Abstract
The rapid evolution of essential developmental genes and their protein products is both intriguing and problematic. The rapid evolution of gene products with simple protein folds and a lack of well-characterized functional domains typically result in a low discovery rate of orthologous genes. Additionally, in the absence of orthologs it is difficult to study the processes and mechanisms underlying rapid evolution. In this study, we have investigated the rapid evolution of centrosomin (cnn), an essential gene encoding centrosomal protein isoforms required during syncytial development in Drosophila melanogaster. Until recently the rapid divergence of cnn made identification of orthologs difficult and questionable because Cnn violates many of the assumptions underlying models for protein evolution. To overcome these limitations, we have identified a group of insect orthologs and present conserved features likely to be required for the functions attributed to cnn in D. melanogaster. We also show that the rapid divergence of Cnn isoforms is apparently due to frequent coding sequence indels and an accelerated rate of intronic additions and eliminations. These changes appear to be buffered by multi-exon and multi-reading frame maximum potential ORFs, simple protein folds, and the splicing machinery. These buffering features also occur in other genes in Drosophila and may help prevent potentially deleterious mutations due to indels in genes with large coding exons and exon-dense regions separated by small introns. This work promises to be useful for future investigations of cnn and potentially other rapidly evolving genes and proteins.
Keywords: Cnn, centrosome, rapid evolution, indels, Drosophila
AS the genomic era advances, molecular biology has shifted from single-gene studies to global analyses of genomes from a broad phylogenetic range of organisms. For single-gene and global studies, a critical first step for molecular biologists is the accurate identification of orthologous genes. The concept of orthology was introduced by Fitch to differentiate between orthologous genes, which have a one-to-one relationship between species, and paralogous genes, which result from a duplication event of the original ortholog within a species (Fitch 1970, 2000). Because orthologs are often more closely related than paralogs, it is generally assumed that gene function is likely to be conserved. Since the functional annotation of uncharacterized genomes is generally computationally based on overall sequence identity with known genes from model organisms, it is essential for orthology assignments to be accurate (Hulsen et al. 2006). The importance of orthology is made evident both by the number of databases reporting orthologous gene sets and by the number of terms introduced to describe relational subcategories in the time since Fitch initially introduced the concept of orthologous and paralogous gene relationships.
Since orthologous genes are related by descent and orthology is based on sequence homology and phylogeny, the process of identification should be relatively simple. This is certainly true for many genes, but for a significant subset of genes several problems confound the process. One method of finding orthologs is to use orthologous gene sets from public databases, but these gene sets are variable depending on the methods used to determine orthology and the number of genomes used to create the database (Altenhoff and Dessimoz 2009). It has also been shown that gene prediction errors (Nagy et al. 2011b) and confusing orthologs, paralogs, and epaktologs, or genes that acquire a conserved domain independently (Nagy et al. 2011a), all confound orthologous gene identification. In both Nagy et al. studies it was found that these errors are common enough to result in erroneous predictions in several global analyses of protein-domain architecture evolution. At the single-gene level, the above problems may be further complicated by rapid evolution and the absence of known curated functional domains. Although it is generally accepted that multi-domain proteins encoded by orthologous genes should share multiple domains and domain architecture, and that low-complexity regions should not be used to determine orthology (Galperin and Koonin 1998; Mushegian et al. 1998), these criteria are easily ignored when molecular data are limited.
One example of how these issues complicate orthologous gene identification is provided by Drosophila melanogaster centrosomin (cnn), a gene whose function is essential for formation of functional centrosomes, organization of the actin and microtubule cytoskeleton, and chromosome segregation during the rapid cleavage divisions in syncytial embryos (Megraw et al. 1999; Vaizel-Ohayon and Schejter 1999; Eisman et al. 2009). Simple searches of orthologous gene sets for cnn from public databases are variable. Based on results from OrthoDB and EggNOG cnn orthologs that are limited to the insects, Homologene predicts that the insects and the vertebrate gene Cyclin Dependent Kinase5 Regulatory Activating Protein2 (CDK5RAP2), but not the paralogous vertebrate gene myomegalin (mmg), and Inparanoid has a complex and somewhat inconsistent mix of putative orthologs. Sequence alignments of these various gene sets suggest that OrthoDB and EggNOG may be the best set for functional inference from D. melanogaster experimental data, but is this a correct assumption?
There are several reasons that may explain the variability in cnn orthologous gene sets. Comparisons of Cnn proteins within the genus Drosophila indicate that the gene and encoded proteins are evolving rapidly and that approximately half the length of all Cnn proteins form simple coiled-coil domains, which are not reliable for determining orthology (Tatusov et al. 1997). Additionally, D. melanogaster cnn is transcriptionally complex, encoding at least a dozen splice variants that form two protein families (Eisman et al. 2009). Possibly because of this complexity and the rapidly evolving sequence, many cnn gene models within the insects are variable, containing either every possible exon in the gene or lacking one or more exons. Finally, with the exception of one small domain (Flory et al. 2002; Sawin et al. 2004; Zhang and Megraw 2007; Fong et al. 2008), cnn encodes no other identified functional protein domains.
The one conserved domain in cnn used to infer orthology across broad phylogenetic distances is found in a protein superfamily and is predicted to be associated with microtubules (Flory et al. 2002; Venkatram et al. 2004). This domain is found in a single gene in fungal and insect species and in two genes in vertebrate species. Although the conservation of the domain predicts that genes in the superfamily are likely to have some functional similarities, it does not necessarily by itself support strong functional inference from any or all characterized genes across the superfamily of proteins. Several experimental studies of genes with this domain from fungi and vertebrates have found that gene function diverges rapidly within these lineages, as well as between the paralogous vertebrate genes (see Discussion). Additionally, in D. melanogaster we have shown that this domain is in a constitutive cnn exon in both long- and short-form families of Cnn proteins (Eisman et al. 2009). However, the localization of short forms to mitotic centrosomes in mutants lacking long forms fails to rescue the mitotic defects (Eisman et al. 2009). This shows that this single domain cannot explain Cnn function and that the domain alone should not determine orthology, but, as mentioned above, in some cases it has.
While the above data suggest known functions for D. melanogaster cnn may be applicable only to other insect species, claims of orthology between cnn and the vertebrate genes CDK5RAP2 and mmg to infer function are becoming common in the literature. Additionally, the Bloomington Stock Center now lists cnn as an ortholog of CDK5RAP2 useful for studying, in Drosophila, the human disease autosomal recessive primary microcephaly. Since both cnn and CDK5RAP2 are centrosomal proteins necessary for the localization of γ-tubulin (Megraw et al. 2001; Fong et al. 2008), it is possible that experimental studies of CDK5RAP2 could provide new insights and directions for future molecular studies of cnn. Alternatively, phenotypic similarities between mutational studies of cnn and CDK5RAP2 may be due to direct and indirect effects associated with loss of essential centrosome function.
To differentiate between these possibilities, it is important to establish a reliable cnn orthologous gene set likely to have similar functions. As mentioned above, cnn is transcriptionally complex and a mixed pool of Cnn-encoded splice variants is present at embryonic centrosomes (Eisman et al. 2009). The identification of all splice variants is difficult at best as gene models in other species predict a single protein and the rapid evolution of cnn make ab initio exon and splicing predictions unreliable. However, the D. melanogaster Cnn-PA isoform spans the entire gene, and this isoform is sufficient to rescue the mutant phenotype provided short isoforms are present (Eisman et al. 2009). Moreover, orthologous genes encoding Cnn-PA-like isoforms will have sequence information for future transcriptional studies and identify the majority of the protein domains necessary for cnn function.
One possible explanation for the fact that gene model predictions for cnn within the insects are often incorrect may be due to the nature of the sequence spanning coding exons and intervening introns. The general assumption is that the reading frame of a coding exon will encounter a stop codon in the adjacent intron. A recent study found that the frequency of in-frame stop codons in the first 40–70 bases of the adjacent downstream intron is 40–70%, respectively (Zhang et al. 2003). However, in this study of insect cnn orthologs, we show that the reading frame of an exon frequently extends well beyond adjacent introns, and in some cases a single reading frame may include multiple coding exons and introns. For discussion purposes, we present the concept of maximum potential open reading frames (MPOs) associated with coding exons. While the MPO for most coding exons is likely to be similar to the actual spliced exon, we have found a subset of genes in Drosophila that contain much longer MPOs and multi-exon MPOs. These extensive MPOs are associated with exon-dense regions containing small introns.
We also show that this type of MPO is associated with frequent nucleotide changes and the rapid accumulation of random small insertions and deletions (indels). Since indels in protein-coding sequences are considered to be rare genomic changes (Rokas and Holland 2000) and indels ranging from a single amino acid to 20 amino acids have been used to determine phylogenies (Gupta 1998; Keeling et al. 2000), why are they apparently common and random in Cnn? The underlying assumption is that these indels are due to codon loss, but the rapid nucleotide sequence divergence adjacent to Cnn indels suggests that some may be nontriplet indels that change the reading frame. In this study, we present evidence that nontriplet indels may be buffered against by extended MPOs, in conjunction with an unknown relaxed splicing mechanism and simple protein folds encoded by the exons. These three features appear to make it possible to retain protein function when normally deleterious mutations occur.
This work presents a detailed analysis of a cnn orthologous gene set from the insects. In addition to the domain mentioned above, we identify three additional highly conserved motifs unique to insects and show that the overall structure of Cnn-PA is conserved across several insect orders. While protein motifs and structure are conserved, the gene and protein diverge considerably between species and orders. The intron–exon structure of cnn is similar within an order but significantly different among the orders investigated, and these changes are correlated with the rapid evolution of Cnn proteins. In addition to gene architecture changes, we show that nucleotide substitution and frequent indels are associated with the rapid evolution of Cnn-PA and that these changes appear to be buffered by the inherent nature of the sequence, simple protein folds, and potentially relaxed splicing mechanisms. While these changes are buffered within a species, we show that the divergence of Cnn-PA has a dominant-negative affect on protein function between species. Finally, we present the concept of MPOs associated with coding exons. The extended MPOs present in cnn appear to be common in rapidly evolving genes in Drosophila and may buffer against potentially deleterious genomic changes. Additionally, we show that comparative studies of changes between homologous MPOs provide a useful tool for the identification of relatively small yet significant changes in coding regions across a broad phylogenetic range. It is our hope that this provides a comprehensive set of orthologs for molecular studies of cnn and a framework for the functional annotation of orthologs, paralogs, and epaktologs that share some degree of homology with Cnn.
Materials and Methods
Identification of centrosomin orthologs
The genus Drosophila orthologs were identified in GBrowse or by BLAST and downloaded from FlyBase (http://flybase.org/) and are taken from the frozen CAF1 assemblies and the Drosophila pseudoobscura Release 2.0. We also retrieved genomic sequences and RNA-Seq data from eight additional sequenced Drosophila genomes done by the Baylor sequencing center for the modENCODE project (http://www.hgsc.bcm.tmc.edu/collaborations/insects/dros_modencode/RNAseq_data/). These were manually annotated by exon-to-exon comparisons after translation based on known D. melanogaster complementary DNA (cDNA) clones available on FlyBase. All non-Drosophila sequences were identified by the orthology links in FlyBase) and downloaded from ENSEMBLE except Bombyx mori, which is based on a full-length cDNA obtained from the silk moth genome project. Sequences were annotated and mapped using MacVector 12.5 (Accelrys Inc.). Electronic versions of the annotated sequences are available on request.
Drosophila stocks
All flies used in this study were grown on standard cornmeal–agar medium at 25°. To express the D. melanogaster GFP::Cnn-PA fusion transgene during embryogenesis, UASpBacNPF::GFP::Cnn-PA homozygous transgenic males from D. melanogaster, Drosophila simulans, Drosophila yakuba, and D. pseudoobscura were crossed to appropriate females homozygous for the pBac(3xP3-EGFPafm)::nos-Gal4::(pW8-) transgene as previously described (Holtzman et al. 2010). Embryos collected from pBac(3xP3-EGFPafm)::nos-Gal4::(pW8-); w; P{w+mC=pUASP- GFP::Cnn-PA} mothers were stained for Cnn, α-tubulin, and DNA to determine the effects of D. melanogaster Cnn-PA chimeric protein on native Cnn function in divergent species.
