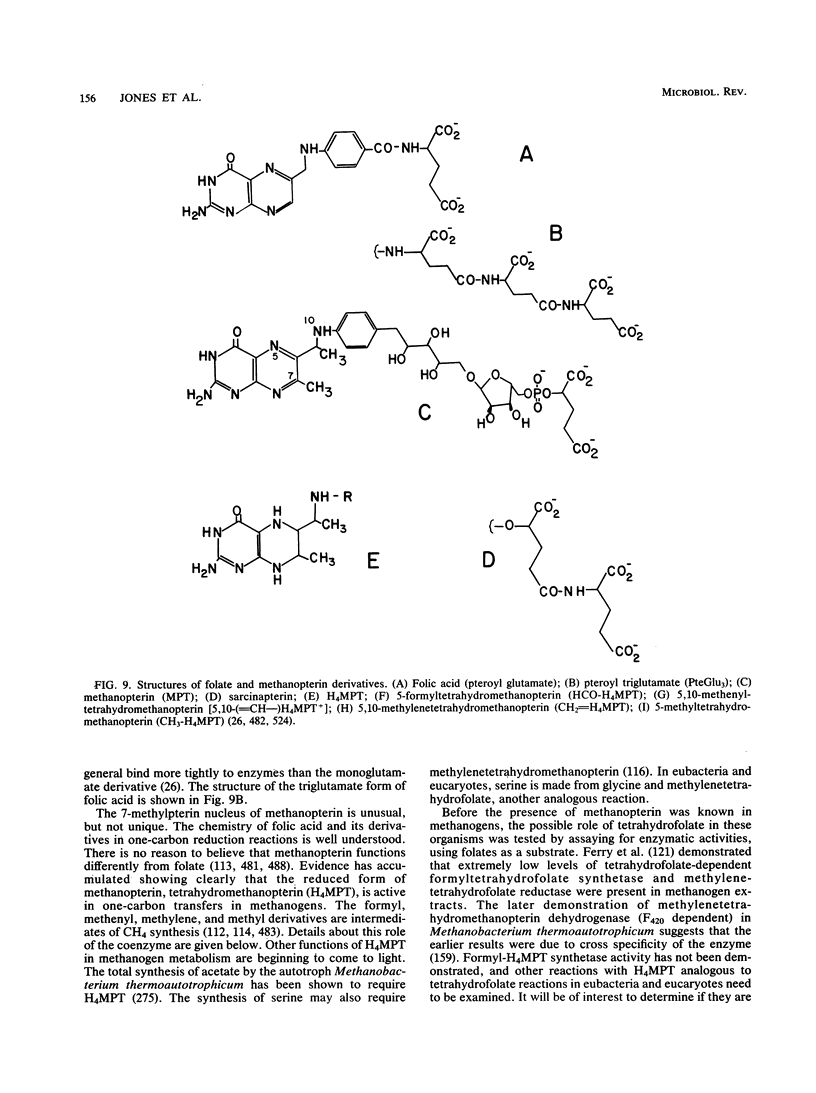
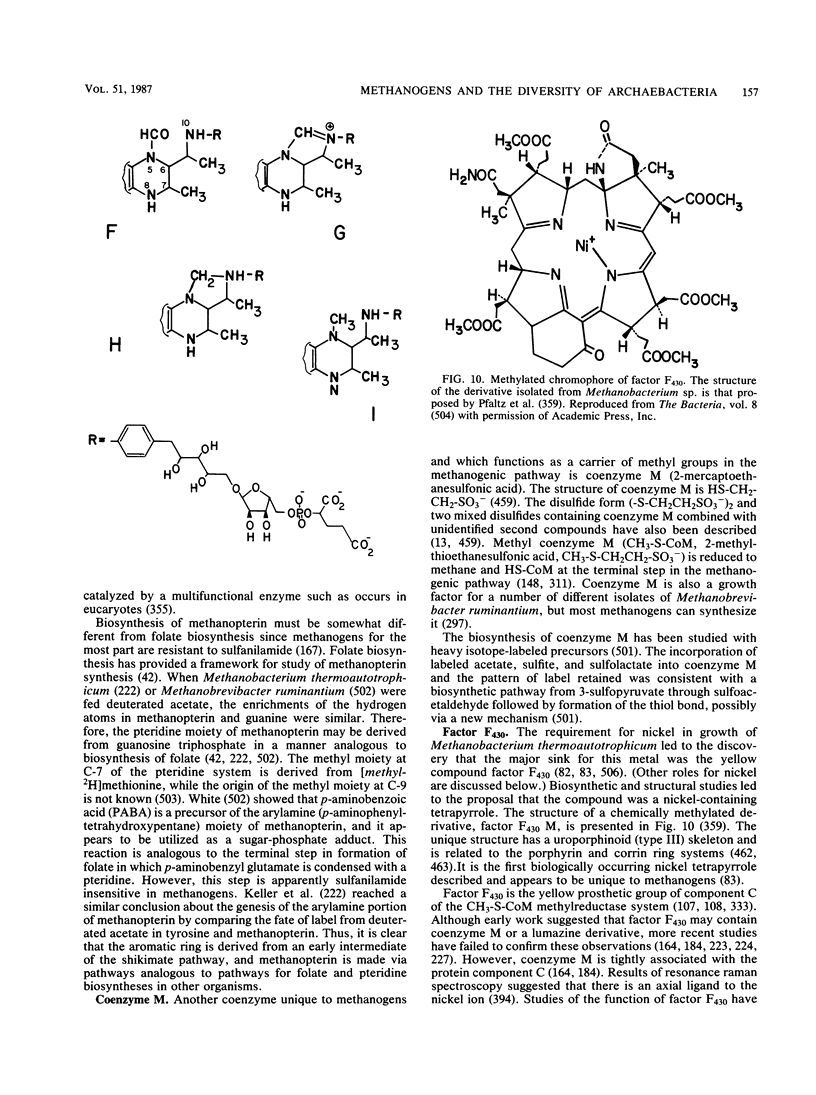
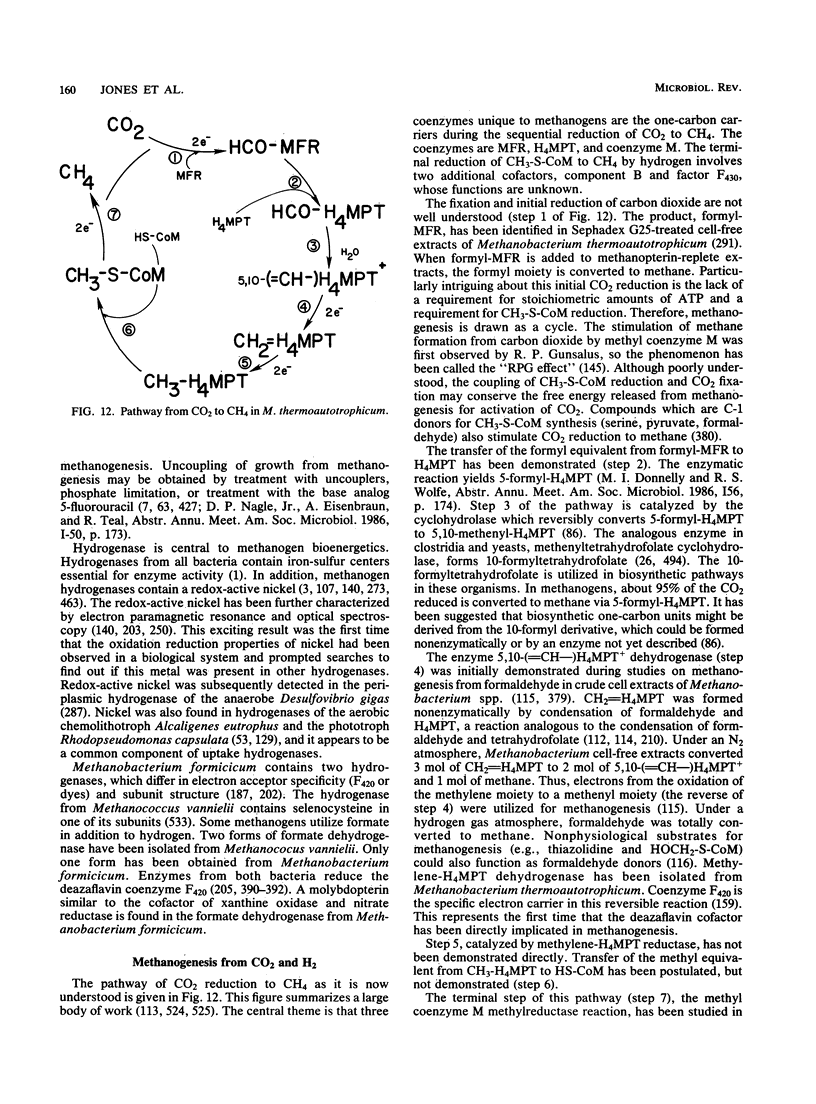
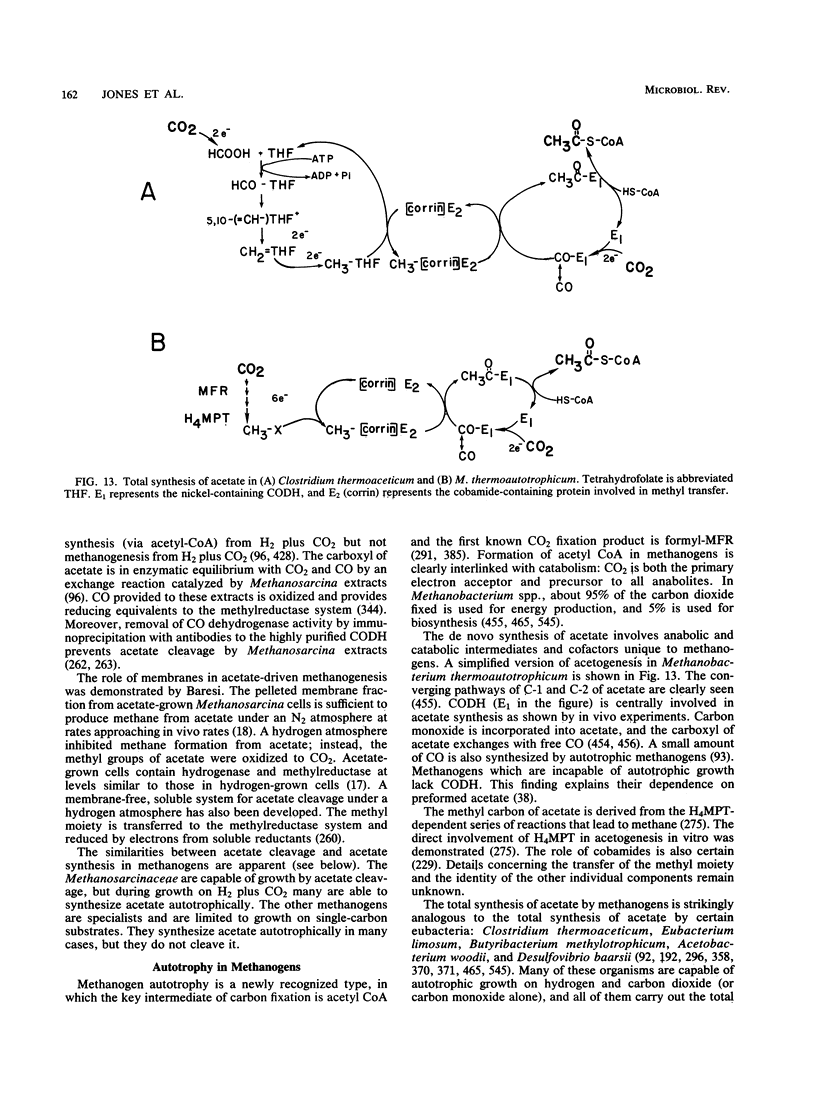
Full text
PDF






























Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. W., Mortenson L. E., Chen J. S. Hydrogenase. Biochim Biophys Acta. 1980 Dec;594(2-3):105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- Albracht S. P., Graf E. G., Thauer R. K. The EPR properties of nickel in hydrogenase from Methanobacterium. FEBS Lett. 1982 Apr 19;140(2):311–313. doi: 10.1016/0014-5793(82)80921-6. [DOI] [PubMed] [Google Scholar]
- Andreesen J. R., Gottschalk G. The occurrence of a modified Entner-doudoroff pathway in Clostridium aceticum. Arch Mikrobiol. 1969;69(2):160–170. doi: 10.1007/BF00409760. [DOI] [PubMed] [Google Scholar]
- Ankel-Fuchs D., Thauer R. K. Methane formation from methyl-coenzyme M in a system containing methyl-coenzyme M reductase, component B and reduced cobalamin. Eur J Biochem. 1986 Apr 1;156(1):171–177. doi: 10.1111/j.1432-1033.1986.tb09563.x. [DOI] [PubMed] [Google Scholar]
- Archer D. B. Uncoupling of Methanogenesis from Growth of Methanosarcina barkeri by Phosphate Limitation. Appl Environ Microbiol. 1985 Nov;50(5):1233–1237. doi: 10.1128/aem.50.5.1233-1237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. Transport of coenzyme M (2-mercaptoethanesulfonic acid) in Methanobacterium ruminantium. J Bacteriol. 1979 Jan;137(1):264–273. doi: 10.1128/jb.137.1.264-273.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderston W. L., Payne W. J. Inhibition of methanogenesis in salt marsh sediments and whole-cell suspensions of methanogenic bacteria by nitrogen oxides. Appl Environ Microbiol. 1976 Aug;32(2):264–269. doi: 10.1128/aem.32.2.264-269.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister J. V., Parker M. W. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci U S A. 1985 Jan;82(1):149–152. doi: 10.1073/pnas.82.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baresi L. Methanogenic cleavage of acetate by lysates of Methanosarcina barkeri. J Bacteriol. 1984 Oct;160(1):365–370. doi: 10.1128/jb.160.1.365-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baresi L., Wolfe R. S. Levels of coenzyme F420, coenzyme M, hydrogenase, and methylcoenzyme M methylreductase in acetate-grown Methanosarcina. Appl Environ Microbiol. 1981 Feb;41(2):388–391. doi: 10.1128/aem.41.2.388-391.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley S. T. Composition of ribosomes of an extremely halophilic bacterium. J Mol Biol. 1966 Feb;15(2):420–427. doi: 10.1016/s0022-2836(66)80117-1. [DOI] [PubMed] [Google Scholar]
- Belay N., Sparling R., Daniels L. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature. 1984 Nov 15;312(5991):286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- Betlach M., Pfeifer F., Friedman J., Boyer H. W. Bacterio-opsin mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1416–1420. doi: 10.1073/pnas.80.5.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar L., Jain M. K., Aubert J. P., Zeikus J. G. Comparison of assimilatory organic nitrogen, sulfur, and carbon sources for growth of methanobacterium species. Appl Environ Microbiol. 1984 Oct;48(4):785–790. doi: 10.1128/aem.48.4.785-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar L., Zeikus J. G., Aubert J. P. Purification and characterization of glutamine synthetase from the archaebacterium Methanobacterium ivanovi. J Bacteriol. 1986 Feb;165(2):638–643. doi: 10.1128/jb.165.2.638-643.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland N. K., Ratkje S. K. Active uptake of glutamate in vesicles of Halobacterium salinarium. Membr Biochem. 1985;6(1):1–17. doi: 10.3109/09687688509065439. [DOI] [PubMed] [Google Scholar]
- Blaut M., Müller V., Fiebig K., Gottschalk G. Sodium ions and an energized membrane required by Methanosarcina barkeri for the oxidation of methanol to the level of formaldehyde. J Bacteriol. 1985 Oct;164(1):95–101. doi: 10.1128/jb.164.1.95-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B. A. Cobamide-dependent methanol-cyanocob(I)alamin methyltransferase of Methanosarcina barkeri. Arch Biochem Biophys. 1968 Mar 20;124(1):314–324. doi: 10.1016/0003-9861(68)90333-0. [DOI] [PubMed] [Google Scholar]
- Blaylock B. A., Stadtman T. C. Methane biosynthesis by Methanosarcina barkeri. Properties of the soluble enzyme system. Arch Biochem Biophys. 1966 Sep 26;116(1):138–152. doi: 10.1016/0003-9861(66)90022-1. [DOI] [PubMed] [Google Scholar]
- Bollschweiler C., Kühn R., Klein A. Non-repetitive AT-rich sequences are found in intergenic regions of Methanococcus voltae DNA. EMBO J. 1985 Mar;4(3):805–809. doi: 10.1002/j.1460-2075.1985.tb03701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts G. P., van der Drift C., Vogels G. D. Stimulation of growth of Bacillus fastidiosus by amino acids. Antonie Van Leeuwenhoek. 1984;50(2):177–182. doi: 10.1007/BF00400179. [DOI] [PubMed] [Google Scholar]
- Brierley C. L., Brierley J. A. A chemoautotrophic and thermophilic microorganism isolated from an acid hot spring. Can J Microbiol. 1973 Feb;19(2):183–188. doi: 10.1139/m73-028. [DOI] [PubMed] [Google Scholar]
- Brock T. D., Brock K. M., Belly R. T., Weiss R. L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84(1):54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Brown G. M., Williamson J. M. Biosynthesis of riboflavin, folic acid, thiamine, and pantothenic acid. Adv Enzymol Relat Areas Mol Biol. 1982;53:345–381. doi: 10.1002/9780470122983.ch9. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Reeve J. N. Polyadenylated, noncapped RNA from the archaebacterium Methanococcus vannielii. J Bacteriol. 1985 Jun;162(3):909–917. doi: 10.1128/jb.162.3.909-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Wolin E. A., Wolin M. J., Wolfe R. S. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Mikrobiol. 1967;59(1):20–31. doi: 10.1007/BF00406313. [DOI] [PubMed] [Google Scholar]
- Byng G. S., Kane J. F., Jensen R. A. Diversity in the routing and regulation of complex biochemical pathways as indicators of microbial relatedness. Crit Rev Microbiol. 1982 May;9(4):227–252. doi: 10.3109/10408418209104491. [DOI] [PubMed] [Google Scholar]
- Cammarano P., Teichner A., Londei P., Acca M., Nicolaus B., Sanz J. L., Amils R. Insensitivity of archaebacterial ribosomes to protein synthesis inhibitors. Evolutionary implications. EMBO J. 1985 Mar;4(3):811–816. doi: 10.1002/j.1460-2075.1985.tb03702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzulo J. J. On the regulatory properties of a halophilic citrate synthase. FEBS Lett. 1973 Mar 15;30(3):339–342. doi: 10.1016/0014-5793(73)80683-0. [DOI] [PubMed] [Google Scholar]
- Cheah K. S. The membrane-bound ascorbate oxidase system of Halobacterium halobium. Biochim Biophys Acta. 1970;205(2):148–160. doi: 10.1016/0005-2728(70)90245-8. [DOI] [PubMed] [Google Scholar]
- Comita P. B., Gagosian R. B. Membrane lipid from deep-sea hydrothermal vent methanogen: a new macrocyclic glycerol diether. Science. 1983 Dec 23;222(4630):1329–1331. doi: 10.1126/science.222.4630.1329. [DOI] [PubMed] [Google Scholar]
- Crider B. P., Carper S. W., Lancaster J. R. Electron transfer-driven ATP synthesis in Methanococcus voltae is not dependent on a proton electrochemical gradient. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6793–6796. doi: 10.1073/pnas.82.20.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Beckler G. S., Reeve J. N., Konisky J. Structure and sequence divergence of two archaebacterial genes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4207–4211. doi: 10.1073/pnas.82.12.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C. J., Gupta R., Doolittle W. F. Transcription and excision of a large intron in the tRNATrp gene of an archaebacterium, Halobacterium volcanii. J Biol Chem. 1985 Mar 10;260(5):3132–3134. [PubMed] [Google Scholar]
- Daniels C. J., Hofman J. D., MacWilliam J. G., Doolittle W. F., Woese C. R., Luehrsen K. R., Fox G. E. Sequence of 5S ribosomal RNA gene regions and their products in the archaebacterium Halobacterium volcanii. Mol Gen Genet. 1985;198(2):270–274. doi: 10.1007/BF00383005. [DOI] [PubMed] [Google Scholar]
- Daniels L., Fuchs G., Thauer R. K., Zeikus J. G. Carbon monoxide oxidation by methanogenic bacteria. J Bacteriol. 1977 Oct;132(1):118–126. doi: 10.1128/jb.132.1.118-126.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels L., Sparling R., Sprott G. D. The bioenergetics of methanogenesis. Biochim Biophys Acta. 1984 Sep 6;768(2):113–163. doi: 10.1016/0304-4173(84)90002-8. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Eisenthal R., Hall S., Kessell S. R., Williams D. L. Dihydrolipoamide dehydrogenase from halophilic archaebacteria. Biochem J. 1984 Mar 15;218(3):811–818. doi: 10.1042/bj2180811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darland G., Brock T. D., Samsonoff W., Conti S. F. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science. 1970 Dec 25;170(3965):1416–1418. doi: 10.1126/science.170.3965.1416. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassarma S., Rajbhandary U. L., Khorana H. G. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc Natl Acad Sci U S A. 1984 Jan;81(1):125–129. doi: 10.1073/pnas.81.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M., De Rosa S., Gambacorta A., Minale L. Caldariellaquinone, a unique benzo(b)thiophen-4,7-quinone from Caldariella acidophila, an extremely thermophilic and acidophilic bacterium. J Chem Soc Perkin 1. 1977;(6):653–657. doi: 10.1039/p19770000653. [DOI] [PubMed] [Google Scholar]
