Abstract
In biology, many quantitative traits are dynamic in nature. They can often be described by some smooth functions or curves. A joint analysis of all the repeated measurements of the dynamic traits by functional quantitative trait loci (QTL) mapping methods has the benefits to (1) understand the genetic control of the whole dynamic process of the quantitative traits and (2) improve the statistical power to detect QTL. One crucial issue in functional QTL mapping is how to correctly describe the smoothness of trajectories of functional valued traits. We develop an efficient Bayesian nonparametric multiple-loci procedure for mapping dynamic traits. The method uses the Bayesian P-splines with (nonparametric) B-spline bases to specify the functional form of a QTL trajectory and a random walk prior to automatically determine its degree of smoothness. An efficient deterministic variational Bayes algorithm is used to implement both (1) the search of an optimal subset of QTL among large marker panels and (2) estimation of the genetic effects of the selected QTL changing over time. Our method can be fast even on some large-scale data sets. The advantages of our method are illustrated on both simulated and real data sets.
Keywords: dynamic traits, Bayesian P-splines, smoothing, variational Bayes (VB) estimation, variable selection
IN quantitative trait loci (QTL) mapping, people are typically interested in finding genomic positions influencing a single quantitative trait. When the repeated measurements over time of a developmental trait (such as body weight, milk production, and mineral density) are available, it is often preferable to analyze all dynamic time points (traits) jointly to obtain a better understanding of the genetic control of the trait over time (Wu and Lin 2006). To analyze such kinds of time course data, one simple possibility is to apply some multiple-trait methods (jointly analyze many unordered correlated traits) based on multivariate regression (Jiang and Zeng 1995; Banerjee et al. 2008). However, the standard multivariate regression often fails to model the specific dependent (order) structure in the dynamic phenotype data. In statistics, the order nature of the time course data (i.e., smoothness property) means that the two nearby measurements should have closer values than the two with farther distances. Regarding the smoothness assumption in the data, the following two different improved statistical approaches have been used for the dynamic trait analysis:
Combining phenotypes: The phenotypic information over time points is combined by using some smoothing and/or data reduction techniques, and the combined data are used as the new response data for mapping QTL. Some examples include Gee et al. (2003), Heuven and Janss (2010), Hurtado et al. (2012), and Sillanpää et al. (2012). They first fitted the repeated measures of the phenotype data at each individual by the logistic growth curve or a high-order polynomial curve, and next they used the estimated curve parameters for all the individuals as the latent trait data in a multivariate Bayesian regression model for mapping QTL. Outside the genetics context, Meier and Bühlmann (2007) proposed a combined-likelihood approach, where they first reweighted the time course response variables by kernel smoothing techniques and then performed univariate regression (with a single response variable) independently on each reweighted response.
Combining genetic effects: In a multiple-trait model, the parameters of the QTL effects (genotypic value) (i.e., time-varying coefficients) are reparameterized by specifying them as a smooth function over time. When the dynamic pattern of the trait is simple, the effects of a QTL over time can be specified as a parametric function (i.e., logistic function). For such cases, Ma et al. (2002) developed a maximum-likelihood-based approach. Alternatively, if the shape of the dynamic trait is complicated, one can fit the nonparametric curve, applying methods such as Legendre polynomials (Lin and Wu 2006; Yang and Xu 2007; Li et al. 2012), wavelets (Zhao et al. 2007), or B-splines (J. Yang et al. 2009; R. Yang et al. 2009; Xiong et al. 2011; Gong and Zou 2012; Xing et al. 2012). In addition, the residual terms of these models were often assumed to share a certain covariance structure such as an autoregressive process (Ma et al. 2002).
In this article, we concentrate on the second approach, which models the smoothness of the marker effects instead of the phenotype data. This might have the advantage of finding the real underlying dynamic pattern of QTL that characterize the developmental traits, so that we can easily interpret the results.
As for a single-trait QTL analysis, variable selection (i.e., choosing a subset of markers that can approximately represent QTL) is an important issue also for mapping dynamic traits. Most of the existing frequentist approaches (Ma et al. 2002; Lin and Wu 2006; Xiong et al. 2011; Gong and Zou 2012) follow single-locus functional mapping, which maps the dynamic traits to each marker one at a time and uses either likelihood methods or least-squares-estimating equations approaches (with independent residual covariance structure). These approaches typically construct a test statistic (e.g., log-likelihood ratio or Wald statistic) to screen the important variables (QTL) through a multiple-testing procedure (e.g., adjusting the P-value by permutation or by Bonferroni correction). In some Bayesian approaches (Yang and Xu 2007; Min et al. 2011; Yang et al. 2011; Li et al. 2012; Xing et al. 2012), the multilocus QTL analysis is performed by assigning shrinkage-inducing priors or spike and slab priors to the marker effects. Wald tests, credible intervals (Li et al. 2011), or Bayes factors can then be used to justify the QTL.
In this article, we develop a Bayesian multivariate regression method with smoothing prior settings for functional QTL mapping. In our model, we choose B-splines to reparameterize the time-varying marker effects. The benefit of using B-splines over Legendre polynomials, which have been intensively used for nonparametric modeling in some earlier functional mapping approaches, was explained in Xing et al. (2012). Although both B-splines and Bayesian modeling have been considered in some earlier works listed above, our approach has the following three new features. First, with B-splines, our focus is on automatic adaptive determination of the degree of smoothness (i.e., number of knots), which is a crucial problem in B-splines modeling. In the earlier works of Xiong et al. (2011) and Gong and Zou (2012), the degree of smoothness of B-splines was chosen explicitly by cross-validation or the Akaike information criterion (AIC). While here we estimate the degree of smoothness implicitly by assigning second-order penalty priors (Fahrmeir and Kneib 2011) to the time-varying parameters. In other words, we estimate the functional forms of the marker effects and infer their degrees of smoothness simultaneously. This could greatly simplify the whole estimation procedure. The original idea of such prior settings was introduced as P-splines (or B-splines with penalty) (Eilers and Marx 1996; Lang and Brezger 2004) and has been widely applied in the estimation problems of the generalized additive models. Second, the above-mentioned Bayesian approaches were based on Markov chain Monte Carlo (MCMC) simulation, which could be computationally inefficient for high-dimensional data with either a large number of markers or a large number of time points. Here, instead, we introduce a fast variational Bayes (Jaakkola and Jordan 2000; Beal 2003) approach for posterior estimation. Variational Bayes is a deterministic approximation method, which has been used in several single-trait QTL mapping studies (Logsdon et al. 2010; Carbonetto and Stephens 2012; Li and Sillanpää 2012). We generalize these ideas in a multivariate regression framework. Third, for model search or variable selection, we adopt a matching pursuit-like algorithm introduced in Nott et al. (2012) for a different context. The idea is similar to the well-known forward/backward selection (Hastie et al. 2009; Segura et al. 2012), but it differs substantially from the Bayesian variable selection procedures used currently for Bayesian QTL mapping (e.g., Yang et al. 2011). The rest of the article is organized as follows: in Methods, the concept of the (functional) multitrait model and B-splines is reviewed, and the prior settings, the variational Bayes algorithms, and the variable selection procedures are introduced; in Example analyses, we show the results of analyzing two types of simulated data sets (including data replicates) and one real mouse data set (Xiong et al. 2011); and finally we summarize some key points of our functional approach in the Discussion.
Methods
Background
A multivariate Gaussian linear regression model for functional QTL mapping can be specified as
| (1) |
for individuals i = 1, … , n and indexes of the time points r = 1, … , k. Here yi(tr) is the measurement of the phenotypic value of individual i at time point tr, and β0(tr) is the intercept term representing the nongenetic additive effect at time point tr. Moreover, xij is the genotype of individual i at marker j coded as 1 for genotype AA, 0 for Aa, and −1 for aa; βj(tr) is the additive genetic effect of marker j at time tr; and ei(tr) is the residual error that is assumed to independently follow a common unknown temporal stochastic process for all i = 1, … , n, with a multivariate normal distributed form ei = [ei(t1), … , ei(tk)] ∼ MVN(0, Σ0). In this article, we further assume that the residual covariance follows (i) an independent diagonal covariance structure and (ii) a stationary first-order autoregressive [AR(1)] structure (Fahrmeir and Kneib 2011), which is defined as (0 < ρ < 1) for indexes of the time points r = 1, … , k, s = 1, … , k.
Remark:
In principle, a QTL usually does not exactly locate at any marker position, but here we consider using a marker only to approximate the true QTL by assuming the high marker density (Xu 2003).
Note that the model (1) differs from some single-locus functional mapping approaches (Ma et al. 2002; Xiong et al. 2011). Those authors specify the model for marker j (j = 1, … , p) as
| (2) |
where ξij1 is the indicator of a particular genotype (e.g., AA) for marker j and individual i, and γj1(tr) and γj2(tr) are the corresponding genotypic values. Model (1) is different from (2). In (1), the multiple loci are included in the same equation, and we assume no dominance effects.
