517 [DOI] [PubMed] [Google Scholar]
- Hsu K. S., Toda T., 2011. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr. Biol. 21: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L. H., Lau L. F., Smith D. L., Mistrot C. A., Hardwick K. G., et al. , 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Hyland K. M., Kingsbury J., Koshland D., Hieter P., 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V. B., Murray A. W., 2007. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 17: 1837–1846 [DOI] [PubMed] [Google Scholar]
- Indjeian V. B., Stern B. M., Murray A. W., 2005. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307: 130–133 [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., et al. , 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11: 673–684 [DOI] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Lechner J., Shevchenko A., Magiera M. M., et al. , 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Tanaka T. U., Lechner J., Schiebel E., 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L. E., Black B. E., Foltz D. R., Cleveland D. W., 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn B., Niedenthal R., Hegemann J. H., 1991. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol. Cell. Biol. 11: 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A. B., van den Broek N. J., Cruijsen C. W., van Osch M. H., et al. , 2008. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 132: 233–246 [DOI] [PubMed] [Google Scholar]
- Jiang W., Lechner J., Carbon J., 1993. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J. Cell Biol. 121: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. W., Fuchs J., Loidl J., 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113(Pt 11): 1903–1912 [DOI] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D., 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Bloom K., Salmon E. D., 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Bachant J. B., Castillo A. R., Giddings T. H., Jr, Winey M., 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell 10: 2377–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., He X., Giddings T. H., Winey M., 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. USA 98: 13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Fowler K. J., Choo K. H., 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14: 2277–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yu H., 2009. Kinase signaling in the spindle checkpoint. J. Biol. Chem. 284: 15359–15363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Chen Y., Zhao Y., Yu H., 2007. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc. Natl. Acad. Sci. USA 104: 20232–20237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yang M., Li B., Qi W., Zhang C., et al. , 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell 32: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K. B., Hyman A. A., Sorger P. K., 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91: 491–500 [DOI] [PubMed] [Google Scholar]
- Kawashima S. A., Tsukahara T., Langegger M., Hauf S., Kitajima T. S., et al. , 2007. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 21: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S. A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y., 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177 [DOI] [PubMed] [Google Scholar]
- Keating P., Rachidi N., Tanaka T. U., Stark M. J., 2009. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J. Cell Sci. 122: 4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. E., Ghenoiu C., Xue J. Z., Zierhut C., Kimura H., et al. , 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., et al. , 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28: 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E., Woehrer S., Peng Y., Mechtler K., Westermann S., 2009. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat. Cell Biol. 11: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Kim S., Yu H., 2011. Mutual regulation between the spindle checkpoint and APC/C. Semin. Cell Dev. Biol. 22: 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Lin D. P., Matsumoto S., Kitazono A., Matsumoto T., 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science 279: 1045–1047 [DOI] [PubMed] [Google Scholar]
- King E. M., Rachidi N., Morrice N., Hardwick K. G., Stark M. J., 2007. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 21: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J., Koshland D., 1991. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell 66: 483–495 [DOI] [PubMed] [Google Scholar]
- Kingston I. J., Yung J. S., Singleton M. R., 2011. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem. 286: 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R., Rose A. M., 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1: 514–521 [DOI] [PubMed] [Google Scholar]
- Kitajima T. S., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y., 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15: 353–359 [DOI] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Kitamura Y., Tanaka T. U., 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 21: 3319–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C., et al. , 2010. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell 18: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M., 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., Yanagida M., 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31: 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P., 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185: 841–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B., O’Toole E., McIntosh J. R., Mellone B., Allshire R., et al. , 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12: 2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockleby J., Vogel J., 2009. The COMA complex is required for Sli15/INCENP-mediated correction of defective kinetochore attachments. Cell Cycle 8: 2570–2577 [DOI] [PubMed] [Google Scholar]
- Koshland D., Hieter P., 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155: 351–372 [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Mitchison T. J., Kirschner M. W., 1988. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 331: 499–504 [DOI] [PubMed] [Google Scholar]