- De Rosa M., Gambacorta A., Gliozzi A. Structure, biosynthesis, and physicochemical properties of archaebacterial lipids. Microbiol Rev. 1986 Mar;50(1):70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M., Gambacorta A., Nicolaus B., Giardina P., Poerio E., Buonocore V. Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem J. 1984 Dec 1;224(2):407–414. doi: 10.1042/bj2240407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange R. J., Green G. R., Searcy D. G. A histone-like protein (HTa) from Thermoplasma acidophilum. I. Purification and properties. J Biol Chem. 1981 Jan 25;256(2):900–904. [PubMed] [Google Scholar]
- DeLange R. J., Williams L. C., Searcy D. G. A histone-like protein (HTa) from Thermoplasma acidophilum. II. Complete amino acid sequence. J Biol Chem. 1981 Jan 25;256(2):905–911. [PubMed] [Google Scholar]
- Dennis P. P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985 Nov 20;186(2):457–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- Diekert G., Klee B., Thauer R. K. Nickel, a component of factor F430 from Methanobacterium thermoautotrophicum. Arch Microbiol. 1980 Jan;124(1):103–106. doi: 10.1007/BF00407036. [DOI] [PubMed] [Google Scholar]
- Diekert G., Konheiser U., Piechulla K., Thauer R. K. Nickel requirement and factor F430 content of methanogenic bacteria. J Bacteriol. 1981 Nov;148(2):459–464. doi: 10.1128/jb.148.2.459-464.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., Claesen C. A., Kell D. B., van der Drift C., Vogels G. D. An adenine nucleotide translocase in the procaryote Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1980 Aug 14;95(3):1288–1293. doi: 10.1016/0006-291x(80)91613-7. [DOI] [PubMed] [Google Scholar]
- Doddema H. J., van der Drift C., Vogels G. D., Veenhuis M. Chemiosmotic coupling in Methanobacterium thermoautotrophicum: hydrogen-dependent adenosine 5'-triphosphate synthesis by subcellular particles. J Bacteriol. 1979 Dec;140(3):1081–1089. doi: 10.1128/jb.140.3.1081-1089.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. I., Escalante-Semerena J. C., Rinehart K. L., Jr, Wolfe R. S. Methenyl-tetrahydromethanopterin cyclohydrolase in cell extracts of Methanobacterium. Arch Biochem Biophys. 1985 Nov 1;242(2):430–439. doi: 10.1016/0003-9861(85)90227-9. [DOI] [PubMed] [Google Scholar]
- Dundas I. E. Ornithine carbamoyltransferase from Halobacterium salinarium. Eur J Biochem. 1972 May 23;27(2):376–380. doi: 10.1111/j.1432-1033.1972.tb01847.x. [DOI] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Carreira L. H., Ljungdahl L. G., Kuo K. C., Gehrke C. W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985 Feb 25;13(4):1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich L. D., Vogels G. D., Wolfe R. S. Distribution of coenzyme F420 and properties of its hydrolytic fragments. J Bacteriol. 1979 Oct;140(1):20–27. doi: 10.1128/jb.140.1.20-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich L. D., Vogels G. D., Wolfe R. S. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978 Oct 31;17(22):4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- Eker A. P., Dekker R. H., Berends W. Photoreactivating enzyme from Streptomyces griseus-IV. On the nature of the chromophoric cofactor in Streptomyces griseus photoreactivating enzyme. Photochem Photobiol. 1981 Jan;33(1):65–72. doi: 10.1111/j.1751-1097.1981.tb04298.x. [DOI] [PubMed] [Google Scholar]
- Ekiel I., Jarrell K. F., Sprott G. D. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur J Biochem. 1985 Jun 3;149(2):437–444. doi: 10.1111/j.1432-1033.1985.tb08944.x. [DOI] [PubMed] [Google Scholar]
- Ekiel I., Smith I. C., Sprott G. D. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983 Oct;156(1):316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson W. L., Whitman W. B. The role of nickel in methanogenic bacteria. Basic Life Sci. 1982;19:403–414. doi: 10.1007/978-1-4684-4142-0_30. [DOI] [PubMed] [Google Scholar]
- Ellefson W. L., Whitman W. B., Wolfe R. S. Nickel-containing factor F430: chromophore of the methylreductase of Methanobacterium. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3707–3710. doi: 10.1073/pnas.79.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson W. L., Wolfe R. S. Component C of the methylreductase system of Methanobacterium. J Biol Chem. 1981 May 10;256(9):4259–4262. [PubMed] [Google Scholar]
- Ellefson W. L., Wolfe R. S. Role of component C in the methylreductase system of Methanobacterium. J Biol Chem. 1980 Sep 25;255(18):8388–8389. [PubMed] [Google Scholar]
- Escalante-Semerena J. C., Leigh J. A., Rinehart K. L., Wolfe R. S. Formaldehyde activation factor, tetrahydromethanopterin, a coenzyme of methanogenesis. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1976–1980. doi: 10.1073/pnas.81.7.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena J. C., Rinehart K. L., Jr, Wolfe R. S. Tetrahydromethanopterin, a carbon carrier in methanogenesis. J Biol Chem. 1984 Aug 10;259(15):9447–9455. [PubMed] [Google Scholar]
- Escalante-Semerena J. C., Wolfe R. S. Formaldehyde oxidation and methanogenesis. J Bacteriol. 1984 May;158(2):721–726. doi: 10.1128/jb.158.2.721-726.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena J. C., Wolfe R. S. Tetrahydromethanopterin-dependent methanogenesis from non-physiological C1 donors in Methanobacterium thermoautotrophicum. J Bacteriol. 1985 Feb;161(2):696–701. doi: 10.1128/jb.161.2.696-701.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. N., Tolman C. J., Kanodia S., Roberts M. F. 2,3-Cyclopyrophosphoglycerate in methanogens: evidence by 13C NMR spectroscopy for a role in carbohydrate metabolism. Biochemistry. 1985 Oct 8;24(21):5693–5698. doi: 10.1021/bi00342a001. [DOI] [PubMed] [Google Scholar]
- Ferguson T. J., Mah R. A. Effect of H(2)-CO(2) on Methanogenesis from Acetate or Methanol in Methanosarcina spp. Appl Environ Microbiol. 1983 Aug;46(2):348–355. doi: 10.1128/aem.46.2.348-355.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castillo R., Rodriguez-Valera F., Gonzalez-Ramos J., Ruiz-Berraquero F. Accumulation of Poly (beta-Hydroxybutyrate) by Halobacteria. Appl Environ Microbiol. 1986 Jan;51(1):214–216. doi: 10.1128/aem.51.1.214-216.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Zillig W., Stetter K. O., Schreiber G. Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature. 1983 Feb 10;301(5900):511–513. doi: 10.1038/301511a0. [DOI] [PubMed] [Google Scholar]
- Fisher J., Spencer R., Walsh C. Enzyme-catalyzed redox reactions with the flavin analogues 5-deazariboflavin, 5-deazariboflavin 5'-phosphte, and 5-deazariboflavin 5'-diphosphate, 5' leads to 5'-adenosine ester. Biochemistry. 1976 Mar 9;15(5):1054–1064. doi: 10.1021/bi00650a016. [DOI] [PubMed] [Google Scholar]
- Forterre P., Elie C., Kohiyama M. Aphidicolin inhibits growth and DNA synthesis in halophilic arachaebacteria. J Bacteriol. 1984 Aug;159(2):800–802. doi: 10.1128/jb.159.2.800-802.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Schneider K., Friedrich B. Nickel in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1982 Oct;152(1):42–48. doi: 10.1128/jb.152.1.42-48.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G., Stupperich E. Evidence for an incomplete reductive carboxylic acid cycle in Methanobacterium thermoautotrophicum. Arch Microbiol. 1978 Jul;118(1):121–125. doi: 10.1007/BF00406084. [DOI] [PubMed] [Google Scholar]
- Fuchs G., Stupperich E., Thauer R. K. Function of fumarate reductase in methanogenic bacteria (Methanobacterium). Arch Microbiol. 1978 Nov 13;119(2):215–218. doi: 10.1007/BF00964276. [DOI] [PubMed] [Google Scholar]
- Geiger B., Mevarech M., Werber M. M. Immunochemical characterization of ferredoxin from Halobacterium of the Dead Sea. Eur J Biochem. 1978 Mar 15;84(2):449–455. doi: 10.1111/j.1432-1033.1978.tb12186.x. [DOI] [PubMed] [Google Scholar]
- Gilles H., Thauer R. K. Uroporphyrinogen III, an intermediate in the biosynthesis of the nickel-containing factor F430 in Methanobacterium thermoautotrophicum. Eur J Biochem. 1983 Sep 1;135(1):109–112. doi: 10.1111/j.1432-1033.1983.tb07624.x. [DOI] [PubMed] [Google Scholar]
- Gradin C. H., Hederstedt L., Baltscheffsky H. Soluble succinate dehydrogenase from the halophilic archaebacterium, Halobacterium halobium. Arch Biochem Biophys. 1985 May 15;239(1):200–205. doi: 10.1016/0003-9861(85)90827-6. [DOI] [PubMed] [Google Scholar]
- Green G. R., Searcy D. G., DeLange R. J. Histone-like protein in the Archaebacterium Sulfolobus acidocaldarius. Biochim Biophys Acta. 1983 Nov 17;741(2):251–257. doi: 10.1016/0167-4781(83)90066-0. [DOI] [PubMed] [Google Scholar]
- Greene R. V., MacDonald R. E. Partial purification and reconstitution of the aspartate transport system from Halobacterium halobium. Arch Biochem Biophys. 1984 Mar;229(2):576–584. doi: 10.1016/0003-9861(84)90190-5. [DOI] [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. ATP activation and properties of the methyl coenzyme M reductase system in Methanobacterium thermoautotrophicum. J Bacteriol. 1978 Sep;135(3):851–857. doi: 10.1128/jb.135.3.851-857.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. Methyl coenzyme M reductase from Methanobacterium thermoautotrophicum. Resolution and properties of the components. J Biol Chem. 1980 Mar 10;255(5):1891–1895. [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. Stimulation of CO2 reduction to methane by methylcoenzyme M in extracts Methanobacterium. Biochem Biophys Res Commun. 1977 Jun 6;76(3):790–795. doi: 10.1016/0006-291x(77)91570-4. [DOI] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Gupta R., Lanter J. M., Woese C. R. Sequence of the 16S Ribosomal RNA from Halobacterium volcanii, an Archaebacterium. Science. 1983 Aug 12;221(4611):656–659. doi: 10.1126/science.221.4611.656. [DOI] [PubMed] [Google Scholar]
