Abstract
The maintenance of genome integrity is essential for organism survival and for the inheritance of traits to offspring. Genomic instability is caused by DNA damage, aberrant DNA replication or uncoordinated cell division, which can lead to chromosomal aberrations and gene mutations. Recently, chromatin regulators that shape the epigenetic landscape have emerged as potential gatekeepers and signalling coordinators for the maintenance of genome integrity. Here, we review chromatin functions during the two major pathways that control genome integrity: namely, repair of DNA damage and DNA replication. We also discuss recent evidence that suggests a novel role for chromatin-remodelling factors in chromosome segregation and in the prevention of aneuploidy.
Throughout the lifespan of an organism, endogenous cellular events and exogenous environmental agents can inflict damage to the cell’s genome and can severely compromise its integrity. To counteract the deleterious effect of these actions and to maintain genomic integrity, three major and evolutionarily conserved cellular pathways have evolved. DNA damage response (DDR) ensures efficient repair of all types of damage, including individual DNA base lesions and breaks1. The chromosome replication pathway governs accurate and unhindered replication of DNA2, and a chromosome segregation pathway preserves the correct number of chromosomes during cell division3. These pathways exhibit crosstalk, forming a network in which disruption of one pathway leads to engagement of the others to protect genome integrity while maintaining cell homeostasis (FIG. 1).
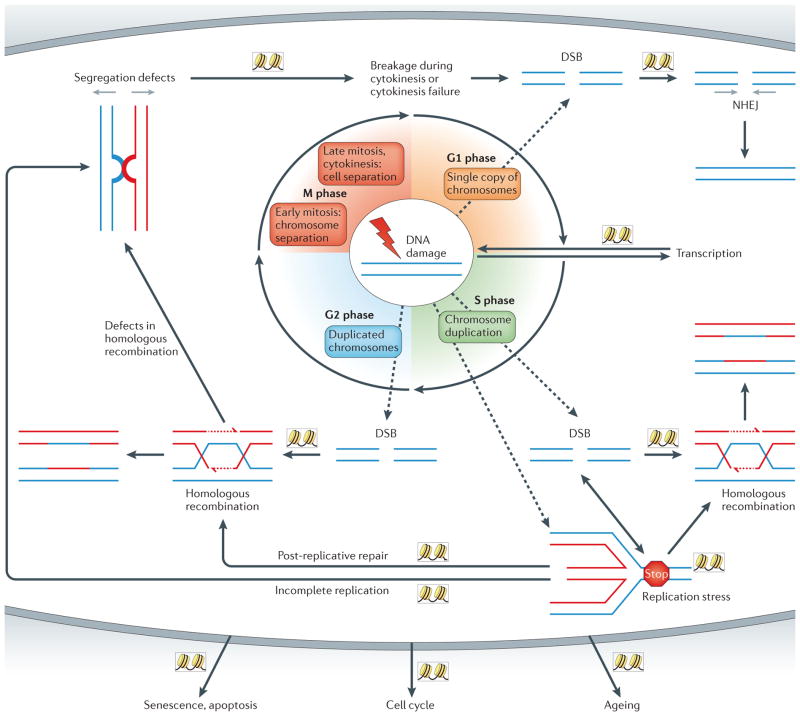
Figure 1. The role of the genome stability network in cell homeostasis.
DNA damage (shown by the lightning bolt) can create double-strand breaks (DSBs) that are repaired by the appropriate pathway, depending on the cell cycle phase. DNA DSBs in G1 phase are preferably repaired by non-homologous end joining (NHEJ), whereas DSBs in S phase or in G2 phase are mainly repaired by homologous recombination. During S phase, replication forks that encounter DNA damage or that undergo other types of replication stress may induce DNA DSBs and/or lead to the formation of aberrant chromatin structures between chromosomes. If they are not resolved, these aberrant structures, which are derived from replication defects or incomplete homologous recombination, can lead to chromosome segregation failure in mitosis or to chromosomal breakage during cytokinesis. To ensure that the most appropriate response is activated, crosstalk between the genome stability pathways is essential. In addition, the genome stability network successfully connects the repair process with other pathways that regulate cell homeostasis. Chromatin has a major role in the different genome stability pathways (as depicted by the nucleosomes in boxes).
As discussed in this Review, the past few years have witnessed a flood of reports that establish a role for chromatin in each of these pathways for genome maintenance. Chromatin structure is subject to at least three regulatory mechanisms. First, covalent histone modifications can alter the physical properties of a chromatin fibre or can regulate the binding of nonhistone proteins. Second, core histones can be replaced by histone variants that provide different biophysical properties to chromatin fibres, that present different opportunities for post-translational modifications or that regulate chromatin-binding partners. Third, chromatin can be remodelled by ATP-dependent chromatin-remodelling enzymes, leading to changes in nucleosome positions, histone eviction or incorporation of histone variants (TABLE 1; Supplementary information S1 (table)). All of these chromatin-regulatory mechanisms have been implicated in the maintenance of genome stability.
Table 1.
Chromatin regulatory factors associated with genome stability pathways
| Name | Proposed molecular function or target | Organism |
|---|---|---|
| ATP-dependent chromatin-remodelling enzymes | ||
| RAD54 | Nucleosome sliding (?) | Saccharomyces cerevisiae, Homo sapiens |
| INO80 | Sliding, H2A.Z dimer eviction | S. cerevisiae, Schizosaccharomyces pombe, Mus musculus, H. sapiens, Arabidopsis thaliana |
| Swr1 | H2A.Z dimer incorporation | S. cerevisiae |
| p400 | H. sapiens | |
| RSC complex | Nucleosome sliding | S. cerevisiae, H. sapiens |
| SWI/SNF complex | Nucleosome sliding, ejection | S. cerevisiae, H. sapiens |
| ISWI complex | Nucleosome sliding, spacing | S. cerevisiae, H. sapiens |
| ALC1 | Nucleosome sliding (?) | H. sapiens |
| SMARCAD1 | Histone dimer exchange | H. sapiens |
| Fft3 | S. pombe | |
| Fun30 | S. cerevisiae | |
| NuRD complex | Nucleosome sliding, ejection | H. sapiens |
| SMARCAL1 | DNA strand annealing (?) | H. sapiens |
| PICH | (?) | H. sapiens |
| Histone-modifying enzymes: kinases | ||
| Tel1, Mec1 | H2A.XS139 | S. cerevisiae |
| ATM, ATR | H. sapiens | |
| WSTF | H2A.XY142 | H. sapiens |
| Cdc7 | H3T45 | S. cerevisiae |
| AURKB | H3S10/S28 | H. sapiens |
| Ipl1 | S. cerevisiae | |
| Ark1 | S. pombe | |
| Bub1 | H2AS121 | S. pombe |
| Histone-modifying enzymes: K-acetyltransferases | ||
| Haspin | H3T3 | S. pombe, Xenopus laevis, H. sapiens |
| Hat1 | H4K5/K12 | S. cerevisiae |
| NuA4 complex | H4K5/K8/K12/K16, H3K14, H2A.ZK3/K8/K10/K14 | S. cerevisiae |
| TIP60 | H4K5/K8/K12/K16, H3K14, H2A.ZK3/K8/K10/K14 | Drosophila melanogaster, H. sapiens |
| GCN5 | H3K9/K14/K18 | S. cerevisiae, H. sapiens |
| CBP, p300 | H3K18, H4K5/K8/K12/K16 | H. sapiens |
| Rtt109 | H3K56 | S. cerevisiae |
| Histone-modifying enzymes: K-deacetylases | ||
| Rpd3 | Amino-tail H3, amino-tail H4 | S. cerevisiae |
| HDAC3 | H. sapiens | |
| Hda1 | H2A.Z, H2B, H3 | S. cerevisiae |
| Sir2 | H4K16 | S. cerevisiae |
| SIRT1, SIRT6 | M. musculus, H. sapiens | |
| HDAC1, HDAC2, HDAC4 | H2A, H2B, H3, H4 | H. sapiens |
| Histone-modifying enzymes: K-methyltransferases | ||
| Set1 | H3K4 | S. cerevisiae |
| SETD8 | H3K20me | H. sapiens |
| MMSET | H4K20me1, H3K20me2 | H. sapiens |
| EZH2 | H3K27 | H. sapiens |
| Dot1 | H3K79 | S. cerevisiae |
| Histone-modifying enzymes: ubiquitylases | ||
| RNF8 | H2A | H. sapiens |
| RNF168 | H2A | H. sapiens |
| Rad6 | H2BK120ub | S. cerevisiae |
| RNF20–RNF40 | H. sapiens | |
| Histone chaperones | ||
| FACT complex | H2A–H2B, H3–H4 | H. sapiens |
| CAF1 | H3.1–H4 | S. cerevisiae, H. sapiens |
| Asf1 | H3–H4 | S. cerevisiae |
| ASF1A, ASF1B | H. sapiens | |
In the ‘Proposed molecular function’ column, the question mark (?) indicates ‘another unknown in vivo function’. A solidus (/) indicates multiple amino acid targets. So, for example, H4K5/K12 means histone H4 at lysine 5 (H4K5) and/or H4K12. References for this table are provided in Supplementary information S1 (table). ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related; AURKB, aurora kinase B; CAF1, also known as CNOT7 in humans and as POP2 in yeast; CBP, CREB-binding protein (also known as CREBBP); Cdc7, cell division control protein 7; Dot1, disrupter of telomere silencing protein 1; FACT complex, facilitates chromatin transcription complex; GCN5, also known as KAT2A in humans; Hat1, histone acetyltransferase 1; HDAC1, histone deacetylase 1; me, methylation; NuRD complex, nucleosome-remodelling and histone deacetylase complex; RNF8, RING finger protein 8; S, serine; SIRT1, sirtuin 1; SMARCAD1, SMARCA containing DEAD/H box 1; SMARCAL1, SMARCA-like 1; T, threonine; TIP60, also known KAT5 in humans; ub, ubiquitylation; Y, tyrosine.