- Krassovsky K., Henikoff J. G., Henikoff S., 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 109: 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A., 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196: 451–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll E. S., Hyland K. M., Hieter P., Li J. J., 1996. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S., Lau D. T., Murray A. W., 2009. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat. Cell Biol. 11: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S., 2010. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Mieck C., Alushin G. M., Nogales E., Westermann S., 2013. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 200: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D. T., Murray A. W., 2012. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Curr. Biol. 22: 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K. S., Salmon E. D., 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., 1994. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 13: 5203–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Carbon J., 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64: 717–725 [DOI] [PubMed] [Google Scholar]
- Lefrancois P., Euskirchen G. M., Auerbach R. K., Rozowsky J., Gibson T., et al. , 2009. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P., Auerbach R. K., Yellman C. M., Roeder G. S., Snyder M., 2013. Centromere-like regions in the budding yeast genome. PLoS Genet. 9: e1003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M., Li Y., Elledge S. J., 2005. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/protein kinase A pathway and a novel subunit required for Ask1 association. Mol. Cell. Biol. 25: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., 1999. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. USA 96: 4989–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A. W., 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531 [DOI] [PubMed] [Google Scholar]
- Li X., Nicklas R. B., 1995. Mitotic forces control a cell-cycle checkpoint. Nature 373: 630–632 [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea C., Mahaffey D., Rechsteiner M., Benezra R., 1997. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl. Acd. Sci. 94: 12431–12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A. A., Wang Y., Qin J., et al. , 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M. J., Lampson M. A., Lens S. M., 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C. B., Hori T., Fukagawa T., et al. , 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochmann B., Ivanov D., 2012. Histone H3 localizes to the centromeric DNA in budding yeast. PLoS Genet. 8: e1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J. A., Biggins S., 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22: 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Xu X., Hall H., Hyland E. M., Boeke J. D., et al. , 2010. Histone h3 exerts a key function in mitotic checkpoint control. Mol. Cell. Biol. 30: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Fang G., Coldiron M., Lin Y., Yu H., et al. , 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7: 224–229 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H., 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9: 59–71 [DOI] [PubMed] [Google Scholar]
- Maddox P. S., Bloom K. S., Salmon E. D., 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., et al. , 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Massimiliano L., Santaguida S., Musacchio A., 2007. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell 131: 730–743 [DOI] [PubMed] [Google Scholar]
- Maresca T. J., 2011. Cell division: aurora B illuminates a checkpoint pathway. Curr. Biol. 21: R557–R559 [DOI] [PubMed] [Google Scholar]
- Maresca T. J., Salmon E. D., 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D. P., Hu X. W., Singleton M. R., 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D. R., Demirel P. B., Stukenberg P. T., Burke D. J., 2012. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26: 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison C. P., Old W. M., Steiner E., Huneycutt B. J., Resing K. A., et al. , 2007. Mps1 activation loop autophosphorylation enhances kinase activity. J. Biol. Chem. 282: 30553–30561 [DOI] [PubMed] [Google Scholar]
- Maure J. F., Kitamura E., Tanaka T. U., 2007. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 17: 2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure J. F., Komoto S., Oku Y., Mino A., Pasqualato S., et al. , 2011. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr. Biol. 21: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., 2011. The CCAN complex: linking centromere specification to control of kinetochore-microtubule dynamics. Semin. Cell Dev. Biol. 22: 946–952 [DOI] [PubMed] [Google Scholar]
- McCarroll R. M., Fangman W. L., 1988. Time of replication of yeast centromeres and telomeres. Cell 54: 505–513 [DOI] [PubMed] [Google Scholar]
- McCleland M. L., Gardner R. D., Kallio M. J., Daum J. R., Gorbsky G. J., et al. , 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B. F., Dong Y., 2010. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell. Mol. Life Sci. 67: 2163–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., 2012. Motors or dynamics: What really moves chromosomes? Nat. Cell Biol. 14: 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., et al. , 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell 135: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., O’Toole E., Zhudenkov K., Morphew M., Schwartz C., et al. , 2013. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 200: 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J. C., Shepperd L. A., Vanoosthuyse V., Lancaster T. C., Sochaj A. M., et al. , 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell 20: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Hailey D. W., Pot I., Givan S. A., Hyland K. M., et al. , 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Wood J. S., Garvik B., Hartwell L. H., 1986. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell 44: 53–63 [DOI] [PubMed] [Google Scholar]
- Megee P. C., Mistrot C., Guacci V., Koshland D., 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4: 445–450 [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D., 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6: 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D., 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11: 3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Yang P., Glowczewski L., Koshland D., Smith M. M., 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613 [DOI] [PubMed] [Google Scholar]