A fundamental principle of functional mapping is that βj(tr)(j = 0, … , p), which is defined at discrete time points tr(r = 1, … , k), actually comes from a continuous function βj(t) with the domain t ∈ [t1, tk]. We may call βj(t) a trend function of the genetic effects since it describes how the effect size of a QTL changes over time. In this trend function, the smoothness property should hold, meaning that the nearby effects should share similar values. For instance, we may expect that the difference between βj(t1) and βj(t2) should be smaller than that between βj(t1) and βj(t3). Introducing smoothness for the time course data may have some advantage over the general methods for modeling multiple traits, where each βj(tr) (r = 1, … , k) is assumed to be independent from the other (Wu and Lin 2006). First, it could provide more biologically meaningful results, where we can directly see a dynamic pattern of a QTL contributing to the development of a trait. Second, when estimating the parameter at a particular time point, the information from the observations of the nearby time points can be shared, and this might be able to increase the statistical power to detect some true signals. One simple parametric way to model the smoothness is to specify a precise functional form to βj(t) over time tr. For example, Ma et al. (2002) specified it as a logistic function , and estimated the parameters aj, bj, and cj by maximum likelihood to determine the exact shape of βj(t). The parametric method is simple and usually has only a small number of parameters to be estimated, but it is able to describe only a quite simple trend such as a monotonically increasing trend or, by other means, to describe a very smooth function with no function values changing abruptly at any time point. To model a more complicated dynamic pattern such as the irregular periodic trend shown in the mouse active-state probability data (Xiong et al. 2011), some more flexible nonparametric or semiparametric methods such as basis expansions and kernels could be good alternatives (Hastie et al. 2009). Many functional mapping approaches are actually intensively based on basis expansions, which represent the additive genetic effect βj(t) as a linear combination of m basis functions as
| (3) |
where the basis functions ψjh(t) are some sort of transformations of the time domain, and αjh are the parameters to be estimated after such a reparameterization. By the given observed time points tr, Equation 3 can be specified in a matrix form,
| (4) |
where βj = [βj(t1), … , βj(tk)]T, αj = [αj1, … , αjm]T, and . The common choice of the basis functions could be, for example, the high-order polynomials, Legendre polynomials, splines (also called piecewise polynomials or truncated power series), or wavelets. Note that, if Ψj is chosen as a k × k identity matrix, then we have βj = αj, which corresponds to the standard nonfunctional multivariate model. Therefore, the nonfunctional multivariate regression could be regarded as a special case of the functional multivariate regression with basis expansions.
We choose B-splines as a univariate basis functions setting in this article. To obtain an intuitive idea of B-splines, it is worth mentioning the spline bases in a general sense. According to Hastie et al. (2009), the spline bases of order s with z (interior) knots are a series of truncated power bases , where the z knots t1 < ζ1 < ζ2 < … < ζz < tk are a sequence of break points defined in the time domain [t1, tk], which divide the domain into z + 1 subintervals. Truncated power bases are not upper bounded, which may cause serious numerical problems during the computation. On the other hand, a B-spline basis, obtained by taking some appropriate differences of the truncated power bases (Fahrmeir and Kneib 2011), has a nice feature that the basis function values are upper bounded by 1, which makes it numerically more stable when fitting the curves. We leave the technical details of the B-splines (e.g., how to generate a B-spline basis by a recursive algorithm) to other authors. Please refer to De Boor (2001) for the general mathematical theory of B-splines and to Ramsay et al. (2009) for the information on some practical implementations and related software.
The total number of basis functions equals the total number of (interior) knot points plus the spline order. For B-splines, the choice of the spline order, the placement of the knots, and especially their number together determine how smooth the curve will be. Based on Hastie et al. (2009), it is usually not necessary to specify the spline order to be higher than 4. Here we just set the order to be 4, which corresponds to the widely used cubic splines. We further simply set the knots to be equally spaced along the time domain.
Prior settings for Bayesian P-spline smoothing
A remaining issue left from the last section is how to appropriately choose the number of knots, which determines the degree of smoothness of the curve. If the trend of the genetic effect is flat and simple, we should push a high degree of smoothness to the curve by using only a small number of knots. On the other hand, if the trend is oscillating and complicated, then the smoothness assumption should be relaxed by specifying a large number of knots. According to Hastie et al. (2009), misspecifying the degree of smoothness (or the number of knots) can easily cause overfitting/underfitting of data. Because a large number of markers may be present in the model, prechoosing an appropriate number of knots for each of them explicitly by using an approach such as cross-validation is an unrealistic task. In Bayesian statistics, a random walk smoothing prior (Lang and Brezger 2004) can be specified for the B-spline parameters αjh to automatically infer the degree of smoothness. The first- and the second-order random walk priors (corresponding to the first- and the second-order difference penalties) can be specified, respectively, as follows:
| (5) |
| (6) |
and
| (7) |
| (8) |
| (9) |
More conveniently, the priors can be written in the matrix form as
| (10) |
where αj = [αj1, … , αjm]T, and K1 and K2 are m × m matrices constructed from the above defined difference penalties. More detailed description of the random walk priors can be found in Appendix A.
The variance parameters , j = 0, … , p, contributing to determine the degree of smoothness for the time-varying marker effects, can be included as a part of the posterior model. For example, we may further assign an inverse gamma prior IG(a, b) with predefined hyperparameters a > 0 and b > 0 to each , so that can be estimated along with other parameters in the posterior. Here the inverse gamma density function is defined as . According to Fahrmeir and Kneib (2011), a typical choice of the hyperparameters is (a, b) = (ε, ε), with ε being a small value, so that the prior for is relatively noninformative. In all of our numerical examples, we choose K2 as the penalty matrix, and we set a = b = 0.0001.
After incorporating smoothness priors into the model, we do not need to be concerned with the choice of knots any more. We may simply specify a global B-spline basis Ψ with a large enough number of equally distributed knots to each marker j. The random walk smoothness priors then play the key role to automatically identify an optimal degree of smoothness of the spline for each individual marker.
We have mentioned earlier that if Ψ is chosen as a k × k identity matrix instead of B-splines, then we exactly obtain a standard nonfunctional multivariate regression model. In this case, we may specify the hierarchical priors as and for parameters αj (= βj). Note that here we assume the coefficients at the same locus j but different time points share a global variance parameter , which makes it differ from those single-trait mapping methods where the coefficients at different time points may be assigned with different variance parameters. The same parameter estimation and variable selection procedure can be applied to both functional and nonfunctional multitrait models, which are described next.
Variational Bayes algorithm
Parameter estimation:
Now everything can be put together. In the basic form of the regression model (1), the parameters βj(tr)(r = 1, … , k) are reparameterized by αjh(h = 1, … , m) defined in (3) and (4) after the B-spline basis expansions, and then we can specify the likelihood function as
| (11) |
where yi = [yi(t1), … , yi(tk)]T, and αj = [αj1, … , αjm]T. The hierarchical smoothness priors for αj (j = 0, … , p) were proposed in the last section. When the residual covariance is assumed to be a diagonal matrix Σ0, each diagonal entry (r = 1, … , k) is assigned a flat Jeffreys’ prior as to make . Let with K = 2(p + 1) + k components. The posterior distribution can be specified as
| (12) |
A variational Bayes (VB) algorithm based on the mean field theory (Jaakkola and Jordan 2000; Beal 2003; Bishop 2006) can be used to efficiently evaluate the unknown parameters in the model. Specifically, the above defined intractable posterior is approximated by a tractable factorized form,
| (13) |
We seek an optimal factorized approximate posterior distribution by minimizing the Kullback–Leibler divergence or, equivalently, maximizing a lower bound of the log-marginal distribution ln p(Y) defined as , where Θ represents the whole parameter space of θ. It can be shown that such a minimization/maximization is reached at
| (14) |
where is the posterior expectation with respect to the factorized approximate distribution with the lth component removed. Since the posterior model belongs to the conjugate exponential family, all the required approximate distributions in (14) can be recognized as standard distributions. Then an iterative coordinate descent algorithm can be easily used to update in (13) for each parameter based on Equations 14 sequentially until convergence. After obtaining the approximate posterior distribution for marker j (j = 1, … , p), interesting quantities such as posterior mean and posterior covariance matrix are directly available.
When the stationary AR(1) residual covariance is used, the covariance matrix Σ0 is actually controlled by two parameters, and ρ (0 < ρ < 1). We may further assign a noninformative Jeffreys’ prior for as and a uniform prior for ρ as p(ρ) = 1[0,1]. A factorized form of the approximate posterior distribution similar to that in Equation 13 can be specified, and the marginal distributions q(⋅|Y) for all the parameters except ρ can be optimized based on Equation 14 as above. The parameter ρ is not conjugate in the posterior and it is difficult to incorporate into the above-mentioned VB updating procedure. To handle this, we can apply the idea of fixed-form VB approximation (Salimans and Knowles 2013), by preassuming q(ρ|Y) to be a Beta(ρ|μ1, μ2) distribution with unknown shape parameters μ1 and μ2. The parameters μ1 and μ2, controlling the shape of the approximate marginal posterior distribution, can be estimated by using the novel Monte Carlo sampling-based method introduced in Salimans and Knowles (2013). Details of these VB algorithms are presented in Appendix B.