Here, we discuss the latest advances in the field of genomic integrity, focusing on inter-connections between chromatin regulators and factors that govern genome stability pathways. As DNA double-strand breaks (DSBs) are the most deleterious form of DNA damage, our discussion will highlight the chromatin response to this type of lesion. We will try to refrain from simply cataloguing all of the chromatin factors that are recruited to DSBs but instead try to focus on factors for which important mechanistic insights have been derived, aiming to address the big question of ‘what do all of these chromatin regulators do?’ In our discussion of DDR, we will first discuss roles for chromatin regulators in the repair process and will follow this with a discussion of their role in cell cycle checkpoints. Roles for chromatin regulators in DNA replication will be followed by a brief discussion of recent evidence that suggests roles for chromatin remodelling in chromosome segregation and in the prevention of aneuploidy.
Chromatin and DNA damage response
Formation of even a single DSB in a cell’s genome can initiate a complex series of events, including cell cycle arrest, recruitment of repair factors to the lesion and orchestration of the actual DSB repair event. In the case of DSB repair, the particular cell cycle phase (that is, G1 versus G2) can determine whether a cell repairs the DSB by either non-homologous end joining (NHEJ) or homologous recombination, both of which are distinct repair pathways that are associated with different chromatin changes4 (FIG. 2). DSB repair by the process of homologous recombination primarily occurs in the S and G2 phases of the cell cycle (FIG. 2). A key feature of homologous recombination is that it requires a homologous DNA duplex that is used as a donor template for DNA-synthesis-dependent repair, providing an explanation for its prevalence after the replication phase of the cell cycle. Typically, a sister chromatid is used as the homologous donor, but DNA sequences located anywhere in the genome represent potential donor templates. By contrast, NHEJ is the most commonly used pathway in G1 cells and appears to be the generally favoured pathway within most mammalian cells, which typically spend most of their lives in G1 phase. During NHEJ, the ends of the broken DNA are essentially re-ligated by molecular machineries recruited to the lesion (FIG. 2). NHEJ requires little processing of the DNA ends, and it can lead to both error-free and error-prone repair1. In both NHEJ and homologous recombination, chromatin is implicated in regulating DSB accessibility and in an additional role as a regulatory platform that helps to coordinate and to integrate the overall complex DDR pathway.
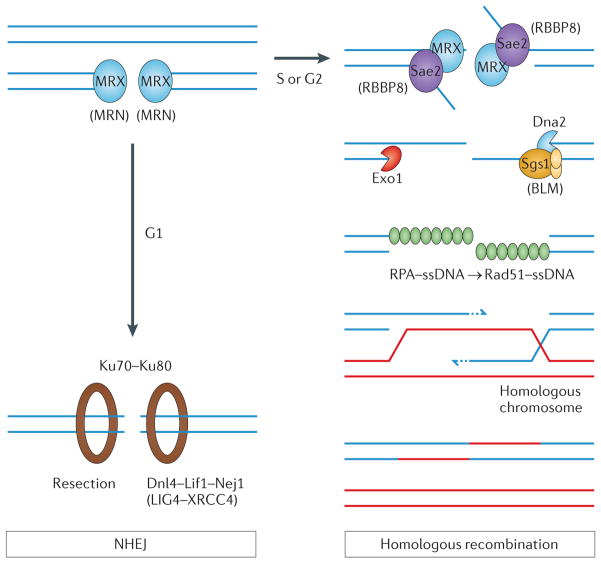
Figure 2. Two primary pathways for double-strand break repair.
Following its formation, a double-strand break (DSB) is initially bound by the yeast Mre11–Rad50–Xrs2 (MRX) or human MRE11–RAD50–NBS1 (MRN) complex. If a DSB is formed in the G1 phase of the cell cycle, the DSB is preferentially repaired by the non-homologous end-joining (NHEJ) pathway (left panel). In this case, binding of the Ku heterodimer (Ku70–Ku80) prevents extensive nucleolytic processing of the DSB and promotes subsequent ligation of the DSB by the Dnl4–Lif1–Nej1 complex in budding yeast and the LIG4–XRCC4 factors in mammals. If a DSB is formed in the S or G2 cell cycle phase, the homologous recombination repair pathway predominates (right panel). Key steps for successful repair by homologous recombination include: nucleolytic processing of the 5′ ends of the dsDNA ends into 3′ single-stranded tails by the initial action of the yeast MRX complex and Sae2 enzyme (known as RBBP8 or CTIP in mammals); extensive processing by either the exodeoxyribonuclease 1 (Exo1) or the Dna2–Sgs1 DNA-end-processing enzymes (the human Bloom’s syndrome protein (BLM) helicase is the homologue of yeast Sgs1); assembly of a recombination protein A (RPA)–ssDNA filament that is subsequently converted into a Rad51–ssDNA filament; completion of a successful homology search and formation of heteroduplex DNA; DNA synthesis that uses the 3′ end of the broken DNA, resolution of the heteroduplex or a double-sided Holliday junction; and, finally, ligation of the ssDNA nicks and termination of the process. Note that formation of the RPA–ssDNA filament is also essential for activation of the DNA damage cell cycle checkpoint. For a recent Review, see REF. 2. Protein or complex names given in brackets in the figure are the human homologues.
Initiating DSB repair in chromatin
Early studies of the role of chromatin in DNA repair led to the ‘access, repair and restore’ model5. This model was based in part on precedents from the transcription field and suggested that chromatin primarily regulates the initial accessibility of the lesion to the repair machinery. Indeed, a large number of the ATP-dependent chromatin-remodelling enzymes that are involved in transcription, as well as histone-modifying enzymes, are recruited to a single DSB soon after their formation (TABLE 1). However, the chromatin rearrangements taking place in response to DNA damage are still not clear, as few studies have demonstrated roles for chromatin regulators in actual chromatin-remodelling events at DSBs. Even the extent of nucleosome loss that occurs within chromatin flanking a DSB is currently under debate (BOX 1). Nonetheless, the RSC chromatin-remodelling complex has been shown to be rapidly recruited to a DSB in Saccharomyces cerevisiae, and this enzyme catalyses the timely eviction or sliding of a few nucleosomes adjacent to the lesion6,7. Similarly, localized disruption of nucleosomes has also been observed in G1-arrested human cells8, supporting an evolutionarily conserved process. In addition, the S. cerevisiae INO80 chromatin-remodelling enzyme has been implicated in nucleosome loss events that occur during the later DNA-processing events of homologous recombination9.
Box 1. How much nucleosome eviction occurs in chromatin surrounding a double-strand break?