Cloning and transformation
To construct the GFP::Cnn-PA fusion protein, we PCR-amplified EGFP (Living Colors by Invitrogen) with the following primers: SpeGfp-5′ (ACT AGT ATG GTG AGC AAG GGC GAG) and GfpMlu-3′ (ACG CGT CTT GTA CAG CTC GTC CAT G). We amplified the Cnn-PA transcript from a previously described Cnn-PA cDNA (Heuer et al. 1995) and added MluI and EcoRI sites with oligos using standard PCR techniques. PCR products were cloned into a Topo-TA pCR2.1 vector (Invitrogen) as per the manufacturer’s protocol and verified by sequencing. All other cloning was done using in-gel ligation techniques (Kalvakolanu and Livingston 1991). PCR products were digested with appropriate enzymes and subcloned into a pBlueskript vector. Following digestion with NotI, the GFP::Cnn-PA fusion was shuttled into the UASpBacNPF vector, and plasmid transformation was done as described (Holtzman et al. 2010).
Fixation and immunostaining
Embryos were collected and fixed in 50% heptane:50% Methanol/EGTA as previously described (Eisman et al. 2006). Embryos were immunostained as previously described (Gorman et al. 1991). Specimens were mounted on glass slides in 90% glycerol and 10% PBS with 0.2 mM n-propyl gallate (Sigma). The following antibodies were used: guinea pig (whole sera) anti-Cnn.Ex1a2 at 1:500 (Megraw et al. 1999) and anti-α-tubulin 84B (Matthews et al. 1989). DNA was stained with TOTO3 (Molecular Probes) at a final concentration of 1:1000. All fluorescent secondary antibodies were used at 1:200 (Jackson ImmunoResearch Labs).
Microscopy and imaging
All images were captured on a Leica TCS confocal microscope with a 63X HCX Plan Apo oil immersion objective, using TCSNT software. Images are all Z-series that range from 2.5 to 6 mm thick, composed of 0.5- to 0.9-mm-thick sections. Projected images were processed and assembled into figures with Adobe Photoshop version 7.0 software.
MPO search, genomic sequences, and gene annotation
To search the D. melanogaster genome for MPOs, we originally determined how far the reading frame of all annotated exons extended beyond the designated 5′ and 3′ ends using Release 4.2. These annotations have been updated and confirmed using a more recent FlyBase release (5.4). From these results we selected genes with at least one MPO extending >3000 nucleotides beyond an annotated end. From this set we selected seven genes and manually annotated the sequence of each gene based on protein isoforms available on FlyBase. The algorithm is available on request. Genomic sequences from FlyBase are from the frozen CAF1 assemblies and the D. pseudoobscura Release 2.0. Sequences were manually annotated as described above. The cnn sequences from a North Carolina population of D. melanogaster (Mackay et al. 2012) were recovered from http://dgrp.gnets.ncsu.edu/. These were annotated as described in the results. Electronic versions of the annotated sequences are available on request.
Results
The coding regions of centrosomin diverge rapidly
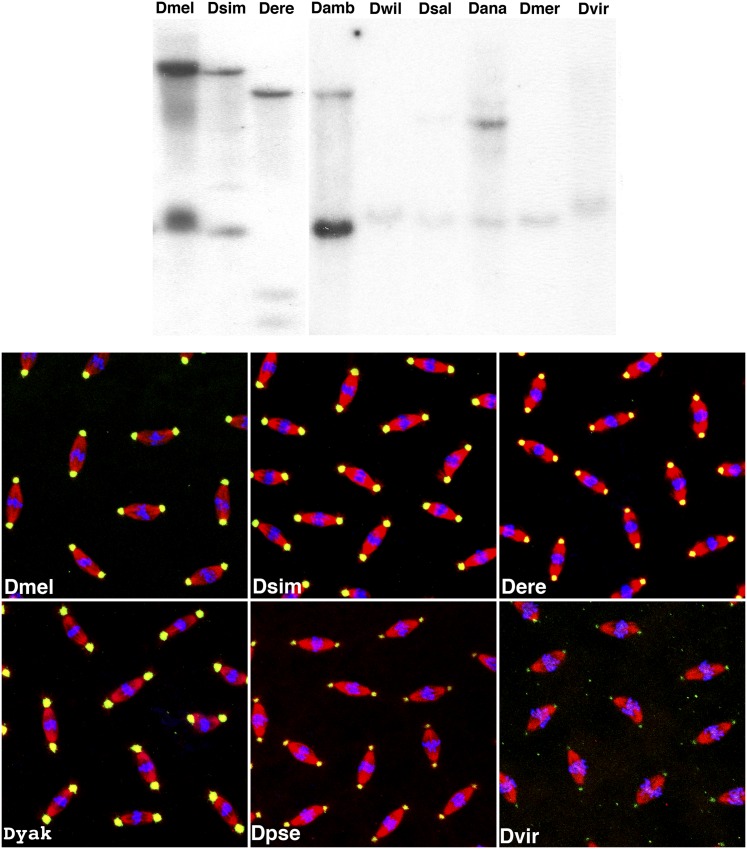
Genetic and molecular analyses of cnn in D. melanogaster suggest that multiple motifs and domains should be conserved in its orthologs. Cnn proteins are required for the initiation of PeriCentriolar Matrix (PCM) biosynthesis in D. melanogaster (Dobbelaere et al. 2008; Conduit and Raff 2010), and this function appears to be conserved within the genus Drosophila as Cnn is required for de novo centrosome formation during parthenogenetic development in Drosophila mercatorum (Eisman and Kaufman 2007). Additionally, we have shown that both long- and short-splice variants or isoforms are present during development and that these two protein families differ significantly at the protein sequence level (Eisman et al. 2009). However, two pieces of evidence suggest that the gene and protein products are evolving at a rapid rate within the genus. First, hybridization efficiency of the Cnn-PA coding sequence to genomic DNA on Southern blots decreases rapidly within the subgenus Sophophora and is not detectable outside the subgenus Drosophila (Figure 1, top). Second, the reactivity of the original Cnn antibody, which covers 67% of the Cnn-PA protein (Heuer et al. 1995), decreases significantly within the genus (Figure 1, bottom).
Figure 1.
The rapid divergence of cnn in Drosophila. (Top) The coding sequence of the centrosomin gene evolves rapidly within the genus Drosophila as shown by the weak hybridization of labeled cnn-RA transcripts with genomic DNA on two Southern blots. The first seven lanes are species in the subgenus Sophophora, and the strongest signals are members of the melanogaster group. The last two lanes are species in the subgenus Drosophila, representing the extent of the phylogenetic range producing detectable signal. (Bottom) The Cnn-PA protein also evolves rapidly, as evidenced by decreased reactivity of the anti-Cnn antibody in immunostained embryos. Cnn staining (green) is strong in Dmel, Dsim, Dere, and Dyak, all members of the melanogaster subgroup, but is weak in Dpse and almost undetectable in Dvir using the same antibody concentrations. The staining in both Dpse and Dvir can be improved by increasing the concentration of the Cnn antibody. Microtubules are shown in red and DNA in blue. Dmel: D. melanogaster; Dsim: D. simulans; Dere: D. erecta; Damb: D. ambigua; Dwil: D. willistoni; Dsal: D. saltans; Dana: D. ananassae; Dmer: D. mercatorum; Dvir: D. virilis; Dyak: D. yakuba; Dpse: D. pseudoobscura.
One possible explanation is that a single relatively small motif (discussed below) found in the protein is responsible for all Cnn function while the remainder of the protein is less important and thus can diverge. Alternatively, since Cnn is apparently essential only during syncytial development and spermatogenesis, the role of the gene in this environment may be relatively novel, and functionally orthologous genes may be present only over a limited phylogenetic range. To differentiate between these two possibilities, we have searched for and analyzed putative orthologs from several Drosophila species and other insects that have sufficient genomic sequence. We first report the results of our analysis of the genus Drosophila.
The D. melanogaster cnn-RA transcript was used to determine orthology as this transcript spans the entire gene, contains both constitutive D. melanogaster cnn exons, and rescues the cnn mutant phenotype in this species provided Cnn short isoforms are present (Eisman et al. 2009). This suggests that the cnn-RA splice variant contains the critical domain or domains necessary for Cnn long-isoform function. Since this transcript spans the entire gene, it also provides landmarks to investigate the conservation, or lack thereof, of the complex splicing previously described for the D. melanogaster cnn gene (Eisman et al. 2009). Based on the D. melanogaster Cnn-PA protein we have identified putative cnn orthologs from 13 Drosophila species.
Conserved motifs identify cnn orthologs
Prior to the identification of the cnn orthologs reported here, we knew that D. melanogaster Cnn-PA had three putative leucine zippers and several coiled-coil regions (Heuer et al. 1995) and that the protein was phosphorylated during syncytial development (Li and Kaufman 1996). To further investigate the rapid divergence of Cnn-PA, we first wanted to identify conserved features in the protein. This was done using the identified putative orthologs and their predicted encoded proteins, which were compared and the conserved sequences identified. It should be noted that we used only orthologs with high-quality genomic sequence that lacked large stretches of ambiguous base calls, or poly-“N” regions in the cnn gene, which unfortunately removed the closely related D. simulans and Drosophila sechellia species from our analyses. Any putative protein orthologs should possess a similar structural architecture and shared motifs. While we have identified many features that promise to be interesting candidates for future molecular experiments, in this study we present only those features likely to be conserved in all cnn orthologs.
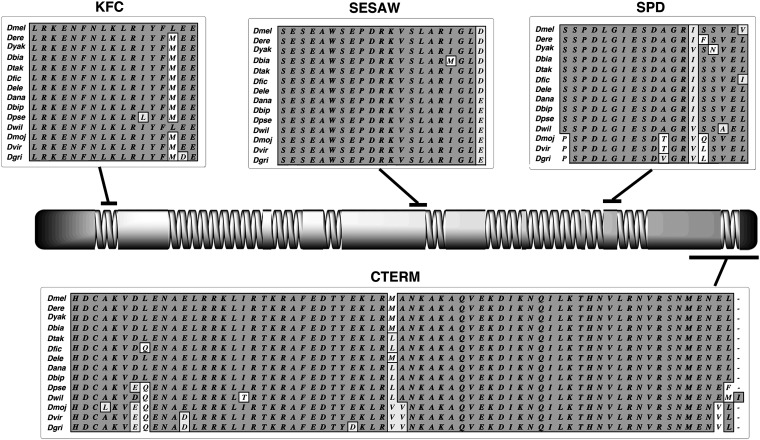
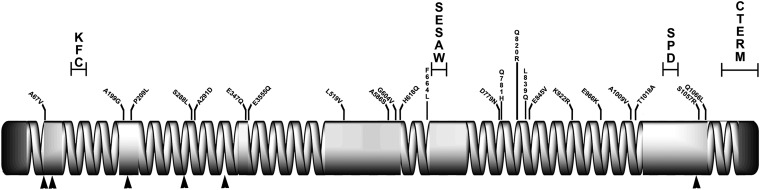
The first conserved Cnn motif is found in all D. melanogaster cnn splice variants and is highly conserved in all Drosophila species investigated here (Figure 2). This motif is a part of a conserved domain in a superfamily of proteins from fungi and animals and is in the amino terminus of these proteins. Experimental evidence shows that several of these proteins are associated with the microtubule cytoskeletal and/or the endomembrane system, but their overall functions are considerably different (see Discussion). The motif has been previously named Centrosomin Motif 1 (Zhang and Megraw 2007), but we prefer the name Ken-Flee Cytoskeleton motif (KFC) motif for two reasons. First, the inclusion of “Centrosomin” in the previous name infers that Cnn function as described for D. melanogaster will be retained in all proteins with this motif. Second, our name is sequence based and implies a more general function for the motif consistent with several experimental studies.
Figure 2.
Conserved motifs and structure of Cnn-PA in Drosophila. Protein alignments of the KFC, SESAW, and SPD motifs and the carboxy terminus are nearly invariant within the genus Drosophila and are present in all long-form splice variants of Cnn. Their position in Cnn-PA is shown aligned to the secondary structure of the protein. Coiled-coil domains are represented as spirals and noncoiled α-helical regions are solid. Dbip: D. biarmipes; Dtak: D. takahashii; Dfic: D. ficusphila; Dele: D. elegans; Dbip: D. bipectinata; Dmoj: D. mojavensis; Dgri: D. grimshawi.