- Merdes A., De Mey J., 1990. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur. J. Cell Biol. 53: 313–325 [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Miller S. A., Johnson M. L., Stukenberg P. T., 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 18: 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C., 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Mirchenko L., Uhlmann F., 2010. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 20: 1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M., 1984. Dynamic instability of microtubule growth. Nature 312: 237–242 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C., 2007. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Molodtsov M. I., Grishchuk E. L., Efremov A. K., McIntosh J. R., Ataullakhanov F. I., 2005. Force production by depolymerizing microtubules: a theoretical study. Proc. Natl. Acad. Sci. USA 102: 4353–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., 2011. A brief history of error. Nat. Cell Biol. 13: 1178–1182 [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W., 1983. Construction of artificial chromosomes in yeast. Nature 305: 189–193 [DOI] [PubMed] [Google Scholar]
- Musacchio A., 2011. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave D. R., Sandman K. M., Reeve J. N., 1991. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl. Acad. Sci. USA 88: 10397–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K., Bloom K. S., 2003. Differential kinetochore protein requirements for establishment vs. propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Tyers R. G., Wong C. C., Yates J. R., 3rd, Drubin D. G., et al. , 2009. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki N., Yoneyama M., Koshiba S., Tochio N., Inoue M., et al. , 2004. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 13: 2089–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V., 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Carbon J., 1987. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 4522–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. M., Waples W. G., Lavoie B. D., Biggins S., 2009. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell 20: 3818–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B., 1997. How cells get the right chromosomes. Science 275: 632–637 [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Koch C. A., 1969. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 43: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K. E., Suzuki A., Hori T., et al. , 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., et al. , 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9: 105–120 [DOI] [PubMed] [Google Scholar]
- Ohkuni K., Abdulle R., Tong A. H., Boone C., Kitagawa K., 2008. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One 3: e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., et al. , 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Oliveira R. A., Nasmyth K., 2010. Getting through anaphase: splitting the sisters and beyond. Biochem. Soc. Trans. 38: 1639–1644 [DOI] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J., 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13: 1140–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Funk C., Schafer A., Lechner J., 2009. Stu1 inversely regulates kinetochore capture and spindle stability. Genes Dev. 23: 2778–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C., Draviam V. M., Marco E., Sorger P. K., De Wulf P., 2009. Roles for the conserved Spc105p/Kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE 4: e7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palframan W. J., Meehl J. B., Jaspersen S. L., Winey M., Murray A. W., 2006. Anaphase inactivation of the spindle checkpoint. Science 313: 680–684 [DOI] [PubMed] [Google Scholar]
- Palmer D. K., O’Day K., Wener M. H., Andrews B. S., Margolis R. L., 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Chen R. H., 2004. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 18: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F., Spencer F., 1996. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell 7: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K., 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Yeh E., Gardner M., Odde D., Salmon E. D., et al. , 2004. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14: 1962–1967 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E., 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302: 2120–2124 [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T., 2011. The ABCs of CENPs. Chromosoma 120: 425–446 [DOI] [PubMed] [Google Scholar]
- Perriches T., Singleton M. R., 2012. Structure of yeast kinetochore Ndc10 DNA-binding domain reveals unexpected evolutionary relationship to tyrosine recombinases. J. Biol. Chem. 287: 5173–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., et al. , 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrasanta L. I., Thrower D., Hsieh W., Rao S., Stemmann O., et al. , 1999. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci. USA 96: 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Tatsutani S. Y., Collins K. A., Biggins S., 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5: 735–745 [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S., 2005. The spindle checkpoint: tension vs. attachment. Trends Cell Biol. 15(9): 486–493 [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S., 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8: 78–83 [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Nelson C. R., Biggins S., 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I., Winston F., 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19: 1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar A., Roy N., Sinha P., 1999. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 31: 349–360 [DOI] [PubMed] [Google Scholar]
- Poddar A., Daniel J. A., Daum J. R., Burke D. J., 2004. Differential kinetochore requirements for establishment and maintenance of the spindle checkpoint are dependent on the mechanism of checkpoint activation in Saccharomyces cerevisiae. Cell Cycle 3: 197–204 [PubMed] [Google Scholar]
- Pohl T. J., Brewer B. J., Raghuraman M. K., 2012. Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet. 8: e1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot I., Measday V., Snydsman B., Cagney G., Fields S., et al. , 2003. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14: 460–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. F., Franck A. D., Gestaut D. R., Cooper J., Gracyzk B., et al. , 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M. R., Venkei Z., Bolanos-Garcia V. M., Debski J., Dadlez M., et al. , 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21: 399–405 [DOI] [PubMed] [Google Scholar]