It is widely believed that nucleosomes are rapidly removed from chromatin that surrounds a double-strand break (DSB), especially following extensive nucleolytic processing of DSBs for homologous recombination. Many chromatin immunoprecipitation (ChIP) studies in yeast have monitored histone loss following the induction of a DSB by the homothallic switching endonuclease (HO). In this case, ChIP studies are carried out in asynchronous cultures at 1–4 hours following DSB formation, time points at which 4–16 kb of ssDNA are formed adjacent to the DSB130. Many studies have shown large decreases in histone levels directly adjacent to an HO-endonuclease-induced DSB, of <500 bp in size, which is consistent with the loss of one or two nucleosomes. Likewise, another study examined histone loss at I-PpoI-induced DSBs in G1-arrested mammalian cells and observed reduced histone levels at 280 bp distal from a DSB8. However, most studies have reported only modest reductions (typically twofold or less) when histone levels are probed at more distal locations, of >500 bp in size, and at time points when kilobases of ssDNA have been formed74,131 (M.P.-C., C.L.P, unpublished observations; for a discussion, see REF. 29). ChIP results between different groups vary for unknown reasons. For instance, one group reported no significant histone loss at an HO-induced DSB at the Saccharomyces cerevisiae mating-type locus MAT when ChIP results were normalized to the input locus74. By contrast, another group reported that histone H3 levels decreased approximately threefold at sites within ~1 kb of the HO-induced DSB, although no decrease was seen at sites ~5 kb distal to this18. Interestingly, this same study reported only a twofold decrease when HO formed a DSB at the S. cerevisiae PDR10 locus18. Recent studies in mammalian cells indicate that nucleosomes are retained at chromatin surrounding a DSB induced by a zinc finger nuclease20, although the extent of DSB processing was not assessed in this work. By comparison, nucleosome loss at gene promoters is commonly associated with a dramatic 5–10-fold decrease in histone levels when assayed by ChIP132,133. RPA, replication protein A.
The functional importance of these chromatin-remodelling events is less clear. In one case in S. cerevisiae, RSC was reported to have a key role in the recruitment of early damage responders, such as the yeast MRX complex (a heterotrimeric complex comprising Mre11, Rad50 and Xrs2) and the Ku heterodimer (that is, Ku70–Ku80)7 (FIG. 3). Additionally, RSC was reported to be important for NHEJ and for timely completion of early steps of the homologous recombination process10. By contrast, another study reported that loss of RSC actually leads to higher levels of NHEJ, to no defect in early steps of homologous recombination and to a surprising defect in the final, ligation step of homologous recombination11. The reasons behind these differing results are unclear. Likewise, chromatin remodelling by the budding yeast INO80 complex does not appear to have an impact on the efficiency or timing of homologous recombination12, even though INO80 promotes the kinetics of DSB processing, and artificial tethering of the yeast INO80 can promote homologous recombination13,14. Likewise, the yeast Fun30 chromatin-remodelling enzyme is required for efficient DSB processing, but this activity appears to be dispensable for the normal kinetics of homologous recombination15,16. By contrast, the Arabidopsis thaliana homologue of INO80 is required for efficient homologous recombination, at least with a chromosomally integrated, inverted repeat luciferase recombination reporter17.
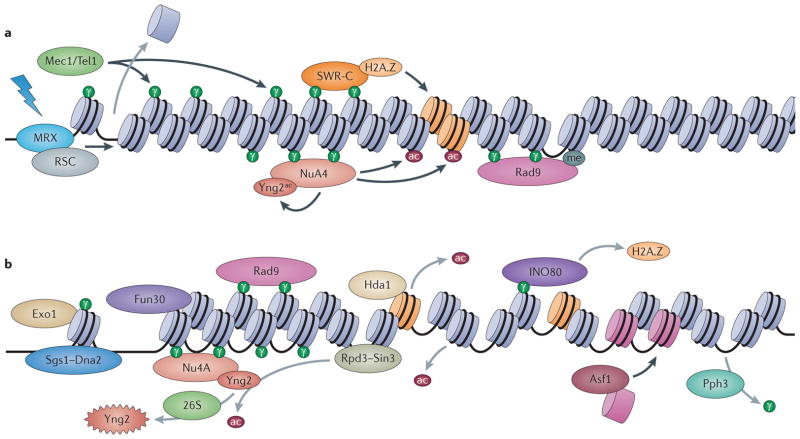
Figure 3. Chromatin dynamics in double-strand break checkpoint response in S. cerevisiae.
a | Formation of a double-strand break (DSB; shown by lightning bolt), initial chromatin alterations and checkpoint activation. Recognition of the broken ends by the Mre11–Rad50–Xrs2 (MRX) complex initiates DNA end processing. The MRX complex recruits the RSC chromatin-remodelling complex, which restructures chromatin at the DSB ends; checkpoint kinases Mec1 and Tel1 (‘Mec1/Tec1’ in the figure) phosphorylate the histone variant H2A.X, leading to the NuA4 complex and SWR complex (SWR-C) recruitment. SWR-C incorporates H2A.Z, which can lead to relocalization of the DSB to the nuclear periphery; NuA4 is self-activated by acetylating its subunit Yng2 (depicted as Yng2ac in the figure) a nd targets histones H2A.Z and H4 for acetylation (abbreviated to ‘ac’ in the figure). Rad9 recruitment is stabilized by its interactions with H2A.X phosphorylated at serine 139 (H2A.XS139ph; known as γH2A.X and abbreviated to ‘γ’ in the figure) and H3 trimethylated at 79 (H3K79me3; abbreviated to ‘me’ in the figure), leading to checkpoint activation. b | The DSB is processed by the Sgs1–Dna2 and exodeoxyribonuclease 1 (Exo1) pathways and is assisted by the chromatin-remodelling complex Fun30, which appears to counteract the inhibitory effect of Rad9 on DNA resection. Initial steps for removing epigenetic marks are also shown: histone deacetylase 1 (Hda1) deacetylates H2A.Z, and Rpd3–Sin3 deacetylates H4 and Yng2. Inactivation of NuA4 occurs by targeted degradation of Yng2 by the 26S proteasome (the degraded Yng2 is represented by the jagged oval). INO80 recruitment leads to eviction of H2A.Z, facilitating checkpoint termination and adaptation. Restoration of chromatin structure is mediated in part by Asf1, which deposits new histones and serine/threonine protein phosphatase 4 catalytic subunit (Pph3) dephosphorylates H2A.X.
In addition to nucleosome loss or mobilization, the dynamic incorporation of histone variants may contribute to the ‘access’ paradigm. The conserved mammalian SWR complex (SWR-C) is recruited to a DSB in yeast18, where it promotes incorporation of the histone variant H2A.Z19 (FIG. 3). (Note that although this complex has many names in yeast, including SWR1, in this Review we use the term SWR-C for the complex in yeast as well as mammals to distinguish it from its catalytic subunit Swr1.) Incorporation of yeast H2A.Z within DSB chromatin promotes processing of DNA ends19, and likewise p400 (also known as EP400), which is the mammalian homologue of yeast Swr1, has been reported to promote nucleosome destabilization in chromatin that is adjacent to an endonuclease-induced DSB20. Thus, incorporation of histone variants appears to support the ‘access’ model.
Several studies indicate that initiation of DSB repair is also associated with unfolding of higher-order, condensed chromatin structures21,22. The mammalian RING finger E3 ubiquitin ligase heterodimer RNF20–RNF40 is rapidly recruited to DSBs, and this enzyme catalyses ubiquitylation of histone H2B at lysine 120 (H2BK120) at DSB chromatin22 (FIG. 4). The H2BK120ub mark disrupts higher-order folding of nucleosomal arrays in vitro23. Interestingly, H2BK120ub appears to regulate recruitment of the chromatin-remodelling enzyme SNF2H (also known as SMARCA5) to DSBs, and SNF2H is required for optimal NHEJ and homologous recombination22,24,25. This suggests that H2BK120ub may promote a more complicated version of the ‘access’ paradigm, controlling local chromatin decompaction. Indeed, chemical-induced relaxation of bulk chromatin appears to alleviate the requirement for RNF20 in DNA repair, supporting the decondensation model22. This model is particularly compelling, as chromatin condensation appears to have an impact on both NHEJ and homologous recombination, as H2BK120ub is required for efficient DSB repair by both pathways22,26
Figure 4. Chromatin dynamics in checkpoint signalling in mammalian cells.