The next two conserved motifs are present in all Drosophila Cnn potential orthologs. The first is centrally located and, based on its sequence, we have named it the SESAW motif (Figure 2). This motif is known to be phosphorylated at a protein kinase A site (http://www.phosphopep.org/) and, based on in silico analyses, also has two casein kinase II and one glycogen synthase site or shaggy kinase site with high probability scores for being phosphorylated. The second Drosophila-conserved sequence we have named the SPD motif (Figure 3). It is located near the carboxy terminus of the protein and has been shown to be phosphorylated at a putative protein kinase C site. Both of these motifs fall in regions predicted to be helical in character but not capable of forming a coiled-coil. Nonetheless, coiled-coil domains bound both. Based on BLAST analyses both motifs are present only in the arthropods. The actual function of these two motifs will be the subject of future studies, but it seems likely that they are involved in regulating Cnn function during the cell cycle.
Figure 3.
Drosophila MPOs confound gene-modeling programs. Splicing predictions for cnn transcripts are difficult due to a lack of strong splicing signals, multi-exon MPOs, and MPOs that extend well beyond the ends of known splice sites. A comparison of MPOs in all Drosophila species in this study (boxed area) showing all MPOs found in cnn reveals that MPOs are variable between species. At the top are all the MPOs in D. melanogaster cnn; the multi-colored boxes are the exons encoding Cnn-PA (below the grey boxes), and the gray and black boxes are coding exons present in other cnn splice variants. The MPO maps show that the transcriptional complexity of D. melanogaster cnn is probably conserved within the genus.
The fourth conserved Cnn motif is at the carboxy terminus (Figure 2) and is nearly invariant in the genus Drosophila. We have previously shown that deletion of this COOH motif and SPD in D. melanogaster results in the mislocalization of truncated Cnn to the region of the chromosomes of dividing nuclei rather than the centrosomes (Eisman et al. 2009). Thus a potential function for one or both of these motifs is to restrict Cnn to the centrosome.
In addition to these four conserved motifs, all Drosophila Cnn-PA proteins are ∼50% coiled-coil, and the arrangement of the coiled-coil regions is similar to D. melanogaster (Figure 2). The three D. melanogaster Cnn-PA leucine zippers are not conserved in the genus Drosophila, and the first and third zippers are absent in most of the proteins described here. The above data predict that true cnn orthologs should encode a coiled-coil protein containing the KFC, SESAW, and SPD motifs, as well as a carboxy terminus similar to the Drosophila tail. It is likely that these proteins will be an important component of the centrosome, but this prediction has yet to be shown experimentally outside Drosophila. Proteins with a subset of these motifs may share some functions attributable to Cnn, but it is not certain that they will be functional orthologs.
Rapid changes in the intron–exon structure of cnn orthologs and MPOs
While conservation of the above motifs is invaluable for the identification of potential cnn orthologs, the motifs provide no insights into the mechanism(s) underlying the rapid evolution of the gene. Since most of the non-Drosophila proteins (discussed below) were based on ab initio gene models, we wanted to know if there were apparent changes in gene architecture or exon–intron structure relative to the D. melanogaster cDNA-based model transcripts.
When we initiated this work the Drosophila genomes were assembled, but there were no GLEAN consensus models available and many of the individual models were highly variable, so we manually annotated the genes. To identify individual exons, we first identified a short coding “word” based in part on the conserved motifs in the presumed exon sequences and then extended the reading frame containing the word upstream and downstream to the first stop codon. We designate these extended reading frames as the MPO of the coding exon. Each MPO was then aligned with the D. melanogaster cnn-RA coding exons to determine a best estimate for splicing. This method was necessary since, based on in silico analysis, cnn has a paucity of strong canonical splicing sequences, which may partially explain the variability seen in individual ab initio models, which has appeared since we started this analysis. This fact combined with the observation that the MPO domains in D. melanogaster and all of the members of the genus examined here are overlapping and in many cases contain multiple exons, extending through regions known to be intronic in D. melanogaster, made the construction of a cogent gene model for the species difficult. A summary of the positions and extent of the MPOs in D. melanogaster and the other 13 species is shown in Figure 3.
To produce reasonable gene models that do not rely solely on ab initio predictions, it is necessary to have some independent assessment of the actual splicing pattern used by the different species. The robust model seen for D. melanogaster has historically relied on a collection of cDNAs, which have been more recently largely substantiated by RNA-Seq data (McQuilton et al. 2012). Unfortunately, with the exception of a single EST in D. pseudoobscura, there is no cDNA evidence available for the other 13 species. Fortunately, however, there are RNA-Seq data for 5 of the 13 species (https://www.hgsc.bcm.edu/content/drosophila-modencode-project). Using these data, it was possible to construct substantiated gene models for the exons encoding the ORF corresponding to Cnn-PA. With these models in hand, it was then possible to infer more precisely models for the orthologs of Cnn-PA in the other 8 species.
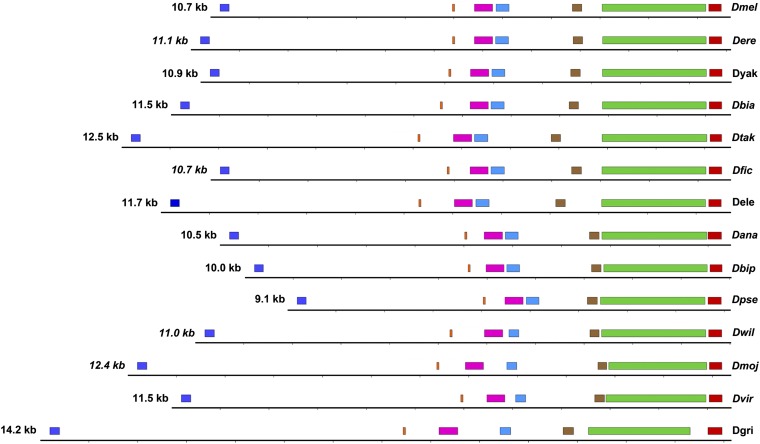
The results of that analysis, shown in Figure 4, reveal that within the genus Drosophila the overall gene architecture is apparently conserved and that putative coding exons are similar in size, but introns and exons have accumulated numerous indels resulting in gene size variability (Figure 4). Unlike the RNA-Seq-substantiated and deduced coding exons, the MPOs are variable in size, generally larger than the spliced exons, and the observed changes do not correlate with the phylogenetic order, suggesting that sequence changes are relatively common and undirected. Much of the variability between MPOs in different species can be accounted for by small internal indels between the conserved motifs, rather than by changes outside of the coding regions, and these indels account for protein size variability (Figure 4). Additionally, both exons 6 and 7 in the D. melanogaster model reside in a single MPO in seven of the Drosophila species investigated. Thus the potential exists to alternatively splice these two exons as a larger single exon retaining the intron between exons 6 and 7 (Figure 3 and Figure 4). However, extensive cDNA data in D. melanogaster show no evidence for the 6↔7 intron being retained during splicing of cnn transcripts, and the available RNA-Seq data indicate that the other species also splice a small intron out of the mature transcript.
Figure 4.
Intronic indels are common and random in Drosophila cnn. Cnn-PA coding exons (color coded as in Fig. 3) span the entire cnn gene. When maps are to scale and aligned at the conserved carboxy terminus, intronic indels are obvious. Because of frequent and random indels, the exon positions are variable as is the size of cnn across the genus. Gene sizes are shown on the left.
Annotation of cnn in other insects
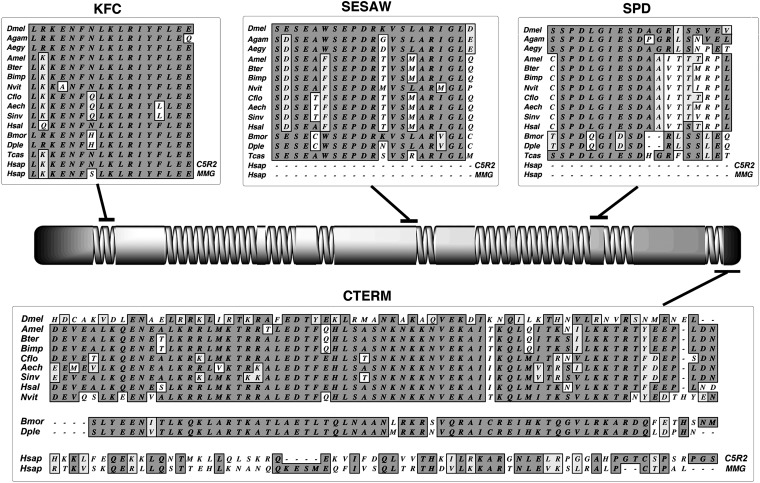
We next identified potential cnn orthologs in several other insects initially based on BLAST analyses of available databases. Our initial probes to these genomes were derived from the conserved domains found in the genus Drosophila. We were able to unambiguously identify KFC, SESAW, and SPD in two mosquitos, eight hymenopterans, two lepidopterans, and Tribolium (Figure 5).
Figure 5.
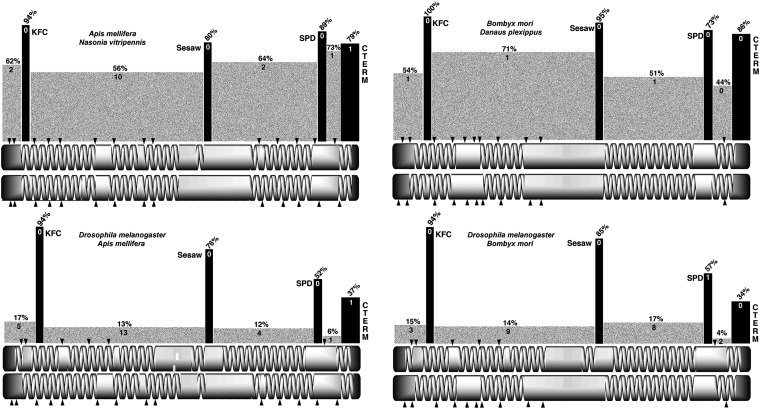
Cnn-PA motifs and structure are conserved within insects. Similar to Figure 2, we show the conserved motifs and carboxy terminus aligned to the structure of Cnn-PA from A. mellifera. Because some databases and the literature assert that the vertebrate genes CDK5RAP2 and mmg are orthologous to cnn, we have included the human co-orthologs in alignments. The human genes have a significantly different structure (not shown), the carboxy terminus has no significant homology to insects, and the terminus is more divergent between human paralogs than it is across the insects (bottom). The SESAW and SPD motifs are not detectable in any vertebrate gene. The Cnn motifs have diverged between orders but are highly conserved within orders. The carboxy terminus (CTERM) is the most divergent of the conserved motifs as shown in alignments between the Hymenoptera (top) and D. melanogaster (top, top line), but is highly conserved within the Hymenoptera and Lepidoptera (bottom). Agam: A. gambiae; Aegy: A. egypti; Amel: A. mellifera; Bter: B. terrestris; Bimp: B. impatiens; Nvit: N. vitripennis; Cflo: C. floridanus; Aech: A. echinatior; Sinv: S. invicta; Hsal: H. salator; Bmor: B. mori; Dple: D. plexippus; Hsap; Homo sapiens.
It was also possible to identify a COOH terminal conserved domain in the mosquitos, Hymenoptera, and Lepidoptera but, while this domain was conserved within those orders, the peptides are divergent between orders. Although the terminus diverges, it is nevertheless recognized by BLAST and Clustal analyses and is always contained in the last coding exon of the annotated genes.
Interestingly, COILS (Lupas et al. 1991) analyses show that all Cnn-PA orthologs investigated have a similar arrangement of coiled-coil domains. Additionally, while Cnn-PA orthologs vary from ∼720 amino acids in the Lepidoptera to almost 1400 amino acids in the Hymenoptera, the spacing of protein motifs is proportionally constant across all species. The second centrally located zipper is present in all insect species examined except the honeybee Apis mellifera. Thus the centrally located leucine zipper may be functionally important in some species, but it is not a good predictor of orthology.
Using these conserved motifs, our knowledge of the gene structure in flies, a cDNA from B. mori, and available gene models for the other species we annotated the cnn gene in these 13 additional insects. For the ant species Harpegnathos salator and Solenopsis invicta we altered the predicted gene models as these two models were missing the first two coding exons present in all other Hymenoptera models and these exons were clearly present in the genomic scaffold used to generate the original models. In the two lepidopterans B. mori and Danaus plexippus, a region similar to D. melanogaster exon 4 has apparently been lost. In the beetle Tribolium castaneum, there are three exons that span the region correlated with D. melanogaster exons 4 and 5, but the peptide sequences for these exons are not similar to any of the other species in this study. The results of this analysis are shown in Figure 6.