Variable selection:
In principle, it is possible to execute the VB algorithm on the multivariate posterior model with a whole set of markers included and then construct the Wald test statistic based on the posterior mean and covariance estimate of αj for the jth marker (see Appendix A) to detect QTL. A similar strategy has been used in some MCMC-based functional mapping studies (Xing et al. 2012). However, it has been shown that although the VB can generally provide accurate posterior mean estimates for the parameters, posterior uncertainties (e.g., posterior variances) are often underestimated (Grimmer 2011; Li and Sillanpää 2012), compared with MCMC. Therefore, the Wald test statistic constructed based on a VB estimate may not be reliable for screening the true positive signals, which motivates us to seek an alternative way to detect QTL under the VB scheme. However, we can nevertheless compute the Wald statistic for each marker and consider them as scores for ranking the markers. Specifically, if we consider a model denoted as M with only a subset of markers included, the corresponding marginal distribution p(Y|M) is called the model evidence or marginal likelihood. The parameter vector in the model M can be defined as . With the marginal likelihoods for all the models computed, we may go in two directions: (i) calculate the posterior distribution p(M|Y) ∝ p(Y|M)p(M) of different models given the data to find which model is preferable [if the prior p(M) is uniform, then p(M|Y) ∝ p(Y|M) (Bishop 2006)], and (ii) compare two models M1 and M2 by the Bayes factor (Kass and Raftery 1995). Although the marginal likelihood p(Y|M) cannot be analytically computed for our problem, in variational Bayes estimation, the above-mentioned lower bound can be treated as an approximation (Bishop 2006). We assume that the prior p(M) is uniformly distributed, and then we can choose an optimal model (the best subset of markers) by satisfying (Beal and Ghahramani 2003). Since it is impractical to perform the VB and compute the lower bounds for all possible combinations of markers, we use a matching pursuit-like greedy algorithm, which is adapted here from Nott et al. (2012), who considered a different model structure. This procedure produces a sequence of candidate models, and we choose the one corresponding to the largest value of the lower bound. Since usually we are interested only in a small number of QTL with large effects, we could stop the algorithm at an early stage by selecting only a small number of variables into the model without considering the whole solution path. In this case, the algorithm will search only through the low-dimensional space, which can be implemented very efficiently. Further details of this algorithm are shown in Appendix C.
Example Analyses
We evaluate performance of our methods with both simulated and real data examples. Our simulation analyses are largely based on the simulated data from the QTLMAS2009 workshop (Coster et al. 2010), and the real data were originally analyzed in Xiong et al. (2011). For parameter estimation and variable selection, we implemented both the functional multitrait VB approach and the nonfunctional multitrait VB approach presented in this article on all these data sets. For simplicity, here we name them as VBfun and VBnonfun, respectively. Furthermore, both diagonal and AR(1) structures were considered in each case to model residual covariance. For the remainder, some of the key features of VBfun and VBnonfun are summarized in Table 1. The methods were implemented in MATLAB on a desktop with an Intel Core 2 2.13 GHz processor and 2 Gb memory. In practice, we used the MATLAB codes (publicly available at http://www.psych.mcgill.ca/misc/fda/software.html) developed by Ramsay et al. (2009) to generate cubic B-spline bases. Our own MATLAB codes for implementing VBfun and VBnonfun methods are available in Supporting Information, File S1.
Table 1. Key features of the functional model and the nonfunctional model.
| Methods | Basis expansions | Priors | Residual covariances |
|---|---|---|---|
| VBfun | βj = Ψk×mαj, where Ψk×m are the B-spline bases | Diagonal or AR(1) | |
| VBnonfun | βj = Ik×kαj, where Ik×k is an identity matrix | Diagonal or AR(1) |
Details are shown in the section Prior settings for Bayesian P-spline smoothing.
Analysis of QTLMAS2009 simulated data
Briefly, the simulated data set includes 453 SNP markers distributed over five chromosomes of 1 M each from 2025 individuals with a certain population structure and the growth traits following logistic curves measured over five time points (0, 132, 265, 397, and 530 days). In total, 18 additive QTL were simulated, with 6 contributing to each of the three parameters φ1 (asymptotic yield), φ2 (inflection point), and φ3 (slope of the curve) of a growth curve. Both genotype (with map information) and phenotype data are publicly available (Coster et al. 2010). The same data set has been analyzed by an MCMC approach of Heuven and Janss (2010) and Sillanpää et al. (2012). A fundamental difference in model strategies between their approaches and ours has been explained in the Introduction. To compare with Heuven and Janss (2010), the same subsample of 1000 individuals is used in our simulation analyses.
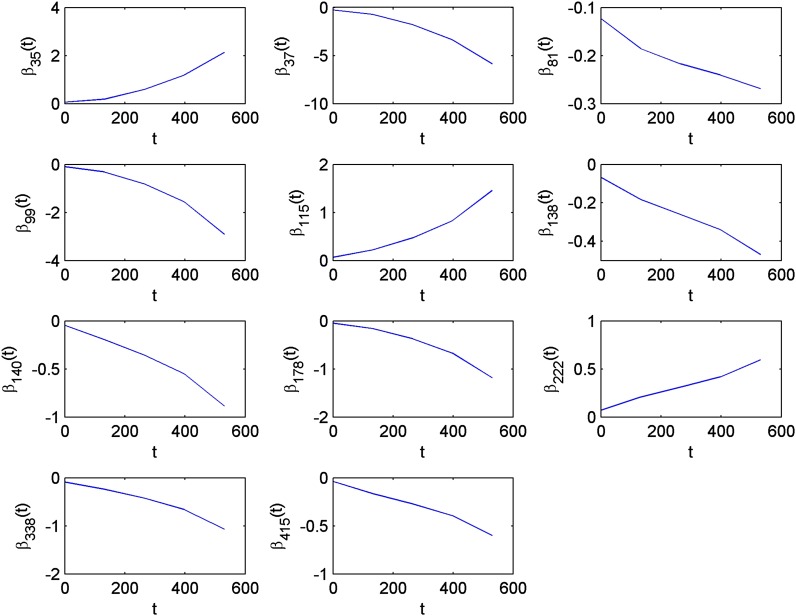
In VBfun, we specified six equidistant knots on the time domain [0, 530]. Note that the number of B-spline bases equals the number of knots plus the spline order, so if the spline order is 4, we obtain 10 B-spline bases in total. For both VBfun and VBnonfun, we ran the VB forward selection for 20 steps that continued with a backward selection procedure (we use the same procedure for the other two data examples). We then identified the best set of variables corresponding to the maximum of the lower bounds. Here we first describe the results by assuming the diagonal residual covariance structure, which might be a more reasonable assumption for the QTLMAS2009 data, since no residual dependence structure was simulated there. The information from the detected QTL is summarized in Table 2. The best model selected by VBfun includes 11 markers, which are all located very close to some of the true simulated QTL. The estimated trend functions of the genetic effects for the 11 selected markers are shown in Figure 1. The trend functions have similar shapes to the logistic growth curves. VBnonfun detects 13 markers that are largely overlapping with the markers found by VBfun, in which all the others are located close to some simulated QTL except marker 452. The estimated trend functions from VBnonfun are shown in Figure S1. In total, VBfun and VBnonfun detected 9 and 11 true simulated QTL, respectively, which is comparable to the results shown in Heuven and Janss (2010), where they reported 9 correctly detected QTL (with false discovery rate <0.05). Similarly to that in Heuven and Janss (2010), our methods are able to detect most of the QTL controlling the parameters φ1 and φ3 of the logistic growth curve, but find fewer that control φ2. In addition, Sillanpää et al. (2012) correctly detected 6 QTL (with QTL inclusion probability >0.5) and 8 QTL (with QTL inclusion probability >0.05). Note that they considered only 500 individuals, which may reduce the statistical power to detect QTL with high probability. Compared to the MCMC approaches of Heuven and Janss (2010) and Sillanpää et al. (2012), one major benefit of our VB method is its computational efficiency. For both VBfun and VBnonfun, the whole computational procedure took only <1 min, whereas the MCMC methods may take hours.
Table 2. The results of QTLMAS2009 data analysis, including the selected markers, their locations (in brackets), the corresponding Wald scores, and the closest true simulated QTL.
| VBfun |
VBnonfun |
|||
|---|---|---|---|---|
| Marker | Wald score | Marker | Wald score | Closest simulated QTL |
| 35 (0.4029) | 320.68 | 35 (0.4029) | 270.29 | (0.4525) 36–37a |
| 37 (0.4447) | 5037.53 | 37 (0.4447) | 4874.49 | (0.4525) 36–37a |
| 81 (0.9137) | 183.05 | 81 (0.9137) | 107.43 | (0.8765) 77–78b |
| 99 (1.0516) | 1114.62 | None | None | (1.0455) 98–99a |
| 115 (1.2566) | 220.05 | 115 (1.2566) | 171.32 | (1.3302) 118–119c |
| 138 (1.4852) | 130.74 | 138 (1.4852) | 95.57 | (1.4889) 138–139b |
| 140 (1.5242) | 143.13 | 140 (1.5242) | 169.91 | (1.4889) 138–139b |
| 178 (1.9011) | 155.87 | 178 (1.9011) | 164.75 | (1.8864) 174–175a |
| 222 (2.3108) | 139.91 | 222 (2.3108) | 127.97 | (2.2622) 216–217b |
| None | None | 288 (3.048) | 126.70 | (3.0962) 293–294b |
| 338 (3.7168) | 248.46 | 338 (3.7168) | 171.26 | (3.6979) 336–337a |
| None | None | 358 (3.8517) | 65.43 | (3.8639) 358–359b |
| 415 (4.6311) | 90.95 | 415 (4.6311) | 70.41 | (4.5971) 411–412c |
| None | None | 452 (4.9494) | 62.84 | (4.7719) 432–433a |
For the Wald tests, the critical value corresponding to the significance level 0.05 is 18.31 for the estimates of VBfun (chi-square distribution with 10 d.f.) and 11.07 for the estimates of VBnonfun (with 5 d.f.), respectively (assuming the diagonal covariance structure in the model).
