Abstract
Purpose
Liver iron quantification by MRI has become routine. Pixelwise (PW) fitting to the iron-mediated signal decay has some advantages but is slower and more vulnerable to noise than region-based techniques. We present a fast, pseudo-pixelwise mapping (PPWM) algorithm.
Materials and methods
The PPWM algorithm divides the entire liver into non-contiguous groups of pixels sorted by rapid relative relaxivity estimates. Pixels within each group of like-relaxivity were binned and fit using a Levenberg-Marquadt algorithm.
Results
The developed algorithm worked about 30 times faster than the traditional PW approach and generated R2* maps qualitatively and quantitatively similar. No systematic difference was observed in median R2* values with a coefficient of variability (CoV) of 2.4%. Intra-observer and inter-observer errors were also under 2.5%. Small systematic differences were observed in the right tail of the R2* distribution resulting in slightly lower mean R2* values (CoV of 4.2%) and moderately lower SD of R2* values for the PPWM algorithm. Moreover, the PPWM provided the best accuracy, giving a lower error of R2* estimates.
Conclusion
The PPWM yielded comparable reproducibility and higher accuracy than the TPWM. The method is suitable for relaxivity maps in other organs and applications.
Keywords: iron overload assessment, fast R2* mapping, liver
INTRODUCTION
Iron overload is a major concern for patients with genetic disorders of iron metabolism as well as with transfusion-dependent anemias [1]. Excess iron is stored in many organs, particularly liver, endocrine glands and heart, where it causes increased morbidity and mortality [2, 3].
The liver is the major site of iron storage and liver iron concentration (LIC) has been used as a measure of total body iron stores [4]. Determination of LIC has become a vital parameter in the management of iron overloaded patients, anchoring the interpretation of trends in serum ferritin. In healthy livers, biopsy remains an accurate test for LIC, but it is invasive, expensive, and subject to sampling errors due to the heterogeneity of iron distribution, with coefficients of variation ranging from 15% to 40% [5–7].
Magnetic Resonance Imaging (MRI) has recently been recognized as a non-invasive alternative to liver biopsy. Moreover, MRI can assess hepatic, cardiac [8, 9], pancreatic [10, 11], and pituitary [12] iron stores. MRI does not measure iron per se, but images water protons as they diffuse near the paramagnetic iron stores that are distorting the magnetic field. These interactions are rendered as a darkening of the images proportional to the iron concentration as a consequence of the increase in relaxation rates R2 and R2* assessed by spin echo and gradient approaches, respectively [13]. R2* techniques are more commonly used in the clinical arena, because they enable faster data acquisition and analysis.
To calculate R2*, the MRI signal is monitored at several gradient echo times (TEs) and a signal decay is mathematically modelled. The fitting can be applied in two different ways. In a region-based fit all the pixels within the region of interest (ROI) are averaged together for each TE and the fitting is performed for this averaged decay curve [8, 9]. This is typically performed in small, peripheral ROIs to avoid contributions from vascular and biliary structures, leaving this approach vulnerable to sampling error in unskilled operators. In the traditional pixelwise mapping (TPWM) approach the fitting is performed for each pixel in the region yielding a complete R2* map. The mean and the median for this distribution are then calculated [14, 15]. The main advantage of the pixelwise approach is that the regions of interest typically encompass all visible liver tissue, removing selection bias, yielding exceedingly low inter-slice and inter-observer variability (<3%). Furthermore, TPWM can expose areas of artifact or pathology that might be missed using a regionbased approach [14, 16].
The primary drawback of traditional R2* mapping is that it is computationally intensive and the signal-to-noise for each individual fit is relatively poor. In this study an automated multiple-ROI approach that simulates R2* mapping (pseudo-pixelwise mapping, or PPWM) is presented. The reproducibility and the accuracy of the PPWM were tested using hepatic MRI data of iron-overloaded patients and simulated hepatic data derived from these images.
MATERIALS AND METHODS
Study population
Baseline data from a phase 2 clinical trial of FBS0701 was used as a test data set because it represented MRI data collected according to local clinical practice from eight major thalassemia centers. Fifty-four patients of mixed race (12 Asian, 2 Black, 4 Thai and 36 White) participated with sites contributing between four and thirteen examinations (median, seven cases). Twenty-two patients were females and mean age was 28.4 ± 8.6 years. Fifty-two patients were diagnosed with thalassemia major and 2 (5.3%) with sickle cell disease; all were regularly transfused and chelated. Mean serum ferritin levels were 3093 ± 2458 ng/ml.