Figure 6.
cnn intron–exon structure changes significantly between insect orders. A comparison of Cnn-PA coding exons relative to D. melanogaster in the Diptera (A), Hymenoptera (B), and Lepidoptera and one coleopteran (C) shows that multiple exon fusions and splitting events have occurred in cnn orthologs. Coding exons are color-coded as in Figure 3 to indicate sequence homology. The gray boxes in T. castaneum have no homology to any other Cnn proteins, but the motifs and structure are conserved, and the lepidopterans have no exons homologous to D. melanogaster exon 4. Although homologous exons are split in the hymenopterans and D. plexippus, the overall arrangement of coding exons is similar to Drosophila. As in Drosophila, gene size (left) is variable.
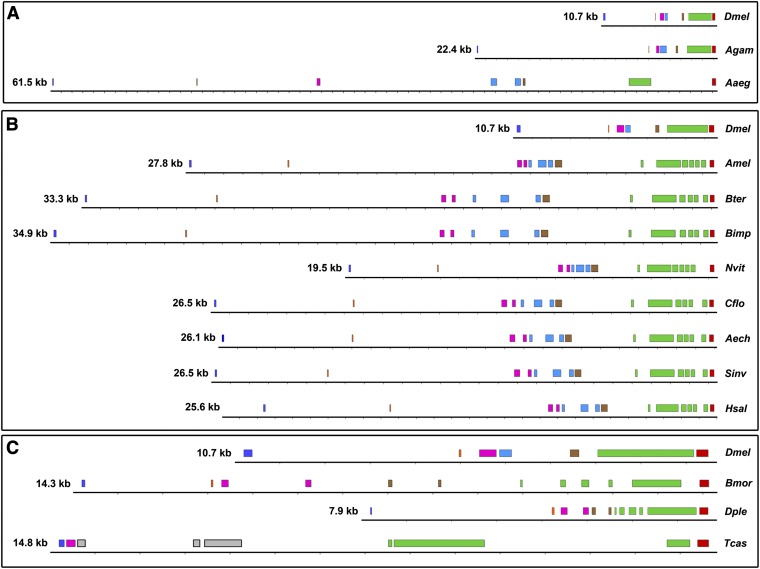
With the exception of the mosquitos, Cnn-PA is apparently encoded by more exons relative to the genus Drosophila in all of the insects investigated. Although fragmented, hymenopteran coding exons are in islands along the length of the gene, and two intronic deletions would result in Drosophila-like maps. In general, the overall gene architecture is conserved within orders but is considerably different among orders. The exception to this pattern is Aedes aegypti, which has an additional intron relative to Anopheles gambiae and the Drosophila species (Figure 6). Of particular interest are the coding sequences encompassing D. melanogaster exons 6 and 7, as this region contains three of the orthologous motifs described above and exon 6 is the largest cnn exon in D. melanogaster. In the non-dipteran species examined, exon 7 is a single terminal exon in all cases, but the homologous exon 6 region appears to be split by three, four, or five introns, depending on the species (Figure 6). Unlike the extensive MPOs found in Drosophila (Figure 2) as well as in the mosquito A. egypti (data not shown), the MPOs in the species shown in Figure 6, with one exception in the Hymenoptera (discussed below), are approximately co-extensive with the coding exons. The presence of multi-exon MPOs in Drosophila sp. and A. gambiae, but not in the species with larger genomes, suggests that multi-exon MPOs may be associated with reduced genome size.
Although the ancestral intron–exon structure of cnn is not known, it seems likely that there have been multiple exon fusions in the dipteran lineages investigated. This is consistent with the finding that the reduction in genome size in Drosophila (Petrov and Hartl 1997, 1998; Petrov et al. 2000; Petrov 2002) and A. gambiae (Nene et al. 2007) is due to deletions of intronic and intergenic regions. Since the immediate gain of a complete intron would likely have deleterious effects on protein function or introduce a premature stop codon, the multi-exon MPOs may represent an intermediate step during the process of exon fusion.
Sequence divergence in Cnn in the genus Drosophila and other insects
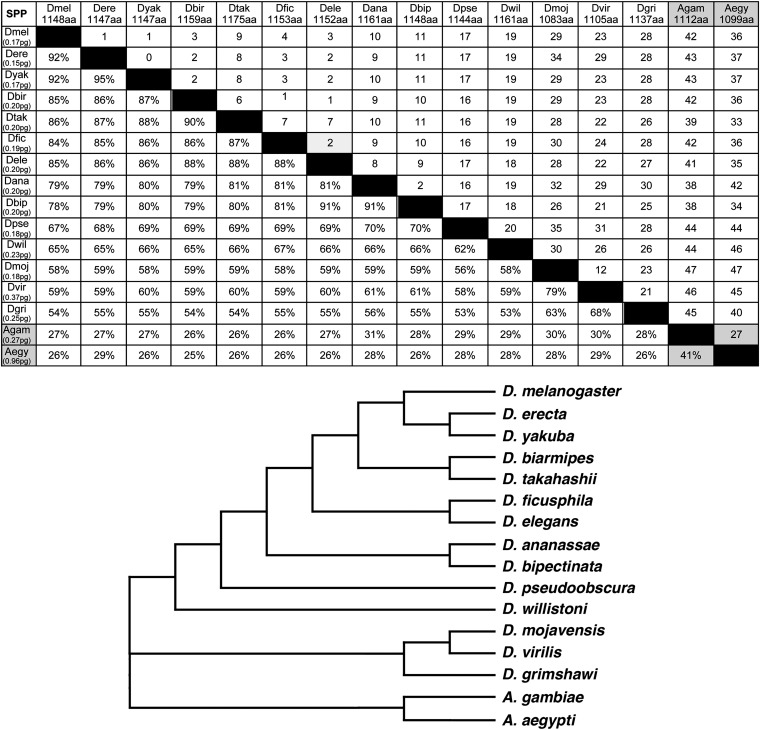
As noted above, there are four highly conserved domains within the amino acid sequence of Cnn-PA in the genus Drosophila. This stands in contrast to the fact that the protein as a whole is evolving rapidly. To determine the nature of this rapid change, we compared the sequences of Cnn-PA in all the dipterans represented in our sample (Figure 7). As can be seen, the percentage of identity of amino acid sequence diverges sharply within the genus Drosophila and is even more dissimilar when comparing the Drosophila species with the two mosquitos. Members of the melanogaster subgroup (Drosophila erecta and D. yakuba) show 92% identity with D. melanogaster and 95% with each other. Additional evidence of similarity is that gapping to produce alignment is minimal. As one moves further from D. melanogaster (D. biarmipis, D. takahashii, D. ficusphila, and D. elegans), identity with D. melanogaster drops to an average of 85% with a concomitant increase in gapping to achieve alignment. The two members of the ananassae group (Drosophila ananassae and Drosophila bipectinata) are further diverged, showing only 78% and 79% identity to D. melanogaster and an increase in gapping. This trend continues into the obscura group (D. pseudoobscura) and, for Drosophila willistoni, 67% and 65% identity to D. melanogaster, respectively, with an additional increment in gapping. It is also interesting to note that comparisons within and between each of the aforementioned groups within the subgenus Sophophora show similar levels of divergence as observed in their comparison to D. melanogaster. The three members of the subgenus Drosophila (Drosophila mojavensis, Drosophila virilis, and Drosophila grimshawi) are all roughly equivalently diverged from D. melanogaster and from each other with a range of 54–68% identity and a significant level of gapping to achieve alignment. Finally, a comparison of the two mosquitos (A. gambiae and A. aegypti) shows that amino acid identity drops to the 25–30% range with a large number of gaps required to find even that level of alignment. Interestingly, a neighbor-joining tree derived solely from the Cnn sequence precisely tracks the consensus phylogeny for the genus Drosophila and its relationship to the mosquitos.
Figure 7.
Sequence divergence and frequent indels are associated with the rapid evolution of Cnn-PA in the Diptera. Pair-wise comparisons of Cnn-PA proteins from Drosophila and mosquitoes (gray boxes) show that the percentage of identity of the protein decreases (section below diagonal solid boxes) and indels accumulate (section above diagonal solid boxes) over relatively short periods of time. A neighbor-joining tree of these Cnn-PA proteins showing the distance based on the number of differences with gaps distributed proportionally shows that the molecular phylogeny for cnn is consistent with the accepted organismal phylogeny for these species. Cnn-PA protein size is shown below species name (top row) and haploid genome sizes are in parentheses (left column).
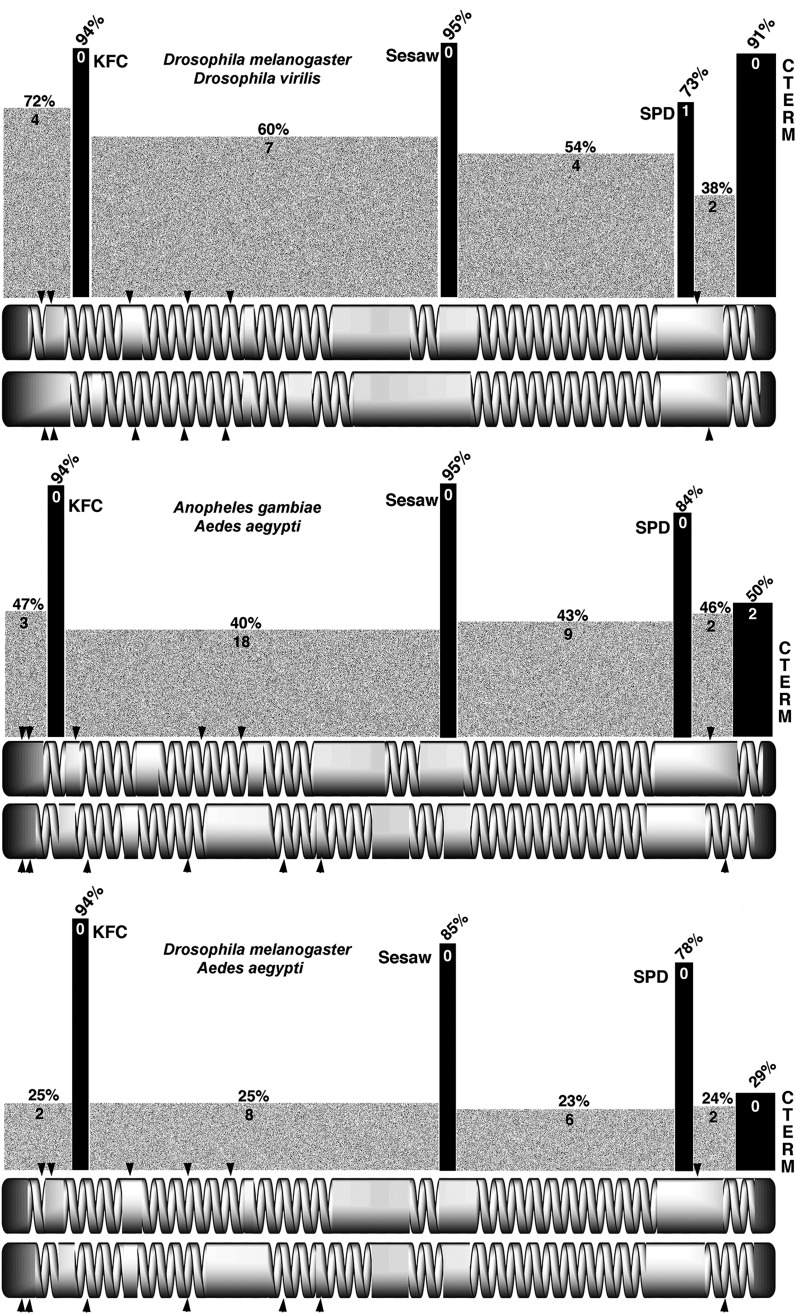
Since Cnn can be seen to diverge rather rapidly even within the genus Drosophila and yet there are four highly conserved motifs, we wanted to see the positions of the domains of conservation relative to those that were diverging. As noted above, Cnn-PA orthologs have a similar coiled-coil domain arrangement and similar spacing of the KFC, SESAW, SPD, and C-terminal motifs allowing for pairwise comparisons between D. melanogaster/D. virilis, A. gambiae/A. aegypti, and D. melanogaster/A. gambiae (Figure 8). The KFC motif is always found near the amino terminus in a coiled-coil domain while the SESAW and SPD motifs are in helical domains that flank a largely coiled-coil domain. The C-terminal motif is separated from the SPD motif by a short domain of mixed coiled-coil and a helical character. The regions between the conserved motifs are much less conserved, and their percentage of identity mirrors that seen in the comparison of all Cnn-PA proteins. It is also the case that essentially all of the gaps needed for an alignment are found in the regions between the conserved motifs.