- Görisch H., Hartl T., Grossebüter W., Stezowski J. J. Archaebacterial malate dehydrogenases. The enzymes from the thermoacidophilic organisms Sulfolobus acidocaldarius and Thermoplasma acidophilum show A-side stereospecificity for NAD+. Biochem J. 1985 Mar 15;226(3):885–888. doi: 10.1042/bj2260885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLMES P. K., HALVORSON H. O. PROPERTIES OF A PURIFIED HALOPHILIC MALIC DEHYDROGENASE. J Bacteriol. 1965 Aug;90:316–326. doi: 10.1128/jb.90.2.316-326.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLMES P. K., HALVORSON H. O. PURIFICATION OF A SALT-REQUIRING ENZYME FROM AN OBLIGATELY HALOPHILIC BACTERIUM. J Bacteriol. 1965 Aug;90:312–315. doi: 10.1128/jb.90.2.312-315.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg C., Baltscheffsky H. Solubilization and separation of two b-type cytochromes from a carotenoid mutant in Halobacterium halobium. FEBS Lett. 1981 Mar 23;125(2):201–204. doi: 10.1016/0014-5793(81)80718-1. [DOI] [PubMed] [Google Scholar]
- Hamilton P. T., Reeve J. N. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol Gen Genet. 1985;200(1):47–59. doi: 10.1007/BF00383311. [DOI] [PubMed] [Google Scholar]
- Harris J. E., Pinn P. A., Davis R. P. Isolation and characterization of a novel thermophilic, freshwater methanogen. Appl Environ Microbiol. 1984 Dec;48(6):1123–1128. doi: 10.1128/aem.48.6.1123-1128.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R., Sickinger H. D., Oesterhelt D. Anaerobic growth of halobacteria. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3821–3825. doi: 10.1073/pnas.77.7.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell P. L., Zvilius G., Escalante-Semerena J. C., Donnelly M. I. Coenzyme F420 dependence of the methylenetetrahydromethanopterin dehydrogenase of Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1985 Dec 31;133(3):884–890. doi: 10.1016/0006-291x(85)91218-5. [DOI] [PubMed] [Google Scholar]
- Hase T., Wakabayashi S., Matsubara H., Kerscher L., Oesterhelt D., Rao K. K., Hall D. O. Complete amino acid sequence of Halobacterium halobium ferredoxin containing an Nepsilon-acetyllysine residue. J Biochem. 1978 Jun;83(6):1657–1670. doi: 10.1093/oxfordjournals.jbchem.a132078. [DOI] [PubMed] [Google Scholar]
- Hase T., Wakabayashi S., Matsubara H., Mevarech M., Werber M. M. Amino acid sequence of 2Fe-2S ferredoxin from an extreme halophile, Halobacterium of the Dead Sea. Biochim Biophys Acta. 1980 May 29;623(1):139–145. doi: 10.1016/0005-2795(80)90016-1. [DOI] [PubMed] [Google Scholar]
- Hatchikian E. C., Bruschi M., Forget N., Scandellari M. Electron transport components from methanogenic bacteria: the ferredoxin from Methanosarcina barkeri (strain Fusaro). Biochem Biophys Res Commun. 1982 Dec 31;109(4):1316–1323. doi: 10.1016/0006-291x(82)91921-0. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P., Moura I., Moura J. J., Xavier A. V., Santos M. H., LeGall J., Howard J. B. Amino acid sequence of a 3Fe:3S ferredoxin from the "archaebacterium" Methanosarcina barkeri (DSM 800). J Biol Chem. 1982 Dec 10;257(23):14192–14197. [PubMed] [Google Scholar]
- Hausinger R. P., Orme-Johnson W. H., Walsh C. Factor 390 chromophores: phosphodiester between AMP or GMP and methanogen factor 420. Biochemistry. 1985 Mar 26;24(7):1629–1633. doi: 10.1021/bi00328a010. [DOI] [PubMed] [Google Scholar]
- Higa A., Cazzulo J. J. Some properties of the citrate synthase from the extreme halophile, Halobacterium cutirubrum. Biochem J. 1975 May;147(2):267–274. doi: 10.1042/bj1470267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein L. I., Dalton B. P. Studies of a halophilic NADH dehydrogenase. I. Purification and properties of the enzyme. Biochim Biophys Acta. 1973 Apr 12;302(2):216–228. doi: 10.1016/0005-2744(73)90150-2. [DOI] [PubMed] [Google Scholar]
- Hochstein L. I. The metabolism of carbohydrates by extremely halophilic bacteria: glucose metabolism via a modified Entner-Doudoroff pathway. Can J Microbiol. 1974 Aug;20(8):1085–1091. doi: 10.1139/m74-170. [DOI] [PubMed] [Google Scholar]
- Hofman J. D., Lau R. H., Doolittle W. F. The number, physical organization and transcription of ribosomal RNA cistrons in an archaebacterium: Halobacterium halobium. Nucleic Acids Res. 1979 Nov 10;7(5):1321–1333. doi: 10.1093/nar/7.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J. S., Miller A. B. Nature of the inactivation of the isocitrate dehydrogenase from an obligate halophile. J Bacteriol. 1970 Jun;102(3):677–681. doi: 10.1128/jb.102.3.677-681.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J. S., Miller A. B. Purification and reversible inactivation of the isocitrate dehydrogenase from an obligate halophile. J Bacteriol. 1969 Jul;99(1):161–168. doi: 10.1128/jb.99.1.161-168.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Schnabel R., Sentenac A., Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui I., Dennis P. P. Characterization of the ribosomal RNA gene clusters in Halobacterium cutirubrum. J Biol Chem. 1985 Jan 25;260(2):899–906. [PubMed] [Google Scholar]
- Hüster R., Gilles H. H., Thauer R. K. Is coenzyme M bound to factor F430 in methanogenic bacteria? Experiments with Methanobrevibacter ruminantium. Eur J Biochem. 1985 Apr 1;148(1):107–111. doi: 10.1111/j.1432-1033.1985.tb08813.x. [DOI] [PubMed] [Google Scholar]
- Iannotti E. L., Kafkewitz D., Wolin M. J., Bryant M. P. Glucose fermentation products in Ruminococcus albus grown in continuous culture with Vibrio succinogenes: changes caused by interspecies transfer of H 2 . J Bacteriol. 1973 Jun;114(3):1231–1240. doi: 10.1128/jb.114.3.1231-1240.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson F. S., Daniels L., Fox J. A., Walsh C. T., Orme-Johnson W. H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982 Apr 10;257(7):3385–3388. [PubMed] [Google Scholar]
- Jaenchen R., Schönheit P., Thauer R. K. Studies on the biosynthesis of coenzyme F420 in methanogenic bacteria. Arch Microbiol. 1984 Apr;137(4):362–365. doi: 10.1007/BF00410735. [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Sprott G. D. Nickel transport in Methanobacterium bryantii. J Bacteriol. 1982 Sep;151(3):1195–1203. doi: 10.1128/jb.151.3.1195-1203.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Sprott G. D. The transmembrane electrical potential and intracellular pH in methanogenic bacteria. Can J Microbiol. 1981 Jul;27(7):720–728. doi: 10.1139/m81-110. [DOI] [PubMed] [Google Scholar]
- Jarsch M., Böck A. DNA sequence of the 16S rRNA/23S rRNA intercistronic spacer of two rDNA operons of the archaebacterium Methanococcus vannielii. Nucleic Acids Res. 1983 Nov 11;11(21):7537–7544. doi: 10.1093/nar/11.21.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. A. Biochemical pathways in prokaryotes can be traced backward through evolutionary time. Mol Biol Evol. 1985 Mar;2(2):92–108. doi: 10.1093/oxfordjournals.molbev.a040338. [DOI] [PubMed] [Google Scholar]
- Johnson M. K., Zambrano I. C., Czechowski M. H., Peck H. D., Jr, DerVartanian D. V., LeGall J. Low temperature magnetic circular dichroism spectroscopy as a probe for the optical transitions of paramagnetic nickel in hydrogenase. Biochem Biophys Res Commun. 1985 Apr 16;128(1):220–225. doi: 10.1016/0006-291x(85)91667-5. [DOI] [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Jones W. J., Donnelly M. I., Wolfe R. S. Evidence of a common pathway of carbon dioxide reduction to methane in methanogens. J Bacteriol. 1985 Jul;163(1):126–131. doi: 10.1128/jb.163.1.126-131.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATES M., YENGOYAN L. S., SASTRY P. S. A DIETHER ANALOG OF PHOSPHATIDYL GLYCEROPHOSPHATE IN HALOBACTERIUM CUTIRUBRUM. Biochim Biophys Acta. 1965 Apr 5;98:252–268. doi: 10.1016/0005-2760(65)90119-0. [DOI] [PubMed] [Google Scholar]
- KUSHNER D. J., BAYLEY S. T., BORING J., KATES M., GIBBONS N. E. MORPHOLOGICAL AND CHEMICAL PROPERTIES OF CELL ENVELOPES OF THE EXTREME HALOPHILE, HALOBACTERIUM CUTIRUBRUM. Can J Microbiol. 1964 Jun;10:483–497. doi: 10.1139/m64-058. [DOI] [PubMed] [Google Scholar]
- Kaine B. P., Gupta R., Woese C. R. Putative introns in tRNA genes of prokaryotes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3309–3312. doi: 10.1073/pnas.80.11.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen R. G., Jencks W. P. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J Biol Chem. 1966 Dec 25;241(24):5851–5863. [PubMed] [Google Scholar]
- Kandler O., König H. Chemical composition of the peptidoglycan-free cell walls of methanogenic bacteria. Arch Microbiol. 1978 Aug 1;118(2):141–152. doi: 10.1007/BF00415722. [DOI] [PubMed] [Google Scholar]
- Kanodia S., Roberts M. F. Methanophosphagen: Unique cyclic pyrophosphate isolated from Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5217–5221. doi: 10.1073/pnas.80.17.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates M., Deroo P. W. Structure determination of the glycolipid sulfate from the extreme halophile Halobacterium cutirubrum. J Lipid Res. 1973 Jul;14(4):438–445. [PubMed] [Google Scholar]