To compare the intrinsic accuracy of the two methods, images were retrieved from 39 patients referring at our Institution in whom complete complex (real and imaginary images) data from their hepatic images had been retained All these patients suffered from thalassemia major and 16 of them were females. Mean age was 18.6 ± 10.6 years.
Informed consent was obtained from prospective data on a protocol approved by their corresponding local institutional review boards; permission for review of retrospective data was also granted from the Committee on Clinical Investigation.
Image acquisition
MRIs were performed on 1.5T scanners at 8 different sites. Table 1 indicates, for each site, the type of scanner and the gradient-echo sequence parameters. All sites collected images according to their local clinical practice. Two centers acquired multiple hepatic slices, but only one slice (midhepatic) was used to compare the two analysis methods. Images at our site were performed on 1.5T GE Signa CVi scanner (GE Healthcare, Waukesha, WI). Complex R2* data were acquired at eight different TEs (1.09–13.38 ms, ΔTE=1.76).
Table 1.
MRI protocol at each MRI site.
| Boston, USA |
Cagliari, Italy |
Genoa, Italy |
Izmir, Turkey |
London, UK |
Oakland, USA |
Siriraj, Thailand |
Torino, Italy |
|
|---|---|---|---|---|---|---|---|---|
| # patients | 4 | 15 | 6 | 6 | 3 | 3 | 10 | 7 |
| Scanner | Philips | Siemens | GE LX | Siemens Symphony | Siemens Avanto | Philips Achieva | Philips Achieva | Philips |
| Software release | 2.6.3.5 | VB15 | 15.0 M4 0910.a | VA30A | VB17 | 2.6.3 | 2.6.3.5 | 2.5.3.3 |
| Coil | 4-channel torso | 8-channel cardiac | 8-channel cardiac | Phased-array body | 4-channel torso | Phased array body | 5-channel cardiac | 4-channel cardiac |
| Min TE (ms) | 1.2 | 0.9 | 1.1 | 1.3 | 0.99 | 1.2 | 1.07 | 1.1 |
| ΔTE (ms) | 1.06 | 1,42 | 2.36 | 1.16 | 1.41 | 2.10 | 0.8 | Variable |
| # echoes | 8 | 12 | 8 | 11 | 12 | 10 | 20 | 9 |
| TR (ms) | 50 | 200 | 200 | 200 | 200 | 17.7 | 80 | 21 |
| FA (°) | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
| Matrix (pixels) | 128X128 | 128X96 | 128X96 | 128X100 | 128X64 | 125X156 | 128X256 | 320X320 |
| Bandwith (Hz/pixel) | 1562 | 1950 | 977 | 1950 | 1950 | 720 | 2480 | 1800 |
| # slices | 1 | 1 | Whole liver | 1 | 1 | 5 | 1 | 1 |
| Slice thk (mm) | 8 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
TE= echo time
TR= repetition time
FA= flip angle
Thk= thickness
Generation of reference data
Noiseless, reference images are necessary to compare the absolute accuracy of post-processing methods. We approximated noiseless images from our complex-valued clinical data by filtering the real and imaginary images using a 5×5 median filtering, raising the signal to noise ratio (SNR) by a factor of five. A reduced noise image relatively free of Rician noise bias was formed by calculating the complex magnitude of the filtered images; pixelwise R2* estimates from these images formed the reference standard. Test images were created by adding Gaussian white noise to the filtered real and imaginary images to degrade their SNR to the originally acquired level. Magnitude images from these data contained similar R2* distribution, noise characteristics, and noise bias as the original source data but were quantifiably comparable to reference images.
R2* mapping: pixelwise and pseudo-pixelwise algorithms
All R2* images were processed centrally using a custom-written software developed in MATLAB (The Mathworks, Natick, MA). For both traditional and pseudo-pixelwise methods, a ROI was drawn around the entire liver boundary, excluding obvious central hilar vessels. Both techniques also used the same intrinsic fitting model consisting of a single exponential with a constant offset model:
| (1) |
S represents the observed signal intensity, S0 is the signal intensity at TE=0, R2* is the transverse relaxation rate, TE represents the echo times and C is a constant value that takes into account the rectification of MRI noise and partial-volume mixing of liver parenchyma with long T2* species such as blood and bile. The fitting was performed using the Levenberg-Marquardt algorithm. Mean, median, and standard deviation (SD) of the multiple R2* values were calculated.