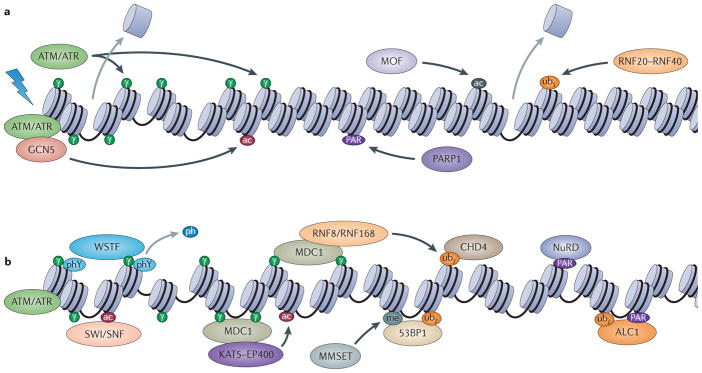
a | DSB formation and initial histone modifications. Ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and RAD3-related (ATR) checkpoint kinases (‘ATM/ATR’ in the figure) phosphorylate the histone variant H2A.X at serine 139 (H2A.XS139ph; termed γH2A.X and depicted as ‘γ’ in the figure) and promote nucleosome eviction. ATM recruits GCN5, which catalyses H3 acetylation (the red ‘ac’ in the figure). Recruitment of MOF catalyses acetylation of histone H4 at lysine 16 (H4K16; the black ‘ac’ in the figure), leading to chromatin decompaction; poly(ADP-ribose) polymerase 1 (PARP1) action leads to PARylation of proteins and histones (‘PAR’). The RING finger heterodimer RNF20–RNF40 ubiquitylates histone H2BK120, which also promotes decompaction (‘ub1’). b | Chromatin decondensation induced by the histone modifications catalysed in panel a leads to subsequent recruitment of WSTF, which dephosphorylates H2A.X at tyrosine 142 (H2A.XY142; phY→ph), leading to mediator of DNA damage checkpoint 1 (MDC1) recruitment by γH2A.X. The SWI/SNF complex is recruited by histone H3 acetylation, and its chromatin-remodelling activity facilitates further spreading of γH2A.X and stabilization of MDC1 on chromatin. MDC1 in turn recruits the KAT5–p400 enzyme, which is activated by trimethylation of H3 at lysine 9 (H3K9; not shown), leading to further histone acetylation, and also recruits RNF8 and RNF168 (‘RNF8/RNF168’ in the figure), which ubiquitylate H2A and/or other proteins (‘ub2’). Ubiquitylation by RNF8 and RNF168 recruits chromodomain helicase DNA-binding protein 4 (CHD4). Histone PARylation (and/or modification by other proteins) leads to recruitment of the nucleosome-remodelling and histone deacetylase (NuRD) complex. Histone H4 is further modified by MMSET-mediated methylation (H4K20me2 or H4K20me3; depicted as ‘me’ in the figure). The combination of ubiquitylation by RNF8 and RNF168 with either H4K20me or PAR-ylation recruits tumour suppressor p53-binding protein 1 (53BP1) or ALC1, respectively.
Chromatin in the later stages of the homologous recombination process
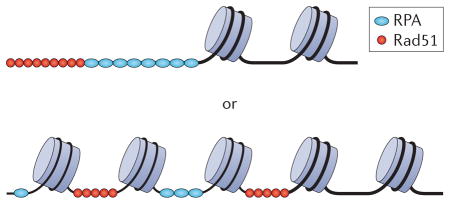
After a DSB has been processed for homologous recombination, the binding of the DNA repair protein Rad51 to ssDNA forms a nucleoprotein filament that carries out a search for a homologous DNA duplex that can be used as a template for repair (FIG. 2). Early studies in both budding yeast and mammalian systems suggested that a homologous duplex could be captured even on an ectopic chromosome, implying that the search might extend to the entire genome if a sister chromatid was not available27,28. Remarkably, in vitro studies with recombinant chromatin fibres and purified budding yeast or human RAD51 demonstrated that a homology search can be completed when the homologous sequence is positioned on the surface of a nucleosome, even in the absence of histone modifications or ATP-dependent remodelling29. Subsequently, however, it was found that the budding yeast SWI/SNF chromatin-remodelling complex can enhance the homology search step but only when the homologous duplex is embedded in a condensed heterochromatin structure11,30.
In the cell nucleus, chromosomes harbour additional layers of higher-order structure, leading to chromosomal domains or more general spatial constraints. Live-cell imaging studies in S. cerevisiae have shown that before DSB formation, a chromosomal locus samples only ~10% of the nuclear volume. However, following formation of an endonuclease-induced DSB in S or G2 cells, the same chromosomal locus moves throughout nearly 40% of the nuclear volume, promoting capture of homology on another chromosome31,32. Surprisingly, formation of a DSB on one chromosome appears to induce an increased mobility of other chromosomes, even when those chromosomes lack homology31. The mechanism by which the entire genome is mobilized by the formation of a single DSB and how much of this phenomenon is evolutionarily conserved is not clear. Studies in mammalian cells have suggested that DNA breaks are actively restricted from moving in the nuclear space21,33, although this may reflect a preference for repair by NHEJ in somatic cells. However, recent observations in yeast may explain earlier studies in mammalian cells that demonstrated a disruption of bulk chromatin structure during DDR8 as well as that showed nuclear repositioning and pairing of the homologous immunoglobulin loci during V(D)J recombination mediated by the DDR machinery34. The dynamic movements of DSBs in yeast nuclei also share interesting similarities to the behaviour of DSBs induced within Drosophila melanogaster heterochromatin35 and the repetitive ribosomal DNA locus of budding yeast36.
It seems likely that the enhanced mobility of DSB chromatin will be highly regulated so as to prevent promiscuous movement of chromosomes that might lead to aberrant recombination events. DSB-induced movements in S. cerevisiae and D. melanogaster heterochromatin require both the exonucleolytic processing of the DSB as well as the Rad51 recombinase, suggesting that formation of a Rad51–ssDNA filament is required for these large-scale movements31,32,35,36. One intriguing model suggests that this nucleoprotein filament directs the recruitment of ATP-dependent chromatin-remodelling enzymes that have a key role in promoting chromosome mobility. Indeed, a recent study found that tethering subunits of the S. cerevisiae INO80 chromatin-remodelling complex to a chromosomal locus is sufficient to enhance nuclear mobility13. Furthermore, inactivation of the Arp8 subunit of INO80 limits the mobility of an endonuclease-induced DSB. On the basis of this evidence, a simple model was suggested in which ATP-dependent nucleosome eviction might catalyse chromosome mobilization. However, tethering other chromatin-remodelling enzymes, such as the SWI/SNF complex, does not seem to share this activity with INO80. These authors also suggest an intriguing possibility that INO80 may disrupt a chromosomal anchor, facilitating DSB mobilization13. Perhaps the orchestration of chromosome movements provides an explanation for why so many chromatin regulators are recruited to a DSB.
A chromatin platform for signal integration
Histone modifications, DSB repair and checkpoint signalling
In addition to the actual repair process, survival after DNA damage requires activation of the DNA damage checkpoint pathway, which halts the cell cycle and coordinates repair of damage with cell cycle transitions. Key initial components of the checkpoint-signalling pathway in humans are two members of the phosphoinositide-3-kinase-related protein kinase (PIKK) family of enzymes, ataxia telangiectasia mutated (ATM; known as Tel1 in S. cerevisiae) and ataxia telangiectasia and Rad3-related (ATR; known as Mec1 in S. cerevisiae)37. ATM binds to unprocessed or minimally processed ends of DSBs and is activated by the Mre11–Rad50–Nbs1 (MRN) complex38. However, recruitment of the human ATR–ATRIP complex (where ATRIP is ATR-interacting protein; in S. cerevisiae, this complex is known as Mec1–Ddc2) at damaged DNA requires extensive ssDNA formation and binding of the single-stranded binding complex replication protein A (RPA). RPA interacts with ATRIP, leading to recruitment of the ATR–ATRIP checkpoint kinase complex at the damaged site38,39. The activation of checkpoint kinases triggers a phosphorylation cascade that has an impact on, among other factors, the human proteins tumour suppressor p53-binding protein 1 (53BP1; in S. cerevisiae, this is known as Rad9) and mediator of DNA damage checkpoint 1 (MDC1). These factors ultimately activate the key transducers checkpoint kinase 2 (CHK2; in S. cerevisiae, this is known as Rad53) and CHK1. The CHK proteins are essential for dispersing the signal to a multitude of downstream targets, arresting the cell cycle and integrating the DNA damage signal with cell metabolism40.