- Kates M. The phytanyl ether-linked polar lipids and isoprenoid neutral lipids of extremely halophilic bacteria. Prog Chem Fats Other Lipids. 1978;15(4):301–342. doi: 10.1016/0079-6832(77)90011-8. [DOI] [PubMed] [Google Scholar]
- Kates M., Wassef M. K., Kushner D. J. Radioisotopic studies on the biosynthesis of the glyceryl diether lipids of Halobacterium cutirubrum. Can J Biochem. 1968 Aug;46(8):971–977. doi: 10.1139/o68-145. [DOI] [PubMed] [Google Scholar]
- Keltjens J. T., Caerteling C. G., Van Kooten A. M., Van Dijk H. F., Vogels G. D. Chromophoric derivatives of coenzyme MF430, a proposed coenzyme of methanogenesis in Methanobacterium thermoautotrophicum. Arch Biochem Biophys. 1983 May;223(1):235–253. doi: 10.1016/0003-9861(83)90589-1. [DOI] [PubMed] [Google Scholar]
- Keltjens J. T., Huberts M. J., Laarhoven W. H., Vogels G. D. Structural elements of methanopterin, a novel pterin present in Methanobacterium thermoautotrophicum. Eur J Biochem. 1983 Feb 15;130(3):537–544. doi: 10.1111/j.1432-1033.1983.tb07183.x. [DOI] [PubMed] [Google Scholar]
- Keltjens J. T., Whitman W. B., Caerteling C. G., van Kooten A. M., Wolfe R. S., Vogels G. D. Presence of coenzyme M derivatives in the prosthetic group (coenzyme MF430) of methylcoenzyme M reductase from Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1982 Sep 30;108(2):495–503. doi: 10.1016/0006-291x(82)90856-7. [DOI] [PubMed] [Google Scholar]
- Kenealy W. R., Thompson T. E., Schubert K. R., Zeikus J. G. Ammonia assimilation and synthesis of alanine, aspartate, and glutamate in Methanosarcina barkeri and Methanobacterium thermoautotrophicum. J Bacteriol. 1982 Jun;150(3):1357–1365. doi: 10.1128/jb.150.3.1357-1365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenealy W., Zeikus J. G. Influence of corrinoid antagonists on methanogen metabolism. J Bacteriol. 1981 Apr;146(1):133–140. doi: 10.1128/jb.146.1.133-140.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keradjopoulos D., Holldorf A. W. Purification and properties of alanine dehydrogenase from Halobacterium salinarium. Biochim Biophys Acta. 1979 Sep 12;570(1):1–10. doi: 10.1016/0005-2744(79)90195-5. [DOI] [PubMed] [Google Scholar]
- Kerscher L., Nowitzki S., Oesterhelt D. Thermoacidophilic archaebacteria contain bacterial-type ferredoxins acting as electron acceptors of 2-oxoacid:ferredoxin oxidoreductases. Eur J Biochem. 1982 Nov;128(1):223–230. doi: 10.1111/j.1432-1033.1982.tb06955.x. [DOI] [PubMed] [Google Scholar]
- Kerscher L., Oesterhelt D., Cammack R., Hall D. O. A new plant-type ferredoxin from halobacteria. Eur J Biochem. 1976 Dec;71(1):101–107. doi: 10.1111/j.1432-1033.1976.tb11094.x. [DOI] [PubMed] [Google Scholar]
- Kerscher L., Oesterhelt D. Purification and properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. Eur J Biochem. 1981 Jun 1;116(3):587–594. doi: 10.1111/j.1432-1033.1981.tb05376.x. [DOI] [PubMed] [Google Scholar]
- Kerscher L., Oesterhelt D. The catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. One-electron transfer at two distinct steps of the catalytic cycle. Eur J Biochem. 1981 Jun 1;116(3):595–600. doi: 10.1111/j.1432-1033.1981.tb05377.x. [DOI] [PubMed] [Google Scholar]
- Kessel M., Klink F. Archaebacterial elongation factor is ADP-ribosylated by diphtheria toxin. Nature. 1980 Sep 18;287(5779):250–251. doi: 10.1038/287250a0. [DOI] [PubMed] [Google Scholar]
- Kessel M., Klink F. Two elongation factors from the extremely halophilic archaebacterium Halobacterium cutirubrum. Assay systems and purification at high salt concentrations. Eur J Biochem. 1981 Mar;114(3):481–486. doi: 10.1111/j.1432-1033.1981.tb05170.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi A., Asai K. Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984 Jun 21;309(5970):677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- Kirby T. W., Lancaster J. R., Jr, Fridovich I. Isolation and characterization of the iron-containing superoxide dismutase of Methanobacterium bryantii. Arch Biochem Biophys. 1981 Aug;210(1):140–148. doi: 10.1016/0003-9861(81)90174-0. [DOI] [PubMed] [Google Scholar]
- Klagsbrun M. An evolutionary study of the methylation of transfer and ribosomal ribonucleic acid in prokaryote and eukaryote organisms. J Biol Chem. 1973 Apr 10;248(7):2612–2620. [PubMed] [Google Scholar]
- Klein A., Schnorr M. Genome complexity of methanogenic bacteria. J Bacteriol. 1984 May;158(2):628–631. doi: 10.1128/jb.158.2.628-631.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak L. J., Grummt F., Burger K. J. Purification and characterization of DNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1985 Jul 25;13(14):5269–5282. doi: 10.1093/nar/13.14.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink F., Schümann H., Thomsen A. Ribosome specificity of archaebacterial elongation factor 2. Studies with hybrid polyphenylalanine synthesis systems. FEBS Lett. 1983 May 2;155(1):173–177. doi: 10.1016/0014-5793(83)80233-6. [DOI] [PubMed] [Google Scholar]
- Kojima N., Fox J. A., Hausinger R. P., Daniels L., Orme-Johnson W. H., Walsh C. Paramagnetic centers in the nickel-containing, deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1983 Jan;80(2):378–382. doi: 10.1073/pnas.80.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzycki J. A., Lehman L. J., Zeikus J. G. Acetate catabolism by Methanosarcina barkeri: evidence for involvement of carbon monoxide dehydrogenase, methyl coenzyme M, and methylreductase. J Bacteriol. 1985 Sep;163(3):1000–1006. doi: 10.1128/jb.163.3.1000-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzycki J. A., Zeikus J. G. Characterization and purification of carbon monoxide dehydrogenase from Methanosarcina barkeri. J Bacteriol. 1984 Apr;158(1):231–237. doi: 10.1128/jb.158.1.231-237.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha S. C., Gochnauer M. B., Kushner D. J., Kates M. Pigments and isoprenoid compounds in extremely and moderately halophilic bacteria. Can J Microbiol. 1974 Feb;20(2):241–245. doi: 10.1139/m74-038. [DOI] [PubMed] [Google Scholar]
- Kushwaha S. C., Kates M. Isolation and identification of "bacteriorhodopsin" and minor C40-carotenoids in Halobacterium cutirubrum. Biochim Biophys Acta. 1973 Aug 23;316(2):235–243. doi: 10.1016/0005-2760(73)90013-1. [DOI] [PubMed] [Google Scholar]
- Kushwaha S. C., Kates M., Sprott G. D., Smith I. C. Novel complex polar lipids from the methanogenic archaebacterium Methanospirillum hungatei. Science. 1981 Mar 13;211(4487):1163–1164. doi: 10.1126/science.7466385. [DOI] [PubMed] [Google Scholar]
- Kwok Y., Wong J. T. Evolutionary relationship between Halobacterium cutirubrum and eukaryotes determined by use of aminoacyl-tRNA synthetases as phylogenetic probes. Can J Biochem. 1980 Mar;58(3):213–218. doi: 10.1139/o80-029. [DOI] [PubMed] [Google Scholar]
- König H., Kandler O., Jensen M., Rietschel E. T. The primary structure of the glycan moiety of pseudomurein from Methanobacterium thermoautotrophicum. Hoppe Seylers Z Physiol Chem. 1983 Jun;364(6):627–636. doi: 10.1515/bchm2.1983.364.1.627. [DOI] [PubMed] [Google Scholar]
- Kühn W., Gottschalk G. Characterization of the cytochromes occurring in Methanosarcina species. Eur J Biochem. 1983 Sep 1;135(1):89–94. doi: 10.1111/j.1432-1033.1983.tb07621.x. [DOI] [PubMed] [Google Scholar]
- LEZIUS A. G., BARKER H. A. CORRINOID COMPOUNDS OF METHANOBACILLUS OMELIANSKII. I. FRACTIONATION OF THE CORRINOID COMPOUNDS AND IDENTIFICATION OF FACTOR 3 AND FACTOR 3 COENZYME. Biochemistry. 1965 Mar;4:510–518. doi: 10.1021/bi00879a021. [DOI] [PubMed] [Google Scholar]
- Lancaster J. R., Jr Membrane-bound flavin adenine dinucleotide in Methanobacterium Bryantii. Biochem Biophys Res Commun. 1981 May 15;100(1):240–246. doi: 10.1016/s0006-291x(81)80088-5. [DOI] [PubMed] [Google Scholar]
- Lancaster J. R., Jr New biological paramagnetic center: octahedrally coordinated nickel(III) in the methanogenic bacteria. Science. 1982 Jun 18;216(4552):1324–1325. doi: 10.1126/science.216.4552.1324. [DOI] [PubMed] [Google Scholar]
- Langworthy T. A. Long-chain diglycerol tetraethers from Thermoplasma acidophilum. Biochim Biophys Acta. 1977 Apr 26;487(1):37–50. doi: 10.1016/0005-2760(77)90042-x. [DOI] [PubMed] [Google Scholar]
- Langworthy T. A., Mayberry W. R., Smith P. F. Long-chain glycerol diether and polyol dialkyl glycerol triether lipids of Sulfolobus acidocaldarius. J Bacteriol. 1974 Jul;119(1):106–116. doi: 10.1128/jb.119.1.106-116.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langworthy T. A., Smith P. F., Mayberry W. R. Lipids of Thermoplasma acidophilum. J Bacteriol. 1972 Dec;112(3):1193–1200. doi: 10.1128/jb.112.3.1193-1200.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Light energy conversion in Halobacterium halobium. Microbiol Rev. 1978 Dec;42(4):682–706. doi: 10.1128/mr.42.4.682-706.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Studies of the electron transport chain of extremely halophilic bacteria. I. Spectrophotometric identification of the cytochromes of Halobacterium cutirubrum. Arch Biochem Biophys. 1968 Dec;128(3):716–724. doi: 10.1016/0003-9861(68)90080-5. [DOI] [PubMed] [Google Scholar]
- LeGall J., Ljungdahl P. O., Moura I., Peck H. D., Jr, Xavier A. V., Moura J. J., Teixera M., Huynh B. H., DerVartanian D. V. The presence of redox-sensitive nickel in the periplasmic hydrogenase from Desulfovibrio gigas. Biochem Biophys Res Commun. 1982 May 31;106(2):610–616. doi: 10.1016/0006-291x(82)91154-8. [DOI] [PubMed] [Google Scholar]