Figure 8.
The divergence of Cnn-PA coiled-coil domains in dipterans. A schematic representation of the divergence of dipteran Cnn-PA conserved motifs (black bars) and the coiled-coil domains (shaded bars) shows the average percentage of identity and the number of gaps needed for alignment across each region. The least-conserved motif is the carboxy terminus. The divergence graph is above structural models for (top pair) D. melanogaster and D. virilis, (middle pair) A. gambiae and A. aegypti, and D. melanogaster and A. gambiae. Arrowheads indicate exon splice sites for each species.
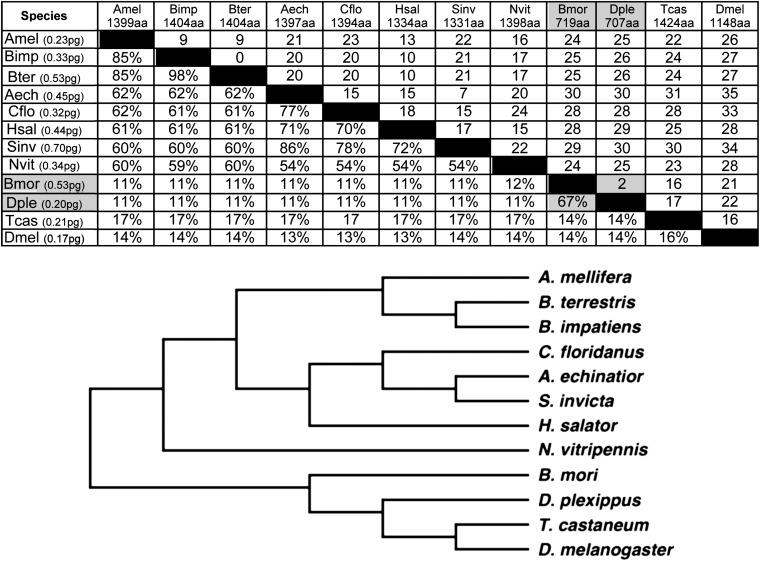
A similar comparison was done for the other insects and the results of that comparison are shown in Figure 9. Since 8 of the 11 species considered were hymenopterans, we based our comparisons on the honeybee A. mellifera. The three species of bee (A. mellifera, Bombus terrestris, and Bombus impatiens) show relatively high levels of sequence identity to A. mellifera at 85%. The alignment does require a modest number of gaps, however. The two bees of the genus Bombus show a high level of sequence identity to each other (98%) and require no gaps to achieve alignment. A comparison with four ant species (Camponotus floridanus, Acromyrmex echinatior, S. invicta, and H. salator) yields sequence identities in the 60–80% range both for a comparison with A. mellifera and within the group of ants, and gaps needed to achieve alignment are moderately high. The last of the Hymenoptera is Nasonia viripenis, a parasitic wasp. It is the most divergent of the group with 50–60% sequence identity when compared to the bees and ants. Gapping is again moderately high. The sequence divergence seen among the Hymenoptera is of the same magnitude as that seen for the comparison of the Drosophila species belonging to the two subgenera (Drosophila and Sophophora). The two lepidopteran species (B. mori and D. plexippus) are diverged at a similar level, showing only 67% identity. When they are compared to the hymenopteran species, the percentage of identity is only 11% with a significant number of gaps needed to achieve alignment due in part to a significant difference in protein size. This same level of divergence is seen in a comparison of the single coleopteran (T. castaneum) and D. melanogaster. This level of divergence when one compares species of different insect orders points up the reason for the difficulty in discovering orthologs of cnn and the reason why molecular probes designed on the D. melanogaster sequence rapidly loose their potency with phylogenetic distance. Despite the rapid divergence of the Cnn protein sequence, the neighbor-joining tree derived from these sequences, as was the case for the Diptera, closely tracks the consensus phylogeny for the species used in this analysis (Figure 9).
Figure 9.
A lower limit for the divergence and number of indels in Cnn-PA orthologs. A table similar to that in Figure 7 showing the divergence of Cnn-PA in the other insects in this study reveals a similar trend in the dipterans. Comparisons between orders suggest that there is a lower limit for sequence divergence and an upper limit for the number of indels tolerated in Cnn-PA.
Pair-wise comparisons of the structure of the Cnn protein models for the hymenopterans and lepidopterans, similar to that for the dipterans, were performed, and selected results are shown in Figure 10. As in the dipterans, the coiled-coil structure and relative positioning of the KFC, SESAW, SPD, and C-terminal motifs are conserved between A. mellifera and N. vitripenis and the two lepidopterans, B. mori and D. plexippus. This conservation of structure is independent of overall protein size as the two lepidopteran proteins are about half the length of the dipteran and hymenopteran examples.
Figure 10.
The divergence of Cnn-PA is consistent in all insects. A schematic representation of the divergence of Cnn-PA, similar to that in Figure 8, showing comparisons between (top left) A. mellifera (bottom) and N. vitripennis (bottom); (top right) B. mori (top) and D. plexippus (bottom); (bottom left) D. melanogaster (top) and A. mellifera (bottom); and (bottom right) D. melanogaster (top) and B. mori (bottom). The rate of change in the Hymenoptera and Lepidoptera is similar to the rate in the Diptera. The comparisons with D. melanogaster show the lower limit of divergence for Cnn-PA across this phylogenetic range. Arrowheads indicate exon splice sites for each species.
As in the Diptera, the KFC motif is found near the amino terminus in a coiled-coil domain, SESAW and SPD are located toward the COOH terminus in helical domains that flank a coiled-coil region, and the C terminus is separated from SPD by a short span of combined coiled-coil/helical character. The regions between the conserved motifs are less well conserved and have the same level of divergence and gapping as seen for the entire protein. These results clearly show that essentially all the divergence occurs in regions of simple coiled-coil domains and helical protein folds. Finally, a comparison of D. melanogaster with A. mellifera and B. mori demonstrates that, despite significant differences in gene structure (exon–intron positions), protein size, and different interspersion of helical and coiled-coil domains, the relative disposition of the conserved motifs and the structure of the protein is maintained. It has been shown that domain architecture is conserved between orthologs and that this high degree of conservation is likely to be necessary for functional conservation (Forslund et al. 2011). Although three of the motifs used in this study are not part of any curated domains, these results suggest that the domain architecture of cnn orthologs will be similar and that more than one motif will be required for functions attributed to Cnn in D. melanogaster.
Sequence divergence in Cnn within a population of D. melanogaster
Clearly, the rapid divergence of Cnn is characterized by sequence changes and multiple insertions and deletions across species. Based on pairwise comparisons of closely related species in this study, it seems likely that there would be minimal allelic variation of cnn within a species. To assess the evolution of Cnn within a species, we used the available sequences from 162 lines from a single North Carolina population of D. melanogaster (http://www.dpgp.org/; http://service004.hpc.ncsu.edu/mackay/Good_Mackay_site/DBRP.html; data not shown). These lines are representative of the haploid genomes present in a single population at the same time point (Mackay et al. 2012). Sixty of the lines were removed from the analysis because of ambiguous base calls or because of heterogeneity at individual bases in the coding sequence of Cnn-PA.
We compared the transcript and protein sequences to the reference, or wild-type Cnn-PA sequence, from FlyBase and found 83 variable nucleotide positions including 23 nonsynonymous substitutions. These changes translate into 29 protein variants plus the wild-type protein and have up to four amino acid changes in Cnn-PA (Table 1). Approximately two-thirds of the substitutions are in coiled-coil regions, and many substitutions are nonsimilar amino acid residues (Figure 11). While many of the variants were present in just one or two genomes, several were more frequent, and one variant with three changes was as frequent as our designated wild-type sequence (14 lines each).
Table 1. Cnn-PA allelic variation in a population.
| North Carolina lines | Variable nucleotide positions | Nonsynonymous positions | Alleles protein variants |
|---|---|---|---|
| 105 | 83 | 23 | 29 + wild type |
| No. of amino acid changes (corresponding no. of alleles) | |||
| 1 aa (9) | 2 aa (6) | 3 aa (11) | 4 aa (3) |
The characterization of the allelic variation from 105 unique strains from a single population (top row) shows the total number of nucleotide substitutions, the number of nonsynonymous substitutions, and the number of protein variants. The number of amino acid differences (one to four) and the number of allelic variants carrying those differences is shown at the bottom of the table. The difference between nonsynonymous substitutions and protein variants is due to unique combinations of one to four nonsynonymous substitutions encoding a total of 29 protein variants.
Figure 11.
Cnn-PA allelic variation within a D. melanogaster population. The 23 amino acid substitutions present within a single population of D. melanogaster are mapped along the structural model of Cnn-PA, showing the amino acid substitution and position above the model. Ten of the 23 amino acid substitutions are not chemically similar residues. In addition to the wild-type protein, unique combinations of one to four of these substitutions produce 29 allelic variants of Cnn-PA in this population. Brackets above the model show the positions of the conserved Cnn motifs, and the positions of the exon splice junctions are indicated by solid triangles below the model.
These results show that there can be a significant protein variation in Cnn-PA within a population, and we assume that these alleles are functionally compatible, although future experimental studies will be required to show that this is true. The above amino acid changes are due to single-nucleotide substitutions and do not explain the rapid sequence divergence that we see in some exons, such as the Cnn-PA exon 4 region. This is the most divergent exon(s) in our orthologs; it has been lost in the lepidopterans and produces multiple gaps in alignments among the species in this study. To investigate the effect of potential deletions within the North Carolina population, we took sequences with single or doublet ambiguous base calls (“n”), removed them from the sequence, and translated the sequence to analyze the resultant peptides. Surprisingly, many of the translated peptides are approximately the same length as the D. melanogaster reference sequence but have diverged nearly 100% from the point of the deletion. We have previously reported on the use of two overlapping reading frames used to produce unique splice variants from the same genomic sequence (Eisman et al. 2009). The result from the North Carolina sequences demonstrates that other exons in cnn have multiple reading frames spanning large portions of the coding exon, potentially buffering against nontriplet insertions or deletions in coding sequence. Obviously, to produce a full-length transcript, splicing would have to be relaxed to accommodate a nontriplet insertion or deletion in an internal coding exon. The lack of strong canonical splicing signals in cnn may allow this to happen, but more extensive sequencing of transcripts from these lines will be required for firm conclusions.
A potential buffering function for MPOs in Drosophila cnn
The above analyses are consistent with the conclusion that cnn is diverging rapidly due to nucleotide substitution and frequent insertions and deletions in coding sequence. Additionally, the intron–exon structure of the gene has changed considerably among insect orders due to intron gain and loss events. The most divergent regions of Cnn-PA orthologs have had multiple exon fusion or intron loss events in Drosophila relative to other orders (arrowheads, Figure 10). Protein alignments between all the species in this study suggest that the short sequences flanking fusion points are divergent between orders and continue to evolve rapidly after the fusion event within orders. Global analyses of Drosophila have shown that most introns are lost by a reverse transcriptase mechanism, resulting in the perfect excision of the intron (Coulombe-Huntington and Majewski 2007; Yenerall et al. 2011). A rare second mechanism of intron loss in Drosophila is precise and imprecise genomic deletion by nonhomologous end joining (NHEJ) during DNA break repair (Llopart et al. 2002; Yenerall et al. 2011). A third mechanism of intron loss in D. melanogaster is genomic deletions that remove 5′ intronic splice sites or shorten the intron below a size capable of being spliced out of the primary transcript, resulting in the retention of the intron (Talerico and Berget 1994). To differentiate among these mechanisms, we analyzed the exon 6/7 boundary in three Drosophila species with known splice sites based on cDNA or RNA-Seq data.
The exon 6/7 splice boundary is present in all species in this study, suggesting that the intron is conserved, but changes in the MPOs in the genus Drosophila (Figure 3) show that the sequence spanning this area is divergent. Consistent with sequence divergence, the protein sequences flanking the splice junction are divergent in Drosophila species. To investigate changes in the intron and flanking coding sequences, we aligned the genomic sequences from the upstream SPD motif to the start of the downstream conserved COOH tail from D. melanogaster, Drosophila biarmipes, and D. pseudoobscura. The splicing is based on cDNA, RNA-Seq, and EST data, respectively.