During the past few years, it has been clear that an efficient checkpoint response in the nucleus induces a host of histone post-translational modifications (TABLE 1), implicating the presence of a regulatory chromatin platform. One of the most intensively studied DSB-induced histone modifications is the histone variant H2A.X phosphorylated at serine 139 (H2A.XS139ph; termed γH2A.X), which is the major form of H2A in budding and fission yeast and appears to be a constitutive component of mammalian chromatin that is harboured by ~10% of nucleosomes41. Phosphorylation of H2A.XS139 by the DDR kinases ATM, ATR and DNA-dependent protein kinase (DNAPK) is one of the earliest events at a DSB, and this mark spreads over at least a megabase of chromatin adjacent to each DSB in mammalian cells (FIG. 4) and up to 50 kb on each side of a DSB in yeast (FIG. 3). Recently, the chromatin-remodelling activity of the human SWI/SNF complex has been implicated in facilitating the formation and maintenance of high levels of γH2A.X42, presumably by enhancing nucleosomal accessibility. This large domain of γH2A.X provides a host of binding sites for the key checkpoint factor MDC1, stabilizing its association with DSB chromatin and thus helping to promote a robust checkpoint response. As discussed above (BOX 1), this chromatin platform may also contain histone–ssDNA complexes and may be intermingled with RPA and RAD51 filaments.
Formation of a γH2A.X domain might not always be sufficient for stable association of MDC1. Disruption of higher-order folding by H4 acetylated at K16 (H4K16ac), a histone mark that unfolds condensed nucleosomal arrays in vivo and in vitro43, has also been suggested to regulate binding of MDC1 to γH2A.X domains (FIG. 4). Conditional knockout of the human histone acetyltransferase MOF (also known as KAT8), which catalyses acetylation of histone H4K16, was found to abrogate MDC1 recruitment44, although a different study using RNA interference to deplete MOF from human cells did not reach the same conclusion45. A recent report shows that H4K16 acetylation undergoes a biphasic response at DNA damage sites, being reduced at early time points by the concerted function of histone deacetylase 1 (HDAC1) and HDAC2 but being increased at later times46. As HDAC1 and HDAC2 promote NHEJ46, these results suggest that early chromatin silencing and compaction might be important in blocking extensive DNA end processing, whereas later relaxation of higher-order chromatin structure may contribute to recruitment of MDC1 to DNA damage foci and to checkpoint regulation.
In addition to its role in checkpoint signalling47,48 and in promoting efficient homologous recombination between sister chromatids49, γH2A.X has recently been shown to have a role in protecting the integrity of DNA ends during V(D)J recombination in B lymphocytes50. Recombination-activating gene 1 (RAG1)- and RAG2-mediated DSBs exclusively occur in the G1 phase of lymphocytes, and these DSBs must be repaired by NHEJ to promote correct V(D)J recombination. In G1-arrested lymphocytes, γH2A.X and MDC1 shield the DNA ends from the DSB-processing enzyme RBBP8 (also known as CTIP), preventing aberrant homologous recombination that can lead to subsequent chromosomal translocations50. This report supports earlier findings showing that H2ax−/− mice are defective in immunoglobulin class-switch recombination51. As mice that lack H2A.X do not exhibit a defect in irradiation-induced cell cycle checkpoints51, these findings suggest that the immune cell function of γH2A.X and MDC1 is likely to be independent of their roles in G1 checkpoint signalling47,48. Likewise, a recent study suggests that γH2A.X and Rad9 also limit DSB processing in S. cerevisiae15. Cumulatively, evidence supports a context-specific role for γH2A.X.
Other histone modifications and histone-modifying enzymes have elaborate roles in the promotion of the MDC1-mediated checkpoint pathway beyond initial recruitment of MDC1 (FIG. 4). MDC1 binds and recruits the E3 ubiquitin ligase RNF8 to DSB chromatin52–54, and RNF8 functions in concert with a second ubiquitin ligase, RNF168, to promote assembly of checkpoint regulators 53BP1 and breast cancer 1, early onset (BRCA1) at damaged chromatin52–56. A recent study suggests that RNF8 may work in part by directing the recruitment of the nucleosome remodelling and histone deacetylase (NuRD) complex (which contains CHD4)57, the recruitment of which also depends on poly(ADP-ribose) polymerase 1 (PARP1)58. Furthermore, recruitment of 53BP1 to DSB chromatin appears to require H4 dimethylated at K20 (H4K20me2)59–61. H4K20me2 is a constitutive histone mark that is increased at laser-irradiation-induced foci and induced at single DSBs62, in contrast to previous reports that did not detect an increase of H4K20me2 signal following DNA damage61. The enzyme responsible for catalysing the deposition of the H4K20me2 and H4K20me3 marks at the site of damage is not known, although downregulation of histone–lysine N-methyltransferase MMSET (also known as NSD2) leads to loss of the H4K20me2 and H4K20me3 DNA-damage-induced marks62. Given that H4K20me1, H4K20me2 and H4K20me3 are histone marks that are associated with transcriptional regulation, an intriguing possibility is that the dual function of MMSET and RNF8 at DSB sites is a part of a genome stability pattern that is essential for the cell to differentiate between transcriptional and DNA damage responses, thereby preventing aberrant chromosomal recruitment of 53BP1 to transcription sites.
Histone modifications integrating DSB repair and apoptosis
In response to genotoxic stress, cells must decide between apoptotic death or repairing the damage and surviving63. Two recent reports in mammalian cells link phosphorylation dynamics of H2A.X at tyrosine 142 (H2A.XY142) in the decision-making process between DNA repair and programmed cell death. H2A.XY142 phosphorylation by WSTF (also known as BAZB1) is a constitutive histone mark that is required for efficient and persistent γH2A.X phosphorylation under DNA damage conditions64,65. Intriguingly, dephosphorylation of H2A.XY142 during the DNA damage response is a requirement for both the activation of the checkpoint response and silencing of the apoptotic death pathway64,65. How the dynamics of H2A.XY142 phosphorylation is controlled is not yet clear, although a recent study implicates the early damage-response protein microcephalin1 (MCPH1)66. These reports bring strong evidence to the notion that post-translational modifications can turn H2A.X into a molecular switch that regulates important cellular decisions.
Histone modifications, repair and transcription
DNA damage can have a direct impact on transcription in cis, and induction of a single DSB in the vicinity of an active promoter in mammalian cells leads to RNA polymerase stalling and ATM-dependent transcriptional silencing67. Silencing requires RNF8- and RNF168-mediated ubiquitylation of H2AK119 (REF. 67), which is a histone mark that has been associated with transcriptional repression68. Whether DSB-induced transcriptional silencing is a molecular mechanism with a defined purpose or whether it is only a consequence of the DSB repair process is still not clear. Interestingly, on completion of DNA repair, transcriptional activity resumes rapidly67. Whether a promoter remains in a poised state for activation with all of the positive histone marks present and transcription cofactors recruited or whether histone marks that mark DSB sites assist in the re-initiation of transcription is an open question. Interestingly, γH2A.X spreading is reciprocally influenced by gene transcription69, suggesting a crosstalk between repair and transcription in mammalian cells.
Histone variants linking DSB repair and checkpoints
Although much of the DNA repair field has focused on histone modifications in the checkpoint response, it is clear not only that the dynamic incorporation of histone variants within DSB chromatin has an impact on the early steps of NHEJ and homologous recombination (see above) but also that specific histone variants are associated with later checkpoint events (FIG. 3). Early during the DDR in budding yeast, the H2A.Z variant is incorporated by SWR-C, replacing either γH2A.X or H2A12,19. Although SWR-C remains associated with DSB chromatin, the presence of H2A.Z at the DSB is only transient18,19: the subsequent recruitment of the INO80 complex is believed to reverse SWR-C action by exchanging nucleosomal H2A.Z–H2B dimers with free H2A.X–H2B12,70 (FIG. 3). H2A.Z dynamics appears to be an important control switch during the DDR pathway — if a DSB is not repaired in a timely fashion, a sumoylated form of H2A.Z has been proposed to mediate anchoring of the DSB to the nuclear periphery, whereas the removal of H2A.Z by INO80 promotes proper checkpoint signalling by an unknown mechanism12,19.