The TPWM method calculates Eq. (1) for each pixel in the region of interest. Since signal-to-noise ratio is relatively modest, spurious R2* values can result, particularly at the boundaries of vascular/biliary structures. The PPWM method overcomes this limitation by using a multistep-step estimation process, described below.
Image smoothing with a 5×5 median filter; this improves point wise estimation in step 2.
- Generation of an approximate R2* map. It is assumed that the signal decay is described by a single exponential model (Eq. (1) 1 without the constant C), and the decay curve is then logarithmically (to base e) scaled, obtaining a straight line:
To eliminate excessive contribution of long T2* species at high iron concentrations, echo times greater than 2 ms are truncated, leaving a 2–3 point R2* estimate in most cases. A non-weighted linear least-squares fit is performed for each pixel. Calculations are vectorized, greatly speeding up calculations in MATLAB (256×256 calculation time around 1 second). While the absolute R2* values are not accurate, relative R2* relationships are generally preserved.(2) All pixels in the traced ROI are sorted according to their approximate R2* values.
Divide the sorted R2* values into N bins, where N is the square root of total number of pixels. Each bin comprises N pixels, not necessarily spatially close, but close in terms of approximated R2* value. From the original, unprocessed images, signal intensity values corresponding to pixels in each bin are averaged together separately for each echo time, creating one signal decay curve for each bin.
The signal decay curve for each bin is fit to equation 1, using Levenberg-Marquadt algorithm, generating N estimates of R2*. A pseudo-R2* map is generated by projecting the R2* value for each bin to each pixel in the bin. Mean, median and standard-deviation R2* values are calculated from the distribution of R2* values.
Reproducibility analysis
Reproducibility analysis was performed for both algorithms. Images were reanalysed by the same observer after at least a 24 - hour delay to evaluate the intra-observer variability. To evaluate the inter-observer variability, the images were presented in random order to another operator, blinded to the results obtained by the first observer.
Accuracy evaluation
The bias, variance, and mean-squared error between R2* maps calculated from the noiseless and noisy magnitude data were used to compare true pixelwise and pseudo-pixelwise accuracy.
Statistical analysis
All data were analyzed using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA) and MedCalc for Windows version 7.2.1.0 (MedCalc Software, Mariakerke, Belgium) statistical packages.
Data were expressed as mean ± SD. Summary data were displayed using scatter plots with regression lines. A coefficient of variation (CoV) was calculated as the ratio of the SD of the half mean square of the differences between the repeated values, to the general mean. The agreement between the algorithms was determined by the Bland-Altman technique, plotting the difference versus the average of the variables. Bias was the mean of the difference between the two methods and agreement was the mean ± 1.96 SDs.
Reproducibility was evaluated using CoV, interclass correlation coefficient (ICC) and Bland-Altman statistics. The ICC was obtained from a two-way random effects model with measures of absolute agreement. An ICC ≥ 0.75 was considered excellent, between 0.40 and 0.75 good, and < 0.40 unsatisfactory.
The accuracy of the individual and median R2* values calculated by the two techniques was assessed in the test images by means of the Bland-Altman analysis and the mean standard error (MSE) with respect to R2* values estimated from the reference images. That is, in each patient a Bland Altman statistics was performed for all the pixels in the ROI and the absolute values of the bias, the lower and the higher limits, and the ranges were compared for the whole population using a paired sample t-test.
RESULTS
Performance of the fast algorithm
All image analyses were performed on a 2.8 GHz Macintosh computer (version 10.6.8). The developed algorithm worked about 30 times faster than the strict approach, proportional to the reduction in subroutine calls to the Levenburg-Marquadt algorithm. To give an example, for a ROI of 6355 pixels, the mean running time after the ROI definition was less than 4 s for the PPWM and 136 s for the TPWM.