Simulated QTL effects on φ1 of the simulated logistic growth curves (traits), respectively.
Simulated QTL effects on φ3 of the simulated logistic growth curves (traits), respectively.
Simulated QTL effects on φ2 of the simulated logistic growth curves (traits), respectively.
Figure 1.
Analysis of QTLMAS2009 data: Estimated trends of genetic effects for the 11 selected markers by using VBfun, which assumed the diagonal residual covariance structure.
Additionally, the results by assuming AR(1) residual covariance structure are shown in Table S1, Figure S2, and Figure S3. Here, the analyses took a longer time, because a stochastic optimization step is built into the VB algorithm for updating the nonconjugate part of the model. The results of both VBfun and VBnonfun here turned out to be slightly worse than in the case of the diagonal covariance structure, in the sense that fewer numbers of QTL are correctly detected. Amazingly, the mean estimate of the parameter ρ, measuring the decline of the correlation with time lag, is 0.91. This indicates that the temporal correlation among the (non-QTL) residual errors is extremely high, although no such residual dependencies were actually simulated. Note that the QTL effects were simulated to the latent trait variables φ1, φ2, and φ3, so that they only indirectly influenced the characteristic of the real phenotypes. Their way of simulation might accidently introduce the high residual correlation to the phenotype data, and the methods assuming AR(1) residual covariance may have difficulties in associating observed dependency into the correct origin. However, the detected QTL between two residual structures partially coincide.
More complicated simulation studies
Here on the basis of the genotype data with 453 markers and 1000 individuals from the QTLMAS2009 workshop, we simulated new phenotype data by having nine additive QTL at markers 35, 52, 78, 98, 118, 174, 216, 358, and 433 together with an intercept term in the simulation model. The trend functions of the genetic and nongenetic effects simulated within the time domain [0, 24] (hr) are given in Table 3. Generally, they were simulated as different curves (linear, logistic, sine …) with various degrees of smoothness. All the other 444 markers were assumed to be inactive over time. The k simulated time points are equally spaced from 0 to 24. The residual terms ei(tr) (r = 1, … , k) were simulated from the AR(1) process. The dynamic phenotype data were then generated based on Equation 1.
Table 3. The simulated trend functions for the intercept and nine QTL.
| Descriptions | Trend functions |
|---|---|
| The intercept with a logistic growth curve | |
| Loci 52 and 358 have constant effects over time | β52 = 2 |
| β358 = 1 | |
| Locus 118 has linear growth effect over time | β118 = 0.1t + 1 |
| Loci 174 and 216 are active only at the early stage | |
| Loci 98 and 433 are active only at the late stage | |
| Locus 78 is active only at the middle stage | |
| Locus 35 has the periodic effects over time |
Again, VBfun and VBnonfun were compared using either diagonal residual covariance structure or AR(1) structure.
Evaluation of parameter estimation:
We first evaluate how accurately our methods can estimate the trend functions of the intercept and the genetic effects without any variable selection. We simulated four data sets with all possible combinations of k = 10, 100 (number of time points) and ρ = 0.5, 0.8 (residual correlation). The noise level was fixed to be 15. The average heritabilities over time points varied from 0.09 to 0.23 for those four data sets. We used both VBfun and VBnonfun with the assumption of AR(1) residual covariance structure to estimate the effects of nine simulated QTL. In VBfun, we specified 16 and 46 equidistant knots (corresponding to 20 and 50 B-spline bases) when k = 10 and k = 100, respectively. Based on our experiments, the results were not sensitive to the number of knots we chose if the number was set to be large enough. However, it is not recommended to use too large a number of knots to save computation time. To measure the accuracy of the parameter estimates, we calculated the mean squared error (MSE) for each simulated QTL (including the intercept term) as
| (15) |
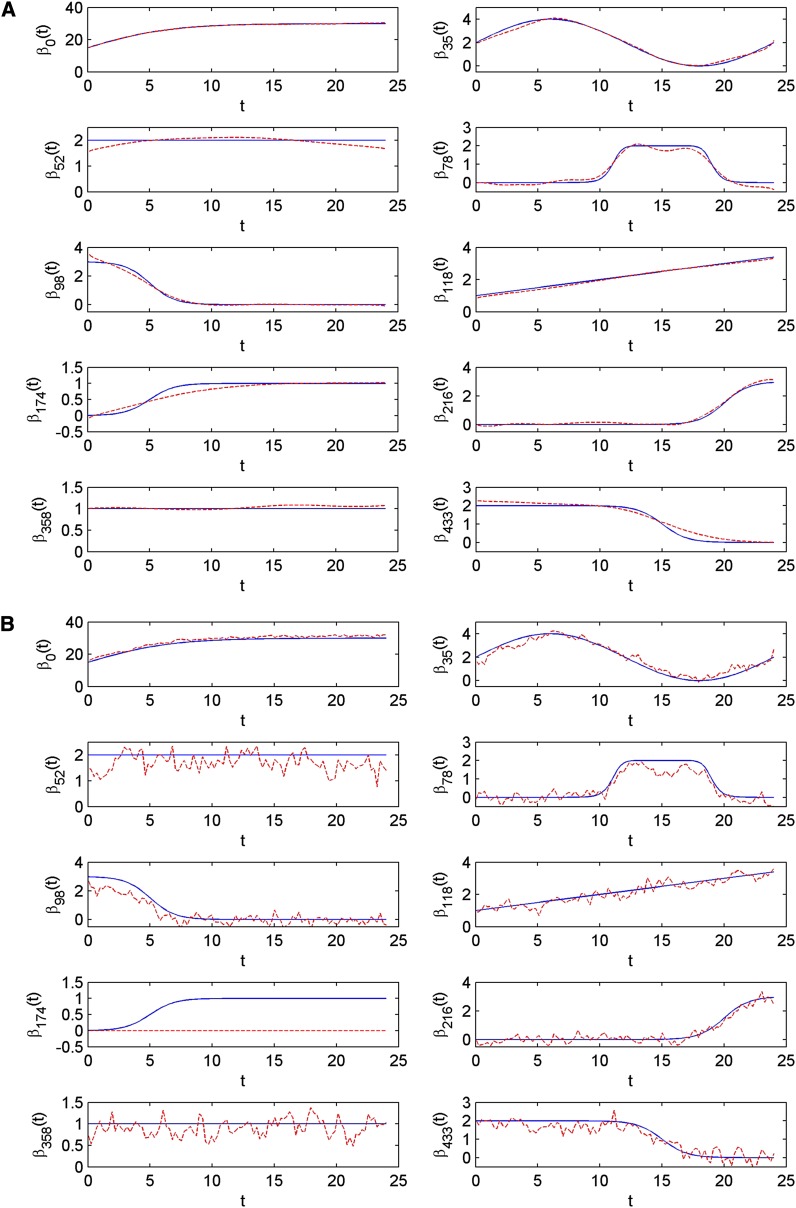
where βj is the simulated trend function for marker j, and is the estimated trend function. The results are summarized in Table 4. Also see Figure 2 including a comparison between the estimated trends and the true simulated trends for the case of k = 100 and ρ = 0.5. Results for the other three cases are presented in Figure S4, Figure S5, and Figure S6. For all the data sets, the posterior mean estimates of the parameters ρ and are not far from their true simulated values, indicating the good performance of the fixed-form VB estimation. Regarding the QTL effects, VBfun tends to provide more accurate estimates than VBnonfun. Taking the case of k = 100 and ρ = 0.5 as an example (see Figure 2), the trend functions estimated by VBfun are almost identical to the true simulated trends. Whereas VBnonfun mistakenly shrinks the estimated effects (over time) of marker 174 toward zero, the estimates of other markers show highly oscillating patterns. VBfun seems to perform best when the number of time points is large (i.e., k = 100) and the residual correlation is not high (i.e., ρ = 0.5).
Table 4. The mean squared error comparing the estimated trend functions to the simulated ones.
| k = 10 | k = 10 | k = 100 | k = 100 | |||||
|---|---|---|---|---|---|---|---|---|
| u = 0.5 | k = 0.8 | u = 0.5 | u = 0.8 | |||||
|
h2 = 0.2265 |
h2 = 0.0922 |
h2 = 0.2347 |
h2 = 0.1259 |
|||||
| QLT effects | F | N | F | N | F | N | F | N |
| β0 | 0.2194 | 2.0375 | 0.4933 | 14.5590 | 0.0246 | 1.7757 | 0.1166 | 19.4684 |
| β35 | 0.0704 | 0.1483 | 0.1138 | 0.5161 | 0.0189 | 0.1267 | 0.0573 | 0.5326 |
| β52 | 0.2535 | 0.7231 | 0.0616 | 3.9968 | 0.0247 | 0.2045 | 0.0324 | 3.9995 |
| β78 | 0.0921 | 0.1037 | 0.0937 | 0.1542 | 0.0248 | 0.1185 | 0.1442 | 1.2072 |
| β98 | 0.1505 | 0.0725 | 0.4961 | 0.5038 | 0.0160 | 0.2116 | 0.4147 | 1.5341 |
| β118 | 0.0127 | 0.0239 | 0.1416 | 0.1495 | 0.0063 | 0.0695 | 0.0185 | 0.1778 |
| β174 | 0.0513 | 0.7201 | 0.1866 | 0.7229 | 0.0129 | 0.7464 | 0.1099 | 0.7473 |
| β216 | 0.0459 | 0.0586 | 0.2224 | 0.0369 | 0.0099 | 0.0606 | 0.0677 | 1.1692 |
| β358 | 0.0069 | 0.0493 | 0.0385 | 0.9960 | 0.0022 | 0.0481 | 0.0085 | 0.9988 |
| β433 | 0.2278 | 0.0278 | 0.2753 | 2.2965 | 0.0332 | 0.1145 | 0.3022 | 2.3295 |
| 14.8034 | 14.7677 | 14.9339 | 14.9758 | 14.9081 | 14.9560 | 15.0325 | 15.1299 | |
| E[ρ] | 0.5088 | 0.5138 | 0.7796 | 0.7882 | 0.4987 | 0.5036 | 0.7779 | 0.7876 |
F and N represent VBfun and VBnonfun, respectively [assuming the AR(1) covariance structure in the model], and h2 is the averaged heritability over time.