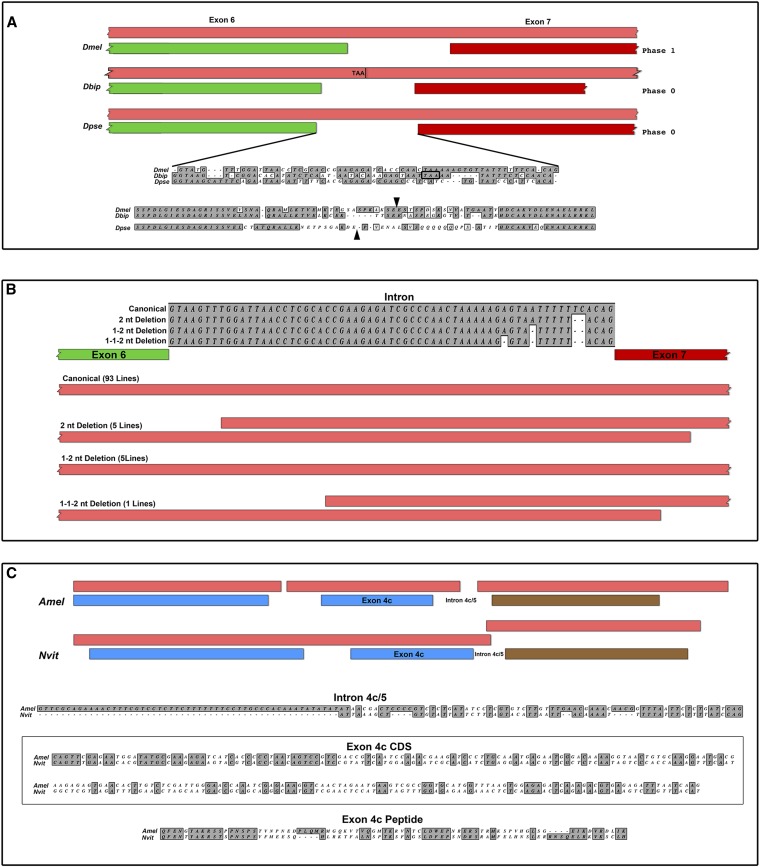
The intron is 61 nucleotides in D. melanogaster and D. pseudoobscura and 57 nucleotides in D. biarmipes. Relative to D. melanogaster, a combination of triplet and nontriplet deletions introduce an in-frame stop codon in D. biarmipes, changing the MPO structure (Figure 12A). Surprisingly, while the D. melanogaster and D. pseudoobscura introns are equal lengths, they are offset by 22 nucleotides with respect to coding sequence and spliced in different phases (Figure 12A). Protein alignments reveal rapid divergence and frequent indels flanking the known splice junction between these three species (arrowheads, Figure 12A). The rapid divergence may be due to an accelerated rate of nonsynonymous substitutions in sequence flanking the intron, nontriplet insertions and deletions that change multiple codons with a single event, or a combination of both mechanisms. While this intron appears to be conserved across a broad phylogenetic range, the sequence and splice sites evolve rapidly.
Figure 12.
Splicing changes and nontriplet indels create new codons in Cnn-PA orthologs. (A) Rapid changes in splicing of the intron between exon 6 (green) and exon 7 (red) in D. melanogaster, D. biarmipes, and D. pseudoobscura due to indels have shortened the intron in D. biarmipes and moved the intron in D. pseudoobscura. The exon and MPO (light red) maps (top) are aligned to the SPD motif (left) and at the start of the carboxy terminus (right) showing the effect of these changes on exon position. A single substitution in D. biarmipes has introduced a stop codon (boxed, middle), changing the MPO. The aligned protein sequences (bottom) showing the two splice sites (arrowheads) show the rapid divergence associated with these changes, which are typical of existing splice sites in orthologs and at sites of exon fusion. The evidence suggests that these changes are due to nontriplet indels and new codon usage in coding sequence and relaxed splicing. (B) An alignment of this same region from the North Carolina lines shows that multiple lines have begun to accumulate nontriplet indels (top). These changes do not change splicing, but they do reveal the buffering capacity of MPOs (light red boxes, bottom) and show how comparisons between MPOs are useful for the detection of very small significant changes in sequence. (C) A schematic (top) comparison of the MPOs (light red) for exon 4c (blue) and exon 5 (brown) between A. mellifera and N. vitripennis suggest that a change has occurred. Comparisons of the aligned introns (top) show a large deletion, and three different nontriplet indels in N. vitripennis have significantly changed the splicing. Alignments of the coding sequence (middle, boxed) and translated peptide encoded by exon 4c show that either nontriplet indels have generated new codons or, for some unknown reason, the nucleotide substitution rate is accelerated in this exon.
The above results show that splicing in Drosophila can evolve rapidly and, in some cases, may be relaxed compared to canonical splicing mechanisms. Additionally, the most parsimonious explanation for rapid protein divergence is that nontriplet deletions have caused frameshifts and the divergent peptides are actually encoded by unique sets of codons. Typically, nontriplet insertions or deletion in coding sequence are predicted to generate deleterious nonsense mutations. One possibility is that the extended reading frames associated with Drosophila MPOs buffer against these events. We have analyzed Drosophila MPOs and have found that there is a paucity of stop codon dinucleotides in both exonic and intronic regions. A paucity of stop codon dinucleotides is also found in the small introns in the other insect species. Perhaps this feature and relaxed splicing mechanisms act as buffers against premature stop codons during intron size reduction prior to the fusion of two exons.
To further investigate the potential buffering capacity of MPOs, we looked at the exon 6/7 intron in the North Carolina D. melanogaster population sequences. The majority of the lines had wild-type sequence, but five lines had a 2-nt deletion, five lines had this deletion and a single-nucleotide deletion, and one line had both deletions and an additional single-nucleotide deletion. These genomic sequences were assembled on the reference D. melanogaster genomic sequence so deletions are represented as “n” in the assemblies. While further sequencing will be required to verify that they are real deletions, the fact that they occur in multiple lines suggests that they are not random sequencing errors. In the two lines with nontriplet deletions, the new reading frames extend well beyond the point of the deletion (Figure 12B) and show that many MPOs have long secondary reading frames overlapping the actual coding sequence. While somewhat surprising, we have previously shown that two overlapping reading frames are differentially used to generate Cnn splice variants (Eisman et al. 2009). These results show how a nontriplet insertion or deletion in coding sequence near a splice junction would result in rapid codon change and be buffered against when coupled with relaxed splicing mechanisms.
The type of MPO discussed above appears to have a limited phylogenetic range and may be restricted to the genus Drosophila. However, we did identify a single multi-exon MPO in the parasitic wasp N. vitripennis that is absent in all other Hymenoptera. The multi-exon MPO spans the last two of three exons homologous to exon 4 in D. melanogaster Cnn-PA and part of the intron between exon 4 and exon 5 (Figure 12C). In the Hymenoptera, both the first two exons homologous to exon 4 and the homologous exon 5 are conserved. The last fragment of exon 4 homology, or exon 4c, is the same length in A. mellifera and N. vitripennis, yet the encoded peptide has diverged significantly (bottom, Figure 12C). The coding sequence has several nontriplet insertions or deletions when gaps are allowed (data not shown) or, because the exons are the same length, an accelerated substitution rate (boxed, Figure 12C). Similar to the above example from Drosophila, the most parsimonious explanation for the rate of change is that nontriplet insertions or deletions create multiple new codons. In addition to coding changes in exon 4c, the N. vitripennis intron between exons 4c and 5 has a large deletion, including the 5′ splice site (Figure 12C), suggesting that splicing can evolve rapidly as it does in Drosophila.
Taken together, these data suggest that multi-exon MPOs, in conjunction with relaxed splicing mechanisms, may buffer small and potentially deleterious insertions and deletions, especially in genes or regions of genes with small introns separating coding exons. These results also show how comparisons of MPOs between species may be a powerful tool for the detection of small, yet significant, changes within genes.
The dominant-negative effect of divergent Cnn-PA between species
The above data identify where Cnn-PA diverges rapidly and provide a possible explanation of some of the processes that underlie these changes, but it does not account for possible effects on protein function. The finding that rapid divergence is associated with coiled-coil regions suggests that these protein folds may buffer against changes since coiled-coil protein folds are composed of simple heptad repeats and have minimal structural constraints (Subbiah 1989). However, if, for example, a protein forms homodimers, sequence divergence and small indels could have a negative effect on protein function. Divergent proteins may repel each other, preventing dimer formation, and numerous indels could shift functional motifs out of register with each other, thus making the coexpression of two heteromorphic peptides possibly antagonistic, resulting in a mutant phenotype.
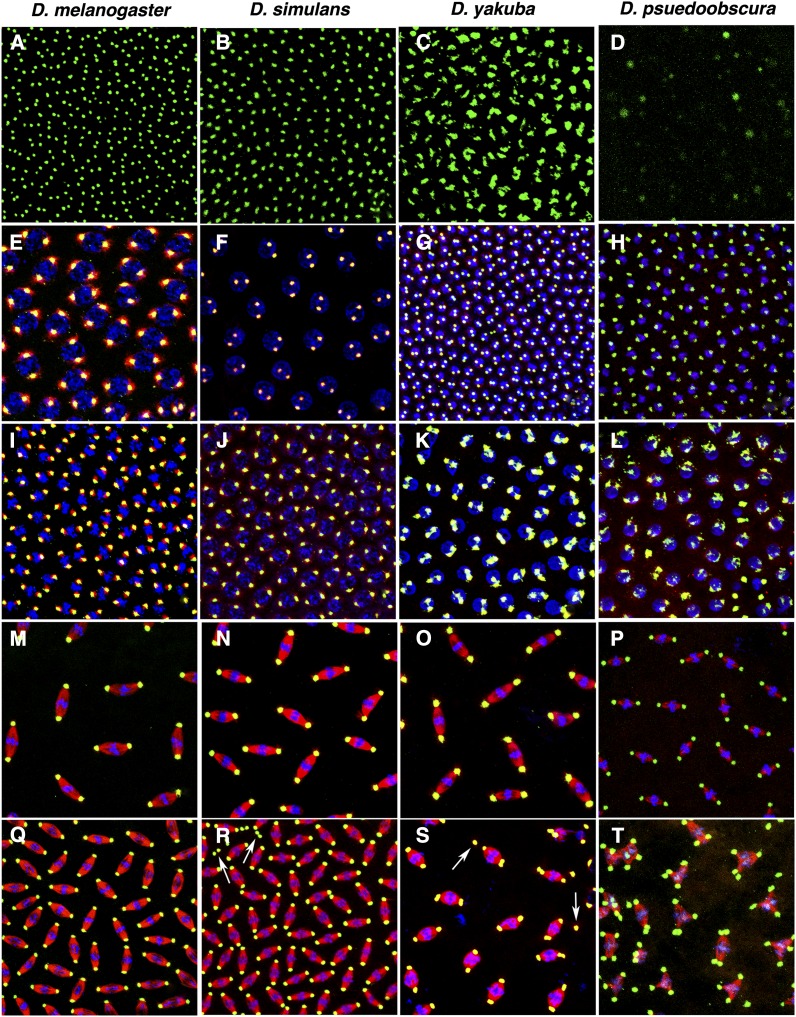
To test this possibility, we have expressed a D. melanogaster GFP::Cnn-PA fusion protein during syncytial development in D. melanogaster, D. simulans, D. yakuba, and D. pseudoobscura. In live embryos, GFP localizes to the centrosome normally in both D. melanogaster and D. simulans, accumulates at relatively high levels in D. yakuba but in an abnormal pattern, and has low and variable accumulation levels in D. pseudoobscura (Figure 13, A-D).
Figure 13.