- Purvis A., Singleton M. R., 2008. Insights into kinetochore-DNA interactions from the structure of Cep3Delta. EMBO Rep. 9: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey V. H., Wang H. W., Nakajima Y., Wong A., Liu J., et al. , 2011. The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol. Biol. Cell 22: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Crispo V., Lucchini G., Piatti S., 2005. Mad3/BubR1 phosphorylation during spindle checkpoint activation depends on both Polo and Aurora kinases in budding yeast. Cell Cycle 4: 972–980 [DOI] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Alexander S. P., 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Schultz A., Cole R., Sluder G., 1994. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. T., Farr K. A., Hoyt M. A., 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 14: 8282–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M. C., Thomas S., Bouck D. C., Kaplan K. B., 2004. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell 15: 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J. S., Cross F. R., Funabiki H., 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell I. D., Grancell A. S., Sorger P. K., 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145: 933–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I. X., Yates J. R., 3rd, Oegema K., et al. , 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 127: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K., Ghosh S. K., Sinha P., 1998. The MCM16 gene of the yeast Saccharomyces cerevisiae is required for chromosome segregation. Mol. Gen. Genet. 260: 242–250 [DOI] [PubMed] [Google Scholar]
- Sarangapani K. K., Akiyoshi B., Duggan N. M., Biggins S., Asbury C. L., 2013. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc. Natl. Acad. Sci. USA 110: 7282–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A. T., van der Waal M. S., Medema R. H., Lens S. M., Kops G. J., 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerling P., Holmberg S., 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24: 4599–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., et al. , 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14: 604–613 [DOI] [PubMed] [Google Scholar]
- Schmitzberger F., Harrison S. C., 2012. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 13: 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. V., Botvinick E., Bonday Z., Furnari F., Berns M., et al. , 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14: 942–952 [DOI] [PubMed] [Google Scholar]
- Shang C., Hazbun T. R., Cheeseman I. M., Aranda J., Fields S., et al. , 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell 14: 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R. H., 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L. A., Meadows J. C., Sochaj A. M., Lancaster T. C., Zou J., et al. , 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero J. H., Koval M., Spencer F., Palmer R. E., Hieter P., et al. , 1991. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 194: 749–773 [DOI] [PubMed] [Google Scholar]
- Shimogawa M. M., Graczyk B., Gardner M. K., Francis S. E., White E. A., et al. , 2006. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr. Biol. 16: 1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa M. M., Wargacki M. M., Muller E. G., Davis T. N., 2010. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle 9: 3619–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M., Camahort R., Mattingly M., Gerton J. L., 2011. Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 286: 12016–12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M., Unruh J. R., Slaughter B. D., Mattingly M., Berman J., et al. , 2012. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. C., Bodor D. L., Stellfox M. E., Martins N. M., Hochegger H., et al. , 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22: 52–63 [DOI] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K. T., et al. , 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ’safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Severin F. F., Hyman A. A., 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127: 995–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Doheny K. F., Hieter P., Kopski K. M., Huffaker T. C., et al. , 1995. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA 92: 12026–12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P., 1990. Mitotic chromosome transmission mutants in Saccharomyces cervisiae. Genetics 124: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Hieter P., 1992. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 8908–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O., Lechner J., 1996. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 15: 3611–3620 [PMC free article] [PubMed] [Google Scholar]
- Stemmann O., Neidig A., Kocher T., Wilm M., Lechner J., 2002. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. USA 99: 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A. D., Haggerty R. A., Vasquez P. A., Vicci L., Snider C. E., et al. , 2013. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J. Cell Biol. 200: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B. M., Murray A. W., 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586 [DOI] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., et al. , 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA 104: 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z., Becker J. S., Talarek N., Kogelsberger S., Pellman D., 2011. Bub1, Sgo1, and Mps1 mediate a distinct pathway for chromosome biorientation in budding yeast. Mol. Biol. Cell 22: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W., 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608 [DOI] [PubMed] [Google Scholar]
- Strunnikov A. V., Kingsbury J., Koshland D., 1995. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G. K., Yen T. J., 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L. J., Guimaraes G. J., DeLuca J. G., 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell 22: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Fukagawa T., 2012. Molecular architecture of vertebrate kinetochores. Exp. Cell Res. 318: 1367–1374 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., et al. , 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Cosma M. P., Wirth K., Nasmyth K., 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98: 847–858 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K., 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2: 492–499 [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., 2010. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 29: 4070–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., et al. , 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H., 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1: 227–237 [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H., 2004. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16: 387–397 [DOI] [PubMed] [Google Scholar]