- Leffers H., Garrett R. A. The nucleotide sequence of the 16S ribosomal RNA gene of the archaebacterium Halococcus morrhua. EMBO J. 1984 Jul;3(7):1613–1619. doi: 10.1002/j.1460-2075.1984.tb02019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leicht W., Werber M. M., Eisenberg H. Purification and characterization of glutamate dehydrogenase from Halobacterium of the Dead Sea. Biochemistry. 1978 Sep 19;17(19):4004–4010. doi: 10.1021/bi00612a020. [DOI] [PubMed] [Google Scholar]
- Leigh J. A. Levels of water-soluble vitamins in methanogenic and non-methanogenic bacteria. Appl Environ Microbiol. 1983 Mar;45(3):800–803. doi: 10.1128/aem.45.3.800-803.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Rinehart K. L., Jr, Wolfe R. S. Methanofuran (carbon dioxide reduction factor), a formyl carrier in methane production from carbon dioxide in Methanobacterium. Biochemistry. 1985 Feb 12;24(4):995–999. doi: 10.1021/bi00325a028. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Wolfe R. S. Carbon dioxide reduction factor and methanopterin, two coenzymes required for CO2 reduction to methane by extracts of Methanobacterium. J Biol Chem. 1983 Jun 25;258(12):7536–7540. [PubMed] [Google Scholar]
- Leps B., Labischinski H., Barnickel G., Bradaczek H., Giesbrecht P. A new proposal for the primary and secondary structure of the glycan moiety of pseudomurein. Conformational energy calculations on the glycan strands with talosaminuronic acid in 1C conformation and comparison with murein. Eur J Biochem. 1984 Oct 15;144(2):279–286. doi: 10.1111/j.1432-1033.1984.tb08461.x. [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Greening R. C., Ferry J. G. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl Environ Microbiol. 1984 Jul;48(1):81–87. doi: 10.1128/aem.48.1.81-87.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., White R. H., Ferry J. G. Identification of methyl coenzyme M as an intermediate in methanogenesis from acetate in Methanosarcina spp. J Bacteriol. 1984 Nov;160(2):521–525. doi: 10.1128/jb.160.2.521-525.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Nicholson D. E., Jr, Fox G. E. Widespread distribution of a 7S RNA in archaebacteria. Curr Microbiol. 1985;12:69–72. doi: 10.1007/BF01567394. [DOI] [PubMed] [Google Scholar]
- Madon J., Leser U., Zillig W. DNA-dependent RNA polymerase from the extremely halophilic archaebacterium Halococcus morrhuae. Eur J Biochem. 1983 Sep 15;135(2):279–283. doi: 10.1111/j.1432-1033.1983.tb07649.x. [DOI] [PubMed] [Google Scholar]
- Mah R. A., Ward D. M., Baresi L., Glass T. L. Biogenesis of methane. Annu Rev Microbiol. 1977;31:309–341. doi: 10.1146/annurev.mi.31.100177.001521. [DOI] [PubMed] [Google Scholar]
- Mankin A. S., Kagramanova V. K., Teterina N. L., Rubtsov P. M., Belova E. N., Kopylov A. M., Baratova L. A., Bogdanov A. A. The nucleotide sequence of the gene coding for the 16S rRNA from the archaebacterium Halobacterium halobium. Gene. 1985;37(1-3):181–189. doi: 10.1016/0378-1119(85)90271-9. [DOI] [PubMed] [Google Scholar]
- Mankin A. S., Teterina N. L., Rubtsov P. M., Baratova L. A., Kagramanova V. K. Putative promoter region of rRNA operon from archaebacterium Halobacterium halobium. Nucleic Acids Res. 1984 Aug 24;12(16):6537–6546. doi: 10.1093/nar/12.16.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Yeats S., Janekovic D., Reiter W. D., Aicher W., Zillig W. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 1984 Sep;3(9):2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson A. T., Yaguchi M., Balch W. E., Wolfe R. S. Sequence homologies in the N-terminal region of the ribosomal 'A' proteins from Methanobacterium Thermoautotrophicum and Halobacterium cutirubrum. Biochim Biophys Acta. 1980 Nov 20;626(1):162–169. doi: 10.1016/0005-2795(80)90207-x. [DOI] [PubMed] [Google Scholar]
- Matheson A. T., Yaguchi M., Christensen P., Rollin C. F., Hasnain S. Purification, properties, and N-terminal amino acid sequence of certain 50S ribosomal subunit proteins from the archaebacterium Halobacterium cutirubrum. Can J Biochem Cell Biol. 1984 Jun;62(6):426–433. doi: 10.1139/o84-058. [DOI] [PubMed] [Google Scholar]
- McBride B. C., Wolfe R. S. A new coenzyme of methyl transfer, coenzyme M. Biochemistry. 1971 Jun 8;10(12):2317–2324. doi: 10.1021/bi00788a022. [DOI] [PubMed] [Google Scholar]
- McConnell D. J., Searcy D. G., Sutcliffe J. G. A restriction enzyme Tha I from the thermophilic mycoplasma Thermoplasma acidophilum. Nucleic Acids Res. 1978 Jun;5(6):1729–1739. doi: 10.1093/nar/5.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P., Hespell R. B., Costerton J. W. Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium. Appl Environ Microbiol. 1981 Apr;41(4):1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar R. C., Shaw K. M., Sprott G. D. Isolation and characterization of a FAD-dependent NADH diaphorase from Methanospirillum hungatei strain GP1. Can J Biochem. 1981 Feb;59(2):83–91. doi: 10.1139/o81-013. [DOI] [PubMed] [Google Scholar]
- Meile L., Kiener A., Leisinger T. A plasmid in the archaebacterium Methanobacterium thermoautotrophicum. Mol Gen Genet. 1983;191(3):480–484. doi: 10.1007/BF00425766. [DOI] [PubMed] [Google Scholar]
- Merkel G. J., Durham D. R., Perry J. J. The atypical cell wall composition of Thermomicrobium roseum. Can J Microbiol. 1980 Apr;26(4):556–559. doi: 10.1139/m80-097. [DOI] [PubMed] [Google Scholar]
- Mevarech M., Eisenberg H., Neumann E. Malate dehydrogenase isolated from extremely halophilic bacteria of the Dead Sea. 1. Purification and molecular characterization. Biochemistry. 1977 Aug 23;16(17):3781–3785. doi: 10.1021/bi00636a009. [DOI] [PubMed] [Google Scholar]
- Mevarech M., Neumann E. Malate dehydrogenase isolated from extremely halophilic bacteria of the Dead Sea. 2. Effect of salt on the catalytic activity and structure. Biochemistry. 1977 Aug 23;16(17):3786–3792. doi: 10.1021/bi00636a010. [DOI] [PubMed] [Google Scholar]
- Mevarech M., Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985 Apr;162(1):461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels M., Bakker E. P. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol. 1985 Jan;161(1):231–237. doi: 10.1128/jb.161.1.231-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J., Conway de Macario E., Macario A. J. Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol. 1982 Jan;43(1):227–232. doi: 10.1128/aem.43.1.227-232.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J. Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol. 1985 Mar;141(2):116–122. doi: 10.1007/BF00423270. [DOI] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J. Oxidation of hydrogen and reduction of methanol to methane is the sole energy source for a methanogen isolated from human feces. J Bacteriol. 1983 Feb;153(2):1051–1055. doi: 10.1128/jb.153.2.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y., Wakabayashi S., Wada K., Matsubara H., Kerscher L., Oesterhelt D. Amino acid sequence of a ferredoxin from thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. Presence of an N6-monomethyllysine and phyletic consideration of archaebacteria. J Biochem. 1985 Mar;97(3):745–753. doi: 10.1093/oxfordjournals.jbchem.a135114. [DOI] [PubMed] [Google Scholar]
- Mitchell R. M., Loeblich L. A., Klotz L. C., Loeblich A. R., 3rd DNA organization of Methanobacterium thermoautotrophicum. Science. 1979 Jun 8;204(4397):1082–1084. doi: 10.1126/science.377486. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969 Jul;99(1):255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Characterization of the deoxyribonucleic acid of various strains of halophilic bacteria. J Bacteriol. 1969 Jul;99(1):248–254. doi: 10.1128/jb.99.1.248-254.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz A., Goebel W. Characterization of the 7S RNA and its gene from halobacteria. Nucleic Acids Res. 1985 Oct 11;13(19):6969–6979. doi: 10.1093/nar/13.19.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz A., Lankat-Buttgereit B., Gross H. J., Goebel W. Common structural features of the genes for two stable RNAs from Halobacterium halobium. Nucleic Acids Res. 1985 Jan 11;13(1):31–43. doi: 10.1093/nar/13.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. C., Searcy D. G., Edwards B. F. Crystallization of a Fe,Zn superoxide dismutase from the archaebacterium Thermoplasma acidophilium. J Mol Biol. 1985 Nov 5;186(1):213–214. doi: 10.1016/0022-2836(85)90273-6. [DOI] [PubMed] [Google Scholar]
- Murray P. A., Zinder S. H. Nutritional Requirements of Methanosarcina sp. Strain TM-1. Appl Environ Microbiol. 1985 Jul;50(1):49–55. doi: 10.1128/aem.50.1.49-55.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Allmansberger R., Klein A. Termination of a transcription unit comprising highly expressed genes in the archaebacterium Methanococcus voltae. Nucleic Acids Res. 1985 Sep 25;13(18):6439–6445. doi: 10.1093/nar/13.18.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle D. P., Jr, Wolfe R. S. Component A of the methyl coenzyme M methylreductase system of Methanobacterium: resolution into four components. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2151–2155. doi: 10.1073/pnas.80.8.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasu S., Kikuchi A. Reverse gyrase; ATP-dependent type I topoisomerase from Sulfolobus. EMBO J. 1985 Oct;4(10):2705–2710. doi: 10.1002/j.1460-2075.1985.tb03990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Kohiyama M. An alpha-like DNA polymerase from Halobacterium halobium. Eur J Biochem. 1985 Oct 15;152(2):293–297. doi: 10.1111/j.1432-1033.1985.tb09197.x. [DOI] [PubMed] [Google Scholar]