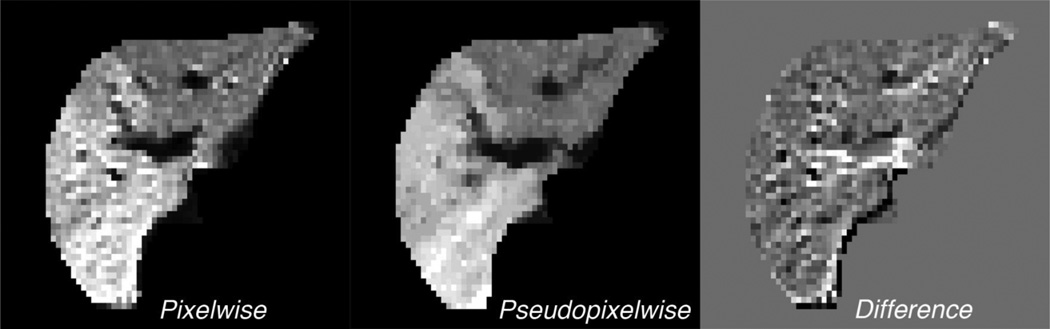
The R2* maps obtained using PPWM were qualitatively and quantitatively similar to those obtained with TPWM assessment. The fast algorithm was able to detect local R2* fluctuations, reflecting changes in iron load or due to the presence of vessels. Figure 1 shows R2* maps calculated for the same patient using TPWM algorithm, PPWM algorithm, and their difference. There were four distinct areas of loading. The vascular and biliary structures had the lowest R2* values, followed by the left lobe, right middle lobe, and right posterior lobe, respectively. The PPWM representation was somewhat smoother, but more clearly distinguished the systematic R2* gradations. The difference image demonstrates the largest deviations in pixels that had partial volumes between biliary structures and hepatic tissue. For the traditional algorithm the median and the mean R2* values were, respectively, 320.6 Hz and 322.0 Hz. For the fast algorithm the median and the mean R2* values were, respectively, 303.4 Hz and 308.5 Hz.
Figure 1.
R2* maps for a patient with thalassemia major calculated using a) traditional and b) fast algorithm and c) ΔR2* map.
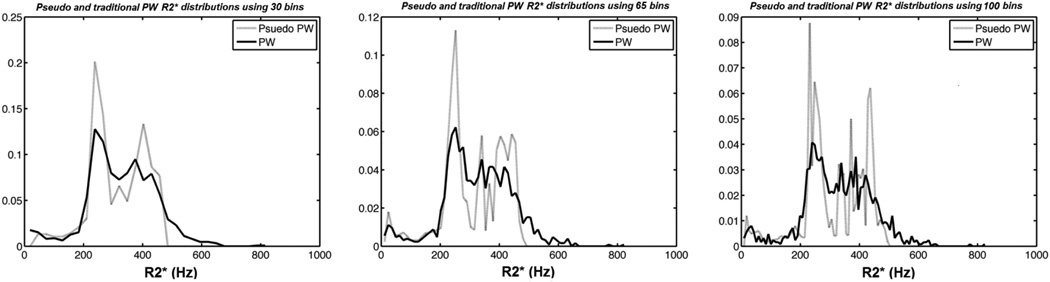
Figure 2 shows frequency histograms associated with the R2* maps presented in Figure 1. The R2* distribution was evaluated using 30, 65, and 100 different bins (since there were 1251 pixels in the region of interest, the default value was 35). Larger bin number yielded finer resolution of map texture but at the price of less noise suppression and greater computation time. Histogram texture was preserved, although the PPWM demonstrated greater contrast of the right posterior lobe. There was less area in the right tail of the PPWM method (even with finer resolution), suggesting that modest averaging suppresses spuriously high R2* estimates generated with true PW methods. In general, there was excellent agreement between the two algorithms, regardless of bin size.
Figure 2.
Frequency histograms associated with the R2* maps presented in Figure 1. The R2* distribution was evaluated for bin lengths of 30, 65, and 100 pixels.