Figure 2.
(A and B) Simulated data with k = 100, u = 0.5: The estimated trend curves (red dashed lines) against the simulated trend curves (blue solid lines).
If the diagonal residual covariance structure is assumed in the model (results are shown in Table S2, Figure S7, Figure S8, Figure S9, and Figure S10), VBfun seems to provide identical estimates compared to the case when AR(1) structure is assumed. On the other hand, VBnonfun performs better together with diagonal covariance structure than with AR(1), in the sense that it does not mistakenly shrink the effects of any QTL toward zero.
Evaluation of variable selection:
Next, to evaluate the quality of the variable selection, we again simulated four data sets. First, we randomly sampled a subset of genotype data with n = 200 or n = 500 (of 1000) individuals. Based on these, we simulated phenotypic measurements with k = 10 or k = 100 time points. For the residuals, the noise level and correlation level ρ were fixed to be 10 and 0.5, respectively. These simulations were repeated 50 times. We then applied the proposed VB methods assuming the AR(1) residual covariance structure on the data and monitored how many times each QTL was correctly identified by VBfun and VBnonfun in each of the four simulated conditions. The results are presented in Table 5. Overall, the VBfun method consistently performed better than VBnonfun. Especially when the number of individuals is small and the number of time points is large (i.e., n = 200 and k = 100), VBfun tends to correctly detect eight of nine QTL with very high frequency, but VBnonfun detected none of them. When n = 500 and k = 100, VBfun is able to correctly identify all nine QTL in almost all 50 replicates, while VBnonfun detects only three. However, when n = 200 and k = 10, VBfun and VBnonfun behave similarly by selecting only two QTL frequently.
Table 5. The number of times each QTL has been correctly selected over 50 replications, together with the average number of wrongly selected markers (false positives).
| n = 200 | n = 200 | n = 500 | n = 500 | |||||
|---|---|---|---|---|---|---|---|---|
|
k = 10 |
k = 100 |
k = 10 |
k = 100 |
|||||
| QTL | F | N | F | N | F | N | F | N |
| 35 | 50 | 47 | 50 | 0 | 50 | 50 | 50 | 50 |
| 52 | 4 | 0 | 49 | 0 | 44 | 0 | 50 | 0 |
| 78 | 0 | 0 | 50 | 0 | 37 | 3 | 50 | 0 |
| 98 | 0 | 0 | 45 | 0 | 36 | 0 | 50 | 0 |
| 118 | 49 | 43 | 50 | 0 | 50 | 50 | 50 | 50 |
| 174 | 0 | 0 | 11 | 0 | 2 | 0 | 46 | 0 |
| 216 | 19 | 1 | 49 | 0 | 47 | 38 | 50 | 0 |
| 358 | 4 | 0 | 50 | 0 | 43 | 0 | 50 | 0 |
| 433 | 0 | 0 | 43 | 0 | 27 | 0 | 50 | 0 |
| Average false positive | 0.04 | 0.04 | 0.32 | 0 | 0.06 | 0.07 | 0.08 | 0 |
F and N represent VBfun and VBnonfun, respectively [assuming the AR(1) covariance structure in the model].
The results of both methods assuming the diagonal residual covariance structure are presented in Table S3. VBfun assuming the residual diagonal covariance structure identifies the correct set of QTL equally as well as the method together with the AR(1) residual covariance, but assuming the diagonal residual covariance results in more false positive QTL. On the other hand, VBnonfun assuming the residual diagonal covariance seems to correctly detect more true QTL than assuming AR(1) covariance.
To further evaluate how well our proposed methods can avoid false positive signals, we simulated another four replicated null data sets, where only an intercept term but no QTL influenced the phenotypes. The sample sizes, number of time points, and residual covariance structures were simulated as above. Results of the average number of false positive QTL over 50 replicates are shown in Table 6. We found that only VBfun together with the diagonal covariance structure may tend to produce a few false positives in some of the replicates, when the nontrivial temporal residual covariance structure indeed exists.
Table 6. Simulation study under the null: the average number of wrongly selected markers (false positives) over 50 replications.
| n = 200 | n = 200 | n = 500 | n = 500 | |||||
|---|---|---|---|---|---|---|---|---|
|
k = 10 |
k = 100 |
k = 10 |
k = 100 |
|||||
| QTL | F | N | F | N | F | N | F | N |
| Diagonal | 0.08 | 0 | 0.38 | 0 | 0.08 | 0 | 0.40 | 0 |
| AR(1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
F and N represent VBfun and VBnonfun [assuming the AR(1) residual covariance structure in the model], respectively.
Analysis of mouse behavioral data
In the real data analysis, we considered a mouse behavioral data set, which has been previously analyzed by Xiong et al. (2011) and is publicly available at QTL Archive (http://qtlarchive.org/db/q?pg=projdetails&proj=xiong_2011). The phenotype data contain active state probabilities (ASP) (y ∈ [0, 1]) with 222 repeated measurements at each consecutive 6-min time interval within 24 hr (from 1:48 pm to 1:54 pm to 11:54 am to12:00 am, with 7 pm to 7 am as the dark period and otherwise as the light period) for 89 backcross mice. The genotype data consist of 233 informative polymorphic SNP markers distributed over 19 chromosomes. Note that the data are quite challenging from the analysis point of view mainly due to the fact that (1) both the numbers of markers and time points are larger than the number of individuals and (2) considerable variations were shown among the active-state probabilities of the mice (Xiong et al. 2011), and the shape of the mean trajectory (see Figure 3A) is also quite complex. For detailed information on phenotyping and genotyping, please refer to Goulding et al. (2008) and Xiong et al. (2011). Before the statistical analysis, we performed the following three preprocessing steps:
The missing genotypes were replaced by their conditional expectations estimated from their flanking markers with known genotypes (Haley and Knott 1992) once before the analysis.
We performed the logit transformation to the phenotypic measurements (Warton and Hui 2011), so that their values were not restricted to the domain [0, 1]. Specially, for those measurements with ASP of 0 and 1, we first changed values to 0.001 and 0.999, respectively, and then performed logit transformation. The mean trajectories of original and transformed phenotypic data are shown in Figure 3, A and B, respectively. Although the logit transformation did not reduce any complexity in the original data, we found that, by directly applying our VB approaches to the original percentage phenotypes without any transformation, the methods would produce some unreasonable results [i.e., the estimated lower bound gave positive values].
To generate B-spline bases, we discretized the 222 time intervals by a set of single time points [1.9, 2, … , 23.9, 24] (e.g., 1.9 represents 1:48 pm, and 24 represents 12:00 am).
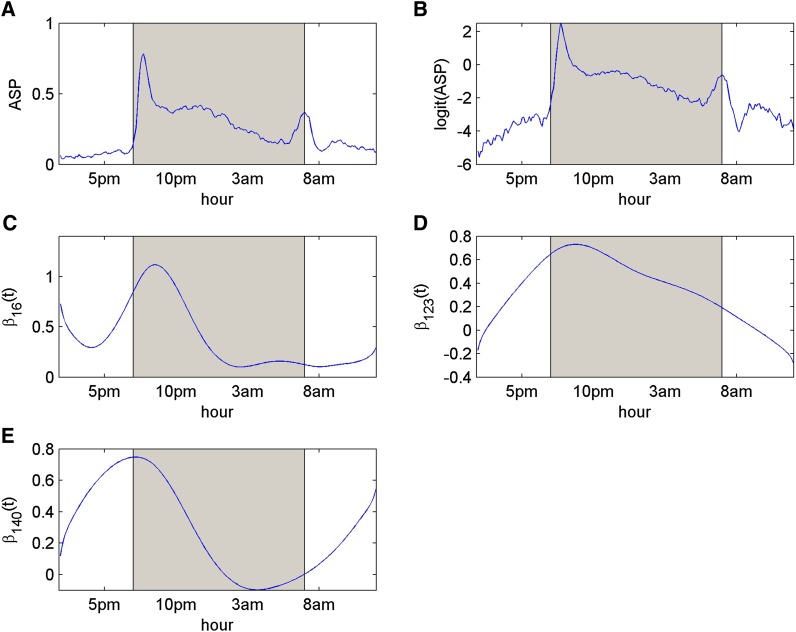
Figure 3.