Minimal divergence between proteins perturbs Cnn-PA function during development. The ectopic expression of a D. melanogaster EGFP::Cnn-PA fusion protein in D. melanogaster, D. simulans, D. yakuba, and D. pseudoobscura during syncytial development shows minimal divergence between proteins expressed in an heterospecific embryo disrupts normal cleavage. Live imaging of EGFP (A–D) shows that the tagged protein localizes to the centrosome, but in D. yakuba the protein is less punctate and more diffuse and in D. pseudoobscura the amount of tagged protein at centrosomes is reduced and variable. Immunostaining of fixed embryos recognizes both the tagged and native Cnn protein and is similar to what is observed in live animals during prophase (compare I–L to A–D). Although centrosomes are more obvious in D. pseudoobscura in fixed material, there is still significant variability among nuclei (L). During metaphase (Q–T) defects are clear in D. simulans (Q) and D. yakuba (S) as indicated by free centrosomes (white arrowheads). Either centrosome replication is precocious or centrosomes are split at many poles in D. yakuba. The phenotype seen in D. pseudoobscura is interesting, as the spindles are similar to spindles in cnn loss-of-function mutant embryos. However, in D. pseudoobscura (T), while Cnn is at centrosomes, spindle poles have multiple centrosomes and many spindles are multipolar. These results show that <10% divergence and a single indel between two coexpressed Cnn proteins is sufficient to significantly perturb normal function. Control animals not expressing the EGFP-tagged Cnn all appear to be normal throughout the cell cycle (prophase: E–H; metaphase: M–P). Fixed embryos (E–T) is stained with anti-Cnn (green) and anti-β-tubulin (red) and DNA is stained with TOTO3 (blue).
In fixed embryos immunostained to detect both the Cnn fusion protein and native Cnn, Cnn staining is very similar to live images during prophase (Figure 13, “D. yakuba” column). During metaphase, defects associated with the accumulation of the fusion protein become apparent. In D. simulans and D. yakuba, centrosomes detach from the spindle and, in D. yakuba, centrosome replication is precocious at some spindles (Figure 13, Q-T). Metaphase spindles in D. pseudoobscura are the most defective, having multiple poles and fusion between spindles (Figure 13, T), similar to the cnn mutant phenotypes described in D. melanogaster (Megraw et al. 1999; Eisman et al. 2009). Consistent with this phenotype, development fails in most of these embryos. The control embryos in these experiments are normal throughout the cell cycle (Figure 13, E-H and M-P) and development proceeds normally.
These data demonstrate that coexpression of divergent alleles of cnn has a dominant-negative effect on protein function during cleavage. The next obvious experiments to conduct are to express cnn orthologs in D. melanogaster and test the rescue ability of divergent alleles when the D. melanogaster native protein is absent, which is beyond the scope of this study.
Multi-exon MPOs are correlated with rapid divergence and alternative splicing
Although every coding exon has an MPO, the prediction is that most MPOs will be co-extensive with the actual coding sequence. Since this is clearly not the case for cnn in Drosophila, we wanted to know if this was a unique phenomenon or if other genes in D. melanogaster contained long MPOs relative to known coding sequences. To identify candidate genes, we queried every annotated exon and asked how much longer the MPO extended beyond the 5′ and 3′ ends of the annotated exon. We found that MPOs in D. melanogaster varied from essentially being the same size as the annotated exon to extending over 14,000 nucleotides in a 5′ or 3′ direction beyond the annotated end of an exon. We identified 398 genes containing at least one MPO extending more than 1000 nucleotides beyond the end of an annotated exon, representing ∼3% of the genes in D. melanogaster. Since we wanted to manually annotate several genes across a range of Drosophila species to identify similarities with the evolution of cnn, we restricted our list to genes with an MPO extending >3000 nucleotides beyond the annotated exon. There were 39 genes in this category from which we chose 7 with traditional Drosophila names, as these genes were likely to be essential developmental genes characterized at the genetic and molecular level. The full extent of this analysis found many changes in our candidate genes, but for this study we report only a few highlights related to our target MPOs identified in the screen (Table 2; Supporting Information: Figure S1, Figure S2, Figure S3, Figure S4, and Figure S5.
Table 2. Divergence of genes with multi-exon MPOs in Drosophila.
| Gene/protein ID | Gene size range (kb) | Protein size range (aa) | Alternative splicing mechanism | Exons annotated |
|---|---|---|---|---|
| Shot-PH | 3070–3498 | Yes (IRE) | Exon 14 only | |
| Dp | Incomplete sequences | Yes (Unknown) | ||
| Pros-PK | 10.8–20.0 | 1403–519 | Yes (IRE) | Entire gene |
| Mud-PL | 10.3–16.5 | 1641–2756 | Yes (IRE) | Exons 2–11 |
| Rno-PB | 11.0–14.8 | 3201–4386 | No | Entire gene |
| Lva-PC | 7.9–9.5 | 2406–2837 | No | Exons 2–8 |
| Futsch-PC | 16.7–21.1 | 4922–6507 | No | Exons 3–8 |
A summary of the comparative analyses of seven genes across nine Drosophila species based on manual annotations. These genes were first identified in D. melanogaster due to the presence of at least one extremely long MPO compared to annotated coding exons. Only the target exon in the SHOT-PH protein is evolving rapidly, and analysis was done only for this exon. Dp sequences were incomplete in most species, hindering a more complete analysis of this gene. For additional details, see File S1 and Figure S1, Figure S2, Figure S3, Figure S4, and Figure S5.
Of the seven genes investigated, six are reported to be rapidly evolving within Drosophila. Only shortstop (shot) is conserved, except for our target MPO, which is evolving rapidly, demonstrating that divergent MPOs can exist in otherwise conserved genes. Target MPOs in dumpy (dp), prospero (pros), short shot, and mushroom body defective (mud) are known to be alternatively spliced by an intron retention/exclusion (IRE) mechanism in all except the uncharacterized splice variants of dp. The target MPOs in rhinocerous (rno), lava lamp (lva), Futsch, and mud are all associated with significant variation in gene models in the genus Drosophila, although our manual annotation of these MPOs finds no reason for this considerable variation (Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, File S1, and Figures 2–5). However, we do find strong evidence in several of the genes studied for the loss or gain of introns outside of our target MPOs, which will require future sequencing to verify these changes. Finally, our target MPOs all encode either coiled-coil or simple repetitive protein domains, similar to the MPOs in cnn.
In general, regions of high conservation separated by regions of high divergence characterize all the peptides encoded by the target MPOs discussed above. The divergent regions introduce multiple small gaps in aligned sequences and result in significant variation in protein length among species similar to what is found in cnn orthologs. Our target MPOs appear to confound gene model algorithms and may be associated with changes in the intron–exon structure of genes. While the mechanisms are not clear, the presence of reading frames extending well beyond the known end of coding exons is apparently indicative of rapid evolution.
Discussion
The rapid divergence of cnn has been both problematic and intriguing for our lab since we first described the gene (Heuer et al. 1995). Surprisingly, the rate of divergence per million years for cnn is more rapid than the rate reported for the vertebrate albumin genes (Doolittle 1995) and more rapid than the rate of 20–30% for cytoskeletal proteins predicted in the same report. The rapid divergence has made it difficult to identify domains and motifs necessary for Cnn function, which is essential for a directed molecular dissection of the protein. Moreover, this rapid evolution and some publicly available orthologous gene sets makes it impossible to completely rule out orthologous relationships among insect, vertebrate, and fungal genes encoding the KFC motif. Although rapid evolution has been problematic, we have always been intrigued by what processes drive the divergence and what compensatory mechanisms exist in Drosophila to offset rapid changes in essential developmental proteins. For these reasons, we have characterized a cnn orthologous gene set from the insects, useful for the identification of additional orthologs, accurate functional annotation, and future investigations of several processes and compensatory mechanisms that potentially underlie the rapid evolution of cnn.
Based on these analyses, cnn orthologs appear to be restricted to the insects, in agreement with OrthoDB and EggNOG orthologous gene sets, although additional genomes may expand this range into other groups. Orthologous Cnn proteins all have the highly conserved KFC, SESAW, and SPD motifs, as well as the more divergent carboxy terminus. The arrangement of these motifs and coiled-coil domains is conserved even though proteins vary in size from 707 amino acids in D. plexippus to 1424 amino acids in T. castaneum. This conservation of structure suggests that function will be conserved (Forslund et al. 2011) and provides a schematic of Cnn domain architecture to direct future studies.
The somewhat limited phylogenetic range of Cnn orthologs also provides new insight into the functional evolution of Cnn. In D. melanogaster, Cnn is required in males during spermatogenesis (Li et al. 1998) and is a maternal-effect embryonic lethal gene in females, required at the centrosome during syncytial development and cellularization (Megraw et al. 1999; Vaizel-Ohayon and Schejter 1999; Eisman et al. 2009). Other than these two stages of development, a maternal supply of Cnn+ in oocytes is sufficient for the development of morphologically normal but sterile adult flies. Since this type of development is common to all species in this study, Cnn may be a novel centrosomal protein that evolved to organize the actin and microtubule cytoskeleton in the absence of cell membranes. The ancestral gene or genes from which cnn is derived remains unknown at this time.
One possibility is that cnn is a chimera derived from multiple genes, as suggested by the superfamily of proteins with the KFC motif. Experimental data for this superfamily exist for the fungal genes Anucleate primary sterigmataB (AspB) in Aspergillus nidulans (Westfall and Momany 2002), microtubule organizer 1 (Mto1p) in Schizosaccharomyces pombe (Sawin et al. 2004), and the vertebrate genes mmg or phosphodiesterase 4d interacting protein (Pde4dip) (Verde et al. 2001) and CDK5RAP2, as well as cnn, in D. melanogaster. The KFC motif in Mto1p (Sawin et al. 2004), CDK5RAP2 (Fong et al. 2008), and Cnn (Megraw et al. 2001; Zhang and Megraw 2007) nucleates γ-tubulin at microtubule-organizing centers although this function is mechanistically different in fungi, vertebrates, and flies (Fong et al. 2008). Unlike these proteins, AspB is involved in nuclear migration during spore formation and colocalizes with F-actin (Westfall and Momany 2002), and Mmg interacts with cyclic nucleotide phosphodiesterase (Verde et al. 2001) and A-kinase (Uys et al. 2011) in vertebrate muscles. Additionally, these proteins share no significant homology outside of the KFC motif and flanking domain, each has unique motifs and domains not present in the other proteins, and removal of the KFC motif for BLAST searches returns a completely different set of “best hits.” Taken together, the diverse functions and limited homology suggest that this superfamily may be epaktologs that have inherited the KFC domain independently in different lineages (Nagy et al. 2011a), rather than orthologs related by descent.
Functional data for the SESAW and SPD motifs and the carboxy terminus are more limited. Global phosphorylation studies of D. melanogaster have shown that the SESAW and SPD motifs are phosphorylated (http://www.phosphopep.org/), suggesting that they are potentially important regulatory motifs used during the cell cycle. The function of the carboxy terminus is mostly unknown, although we have shown that loss of the SPD motif and carboxy terminus results in very low levels of truncated Cnn at the centrosome and in high levels at the metaphase plate (Eisman et al. 2009). A recent study reported that a motif in the Cnn carboxy terminus is conserved in mouse CDK5RAP2 (Wang et al. 2010), but a PsiBLAST with the D. melanogaster motif found minimal homology, with an E-value of 7.3 between fly and mouse. Clearly, future studies of Cnn are required to define the molecular basis of Cnn function.
The conservation of Cnn domain architecture is in striking contrast to the divergence of orthologous gene architecture, gene size, and coding sequences. In general, the intron–exon structure of cnn is conserved within insect orders but is strikingly different between orders. Relative to the dipterans, Cnn is encoded by more exons in all species investigated here with a trend toward exon fusion in the dipterans. The most common mechanism for intron loss is mediated by reverse transcriptase (Coulombe-Huntington and Majewski 2007; Yenerall et al. 2011), but our results suggest that cnn introns are lost by genomic deletions (Llopart et al. 2002; Yenerall et al. 2011). Our comparison of the exon structure of cnn orthologs shows a potential intermediate step in this process, especially within the hymenopterans. Initially, intronic deletions bring multiple exons into close proximity with each other, forming exon islands. These data do not resolve how multiple exons fuse, but imprecise NHEJ repair (Llopart et al. 2002; Yenerall et al. 2011) or intronic deletions resulting in intron retention (Talerico and Berget 1994) are plausible mechanisms. These two mechanisms, as well as the loss of coding exons in the lepidopterans and the incorporation of nonhomologous coding sequence in T. castaneum, would account for some of the divergence of Cnn-PA proteins.
The divergence of Cnn-PA proteins appears to be due to several types of changes. Imprecise exon fusion events, nonsynonymous nucleotide substitution, frequent small indels within coding exons, and, based on our analyses, nontriplet indels resulting in frameshifts that generate new codons all play a role in the divergence of Cnn-PA. Within orders, Cnn-PA divergence is rapid but uniform across the phylogeny, whereas comparisons between orders show that Cnn-PA has minimal homology, which may represent the minimal conservation necessary to retain Cnn-PA function. Analysis of the D. melanogaster North Carolina lines suggests that nucleotide substitution is responsible for allelic variation within a population and that the accumulation of indels occurs after speciation. However, as presented in the Results, we found evidence for nontriplet indels in exon 4, suggesting that these types of changes could exist within a single population. Future RNA-seq analyses of the lines in the North Carolina population will be required to verify the validity of this cause of the apparent rapid changes in coding sequence.