- Nathans J., Thomas D., Hogness D. S. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986 Apr 11;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Willick G. E., Matheson A. T. The 5 S RNA.protein complex from an extreme halophile, Halobacterium cutirubrum. Studies on the RNA-protein interaction. J Biol Chem. 1979 Mar 10;254(5):1506–1512. [PubMed] [Google Scholar]
- Nelson M. J., Brown D. P., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by hydrogenase from Methanobacterium formicicum. Biochem Biophys Res Commun. 1984 May 16;120(3):775–781. doi: 10.1016/s0006-291x(84)80174-6. [DOI] [PubMed] [Google Scholar]
- Nelson M. J., Ferry J. G. Carbon monoxide-dependent methyl coenzyme M methylreductase in acetotrophic Methosarcina spp. J Bacteriol. 1984 Nov;160(2):526–532. doi: 10.1128/jb.160.2.526-532.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll K. M., Rinehart K. L., Jr, Tanner R. S., Wolfe R. S. Structure of component B (7-mercaptoheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4238–4242. doi: 10.1073/pnas.83.12.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda G., Strom A. R., Visentin L. P., Yaguchi M. An acidic, alanine-rich 50 S ribosomal protein from Halobacterium cutirubrum: amino acid sequence homology with Escherichia coli proteins L7 and L12. FEBS Lett. 1974 Jul 15;43(2):127–130. doi: 10.1016/0014-5793(74)80983-x. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Pace N. R., Nuell M., Kaine B. P., Gupta R., Woese C. R. Sequence of the 16S rRNA gene from the thermoacidophilic archaebacterium Sulfolobus solfataricus and its evolutionary implications. J Mol Evol. 1985;22(4):301–307. doi: 10.1007/BF02115685. [DOI] [PubMed] [Google Scholar]
- Pappenheimer A. M., Jr, Dunlop P. C., Adolph K. W., Bodley J. W. Occurrence of diphthamide in archaebacteria. J Bacteriol. 1983 Mar;153(3):1342–1347. doi: 10.1128/jb.153.3.1342-1347.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterek J. R., Smith P. H. Isolation and characterization of a halophilic methanogen from great salt lake. Appl Environ Microbiol. 1985 Oct;50(4):877–881. doi: 10.1128/aem.50.4.877-881.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukert J. L., Straus L. D., Rabinowitz J. C. Formyl-methyl-methylenetetrahydrofolate synthetase-(combined). An ovine protein with multiple catalytic activities. J Biol Chem. 1976 Aug 25;251(16):5104–5111. [PubMed] [Google Scholar]
- Pauling C. Bacteriophages of Halobacterium halobium: isolated from fermented fish sauce and primary characterization. Can J Microbiol. 1982 Aug;28(8):916–921. doi: 10.1139/m82-138. [DOI] [PubMed] [Google Scholar]
- Paynter M. J., Hungate R. E. Characterization of Methanobacterium mobilis, sp. n., isolated from the bovine rumen. J Bacteriol. 1968 May;95(5):1943–1951. doi: 10.1128/jb.95.5.1943-1951.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezacka E., Wood H. G. The autotrophic pathway of acetogenic bacteria. Role of CO dehydrogenase disulfide reductase. J Biol Chem. 1986 Feb 5;261(4):1609–1615. [PubMed] [Google Scholar]
- Pfeifer F., Betlach M. Genome organization in Halobacterium halobium: a 70 kb island of more (AT) rich DNA in the chromosome. Mol Gen Genet. 1985;198(3):449–455. doi: 10.1007/BF00332938. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981 Jan;145(1):369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Genetic variability in Halobacterium halobium. J Bacteriol. 1981 Jan;145(1):375–381. doi: 10.1128/jb.145.1.375-381.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A., van der Drift C., Vogels G. D. Corrinoids from Methanosarcina barkeri: structure of the alpha-ligand. Biochem Biophys Res Commun. 1982 Sep 30;108(2):731–737. doi: 10.1016/0006-291x(82)90890-7. [DOI] [PubMed] [Google Scholar]
- Pond J. L., Langworthy T. A., Holzer G. Long-chain diols: a new class of membrane lipids from a thermophilic bacterium. Science. 1986 Mar 7;231(4742):1134–1136. doi: 10.1126/science.231.4742.1134. [DOI] [PubMed] [Google Scholar]
- Postgate J. R. Methane as a minor product of pyruvate metabolism by sulphate-reducing and other bacteria. J Gen Microbiol. 1969 Aug;57(3):293–302. doi: 10.1099/00221287-57-3-293. [DOI] [PubMed] [Google Scholar]
- Powers S. G., Snell E. E. Ketopantoate hydroxymethyltransferase. II. Physical, catalytic, and regulatory properties. J Biol Chem. 1976 Jun 25;251(12):3786–3793. [PubMed] [Google Scholar]
- Prangishvili D. A., Vashakidze R. P., Chelidze M. G., Gabriadze IYu A restriction endonuclease SuaI from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1985 Nov 11;192(1):57–60. doi: 10.1016/0014-5793(85)80042-9. [DOI] [PubMed] [Google Scholar]
- Prangishvilli D., Zillig W., Gierl A., Biesert L., Holz I. DNA-dependent RNA polymerase of thermoacidophilic archaebacteria. Eur J Biochem. 1982 Mar 1;122(3):471–477. doi: 10.1111/j.1432-1033.1982.tb06461.x. [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W., Ljungdahl L. G., DerVartanian D. V. Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J Bacteriol. 1983 Sep;155(3):1224–1237. doi: 10.1128/jb.155.3.1224-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W., Wood H. G. Acetate biosynthesis by acetogenic bacteria. Evidence that carbon monoxide dehydrogenase is the condensing enzyme that catalyzes the final steps of the synthesis. J Biol Chem. 1985 Apr 10;260(7):3970–3977. [PubMed] [Google Scholar]
- Rauhut R., Gabius H. J., Engelhardt R., Cramer F. Archaebacterial phenylalanyl-tRNA synthetase. Accuracy of the phenylalanyl-tRNA synthetase from the archaebacterium Methanosarcina barkeri, Zn(II)-dependent synthesis of diadenosine 5',5'''-P1,P4-tetraphosphate, and immunological relationship of OFFnylalanyl-tRNA synthetases from different urkingdoms. J Biol Chem. 1985 Jan 10;260(1):182–187. [PubMed] [Google Scholar]
- Rauhut R., Gabius H. J., Kühn W., Cramer F. Phenylalanyl-tRNA synthetase from the archaebacterium Methanosarcina barkeri. J Biol Chem. 1984 May 25;259(10):6340–6345. [PubMed] [Google Scholar]
- Rivard C. J., Henson J. M., Thomas M. V., Smith P. H. Isolation and Characterization of Methanomicrobium paynteri sp. nov., a Mesophilic Methanogen Isolated from Marine Sediments. Appl Environ Microbiol. 1983 Aug;46(2):484–490. doi: 10.1128/aem.46.2.484-490.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A. M., Wolfe R. S. ATP requirement for methanogenesis in cell extracts of methanobacterium strain M.o.H. Biochim Biophys Acta. 1969 Dec 30;192(3):420–429. doi: 10.1016/0304-4165(69)90391-2. [DOI] [PubMed] [Google Scholar]
- Romesser J. A., Wolfe R. S. Coupling of methyl coenzyme M reduction with carbon dioxide activation in extracts of Methanobacterium thermoautotrophicum. J Bacteriol. 1982 Nov;152(2):840–847. doi: 10.1128/jb.152.2.840-847.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romesser J. A., Wolfe R. S. Interaction of coenzyme M and formaldehyde in methanogenesis. Biochem J. 1981 Sep 1;197(3):565–571. doi: 10.1042/bj1970565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C. S., Pirt S. J. Conversion of glucose to fatty acids and methane: roles of two mycoplasmal agents. J Bacteriol. 1981 Jul;147(1):248–254. doi: 10.1128/jb.147.1.248-254.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière P. E., Escalante-Semerena J. C., Wolfe R. S. Component A2 of the methylcoenzyme M methylreductase system from Methanobacterium thermoautotrophicum. J Bacteriol. 1985 Apr;162(1):61–66. doi: 10.1128/jb.162.1.61-66.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEHGAL S. N., KATES M., GIBBONS N. E. Lipids of Halobacterium cutirubrum. Can J Biochem Physiol. 1962 Jan;40:69–81. [PubMed] [Google Scholar]
- SMITH P. H., HUNGATE R. E. Isolation and characterization of Methanobacterium ruminantium n. sp. J Bacteriol. 1958 Jun;75(6):713–718. doi: 10.1128/jb.75.6.713-718.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Sauer F. D., Erfle J. D., Mahadevan S. Evidence for an internal electrochemical proton gradient in Methanobacterium thermoautotrophicum. J Biol Chem. 1981 Oct 10;256(19):9843–9848. [PubMed] [Google Scholar]
- Sauer F. D., Mahadevan S., Erfle J. D. Methane synthesis by membrane vesicles and a cytoplasmic cofactor isolated from Methanobacterium thermoautotrophicum. Biochem J. 1984 Jul 1;221(1):61–69. doi: 10.1042/bj2210061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1986 Feb;165(2):405–411. doi: 10.1128/jb.165.2.405-411.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1983 Aug;155(2):467–472. doi: 10.1128/jb.155.2.467-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Properties of formate dehydrogenase in Methanobacterium formicicum. J Bacteriol. 1982 Apr;150(1):1–7. doi: 10.1128/jb.150.1.1-7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid G., Böck A. Immunological comparison of ribosomal proteins from archaebacteria. J Bacteriol. 1981 Aug;147(2):282–288. doi: 10.1128/jb.147.2.282-288.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid G., Strobel O., Stöffler-Meilicke M., Stöffler G., Böck A. A ribosomal protein that is immunologically conserved in archaebacteria, eubacteria and eukaryotes. FEBS Lett. 1984 Nov 19;177(2):189–194. doi: 10.1016/0014-5793(84)81281-8. [DOI] [PubMed] [Google Scholar]