The results of mouse behavioral data analysis by using VBfun, which assumed the AR(1) residual covariance structure: (A–E) The mean trajectory of the original ASPs (A), the mean trajectory after logit transformation (B), and estimated trends of genetic effects (in logit scale) (C–E) for markers 16, 123, and 140, respectively. The shaded area represents the dark period (7 pm to 7 am).
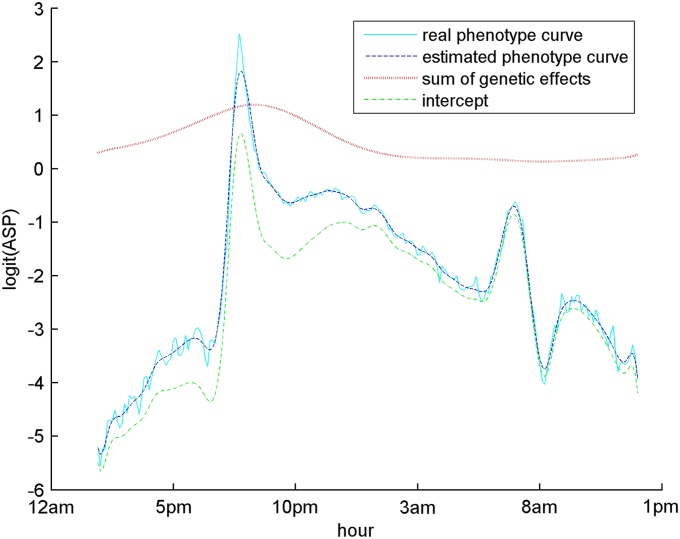
Here we first describe the results of VBfun when the AR(1) residual covariance structure is assumed. As in the last simulated example, we generated 50 B-spline bases needed in VBfun as an upper limit for complexity. VBfun then suggested the best model with three putative QTL, whose positions are shown in Table 7. The posterior mean estimates of AR(1) parameters and ρ are 1.79 and 0.66, respectively. Among these three putative QTL, loci 16 (rs3689947) and 123 (rs6207781), with the largest Wald scores, are located at 81.40 cM on chromosome 1 and at 20.74 cM on chromosome 9, respectively. The estimating equations approach of Xiong et al. (2011) detected two major QTL, which are located at 75 cM (between loci 15 and 16) on chromosome 1 and at 10 cM (at locus 119) on chromosome 9 [S. Sen (University of California, San Francisco), personal communication]. Note that in the genotype data some adjacent markers are very highly correlated with each other, and our greedy search algorithm tends to select only a single marker from a group of highly correlated markers. This might explain why the positions found by our method are slightly different from theirs. In addition, our method detected another interesting locus 140, on chromosome 10. An overall genetic effect trend of the three putative QTL is calculated by (), and the curve is shown in Figure 4. The overall trend of the genetic effects shows a peak between 5 am and 2 pm, during which time the mean trajectory of the phenotypes also shows a clear peak. This indicates that the three putative QTL detected by our method may contribute to the phenotypic variation during the time period when the mice are highly activated. We may further reestimate the phenotypes by summing the estimated intercept and genetic effects of the selected loci. The mean trajectory of the reestimated phenotypes is also shown in Figure 4, which provides a smooth description of the original mean phenotype curve.
Table 7. The results of mouse behavioral data analysis by using VBfun, which assumed the AR(1) residual covariance structure, including the selected markers, their chromosomes, locations, and the corresponding Wald scores.
| Marker | Chromosome | Location (cM) | Wald score |
|---|---|---|---|
| 16 (rs13476259) | 1 | 81.40 | 148.6427 |
| 123 (rs6207781) | 9 | 20.74 | 122.1237 |
| 140 (rs3654717) | 10 | 55.93 | 110.5854 |
For the Wald tests, the critical value corresponding to the significance level 0.05 is 67.50 for the estimates (chi-square distribution with 50 d.f.).
Figure 4.
Cyan solid line is the mean trajectory of the real phenotypes in logit scale, the same as in Figure 3. Red dotted line represents an overall trend of the genetic effects of the three putative QTL. Dark green dotted-dashed line is the estimated intercept. Blue dashed line represents the mean of the reestimated phenotypic trajectory (the genetics effects plus the intercept) in the red dashed line.
On the other hand, when the diagonal covariance structure is assumed (results are shown in Table S4, Figure S11, and Figure S12), VBfun is able to detect loci 18, 123, and 137 (note that loci 18 and 137 are near to loci 16 and 140), which were also found in the AR(1) case. However, additionally, VBfun tends to find many other markers with relatively small effects that are false positives. Thus, we can conclude that assuming a time-dependent AR(1) residual covariance seems to effectively control false positive signals.
Finally, VBnonfun and two different residual covariance structures were applied on the mouse data as well, but they did not detect any signals (results not shown).
Discussion
We have developed here an efficient Bayesian nonparametric estimation procedure for mapping dynamic traits. Compared to several earlier approaches such as the estimating equation approach of Xiong et al. (2011), one major benefit of our Bayesian approach is that some prior information for achieving smoothness can be easily built into the model, which is helpful to simplify the estimation procedure (i.e., without the need to explicitly determine the optimal number of B-spline bases). Our nonparametric method should be generally applicable to many kinds of dynamic traits whether their trajectories are smooth or rather complicated. In this article, we have not considered yet the possible impact of some environmental covariates such as temperature and age variables and their interactions with QTL, as well as the QTL–QTL interactions on the target dynamic traits. Following works such as Zhang and Xu (2005), Yi and Banerjee (2009), Min et al. (2011), and Li and Sillanpää (2012), it is not difficult to extend our current marker set by including the environmental covariates or the pairwise marker–environment or pairwise marker–marker interaction terms as new “marker” variables for variable selection.
Our model specification is largely based on the Bayesian P-splines, which has been applied in various nonparametric modeling fields such as structured additive models (Fahrmeir et al. 2010) and time-varying coefficient models (Lee and Shaddick 2007). One common problem of such a model specification is that a relatively large number of B-spline bases need to be used in the model, which makes the simulation-based MCMC algorithm infeasible for large-scale data sets with hundreds of markers and time points. This motivates us to alternatively use a fast deterministic variational Bayes algorithm for computation. The VB approximation method can not only provide accurate posterior mean estimates to the parameters of genetic effects, but also provides a lower bound estimate of the model evidence that can be used to guide variable selection (Beal and Ghahramani 2003). The matching pursuit-like algorithm adapted here from Nott et al. (2012) is a simple and efficient method for searching an “optimal subset ” of markers that roughly maximizes lower bounds. However, since such a greedy algorithm does not fully explore the whole model space, it usually cannot find a “perfect model ” that corresponds to the global maximum of the lower bounds especially in the case of polygenic traits. We recommend using such a procedure only in the case of sparse genetic architecture to seek a small number of markers with relatively large genetic effects.
An important goal of our data analyses was to compare the functional (multivariate) mapping approach (i.e., VBfun) and the general nonfunctional multiple-trait approach (i.e., VBnonfun) for analyzing the functional valued dynamic traits. Overall, the results from those three examples showed that the functional approach performs better than or at least equally as well as the nonfunctional approach, from the perspective of both parameter estimation and variable selection. Especially when the number of time points is relatively large and the number of individuals is relatively small, the functional approach tends to show much higher statistical power to detect QTL than the nonfunctional approach, indicating that the functional approach owns the advantage of combining the information from different repeated measurement points by specifying basis expansions and smoothing priors for genetic effects in the model. Dynamic phenotype data measured at a large number of time points are often available from some high-throughput automated phenotyping platforms (e.g., Eberius and Lima-Guerra 2009). On the other hand, when the number of time points is small, the benefit of using the functional approach is reduced. Note that our VB framework is quite flexible in the sense that the B-splines can be substituted by many other possible methods of basis expansions and corresponding priors as well. In practice, it is useful to compare different methods and choose the most preferable one.
The primary aim of our nonparametric method is to determine a subset of important markers approximating QTL and estimate the trends of their genetic effects over time. We also tested two possible residual covariance structures, (i) a nonstationary diagonal covariance structure and (ii) a stationary AR(1) covariance structure, and evaluated their impacts on the QTL mapping. In most of our analyses, we found that compared to the AR(1) covariance structure, the simple diagonal structure assuming time independence of residual errors does not significantly affect the accuracy of the parameter estimation, but tends to significantly underestimate the uncertainty (i.e., the posterior covariance for each marker), which may result in including some false positive QTL into the variable selection procedure. Therefore, even though the computation with AR(1) covariance structure is more expensive due to the presence of nonconjugacy in the posterior, it might be a more suitable choice especially when the heritabilities of the dynamic traits under study are low. Other more complicated covariance structures such as some nonstationary parametric structures (Liu and Wu 2009) or nonparametric structures (Yap et al. 2009) can be possibly incorporated if needed, but they require the development of more specific algorithms for the computation of those newly involved parameters. Furthermore, it is necessary to point out that due to the approximative nature of the VB algorithm, the uncertainty estimates for markers may still be underestimated even by using an appropriate residual covariance structure or, by other means, the estimated Wald statistic might be upward biased. On the other hand, the MCMC method is able to provide more accurate uncertainty estimates than the VB methods. Thus, after obtaining a small subset of markers from our VB variable selection procedure, we may then apply a MCMC algorithm to more accurately estimate the Wald statistics and perform the formal hypothesis testing. These can be taken as topics for future research.