- Tien J. F., Umbreit N. T., Gestaut D. R., Franck A. D., Cooper J., et al. , 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J. D., Sorger P. K., 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. S., Takagaki K., Kumada K., Hirayama Y., Noda T., et al. , 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit N. T., Davis T. N., 2012. Mitosis puts sisters in a strained relationship: force generation at the kinetochore. Exp. Cell Res. 318: 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M. S., Saurin A. T., Vromans M. J., Vleugel M., Wurzenberger C., et al. , 2012. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13: 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K. G., 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Chandrasekaran S., Sundin L. J., Reidy K. T., Wan X., et al. , 2012. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat. Cell Biol. 14: 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J. S., Gardner R., Stephens A. D., Yeh E., Bloom K., 2012. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol. Biol. Cell 23: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V. A., Zaytsev A. V., Gudimchuk N., Grissom P. M., Gintsburg A. L., et al. , 2013. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc. Natl. Acad. Sci. USA 110: 7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Cai S., Khodjakov A., 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. W., Ramey V. H., Westermann S., Leschziner A. E., Welburn J. P., et al. , 2007. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol. 14: 721–726 [DOI] [PubMed] [Google Scholar]
- Wang H. W., Long S., Ciferri C., Westermann S., Drubin D., et al. , 2008. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol. 383: 894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Burke D. J., 1995. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6838–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. J., Huffaker T. C., 1997. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargacki M. M., Tay J. C., Muller E. G., Asbury C. L., Davis T. N., 2010. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle 9: 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. D., Brady D. M., Johnston R. C., Hanna J. S., Hardwick K. G., et al. , 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13: 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. A., Gerton J. L., Polancic J. E., DeRisi J. L., Koshland D., et al. , 2004. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2: E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. R., Sorger P. K., Harrison S. C., 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA 102: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. R., Schnell J. R., Larsen N. A., Sorger P. K., Chou J. J., et al. , 2006. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure 14: 1003–1009 [DOI] [PubMed] [Google Scholar]
- Wei R. R., Al-Bassam J., Harrison S. C., 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Weiss E., Winey M., 1996. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J. P., Cheeseman I. M., 2008. Toward a molecular structure of the eukaryotic kinetochore. Dev. Cell 15: 645–655 [DOI] [PubMed] [Google Scholar]
- Welburn J. P., Vleugel M., Liu D., Yates J. R., III, Lampson M. A., et al. , 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells W. A. E., Murray A. W., 1996. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J. Cell Biol. 133: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., et al. , 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., et al. , 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H. W., Avila-Sakar A., Drubin D. G., Nogales E., et al. , 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G., 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76: 563–591 [DOI] [PubMed] [Google Scholar]
- Widlund P. O., Lyssand J. S., Anderson S., Niessen S., Yates J. R., 3rd, et al. , 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell 17: 1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A., Kilmartin J. V., 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A., Jensen O. N., Holmes S., Soues S., Mann M., et al. , 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windecker H., Langegger M., Heinrich S., Hauf S., 2009. Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin. EMBO Rep. 10: 1022–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Bloom K., 2012. Mitotic spindle form and function. Genetics 190: 1197–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B., 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mamay C. L., O’Toole E. T., Mastronarde D. N., Giddings T. H., Jr, et al. , 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., et al. , 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Mizuguchi G., Wisniewski J., Huang Y., Wei D., et al. , 2011. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell 43: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhu S., Wang W., Zhang X., Old W., et al. , 2009. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol. Biol. Cell 20: 10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y., 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330: 239–243 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C. H., Tanno Y., Watanabe Y., 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14: 746–752 [DOI] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D. R., Machius M., Rizo J., et al. , 2007. p31comet blocks Mad2 activation through structural mimicry. Cell 131: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., et al. , 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Carbon J., 1999. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA 96: 13208–13213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Kahana J. A., Silver P. A., Morphew M. K., McIntosh J. R., et al. , 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Kelstrup C. D., Hu X. W., Kaas Hansen M. J., Singleton M. R., et al. , 2012. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J. Cell Sci. 125: 3243–3253 [DOI] [PubMed] [Google Scholar]
- Zhou P., Howley P. M., 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2: 571–580 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Feng H., Zhou B. R., Ghirlando R., Hu K., et al. , 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zich J., Hardwick K. G., 2010. Getting down to the phosphorylated ’nuts and bolts’ of spindle checkpoint signalling. Trends Biochem. Sci. 35: 18–27 [DOI] [PubMed] [Google Scholar]
- Zich J., Sochaj A. M., Syred H. M., Milne L., Cook A. G., et al. , 2012. Kinase activity of fission yeast Mph1 is required for Mad2 and Mad3 to stably bind the anaphase promoting complex. Curr. Biol. 22: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]