- Schmid K., Thomm M., Laminet A., Laue F. G., Kessler C., Stetter K. O., Schmitt R. Three new restriction endonucleases MaeI, MaeII and MaeIII from Methanococcus aeolicus. Nucleic Acids Res. 1984 Mar 26;12(6):2619–2628. doi: 10.1093/nar/12.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H. An immune strain of Halobacterium halobium carries the invertible L segment of phage PhiH as a plasmid. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1017–1020. doi: 10.1073/pnas.81.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H., Palm P., Dick K., Grampp B. Sequence analysis of the insertion element ISH1.8 and of associated structural changes in the genome of phage PhiH of the archaebacterium Halobacterium halobium. EMBO J. 1984 Aug;3(8):1717–1722. doi: 10.1002/j.1460-2075.1984.tb02037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H., Zillig W., Pfäffle M., Schnabel R., Michel H., Delius H. Halobacterium halobium phage øH. EMBO J. 1982;1(1):87–92. doi: 10.1002/j.1460-2075.1982.tb01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R., Thomm M., Gerardy-Schahn R., Zillig W., Stetter K. O., Huet J. Structural homology between different archaebacterial DNA-dependent RNA polymerases analyzed by immunological comparison of their components. EMBO J. 1983;2(5):751–755. doi: 10.1002/j.1460-2075.1983.tb01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönheit P., Beimborn D. B. ATP synthesis in Methanobacterium thermoautotrophicum coupled to CH4 formation from H2 and CO2 in the apparent absence of an electrochemical proton potential across the cytoplasmic membrane. Eur J Biochem. 1985 May 2;148(3):545–550. doi: 10.1111/j.1432-1033.1985.tb08874.x. [DOI] [PubMed] [Google Scholar]
- Searcy D. G., Stein D. B. Nucleoprotein subunit structure in an unusual prokaryotic organism: Thermoplasma acidophilum. Biochim Biophys Acta. 1980 Aug 26;609(1):180–195. doi: 10.1016/0005-2787(80)90211-7. [DOI] [PubMed] [Google Scholar]
- Searcy K. B., Searcy D. G. Superoxide dismutase from the Archaebacterium Thermoplasma acidophilum. Biochim Biophys Acta. 1981 Aug 28;670(1):39–46. doi: 10.1016/0005-2795(81)90046-5. [DOI] [PubMed] [Google Scholar]
- Seely R. J., Fahrney D. E. Levels of cyclic-2,3-diphosphoglycerate in Methanobacterium thermoautotrophicum during phosphate limitation. J Bacteriol. 1984 Oct;160(1):50–54. doi: 10.1128/jb.160.1.50-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seely R. J., Krueger R. D., Fahrney D. E. Cyclic-2,3-diphosphoglycerate levels in Methanobacterium thermoautotrophicum reflect inorganic phosphate availability. Biochem Biophys Res Commun. 1983 Nov 15;116(3):1125–1128. doi: 10.1016/s0006-291x(83)80259-9. [DOI] [PubMed] [Google Scholar]
- Shapiro S., Wolfe R. S. Methyl-coenzyme M, an intermediate in methanogenic dissimilation of C1 compounds by Methanosarcina barkeri. J Bacteriol. 1980 Feb;141(2):728–734. doi: 10.1128/jb.141.2.728-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek M., DasSarma S., RajBhandary U. L., Khorana H. G. A transposable element from Halobacterium halobium which inactivates the bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7268–7272. doi: 10.1073/pnas.79.23.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr U. B., Messner P. Crystalline surface layers on bacteria. Annu Rev Microbiol. 1983;37:311–339. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- Smith M. R., Lequerica J. L., Hart M. R. Inhibition of methanogenesis and carbon metabolism in Methanosarcina sp. by cyanide. J Bacteriol. 1985 Apr;162(1):67–71. doi: 10.1128/jb.162.1.67-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Mah R. A. Growth and methanogenesis by Methanosarcina strain 227 on acetate and methanol. Appl Environ Microbiol. 1978 Dec;36(6):870–879. doi: 10.1128/aem.36.6.870-879.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N., Matheson A. T., Yaguchi M., Willick G. E., Nazar R. N. The 5-S RNA . protein complex from an extreme halophile, Halobacterium cutirubrum. Purification and characterization. Eur J Biochem. 1978 Sep 1;89(2):501–509. doi: 10.1111/j.1432-1033.1978.tb12554.x. [DOI] [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. Methanosarcina acetivorans sp. nov., an Acetotrophic Methane-Producing Bacterium Isolated from Marine Sediments. Appl Environ Microbiol. 1984 May;47(5):971–978. doi: 10.1128/aem.47.5.971-978.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. Isolation and Characterization of a Methylotrophic Marine Methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol. 1983 Feb;45(2):684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R. W., Daniels L., Fulton G., Orme-Johnson W. H. Product isotope effects on in vivo methanogenesis by Methanobacterium thermoautotrophicum. Biochemistry. 1980 Aug 5;19(16):3678–3683. doi: 10.1021/bi00557a007. [DOI] [PubMed] [Google Scholar]
- Sprott G. D., McKellar R. C., Shaw K. M., Giroux J., Martin W. G. Properties of malate dehydrogenase isolated from Methanospirillum hungatii. Can J Microbiol. 1979 Feb;25(2):192–200. doi: 10.1139/m79-030. [DOI] [PubMed] [Google Scholar]
- Sprott G. D., Shaw K. M., Jarrell K. F. Ammonia/potassium exchange in methanogenic bacteria. J Biol Chem. 1984 Oct 25;259(20):12602–12608. [PubMed] [Google Scholar]
- Stackebrandt E., Ludwig W., Schubert W., Klink F., Schlesner H., Roggentin T., Hirsch P. Molecular genetic evidence for early evolutionary origin of budding peptidoglycan-less eubacteria. Nature. 1984 Feb 23;307(5953):735–737. doi: 10.1038/307735a0. [DOI] [PubMed] [Google Scholar]
- Steber J., Schleifer K. H. Halococcus morrhuae: a sulfated heteropolysaccharide as the structural component of the bacterial cell wall. Arch Microbiol. 1975 Oct 27;105(2):173–177. doi: 10.1007/BF00447133. [DOI] [PubMed] [Google Scholar]
- Stoeckenius W., Bogomolni R. A. Bacteriorhodopsin and related pigments of halobacteria. Annu Rev Biochem. 1982;51:587–616. doi: 10.1146/annurev.bi.51.070182.003103. [DOI] [PubMed] [Google Scholar]
- Stoeckenius W. The purple membrane of salt-loving bacteria. Sci Am. 1976 Jun;234(6):38–46. doi: 10.1038/scientificamerican0676-38. [DOI] [PubMed] [Google Scholar]
- Stoeckenius W. The rhodopsin-like pigments of halobacteria: light-energy and signal transducers in an archaebacterium. Trends Biochem Sci. 1985 Dec;10(12):483–486. doi: 10.1016/0968-0004(85)90210-5. [DOI] [PubMed] [Google Scholar]
- Storer A. C., Sprott G. D., Martin W. G. Kinetic and physical properties of the L-malate-NAD+ oxidoreductase from Methanospirillum hungatii and comparison with the enzyme from other sources. Biochem J. 1981 Jan 1;193(1):235–244. doi: 10.1042/bj1930235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A. R., Visentin L. P. Acidic ribosomal proteins from the extreme halophile, Halobacterium cutirubrum. The simultaneous separation, identification and molecular weight determination. FEBS Lett. 1973 Dec 1;37(2):274–280. doi: 10.1016/0014-5793(73)80477-6. [DOI] [PubMed] [Google Scholar]
- Stupperich E., Hammel K. E., Fuchs G., Thauer R. K. Carbon monoxide fixation into the carboxyl group of acetyl coenzyme A during autotrophic growth of Methanobacterium. FEBS Lett. 1983 Feb 7;152(1):21–23. doi: 10.1016/0014-5793(83)80473-6. [DOI] [PubMed] [Google Scholar]
- Taylor C. D., Wolfe R. S. Structure and methylation of coenzyme M(HSCH2CH2SO3). J Biol Chem. 1974 Aug 10;249(15):4879–4885. [PubMed] [Google Scholar]
- Thauer R. K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977 Mar;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Altenbuchner J., Stetter K. O. Evidence for a plasmid in a methanogenic bacterium. J Bacteriol. 1983 Feb;153(2):1060–1062. doi: 10.1128/jb.153.2.1060-1062.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornabene T. G., Langworthy T. A. Diphytanyl and dibiphytanyl glycerol ether lipids of methanogenic archaebacteria. Science. 1979 Jan 5;203(4375):51–53. doi: 10.1126/science.758677. [DOI] [PubMed] [Google Scholar]
- Tornabene T. G., Wolfe R. S., Balch W. E., Holzer G., Fox G. E., Oro J. Phytanyl-glycerol ethers and squalenes in the archaebacterium Methanobacterium thermoautotrophicum. J Mol Evol. 1978 Aug 2;11(3):259–266. doi: 10.1007/BF01734487. [DOI] [PubMed] [Google Scholar]
- Torsvik T., Dundas I. D. Bacteriophage of Halobacterium salinarium. Nature. 1974 Apr 19;248(5450):680–681. doi: 10.1038/248680a0. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Fukuyama K., Wakabayashi S., Wada K., Matsubara H., Kerscher L., Oesterhelt D. Preliminary X-ray diffraction studies on a ferredoxin from the thermophilic archaebacterium, Thermoplasma acidophilum. J Mol Biol. 1985 Nov 20;186(2):481–482. doi: 10.1016/0022-2836(85)90122-6. [DOI] [PubMed] [Google Scholar]
- Tu J. K., Prangishvilli D., Huber H., Wildgruber G., Zillig W., Stetter K. O. Taxonomic relations between archaebacteria including 6 novel genera examined by cross hybridization of DNAs and 16S rRNAs. J Mol Evol. 1982;18(2):109–114. doi: 10.1007/BF01810829. [DOI] [PubMed] [Google Scholar]
- Tu J., Zillig W. Organization of rRNA structural genes in the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res. 1982 Nov 25;10(22):7231–7245. doi: 10.1093/nar/10.22.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng S. F., Wolfe R. S., Bryant M. P. Factor 420-dependent pyridine nucleotide-linked hydrogenase system of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):184–191. doi: 10.1128/jb.121.1.184-191.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzing S. F., Bryant M. P., Wolfe R. S. Factor 420-dependent pyridine nucleotide-linked formate metabolism of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):192–196. doi: 10.1128/jb.121.1.192-196.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin L. P., Chow C., Matheson A. T., Yaguchi M., Rollin F. Halobacterium cutirubrum ribosomes. Properties of the ribosomal proteins and ribonucleic acid. Biochem J. 1972 Nov;130(1):103–110. doi: 10.1042/bj1300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossbrinck C. R., Woese C. R. Eukaryotic ribosomes that lack a 5.8S RNA. Nature. 1986 Mar 20;320(6059):287–288. doi: 10.1038/320287a0. [DOI] [PubMed] [Google Scholar]