A key question regarding protein insertions and deletions is whether the underlying nucleotide indels are triplet or nontriplet changes. The former presumably removes a single codon whereas the latter causes a frameshift. The assumption of global analyses is that protein indels are the result of the perfect insertion or deletion of codons, but the complete divergence of multiple amino acids surrounding indels in Cnn-PA alignments suggests that this is not always true. The most parsimonious explanation for the divergence of coding regions flanking the exon 6/7 splice site in Drosophila and the exon 4c/5 splice site in the hymenopterans is nontriplet indels. In both examples, upstream and downstream conserved regions flank the divergent regions so that indels must eventually total triplet combinations or the splicing machinery must restore the downstream reading frame. Although more data are required to address these possibilities, it seems probable that both mechanisms resolve nontriplet indels in coding sequences.
The types of changes driving the rapid evolution of Cnn-PA suggest that there must be buffering mechanisms at the sequence and protein level. In this study, we have presented MPOs as a possible buffering mechanism at the sequence level. The buffering capacity of MPOs appears to be inherent in the sequence that they span, as both the coding and small introns have a paucity of dinucleotides needed to introduce stop codons. This results in multiple overlapping reading frames in the same genomic region, as demonstrated by the intron between exons 6 and 7 in the North Carolina lines. In this example, splicing is not changed, but, if similar changes occur in coding sequence, an as-yet-unknown but apparently present splicing mechanism could retain protein function. Perhaps at the sequence level evolution selects for a paucity of stop codon dinucleotides to buffer against the inevitable deleterious deletions that will occur when genome size is significantly reduced.
Although buffering at the sequence level makes it possible to transcribe a similar primary sequence, the resultant protein must fold properly to retain function. Based on Cnn-PA orthologs and the proteins in the MPO screen, rapid divergence of primary sequence may be restricted to structural proteins. The most frequent motif found in this study is the coiled-coil domain, an α-helical fold composed of simple heptad repeats and a common component of structural proteins. Since many residues can substitute at the heptad positions, in this protein motif it is not surprising that this fold can diverge rapidly and retain function. The rapid divergence of Futsch suggests that a second fold with buffering capacity is high-copy-number simple repeats. Although we did not analyze the evolution of Dp, it has been shown that the repetitive PIGFEAST region is undergoing concerted evolution in Drosophila due to unequal crossing (Carmon et al. 2007, 2010), similar to what we see in Futsch. While other protein folds, such as disordered regions, are likely to have a buffering capacity, the rapid divergence associated with Cnn-PA orthologs is likely to be restricted to a subset of protein types. An additional and somewhat surprising buffering capacity may be inherent in the insects in that they can tolerate significant differences in protein size between closely related species. This suggests that the molecular machinery in these animals is relatively plastic with a wide range of size tolerances.
While rapid divergence may be buffered at several levels, it seems reasonable to expect some constraints on protein function, especially if coiled-coil domains interact to form oligomers. Although we do not know the oligomerization state of Cnn-PA, in the simplest state Cnn-PA forms a parallel homodimer, so divergent Cnn-PA orthologs should interfere with protein folding and domain alignment. To test this hypothesis, we ectopically expressed a D. melanogaster GFP::Cnn-PA fusion protein in D. simulans, D. yakuba, and D. pseudoobscura during syncytial development. These experiments showed that this fusion protein interferes with native Cnn-PA function in other species and that the intensity of the dominant-negative effect increases as the divergence of the native protein increases. Since the GFP::Cnn-PA fusion protein rescues the cnnmfs mutant phenotype in D. melanogaster, and the D. simulans and D. yakuba Cnn-PA proteins are very similar, it is unlikely that these results are due to the GFP tag or to functional changes between species. This suggests that, for a functional homodimer to form, the degree of protein divergence tolerated is low and indels are rare within a species. However, since we see effects in D. yakuba between Cnn-PA orthologs and the same type of divergence is taking place in other essential developmental proteins, it is tempting to propose that the combined effect may be associated with speciation, specifically in light of the data presented here in postfertilization isolation.
We have presented a comprehensive set of cnn orthologs and describe several features likely to be necessary for Cnn function associated with D. melanogaster. While proteins that have a subset of these features may have some shared functions, it seems unwise to transfer the full function of cnn based on mutational analyses in D. melanogaster onto genes with limited homology. This orthologous gene set provides valuable information for future molecular and transcriptional studies. We realize much of the divergence in cnn does not fit neatly into current models, in part because in silico global analyses of multiple genomes cannot “see” the small-yet-significant changes described. One way to detect these changes would be to incorporate MPOs into computer programs as comparisons of MPO changes between species as a means to detect even a single-nucleotide deletion. It is our hope that this work provides a basis for continued investigations into how rare genomic changes and presumably deleterious mutations can drive the rapid evolution of essential genes.
Supplementary Material
Acknowledgments
The authors thank Melissa Phelps and Stacy Holtzman for outstanding technical assistance and enduring moral support; FlyBase for easy access to genetic and genomic information; the Bloomington Drosophila Stock Center for fly stocks; the Center for Genomics and Bioinformatics for help with the algorithm to search for MPOs; the Baylor Sequencing Center and Fringy Richards for access to the recently sequenced modENCODE Drosophila genomes and the associated RNAseq data; Josh Goodman for help in retrieving the relevant sequences from the Mackay Raleigh population sequences; and, finally, Matt Hahn for his evisceration of a previous incarnation of this article. His comments were helpful, insightful, and all remaining deficiencies are due to our pigheaded refusal to take his well-proffered advice. Supported by the Indiana Genomics Initiative.
Footnotes
Communicating editor: N. Perrimon
Literature Cited
- Altenhoff A. M., Dessimoz C., 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLOS Comput. Biol. 5: e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon A., Wilkin M., Hassan J., Baron M., MacIntyre R., 2007. Concerted evolution within the Drosophila dumpy gene. Genetics 176: 309–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon A., Guertin M. J., Grushko O., Marshall B., MacIntyre R., 2010. A molecular analysis of mutations at the complex dumpy locus in Drosophila melanogaster. PLoS ONE 5: e12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P. T., Raff J. W., 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20: 2187–2192 [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J., Majewski J., 2007. Intron loss and gain in Drosophila. Mol. Biol. Evol. 24: 2842–2850 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., et al. , 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64: 287–314 [DOI] [PubMed] [Google Scholar]
- Eisman R., Kaufman T. C., 2007. Cytological investigation of the mechanism of parthenogenesis in Drosophila mercatorum. Fly (Austin) 1: 317–329 [DOI] [PubMed] [Google Scholar]
- Eisman R. C., Stewart N., Miller D., Kaufman T. C., 2006. centrosomin’s beautiful sister (cbs) encodes a GRIP-domain protein that marks Golgi inheritance and functions in the centrosome cycle in Drosophila. J. Cell Sci. 119: 3399–3412 [DOI] [PubMed] [Google Scholar]
- Eisman R. C., Phelps M. A., Kaufman T. C., 2009. Centrosomin: a complex mix of long and short isoforms is required for centrosome function during early development in Drosophila melanogaster. Genetics 182: 979–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–113 [PubMed] [Google Scholar]
- Fitch W. M., 2000. Homology a personal view on some of the problems. Trends Genet. 16: 227–231 [DOI] [PubMed] [Google Scholar]
- Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N., 2002. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13: 47–58 [PubMed] [Google Scholar]
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z., 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Pekkari I., Sonnhammer E. L., 2011. Domain architecture conservation in orthologs. BMC Bioinformatics 12: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Koonin E. V., 1998. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1: 55–67 [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Donatelli I., Guo Y. J., et al. , 1991. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J. Virol. 65: 3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62: 1435–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer J. G., Li K., Kaufman T. C., 1995. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development 121: 3861–3876 [DOI] [PubMed] [Google Scholar]
- Holtzman S., Miller D., Eisman R., Kuwayama H., Niimi T., et al. , 2010. Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly (Austin) 4: 349–362 [DOI] [PubMed] [Google Scholar]
- Hulsen T., Huynen M. A., de Vlieg J., Groenen P. M., 2006. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 7: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvakolanu D. V., Livingston W. H., III, 1991. Rapid and inexpensive protocol for generating greater than 95% recombinants in subcloning experiments. Biotechniques 10: 176–177 [PubMed] [Google Scholar]
- Keeling P. J., Luker M. A., Palmer J. D., 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 17: 23–31 [DOI] [PubMed] [Google Scholar]
- Li K., Kaufman T. C., 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85: 585–596 [DOI] [PubMed] [Google Scholar]
- Li K., Xu E. Y., Cecil J. K., Turner F. R., Megraw T. L., et al. , 1998. Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J. Cell Biol. 141: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A., Comeron J. M., Brunet F. G., Lachaise D., Long M., 2002. Intron presence-absence polymorphism in Drosophila driven by positive Darwinian selection. Proc. Natl. Acad. Sci. USA 99: 8121–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J., 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. A., Miller D. F., Kaufman T. C., 1989. Developmental distribution of RNA and protein products of the Drosophila alpha-tubulin gene family. Dev. Biol. 132: 45–61 [DOI] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J.; FlyBase Consortium, 2012. FlyBase 101: the basics of navigating FlyBase. Nucleic Acids Res. 40: D706–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C., 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126: 2829–2839 [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Kao L. R., Kaufman T. C., 2001. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11: 116–120 [DOI] [PubMed] [Google Scholar]
- Mushegian A. R., Garey J. R., Martin J., Liu L. X., 1998. Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 8: 590–598 [DOI] [PubMed] [Google Scholar]
- Nagy A., Bányai L., Patthy L., 2011a Reassessing domain architecture evolution of metazoan proteins: major impact of errors caused by confusing paralogs and epaktologs. Genes 2: 516–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Szláma G., Szarka E., Trexler M., Bányai L., et al. , 2011b Correction: Nagy, et al. Reassessing domain architecture evolution of metazoan proteins: major impact of gene prediction errors. Genes 2011, 2, 449–501. Genes 2: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V., Wortman J. R., Lawson D., Haas B., Kodira C., et al. , 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. A., 2002. DNA loss and evolution of genome size in Drosophila. Genetica 115: 81–91 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Hartl D. L., 1997. Trash DNA is what gets thrown away: high rate of DNA loss in Drosophila. Gene 205: 279–289 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Hartl D. L., 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol. Biol. Evol. 15: 293–302 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Sangster T. A., Johnston J. S., Hartl D. L., Shaw K. L., 2000. Evidence for DNA loss as a determinant of genome size. Science 287: 1060–1062 [DOI] [PubMed] [Google Scholar]
- Rokas A., Holland P. W., 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15: 454–459 [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Lourenco P. C., Snaith H. A., 2004. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14: 763–775 [DOI] [PubMed] [Google Scholar]
- Subbiah S., 1989. Are coiled-coil proteins evolutionarily related? J. Mol. Biol. 206: 689–692 [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M., 1994. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 14: 3434–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Koonin E. V., Lipman D. J., 1997. A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Uys G. M., Ramburan A., Loos B., Kinnear C. J., Korkie L. J., et al. , 2011. Myomegalin is a novel A-kinase anchoring protein involved in the phosphorylation of cardiac myosin binding protein C. BMC Cell Biol. 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon D., Schejter E. D., 1999. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9: 889–898 [DOI] [PubMed] [Google Scholar]
- Venkatram S., Tasto J. J., Feoktistova A., Jennings J. L., Link A. J., et al. , 2004. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 15: 2287–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., Pahlke G., Salanova M., Zhang G., Wang S., et al. , 2001. Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276: 11189–11198 [DOI] [PubMed] [Google Scholar]
- Wang Z., Wu T., Shi L., Zhang L., Zheng W., et al. , 2010. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol. Chem. 285: 22658–22665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P. J., Momany M., 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13: 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenerall P., Krupa B., Zhou L., 2011. Mechanisms of intron gain and loss in Drosophila. BMC Evol. Biol. 11: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Megraw T. L., 2007. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell 18: 4037–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lee J., Chasin L. A., 2003. The effect of nonsense codons on splicing: a genomic analysis. RNA 9: 637–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.