Agreement between algorithms: median R2* values
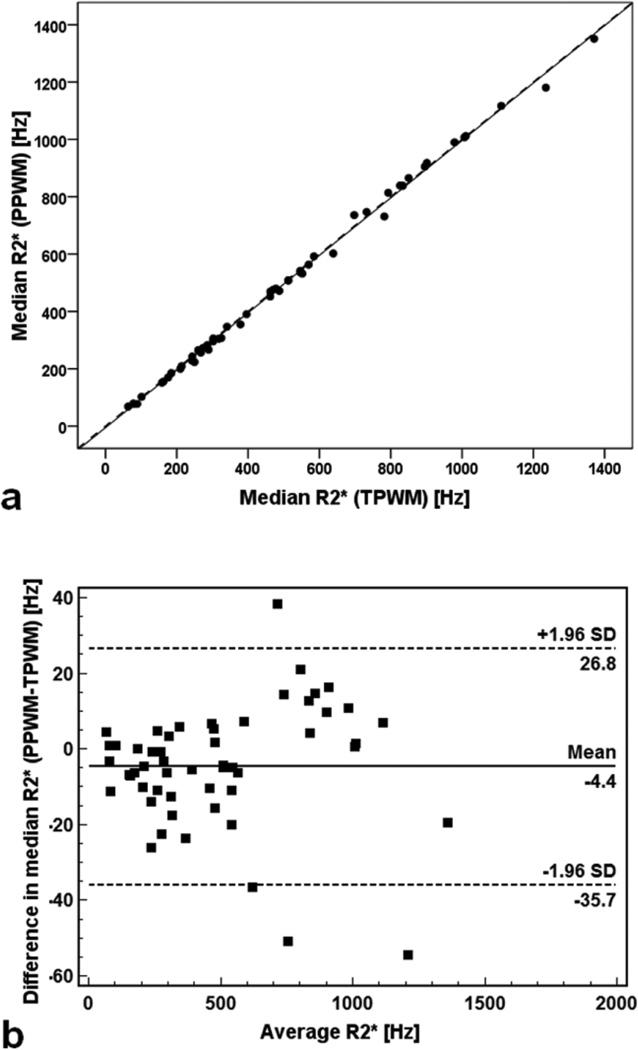
Good agreement was observed across the entire study population. Median R2* values obtained with TPWM and PPWM were 496.9 ± 321.9 Hz (range: 63.5 – 1370.5 Hz) and 492.5 ± 322.9 Hz (range: 68.1 – 1350.9 Hz), respectively. Figure 3a shows median R2* obtained with the PPWM algorithm as a function of values obtained with the TPWM algorithm. The line of best fit had a slope of 1.002 ± 0.007 and an intercept of −5.339 ± 4.053 Hz. The R-squared value for the fit was 0.998. Figure 3b shows the Bland-Altman plot. The mean difference was −4.4 ± 15.9 Hz. The error was proportional to the average value, with a CoV of 2.3%.
Figure 3.
Comparison between the median R2* values obtained with the PPWM algorithm and the correspondent median R2* values obtained with the TPWM algorithm. a) Scatter plot with regression line. The dotted line represents the unity line. b) Bland–Altman plot of absolute differences. Dotted lines indicate limits of agreement
Agreement between algorithms: mean R2* values
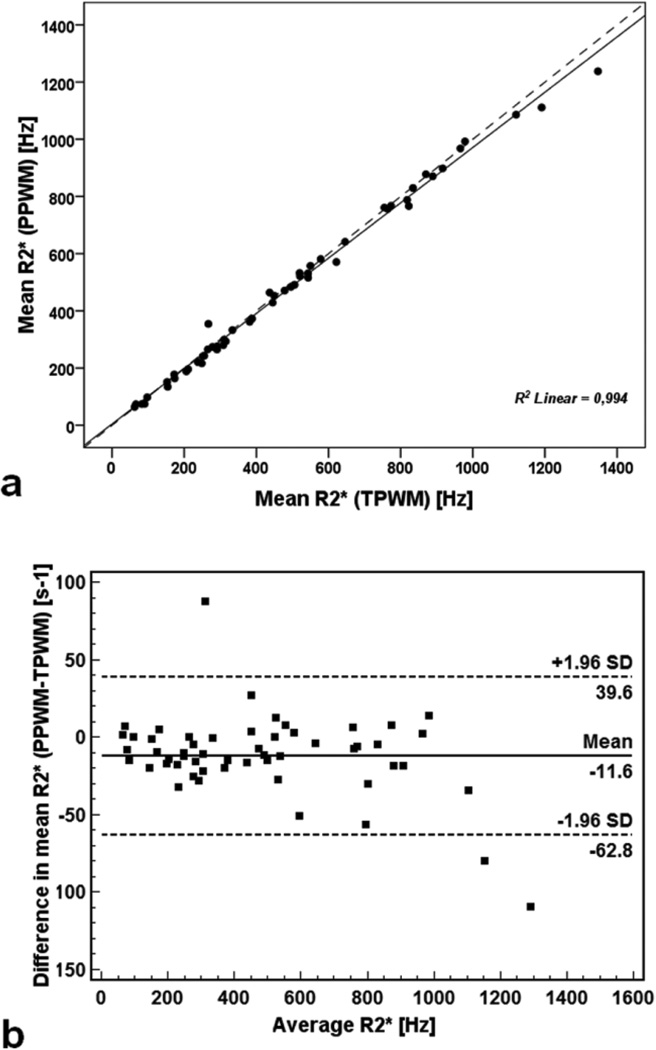