- Wais A. C., Kon M., MacDonald R. E., Stollar B. D. Salt-dependent bacteriophage infecting Halobacterium cutirubrum and H. halobium. Nature. 1975 Jul 24;256(5515):314–315. doi: 10.1038/256314a0. [DOI] [PubMed] [Google Scholar]
- Wasserman G. F., Benkovic P. A., Young M., Benkovic S. J. Kinetic relationships between the various activities of the formyl-methenyl-methylenetetrahydrofolate synthetase. Biochemistry. 1983 Mar 1;22(5):1005–1013. doi: 10.1021/bi00274a002. [DOI] [PubMed] [Google Scholar]
- Weidinger G., Klotz G., Goebel W. A large plasmid from Halobacterium halobium carrying genetic information for gas vacuole formation. Plasmid. 1979 Jul;2(3):377–386. doi: 10.1016/0147-619x(79)90021-0. [DOI] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate assimilation pathway of Methanosarcina barkeri. J Bacteriol. 1979 Jan;137(1):332–339. doi: 10.1128/jb.137.1.332-339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. L. Subunit cell wall of Sulfolobus acidocaldarius. J Bacteriol. 1974 Apr;118(1):275–284. doi: 10.1128/jb.118.1.275-284.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werber M. M., Mevarech M. Purification and characterization of a highly acidic 2Fe-ferredoxin from Halobacterium of the dead sea. Arch Biochem Biophys. 1978 Apr 30;187(2):447–456. doi: 10.1016/0003-9861(78)90056-5. [DOI] [PubMed] [Google Scholar]
- White R. H. Biosynthesis of the 7-methylated pterin of methanopterin. J Bacteriol. 1986 Jan;165(1):215–218. doi: 10.1128/jb.165.1.215-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Wolfe R. S. Activation of the methylreductase system from Methanobacterium bryantii by ATP. J Bacteriol. 1983 May;154(2):640–649. doi: 10.1128/jb.154.2.640-649.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Wolfe R. S. Activation of the methylreductase system from Methanobacterium bryantii by corrins. J Bacteriol. 1985 Oct;164(1):165–172. doi: 10.1128/jb.164.1.165-172.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Wolfe R. S. Presence of nickel in factor F430 from Methanobacterium bryantii. Biochem Biophys Res Commun. 1980 Feb 27;92(4):1196–1201. doi: 10.1016/0006-291x(80)90413-1. [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Wolfe R. S. Purification and analysis of cobamides of Methanobacterium bryantii by high-performance liquid chromatography. Anal Biochem. 1984 Feb;137(1):261–265. doi: 10.1016/0003-2697(84)90380-4. [DOI] [PubMed] [Google Scholar]
- Wich G., Jarsch M., Böck A. Apparent operon for a 5S ribosomal RNA gene and for tRNA genes in the archaebacterium Methanococcus vannielii. Mol Gen Genet. 1984;196(1):146–151. doi: 10.1007/BF00334107. [DOI] [PubMed] [Google Scholar]
- Wieland F., Dompert W., Bernhardt G., Sumper M. Halobacterial glycoprotein saccharides contain covalently linked sulphate. FEBS Lett. 1980 Oct 20;120(1):110–114. doi: 10.1016/0014-5793(80)81058-1. [DOI] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Effect of sulfate on carbon and electron flow during microbial methanogenesis in freshwater sediments. Appl Environ Microbiol. 1977 Feb;33(2):275–281. doi: 10.1128/aem.33.2.275-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Microbial methanogenesis and acetate metabolism in a meromictic lake. Appl Environ Microbiol. 1979 Feb;37(2):213–221. doi: 10.1128/aem.37.2.213-221.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gupta R., Hahn C. M., Zillig W., Tu J. The phylogenetic relationships of three sulfur dependent archaebacteria. Syst Appl Microbiol. 1984;5:97–105. doi: 10.1016/s0723-2020(84)80054-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Fox G. E. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]
- Wood A. G., Whitman W. B., Konisky J. A newly-isolated marine methanogen harbors a small cryptic plasmid. Arch Microbiol. 1985 Aug;142(3):259–261. doi: 10.1007/BF00693400. [DOI] [PubMed] [Google Scholar]
- Wood J. M., Moura I., Moura J. J., Santos M. H., Xavier A. V., LeGall J., Scandellari M. Role of vitamin B12 in methyl transfer for methane biosynthesis by Methanosarcina barkeri. Science. 1982 Apr 16;216(4543):303–305. doi: 10.1126/science.7063887. [DOI] [PubMed] [Google Scholar]
- Xu W. L., Doolittle W. F. Structure of the archaebacterial transposable element ISH50. Nucleic Acids Res. 1983 Jun 25;11(12):4195–4199. doi: 10.1093/nar/11.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Morisaki M., Kumaoka H. Different biosynthetic pathways of the pyrimidine moiety of thiamin in procaryotes and eucaryotes. Biochim Biophys Acta. 1983 Mar 15;756(1):41–48. doi: 10.1016/0304-4165(83)90022-3. [DOI] [PubMed] [Google Scholar]
- Yamazaki S. A selenium-containing hydrogenase from Methanococcus vannielii. Identification of the selenium moiety as a selenocysteine residue. J Biol Chem. 1982 Jul 25;257(14):7926–7929. [PubMed] [Google Scholar]
- Yamazaki S., Tsai L., Stadtman T. C. Analogues of 8-hydroxy-5-deazaflavin cofactor: relative activity as substrates for 8-hydroxy-5-deazaflavin-dependent NADP+ reductase from Methanococcus vannielii. Biochemistry. 1982 Mar 2;21(5):934–939. doi: 10.1021/bi00534a019. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Tsai L., Stadtman T. C., Jacobson F. S., Walsh C. Stereochemical studies of 8-hydroxy-5-deazaflavin-dependent NADP+ reductase from Methanococcus vannielii. J Biol Chem. 1980 Oct 10;255(19):9025–9027. [PubMed] [Google Scholar]
- Yeats S., McWilliam P., Zillig W. A plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J. 1982;1(9):1035–1038. doi: 10.1002/j.1460-2075.1982.tb01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder A. J., Huser B. A., Brock T. D., Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980 Jan;124(1):1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Fuchs G., Kenealy W., Thauer R. K. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J Bacteriol. 1977 Nov;132(2):604–613. doi: 10.1128/jb.132.2.604-613.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus J. G., Henning D. L. Methanobacterium arbophilicum sp.nov. An obligate anaerobe isolated from wetwood of living trees. Antonie Van Leeuwenhoek. 1975;41(4):543–552. doi: 10.1007/BF02565096. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Kerby R., Krzycki J. A. Single-carbon chemistry of acetogenic and methanogenic bacteria. Science. 1985 Mar 8;227(4691):1167–1173. doi: 10.1126/science.3919443. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G. The biology of methanogenic bacteria. Bacteriol Rev. 1977 Jun;41(2):514–541. doi: 10.1128/br.41.2.514-541.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus J. G., Wolfe R. S. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972 Feb;109(2):707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Stetter K. O., Janeković D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur J Biochem. 1979 Jun 1;96(3):597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x. [DOI] [PubMed] [Google Scholar]
- Zillig W., Stetter K. O., Tobien M. DNA-dependent RNA polymerase from Halobacterium halobium. Eur J Biochem. 1978 Nov 2;91(1):193–199. doi: 10.1111/j.1432-1033.1978.tb20951.x. [DOI] [PubMed] [Google Scholar]
- Zinder S. H., Cardwell S. C., Anguish T., Lee M., Koch M. Methanogenesis in a Thermophilic (58 degrees C) Anaerobic Digestor: Methanothrix sp. as an Important Aceticlastic Methanogen. Appl Environ Microbiol. 1984 Apr;47(4):796–807. doi: 10.1128/aem.47.4.796-807.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Elias A. F. Growth substrate effects on acetate and methanol catabolism in Methanosarcina sp. strain TM-1. J Bacteriol. 1985 Jul;163(1):317–323. doi: 10.1128/jb.163.1.317-323.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Mah R. A. Isolation and Characterization of a Thermophilic Strain of Methanosarcina Unable to Use H(2)-CO(2) for Methanogenesis. Appl Environ Microbiol. 1979 Nov;38(5):996–1008. doi: 10.1128/aem.38.5.996-1008.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beelen P., Labro J. F., Keltjens J. T., Geerts W. J., Vogels G. D., Laarhoven W. H., Guijt W., Haasnoot C. A. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur J Biochem. 1984 Mar 1;139(2):359–365. doi: 10.1111/j.1432-1033.1984.tb08014.x. [DOI] [PubMed] [Google Scholar]
- van Beelen P., Stassen A. P., Bosch J. W., Vogels G. D., Guijt W., Haasnoot C. A. Elucidation of the structure of methanopterin, a coenzyme from Methanobacterium thermoautotrophicum, using two-dimensional nuclear-magnetic-resonance techniques. Eur J Biochem. 1984 Feb 1;138(3):563–571. doi: 10.1111/j.1432-1033.1984.tb07951.x. [DOI] [PubMed] [Google Scholar]
- van der Meijden P., Heythuysen H. J., Pouwels A., Houwen F., van der Drift C., Vogels G. D. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch Microbiol. 1983 Jun;134(3):238–242. doi: 10.1007/BF00407765. [DOI] [PubMed] [Google Scholar]
- van der Meijden P., van der Lest C., van der Drift C., Vogels G. D. Reductive activation of methanol: 5-hydroxybenzimidazolylcobamide methyltransferase of Methanosarcina barkeri. Biochem Biophys Res Commun. 1984 Feb 14;118(3):760–766. doi: 10.1016/0006-291x(84)91460-8. [DOI] [PubMed] [Google Scholar]