The final steps of repair
Repair of the DSB, either by NHEJ or homologous recombination, signals the return of the cell to homeostasis. DNA repair factors and chromatin regulators are removed from the site of repair, the checkpoint response is inactivated and the epigenetic landscape is returned to its original state. Several studies have shown that γH2A.X is targeted for dephosphorylation following repair in both yeast and mammalian cells, and its removal is key for downregulating the DNA damage checkpoint response71–73. The final steps of repair also lead to assembly of new nucleosomes that contain the histone mark H3K56ac, which is required for inactivation of the DNA damage checkpoint74. H3K56ac is known to stimulate histone turnover at gene promoter regions, perhaps providing a molecular explanation for why newly repaired chromatin is more labile20.
Chromatin and the fidelity of DNA replication
Throughout this Review, we have focused our discussion on the DDR pathway during G1 or G2 cells, but what happens when DNA damage occurs in S phase? Replication of an ssDNA nick will directly generate a DSB. Moreover, replication fork progression can be stalled by encounters with small or bulky DNA base adducts, DNA-crosslinking agents or by depletion of nucleotide pools, and this can lead to DSBs. There are natural replication fork barriers, such as repetitive DNA sequences, and stable protein–DNA chromosomal structures, such as heterochromatinized domains or actively transcribed sequences, that can lead to topological alterations and formation of secondary DNA structures in the context of chromatin. These have to be resolved before the cell can enter mitosis2.
Histone modifications and the intra-S-phase checkpoint
The intra-S-phase checkpoint induced by fork stalling serves to block further replication initiation events, to slow S phase progression, to regulate transcription of genes and to keep the stalled replication forks in a competent state, thereby promoting a rapid restart following repair75,76. An early step in the intra-S-phase checkpoint in humans and S. cerevisiae is the activation of the ATR checkpoint kinase, which generates a chromatin domain of γH2A.X77. The γH2A.X does not appear to spread to the great extents seen for DSBs in G1 or G2 cells. In addition, in S. cerevisiae, histone H3 at threonine 45 (H3T45) appears to be exclusively phosphorylated inside S phase by the histone kinase complex Cdc7–Dbf4 (where Cdc7 is cell division control protein 7, and Dbf4 is dumb-bell-forming protein 4), promoting DNA replication under replication stress conditions78. Currently, it is unclear how phosphorylation of H3T45 integrates with the intra-S-phase checkpoint. In an interesting parallel with the checkpoint response, chromatin dynamics in S. cerevisiae also appears to be involved in pathway choice of replication-coupled DNA damage repair. Trimethylation of H3K79 by disrupter of telomere silencing protein 1 (Dot1) inhibits the error-prone translesion synthesis (TLS) pathway79,80 most probably by recruiting Rad9 (REF. 81), which is a negative regulator of TLS82. Thus, the reported data point to the direction that histone modifications, as in the DDR pathway, may have a similar role in integrating signals into replication-coupled, genome stability networks.
Chromatin in fork progression and genome stability
The process of nucleosome assembly plays a key part in genome stability during DNA replication by regulating fork stability and progression. Histone chaperones such as the CAF1 complex and anti-silencing function protein 1 (Asf1) have been shown to be crucial in the establishment of nascent chromatin and therefore in the maintenance of genome stability during DNA replication. They may function either by depositing newly synthesized histones83 onto the daughter DNA strands or by transferring parental histones from parental to nascent strands84. Nucleosome assembly also appears to be coupled to Okazaki fragment maturation, suggesting a novel level of regulation by chromatin85. Chaperone function is regulated by several histone modifications, including acetylation of H3K56 by the Rtt109 acetyltransferase and acetylation of H3 amino-terminal lysines by Gcn5, both of which occur in S. cerevisiae83,86. Although the function of chaperones inside S phase has been extensively discussed elsewhere87, one major theme that has arisen from these studies is the idea that nucleosome assembly on the daughter strands can have a major impact on both fork progression and the stability of stalled forks.
In S. cerevisiae, the INO80 chromatin-remodelling enzyme is associated with the replication fork, and stalling of a fork leads to additional recruitment of INO80 (REF. 88). In the absence of INO80, fork progression is slowed but, more importantly, forks that stall because of replication stress are destabilized and collapse, leading to an inability to restart forks and a loss of cell viability89,90. In an ino80 mutant, the H2A.Z histone variant accumulates to aberrant levels, and genetic studies indicate that this misincoporated H2A.Z destabilizes stalled forks, leading to fork collapse70. Although it is unknown whether INO80 remodels parental or newly deposited H2A.Z-containing nucleosomes, we favour a model in which INO80 moves with the fork, evicting improperly deposited H2A.Z (FIG. 5). Interestingly, the misincorporated H2A.Z is hypoacetylated, and expression of a version of H2A.Z that mimics a hyperacetylated state alleviates many of the phenotypes of an ino80 mutant70. Depletion of human INO80 also impairs DNA synthesis in mammalian cells, suggesting a highly conserved role for INO80 during DNA replication91. Exactly how INO80 stabilizes stalled forks and why hypoacetylated H2A.Z is detrimental to fork progression is unclear. The DNA damage tolerance pathway has also been suggested to be compromised in the absence of yeast INO80 (REF. 92), although this may simply be due to extensive fork collapse, which occurs in the absence of INO80, and to the inability of the fork to retain replisome subunits under stress conditions89.
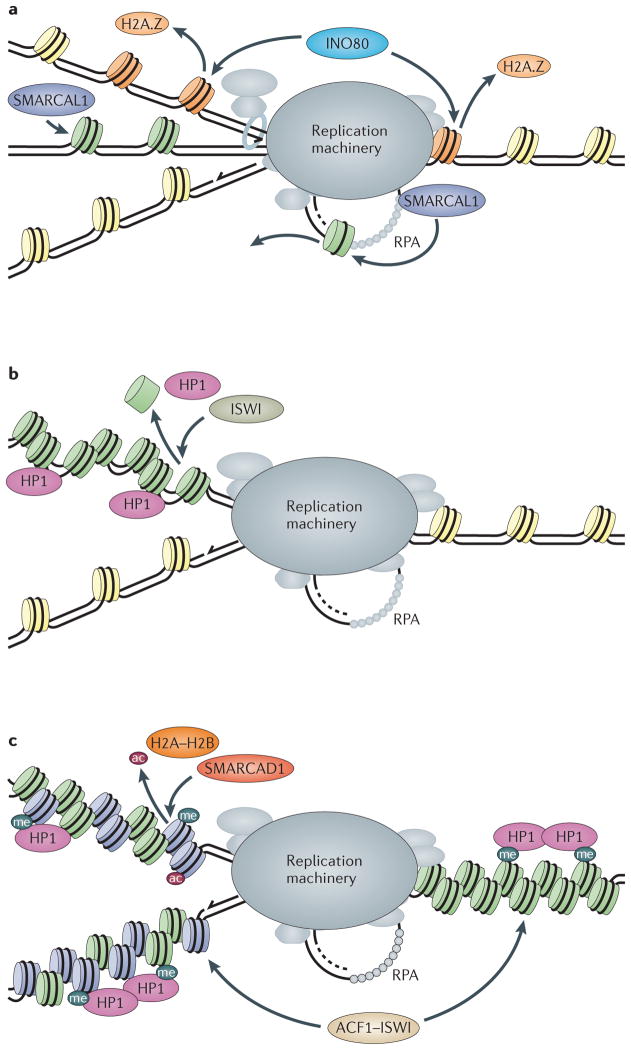
Figure 5. Chromatin-remodelling activities at the replication fork.
a | INO80 may remove H2A.Z–H2B dimers (‘H2A.Z’ in the figure) from nucleosomes (H2A.Z nucleosomes depicted in orange) ahead of the fork or from newly formed nucleosomes on replicated DNA (orange nucleosomes). SMARCA-like protein 1 (SMARCAL1) catalyses fork regression and Holliday junction migration and may remove nucleosomes from regressed dsDNA or from ssDNA. b | The ISWI complex may protect newly replicated DNA from unwarranted heterochromatinization (green nucleosomes) and aberrantly recruited heterochromatin protein 1 (HP1) by repositioning nucleosomes behind the fork. c | SMARCA-containing DEAD/H box 1 (SMARCAD1) may protect heterochromatin (depicted as HP1-associated nucleosomes) during replication by deacetylating newly synthesized histones (purple nucleosomes), promoting histone H3 trimethylated at lysine 9 (H3K9me3; ‘me’ in the figure) and exchanging H2A–H2B histone dimers. Human ACF1–ISWI complex promotes replication of late-replicating heterochromatin regions, which act either in front or behind the fork. RPA, replication protein A.