Supplementary Material
Acknowledgments
We are grateful to the three anonymous referees for their valuable comments, which helped us to substantially improve the contents of this article. This work was supported by the Finnish Graduate School of Population Genetics and by a research grant from the Academy of Finland.
Appendix A
Our modeling strategy is largely based on an idea of a combination of B-splines with a difference penalty added on the parameters αjh defined in Equation 3 or so called P-splines (Eilers and Marx 1996). An important property of B-splines is that if all the parameters are the same, then the fitted curve is a horizontal line (a constant value). Inspired by this fact, the frequentist P-splines with the first- and second-order penalties are defined as
| (A1) |
and
| (A2) |
respectively, where λj > 0 is a tuning parameter. The difference penalties or play a role to push the adjacent parameters of B-splines to share similar values to induce the smoothness to the curve. The tuning parameter λj > 0 for marker j determines how smooth a curve will be. Note that the higher-order penalties can be specified as well, but only the first-order and second-order penalties are widely used in practice (Fahrmeir and Kneib 2011). From the perspective of the Bayesian statistics, the difference penalties above can be interpreted as the random walk smoothing priors (Lang and Brezger 2004), which have been introduced previously in Equations 5–9.
Next, we explain briefly how to convert the random walk smoothing priors (5)–(9) to the matrix form, which is defined in (10). For the first-order case, from (5) and (7), we can obtain . It is not difficult to show that
| (A3) |
where
Then we have with . Similarly for the second-order case, we obtain with , where
Finally, note that, in the original article on Bayesian P-splines (Lang and Brezger 2004), the prior specifications in (5) or (7) and (8) were replaced by p(αj1) ∝ 1 or p(αj1) ∝ 1 and p(αj2) ∝ 1, which results in a rank deficient penalty matrix K1 or K2. Our prior setting given above, proposed by Chib and Jeliazkov (2008), guarantees K1 or K2 to be full rank, which is required to proceed to the variational Bayes estimation that is introduced in Appendixes B and C.
Appendix B
VB Estimation
As mentioned earlier, the mean field variational Bayes algorithm can be applied to compute a factorized approximate posterior distribution , by minimizing the Kullback–Leibler divergence between it and the true posterior distribution. It is known that the minimization of the KL divergence with respect to each q(θl|Y) is reached at , defined by formula (14). If the posterior belongs to the conjugate exponential family so that for l = 1, … , K can be derived as standard parametric distributions, then a simple coordinate descent algorithm can be used for sequentially updating each . However, if for one parameter , the marginal distribution defined by (14) is not recognized as any standard distribution, then proceeding with the VB algorithm based on (14) is no longer straightforward, since it might not be possible to derive an analytical form of the expectation . Instead, we may assume that the marginal approximate distribution of is in a fixed form belonging to the exponential family, where η is a u × 1 vector of natural parameters that determines the shape of the approximate distribution, is a 1 × u vector of statistics, a(η) defines the normalizing constant, and is a base measure. The minimization of the above-mentioned KL divergence with respect to is now equivalent to the optimization problem
| (B1) |
where the expectation can be easily computed, if defined in (14) for all other parameters expect are standard parametric distributions.
Salimans and Knowles (2013) demonstrated that the minimization is given at
| (B2) |
where , and . If we can sample from , then the two expectations in (B2) can be evaluated by the Monte Carlo simulation methods. Based on these ideas, Salimans and Knowles (2013) designed a stochastic optimization algorithm to estimate or, alternatively, an optimal marginal approximate distribution . We use Algorithm 1 in their article for evaluating the approximate distribution of ρ, which is a key parameter defining the AR(1) covariance matrix.
Next, following the above principles, we present the specific VB algorithms for estimating the marker effects. The variable selection part is then explained in Appendix C.
First, let us focus on the case of the diagonal covariance structure, which is for r = 1, … , k. The logarithm of the joint distribution is
| (B3) |
where a = b = 0.0001. By using formula (14), we can derive the analytical form of q(•|Y) for each parameter in θ as follows.
I. Derivation of
By keeping only terms containing α0, we obtain
| (B4) |
where C represents those terms that do not contain α0, and represents a k × k diagonal matrix with for r = 1, … , k. Here the notation E[•] represents the posterior expectation (first moment) of the parameter • with respect to its approximate marginal distribution . We can recognize from (B4) that is a multivariate normal distribution with mean
| (B5) |
and covariance matrix
| (B6) |
Furthermore, the second moment can be calculated as .
II. Derivation of (j = 1, … , p)
| (B7) |
| (B8) |
We recognize as a multivariate normal distribution with mean (or the first moment)
| (B9) |
and covariance matrix
| (B10) |
Furthermore, the second moment can be calculated as .
III. Derivation of (j = 1, … , p)
We have
| (B11) |
where
| (B12) |
and
| (B13) |
is recognized as an inverse gamma distribution, IG(A1j,B1j). The moment function can be computed as .
IV. Derivation of (r = 1, … , k)
We have
| (B14) |
where
| (B15) |
and
| (B16) |
where ψr represents the rth row vector of Ψ. is recognized as an inverse gamma distribution IG(A2r, B2r). In addition, the moment function needs to be computed.
Furthermore, the lower bound can be evaluated as
| (B17) |
To proceed with the VB algorithm, we need to update these approximate marginal posterior distributions for each parameter sequentially or, by other means, we just need to update the values of the quantities including E[α0], COV[α0], , E[αj], COV[αj], , A2r, B2r, (for r = 1, … , k), A1j, B1j, and (for j = 1, … , p) in turn, until convergence. The convergence can be checked by the lower bound. In step t, we calculate . If it is smaller than some predefined threshold (some small positive value such as ), then the algorithm can stop.
On the other hand, if an AR(1) structure for r = 1, … , k, s = 1, … , k is used, the above steps I–III can be used to update and for j = 0, … , p here as well. Next, we add two extra steps (IV* and V*) for updating and , respectively. Note that is updated based on formula (14), whereas is estimated by the above-mentioned fixed-form variational method of Salimans and Knowles (2013).
IV*. Derivation of
We have
| (B18) |
where
| (B19) |
and
| (B20) |
where
| (B21) |
and
is recognized as an inverse gamma distribution IG(A3, B3). In addition, the moment function needs to be computed.
V*. Derivation of
Since 0 < ρ < 1, it is reasonable to preassume q(ρ|Y) to be a beta(ρ|μ1, μ2) (μ1 > 0, μ2 > 0) distribution. It is well known that the beta distribution belongs to the exponential family, with shape parameters η = [μ1, μ2]T and sufficient statistics t(ρ) = [ln(ρ), ln(1 − ρ)]. Then it is straightforward to apply the above-mentioned stochastic optimization algorithm to estimate optimal values of and and obtain , with mean , variance , and the second moment E[ρ2] = E[ρ2] + VAR[ρ]. Then we need to evaluate the expectation , where
which is required in steps I and II.
The lower bound is computed as
| (B22) |
where Be(⋅) and ψ(⋅) represent the beta function and the digamma function (Bishop 2006), respectively. The expectation can be computed by Monte Carlo integration. Alternatively, we may do the approximation , which should not affect the results significantly based on our own empirical experimentation.
After obtaining the posterior mean and covariance estimate of αj, the Wald test statistic (score), needed in the variable selection, can be obtained as
| (B23) |
which asymptotically follows a chi-square distribution with m d.f.
Appendix C
VB Variable Selection
The aim of the model (variable) selection is to seek a model with the highest value of a lower bound. A model search algorithm adopted from Nott et al. (2012) is used here. Starting from a null model with the intercept term only, the algorithm adds one marker into the model at a time, so that it produces a sequence of candidate models. Conventionally, when a model M with a subset of selected markers as well as the corresponding approximate distribution and the lower bound have been obtained, we may pick up a new marker from Mc, the complementary set of M, which satisfies
| (C1) |
where the lower bounds can be computed by the VB algorithm introduced in Appendix B. In practice, this involves a repeated computation of the approximate distribution for all possible new models M ∪ jnew (jnew ∈ Mc), which will be inefficient for a data set with a large number of markers. This motivates us to find a faster way to rank the markers in Mc instead of using (C1). It is easy to see that, for each M ∪ jnew, the computational burden appears if we try to update all the components in as we have done in the regular VB computation. Instead, here we consider the following approximation:
| (C2) |
By other means, we update only the approximate distributions for two parameters and of the new marker jnew, by fixing the other part to the approximate distribution , which should have already been evaluated for the model M in the earlier round. This procedure is roughly equivalent to using VB to approximate the following posterior distribution,
| (C3) |
where , for i = 1, … , n, and , E[α0 | M], and E[αj | M] (j ∈ M) are moment functions of the approximate distribution , which are considered as fixed values here. By applying the VB algorithm on the above posterior, we obtain an approximate distribution for ,
| (C4) |
with mean
| (C5) |
and covariance matrix
| (C6) |
and for , we have
| (C7) |
with
| (C8) |
and
| (C9) |
Other required moment functions can be computed by
| (C10) |
and
| (C11) |
The corresponding lower bound is
| (C12) |
where . After computing the lower bound L* for each marker in Mc, we choose the marker that corresponds to the maximum of L*. It can be interpreted as a marker that contributes the most to the increment of the lower bound of the model M. We then add into the model M to obtain a new , and at the same time should be deleted from the set Mc. In addition, the approximate distribution and the lower bound can be fully evaluated by the standard VB algorithm now and can be used for the next round. These forward selection steps can be iteratively implemented until the maximum number of markers (say T) is reached, and a sequence of candidate models (i = 1, … , T) with their lower bound estimates can be obtained. Since such a selection strategy is greedy, it may pick up some wrong markers into the model. To improve the reliability of model selection, we may further add a backward elimination procedure. In reverse, starting from the final model constructed by the forward selection, one marker was dropped from the model at a time until the null model is reached. This procedure produces another sequence of models (i = T − 1, … , 1). Specifically, at the stage of model (i > 1), the standard VB algorithm is used to estimate the approximate posterior distribution , and then the Wald test statistic (see Appendix B) can be calculated for every marker in the model. The Wald test statistic here is interpreted as a score for measuring the importance of one marker. Then we drop the marker corresponding to the smallest Wald test statistic out of the current model and obtain the next model . After completing both the forward and the backward selection procedures, we then select an optimal model with the maximum value of the lower bounds from all the candidate models.