In mammalian cells, a putative chromatin-remodelling enzyme called SMARCA-like protein 1 (SMARCAL1; also known as HARP), which is responsible for Schimke’s immuno-osseous dysplasia93, also appears to stabilize stalled forks94–97. SMARCAL1 has annealing helicase activity, interacts with RPA94,97,98 and could act in various stages of DNA synthesis by limiting the amount of ssDNA at the fork99,100. Whether SMARCAL1 has ATP-dependent chromatin-remodelling activity is not yet known; however, this is highly possible given its close relation to other ATP-dependent chromatin-remodelling enzymes. One hypothesis is that SMARCAL1 might promote DNA replication by removing unproductive histone–ssDNA intermediates from the fork, combining its annealing and putative chromatin-remodelling activities (FIG. 5a).
Chromatin regulators, genome integrity and heterochromatin
A major source of genome instability arises from illicit recombination between the vast amounts of repetitive DNA found within most eukaryotic genomes. Such events are inhibited by the packaging of repetitive DNA into condensed heterochromatic structures. During S phase, these heterochromatic structures must be faithfully replicated to ensure genome stability. Notably, several chromatin-remodelling enzymes have been implicated in the efficient replication of heterochromatin domains. For instance, both S. cerevisiae Isw2 and its mammalian homologue SNF2H have been implicated in facilitating DNA replication through late replicating regions and within heterochromatin, respectively101,102. In addition, SNF2H also appears to protect newly replicated DNA from heterochromatinization during S phase as a part of a complex with the WSTF protein103. However, given the preferred function of the ISWI family members to deposit and to reposition nucleosomes across the genome104,105, it remains unclear whether members of the ISWI family disrupt nucleosomes in front of the fork or whether they control nucleosome assembly on the daughter chromatids.
In human cells, the SMARCA-containing DEAD/H box 1 (SMARCAD1) chromatin-remodelling enzyme is recruited to replication foci by the proliferating cell nuclear antigen (PCNA) sliding clamp, and it facilitates the maintenance of heterochromatin inside S phase106. SMARCAD1 appears to function by promoting removal of acetyl marks on newly synthesized and deposited H3 and H4 histones106 and promoting the resultant H3K9me3 mark, which is essential for binding of heterochromatin protein (HP1) and transcription intermediary factor 1β (TIF1β; also known as KAP1). Interestingly, the Schizosaccharomyces pombe SMARCAD1 homologue, Fft3, also protects silent chromatin domains from euchromatic assembly, suggesting an evolutionarily conserved function107.
Chromosomal stability in mitosis
Repair of DNA damage or accurate DNA replication would be meaningless if chromosomes were not properly segregated and equally inherited during cell division. Defects in chromosome segregation increase the rate of chromosomal instability (CIN) during mitosis, leading to aneuploidy. Similarly, CIN can lead to failure of cytokinesis owing to unresolved sister chromatids or to the presence of chromatin bridges at the intercellular canal that connects the two daughter cells, resulting in polyploidy108.
Specialized chromatin structures and histone modifications — mainly phosphorylation events — have long been known to be crucial for chromosome segregation and CIN prevention. For example, CenH3 is the centromere-specific H3-type histone variant that is essential for the assembly of kinetochores109,110. The histone variant H2A.Z is also found to be enriched at kinetochores and promotes chromosome segregation, albeit with an unknown mechanism111,112. Finally, phosphorylation of H3 at serine 10 (H3S10) and of H3S28 by aurora kinase B (AURKB) is important for chromosome condensation during mitosis in mammals113.
Recently, it was shown in S. pombe that phosphorylation of H2AS121 by Bub1 recruits the shugoshin proteins Sgo1 and Sgo2 to centromeres114. Sgo1 protects sister chromatid cohesion, and Sgo2 directs recruitment of the serine/theronine protein kinase Ark1 (S. pombe homologue of AURKB), which is a subunit of the chromosomal passenger complex (CPC) that regulates the spindle assembly checkpoint and microtubule– kinetochore attachment. Three independent groups have shown that phosphorylation of histone H3 at tyrosine 3 (H3T3ph) by haspin is an additional mark that promotes AURKB localization at human centromeres. The H3T3ph mark is recognized by the CPC subunit survivin115, and it cooperates with H2AS121ph to recruit the CPC to the centromere116. Interestingly, phosphorylation of condensin by Ark1 during mitosis leads to interaction of condensin with histones H2A and H2A.Z as well as to its recruitment to kinetochores and chromosome arms117. Whether H2A and H2A.Z differentially regulate the binding of condensin is still an open question. Cumulatively, these studies establish chromatin as an essential regulatory structure for the establishment of the mitotic apparatus and hence for the maintenance of genome stability throughout division.
The ATP-dependent chromatin-remodelling enzymes RSC118, INO80 (REF. 119) and SWI/SNF120 have also been shown to be recruited to centromeres and have been implicated in chromosome segregation in S. cerevisiae, although the molecular mechanism of their function at the centromere is unknown. Moreover, S. cerevisiae SWI/SNF also appears to have a role in removing the centromere-specific histone H3-like protein Cse4 from non-centromeric sites121. Interestingly, human INO80 was also found to localize at the midbody91, and depletion of INO80 or some subunits of RSC leads to polyploidy in yeast91,122,123. As cytokinesis is tightly coordinated with completion of chromosome segregation124,125, these observations suggest a function for INO80 during cytokinesis, possibly coordinating chromatin dynamics with cell division. Interestingly, a similar role has been proposed for human PICH (also known as ERCC6L), a SNF2-like ATPase that colocalizes with Bloom’s syndrome protein (BLM) at anaphase bridges126,127. As defects in DNA replication at the centromeres or fragile sites can lead to the prevalence of sister chromatid bridges in mitosis128, it has been suggested that BLM and PICH promote the resolution of the bridges during anaphase127.
Future directions
Although initial thoughts on the role of chromatin in genome stability were driven by simple paradigms derived from transcriptional studies (such as ‘access, repair and restore’), it is clear that chromatin dynamics orchestrates a more complex and intriguing set of roles in many nuclear events. In this Review, we have highlighted several recent studies that pinpoint a function for chromatin in promoting integration of different nuclear events, such as DNA repair, DNA replication and transcription. As coordination of different genome stability pathways is essential for normal cell proliferation (FIG. 1), an open question in the field is how this crosstalk is accomplished at a molecular level. We envisage that chromatin will have a major role in this crucial, cell-homeostasis-regulating function, and this role is exemplified by the number of chromatin regulators implicated in various diseases.
For each of the genome stability pathways discussed here, studies need to continue their focus on mechanism and on how integration of chromatin regulation is accomplished in each pathway. Why are so many enzymes brought to a DSB? Why does the DDR pathway require multiple ATP-dependent chromatin-remodelling enzymes? How are chromosomes mobilized within the nucleus? Why are chromatin-remodelling enzymes localized to centrosomes129, and what role do they have here in the absence of detectable histones? Are there mitotic roles for INO80 and other chromatin-remodelling enzymes that take advantage of their ability to induce large-scale chromosome movements, as proposed for DNA repair? Do histone-modifying enzymes target only chromatin substrates, or are non-histone proteins equally or more important? This final question is important for studies in mammalian cells, where it is more difficult directly to assess the functional consequences of histone modifications.
Given that the mechanism of DNA repair changes as a function of cell cycle position, in vivo studies must continue to move away from studies in asynchronous cells. Likewise, studies in yeast typically look at the DDR pathway under conditions in which the DSB cannot be repaired; such studies are useful for analysis of factor recruitment, but more work needs to focus on the actual repair events to provide mechanistic insight. New focus on later steps of the homologous recombination process may also yield new insights into chromatin regulation. For instance, it is not clear whether the entire host of histone modifications survives the repair event and thus must be actively removed or whether the final, replication-dependent steps of homologous recombination lead to displacement of the modified, parental nucleosomes. We anticipate that biochemical studies will continue to recapitulate genome stability pathways with chromatin templates, providing insights into mechanism as well as instructing new models to be tested in vivo.
Acknowledgments
We thank L. Prendergast and members of the Peterson laboratory for helpful comments and discussion.
Glossary
- Holliday junction
An intermediate in homologous recombination comprised of four DNA strands
- V(D)J recombination
A somatic recombination event in lymphoid cells in which different variable, diverse and joining gene segments are combined as a part of the process to form diverse immunoglobulins and T cell receptors
- Resection
Exonucleolytic processing of the 5′ DNA strand at double-strand breaks, resulting in a 3 ssDNA ‘tail’
- DNA base adducts
DNA bases that contain a covalently bound chemical, often induced by cellular exposure to carcinogens
- Translesion synthesis
(TLS). A DNA tolerance pathway that allows replication to proceed through DNA lesions. This pathway involves fork-associated switching of a normal polymerase for a specialized translesion polymerase
- Chromosomal instability
(CIN). A cellular phenotype characterized by high rates of chromosome mis-segregation, leading to loss or gain of whole chromosomes
- Cytokinesis
A late stage of mitosis in which a cell divides to form two daughter cells
- Midbody
A transient structure found in mammalian cells that connects two daughter cells at the end of cytokinesis. The midbody is important for the final abscission (cleavage) event
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 3.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nature Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 4.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 5.Green CM, Almouzni G. When repair meets chromatin. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 7.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. This work demonstrates a role for the RSC complex in chromatin-remodelling nucleosomes that are proximal to a DSB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 9.Tsukuda T, et al. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair. 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and SWI/SNF ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. In this paper, it is shown that the SWI/SNF complex is essential for homologous recombination in heterochromatin in vivo, and a late role for the RSC enzyme during homologous recombination is suggested. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between INO80 and SWR1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann FR, et al. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 2012;26:369–383. doi: 10.1101/gad.176156.111. In this intriguing work, it is indicated that the INO80 chromatin-remodelling enzyme can promote large-scale chromosome movements when tethered to a DNA locus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end-resection and checkpoint deactivation. Mol Cell Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell. 2004;16:479–485. doi: 10.1016/j.molcel.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 18.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, et al. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Fierz B, et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nature Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. This elegant biochemical study demonstrates that H2Bub disrupts chromatin higher-order folding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogiwara H, et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Molina S, et al. Role for hACF1 in the G2/M damage checkpoint. Nucleic Acids Res. 2011;39:8445–8456. doi: 10.1093/nar/gkr435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinha M, Peterson CL. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics. 2009;1:371–385. doi: 10.2217/epi.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. In this paper, a reconstitution of a homologous recombination reaction with heterochromatin arrays defines a new role for the SWI/SNF complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nature Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. This outstanding study is the first to demonstrate DSB-induced chromosome mobility and its role in the homology search process. [DOI] [PubMed] [Google Scholar]
- 32.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nature Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 33.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nature Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nature Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. In this study, regulated DSB mobilization suggests a new level of regulation for recombinational repair in heterochromatin domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres-Rosell J, et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nature Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 37.Abraham RT. Cell cycle checkpoint signaling through the ATM & ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 38.Durocher D. In: The DNA Damage Response: Implications on Cancer Formation and Treatment. Khanna KK, Yosef S, editors. Ch 1. Springer; 2009. pp. 1–24. [Google Scholar]
- 39.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 40.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 41.West MH, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- 42.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 44.Li X, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma GG, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nature Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammet A, Magill C, Heierhorst J, Jackson SP. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Javaheri A, et al. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci USA. 2006;103:13771–13776. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie A, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helmink BA, et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 55.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 56.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 57.Luijsterburg MS, et al. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 61.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 64.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2008;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh N, et al. Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1. Proc Natl Acad Sci USA. 2012;109:14381–14386. doi: 10.1073/pnas.1212366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. The elegant approach used here provides molecular insight into the crosstalk between DDR and transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 69.Iacovoni JS, et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. This paper defines a new histone variant exchange activity for INO80 and provides evidence that aberrant distribution of H2A.Z has a negative impact on genome stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chowdhury D, et al. Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–1026. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keogh MC, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 74.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 76.De Piccoli G, et al. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 77.Cobb JA, et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baker SP, et al. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nature Cell Biol. 2010;12:294–298. doi: 10.1038/ncb2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levesque N, Leung GP, Fok AK, Schmidt TI, Kobor MS. Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway. J Biol Chem. 2010;285:35113–35122. doi: 10.1074/jbc.M110.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conde F, San-Segundo PA. Role of Dot1 in the response to alkylating DNA damage in Saccharomyces cerevisiae: regulation of DNA damage tolerance by the error-prone polymerases Polzeta/Rev1. Genetics. 2008;179:1197–1210. doi: 10.1534/genetics.108.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wysocki R, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murakami-Sekimata A, Huang D, Piening BD, Bangur C, Paulovich AG. The Saccharomyces cerevisiae RAD9, RAD17 and RAD24 genes are required for suppression of mutagenic post-replicative repair during chronic DNA damage. DNA Repair. 2010;9:824–834. doi: 10.1016/j.dnarep.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 85.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 88.Au TJ, Rodriguez J, Vincent JA, Tsukiyama T. ATP-dependent chromatin remodeling factors tune S phase checkpoint activity. Mol Cell Biol. 2011;31:4454–4463. doi: 10.1128/MCB.05931-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papamichos-Chronakis M, Peterson CL. The INO80 chromatin-remodeling enzyme regulates replisome function and stability. Nature Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 90.Shimada K, et al. INO80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 91.Hur SK, et al. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell Mol Life Sci. 2010;67:2283–2296. doi: 10.1007/s00018-010-0337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Falbo KB, et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nature Struct Mol Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boerkoel CF, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nature Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 94.Ciccia A, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Driscoll R, Cimprich KA. HARPing on about the DNA damage response during replication. Genes Dev. 2009;23:2359–2365. doi: 10.1101/gad.1860609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Betous R, et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nature Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Collins N, et al. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nature Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 103.Poot RA, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nature Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 104.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 105.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rowbotham SP, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 107.Stralfors A, Walfridsson J, Bhuiyan H, Ekwall K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011;7:e1001334. doi: 10.1371/journal.pgen.1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nature Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 109.Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nature Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 110.Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nature Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim HS, et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nature Struct Mol Biol. 2009;16:1286–1293. doi: 10.1038/nsmb.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krogan NJ, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez-Cadahia B, Drobic B, Davie JR. H3 phosphorylation: dual role in mitosis and interphase. Biochem Cell Biol. 2009;87:695–709. doi: 10.1139/O09-053. [DOI] [PubMed] [Google Scholar]
- 114.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 115.Kelly AE, et al. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 117.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 118.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ogiwara H, Enomoto T, Seki M. The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle. 2007;6:1090–1095. doi: 10.4161/cc.6.9.4130. [DOI] [PubMed] [Google Scholar]
- 120.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast RSC and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gkikopoulos T, et al. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu S, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nature Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Campsteijn C, Wijnands-Collin AM, Logie C. Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS Genet. 2007;3:e92. doi: 10.1371/journal.pgen.0030092. This comprehensive genetic analysis of RSC led to the discovery of new role for RSC in the control of ploidy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 125.Norden C, et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 126.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 127.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 129.Sillibourne JE, Delaval B, Redick S, Sinha M, Doxsey SJ. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol Biol Cell. 2007;18:3667–3680. doi: 10.1091/mbc.E06-07-0604. This intriguing work suggests a novel role for chromatin-remodelling enzymes in centrosome assembly and function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 131.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Korber P, Luckenbach T, Blaschke D, Horz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol Cell Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klopf E, et al. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2009;29:4994–5007. doi: 10.1128/MCB.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]