Footnotes
Communicating editor: N. Yi
Literature Cited
- Banerjee S., Yandell B. S., Yi N., 2008. Bayesian QTL mapping for multiple traits. Genetics 179: 2275–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal, M. J., 2003 Variational algorithms for approximate Bayesian inference. Ph.D. Thesis, University of London, London. [Google Scholar]
- Beal M. J., Ghahramani Z., 2003. The variational Bayesian EM algorithm for incomplete data: with application to scoring graphical model structures. Bayesian Stat. 7: 453–464 [Google Scholar]
- Bishop C. M., 2006. Pattern Recognition and Machine Learning. Springer-Verlag, New York [Google Scholar]
- Carbonetto P., Stephens M., 2012. Scalable variational inference for Bayesian variable selection in regression, and its accuracy in genetic association studies. Bayesian Anal. 7: 73–108 [Google Scholar]
- Chib S., Jeliazkov I., 2008. Inference in semiparametric dynamic models for binary longitudinal data. J. Am. Stat. Assoc. 101: 685–700 [Google Scholar]
- Coster, A., J. W. M. Bastiaansen, M. P. L. Calus, C. Maliepaard, and M. C. A. M. Bink, 2010 QTLMAS 2009: simulated dataset. BMC Proc. 4: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boor C. M., 2001. A Practical Guide to Splines. Springer-Verlag, New York [Google Scholar]
- Eberius M., Lima-Guerra J., 2009. High-throughput plant phenotyping: data acquisition, transformation, and analysis, pp. 259–278 in Bioinformatics, edited by Edwards D., Stajich J., Hansen D. Springer-Verlag, New York [Google Scholar]
- Eilers P. H. C., Marx B. D., 1996. Flexible smoothing using B-splines and penalized likelihood. Stat. Sci. 11: 89–121 [Google Scholar]
- Fahrmeir L., Kneib T., 2011. Bayesian Smoothing and Regression for Longitudinal, Spatial and Event History Data. Oxford University Press, New York [Google Scholar]
- Fahrmeir L., Kneib T., Konrath S., 2010. Bayesian regularisation in structured additive regression: a unifying perspective on shrinkage, smoothing and predictor selection. Stat. Comput. 20: 203–219 [Google Scholar]
- Gee C., Morrison J. L., Thomas D. C., Gauderman W. J., 2003. Segregation and linkage analysis for longitudinal measurements of a quantitative trait. BMC Genet. 4: S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Zou F., 2012. Varying coefficient models for mapping quantitative trait loci using recombinant inbred intercrosses. Genetics 190: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding E. H., Schenk A. K., Juneja P., MacKay A. W., Wade J. M., et al. , 2008. A robust automated system elucidates mouse home cage behavioral structure. Proc. Natl. Acad. Sci. USA 105: 20575–20582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer J., 2011. An introduction to Bayesian inference via variational approximations. Polit. Anal. 19: 32–47 [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J., 2009. Elements of Statistical Learning, Ed. 2 Springer-Verlag, New York [Google Scholar]
- Heuven, H. C. M., and L. L. G. Janss, 2010 Bayesian multi-QTL mapping for growth curve parameters. BMC Proc. 4: S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado P. X., Schnabel S. K., Zaban A., Veteläinen M., Virtanen E., et al. , 2012. Dynamics of senescence-related QTLs in potato. Euphytica 183: 289–302 [Google Scholar]
- Jaakkola T. S., Jordan M. I., 2000. Bayesian parameter estimation via variational methods. Stat. Comput. 10: 25–37 [Google Scholar]
- Jiang C., Zeng Z.-B., 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795 [Google Scholar]
- Lang S., Brezger A., 2004. Bayesian P-splines. J. Comput. Graph. Stat. 13: 183–212 [Google Scholar]
- Lee D., Shaddick G., 2007. Time-varying coefficient models for the analysis of air pollution and health outcome data. Biometrics 63: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Li J., Das K., Fu G., Li R., Wu R., 2011. The Bayesian LASSO for genome-wide association studies. Bioinformatics 27: 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., R. Li, and R. Wu, 2012 Bayesian group lasso for nonparametric varying coefficient models. Available at: http://www.personal.psu.edu/jzl185/jonathan/paper/bayesian_lasso_fi.pdf [DOI] [PMC free article] [PubMed]
- Li Z., Sillanpää M. J., 2012. Estimation of quantitative trait locus effects with epistasis by variational Bayes algorithms. Genetics 190: 231–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Wu R., 2006. A joint model for nonparametric functional mapping of longitudinal trajectory and time-to-event. BMC Bioinformatics 7: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wu R., 2009. A Bayesian algorithm for functional mapping of dynamic complex traits. Algorithms 2: 667–691 [Google Scholar]
- Logsdon B. A., Hoffman G. E., Mezey J. G., 2010. A variational Bayes algorithm for fast and accurate multiple locus genome-wide association analysis. BMC Bioinformatics 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Casella G., Wu R., 2002. Functional mapping of quantitative trait loci underlying the character process: a theoretical framework. Genetics 161: 1751–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier L., Bühlmann P., 2007. Smoothing ℓ1-penalized estimators for high-dimensional time-course data. Electron. J. Stat. 1: 597–615 [Google Scholar]
- Min L., Yang R., Wang X., Wang B., 2011. Bayesian analysis for genetic architecture of dynamic traits. Heredity 106: 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott D. J., Tran M. N., Leng C., 2012. Variational approximation for heteroscedastic linear models and matching pursuit algorithms. Stat. Comput. 22: 497–512 [Google Scholar]
- Ramsay J., Hooker G., Graves S., 2009. Functional Data Analysis with R and MATLAB. Springer-Verlag, New York [Google Scholar]
- Salimans, T., and D. A. Knowles, 2013 Fixed-form variational posterior approximation through stochastic linear regression. Available at: http://arxiv.org/abs/1206.6679
- Segura V., Vilhjálmsson B. J., Platt A., Korte A., Seren Ü., et al. , 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää M. J., Pikkuhookana P., Abrahamsson S., Knürr T., Fries A., et al. , 2012. Simultaneous estimation of multiple quantitative trait loci and growth curve parameters through hierarchical Bayesian modeling. Heredity 108: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton D. I., Hui F. K. C., 2011. The arcsine is asinine: the analysis of proportions in ecology. Ecology 92: 3–10 [DOI] [PubMed] [Google Scholar]
- Wu R., Lin M., 2006. Functional mapping-how to map and study the genetic architecture of dynamical complex traits. Nat. Rev. Genet. 7: 229–237 [DOI] [PubMed] [Google Scholar]
- Xing J., Li J., Yang R., Zhou X., Xu S., 2012. Bayesian B-spline mapping for dynamic quantitative traits. Genet. Res. 94: 85–95 [DOI] [PubMed] [Google Scholar]
- Xiong H., Goulding E. H., Carlson E. J., Tecott L. H., McCulloch C. E., et al. , 2011. A flexible estimating equations approach for mapping function valued traits. Genetics 189: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., 2003. Estimating polygenic effects using markers of the entire genome. Genetics 163: 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wu R., Casella G., 2009. Nonparametric functional mapping of quantitative trait loci. Biometrics 65: 30–39 [DOI] [PubMed] [Google Scholar]
- Yang R., Xu S., 2007. Bayesian shrinkage analysis of quantitative trait loci for dynamic traits. Genetics 176: 1169–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Li J., Wang X., Zhou X., 2011. Bayesian functional mapping of dynamic quantitative traits. Theor. Appl. Genet. 123: 483–492 [DOI] [PubMed] [Google Scholar]
- Yap J. S., Fan J., Wu R., 2009. Nonparametric modeling of longitudinal covariance structure in functional mapping of quantitative trait loci. Biometrics 65: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Banerjee S., 2009. Hierarchical generalized linear models for multiple quantitative trait locus mapping. Genetics 181: 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-M., Xu S., 2005. A penalized maximum likelihood method for estimating epistatic effects of QTL. Heredity 95: 96–104 [DOI] [PubMed] [Google Scholar]
- Zhao W., Li H., Hou W., Wu R., 2007. Wavelet-based parametric functional mapping of developmental trajectories with high-dimensional data. Genetics 176: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.