Mean R2* values were systematically slightly (2.7%) higher using the TPWM than the PPWM method, measuring 486.5 ± 314.1 Hz (range: 62.4 – 1346.7 Hz) and 474.9 ± 304.5 Hz (range: 64.2 – 1237.6 Hz), respectively. Figure 4a shows mean R2* obtained with the PPWM algorithm as a function of values obtained with the traditional algorithm. The line of best fit had a slope of 0.967 ± 0.011 and an intercept of 4.641± 6.099 Hz. The R-squared value for the fit was 0.994. Figure 4b shows the Bland-Altman plot. The mean difference was 11.6 ± 26.1 Hz. Differences also scaled with average R2* value, having a CoV of 4.2%.
Figure 4.
Comparison between the mean R2* values obtained with the PPWM algorithm and the correspondent mean R2* values obtained with the TPWM algorithm. a) Scatter plot with regression line. The dotted line represents the unity line. b) Bland–Altman plot of absolute differences. Dotted lines indicate limits of agreement.
Comparison of Standard Deviation
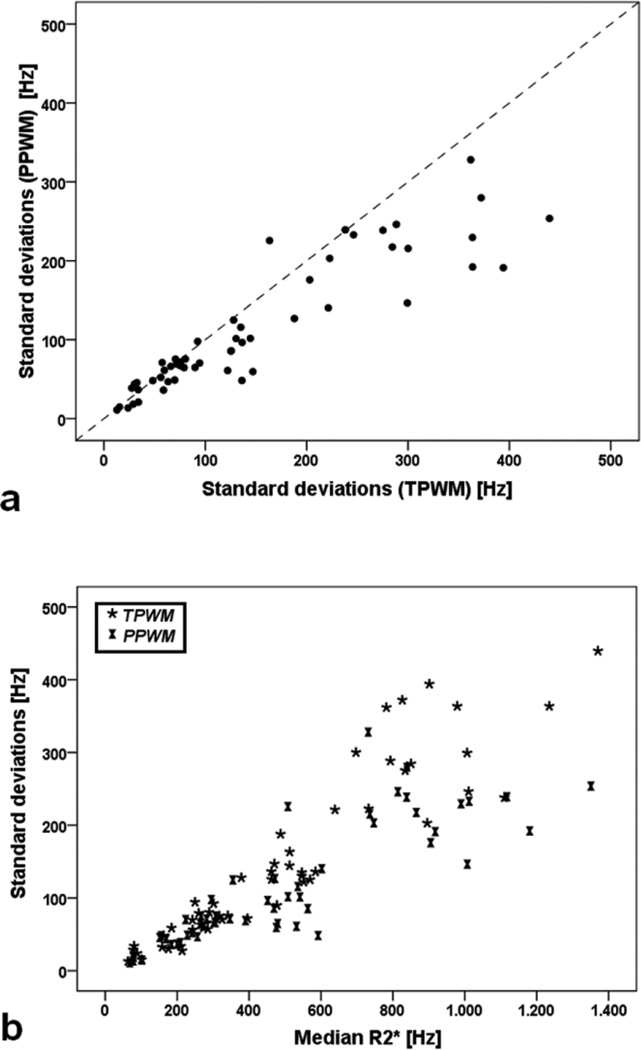
Figure 5a shows SD obtained with the fast algorithm as a function of SD obtained with the traditional algorithm. The relationship was not linear with the PPWM generating systematically lower SD estimates for R2* values > 400 Hz (Figure 5b). For both algorithms, SD rises with median R2*; this relationship is concave-up for the traditional method and more linear with the PPWM method. Bland Altman analysis demonstrates that PPWM methods were 22% lower than using TPWM analysis.
Figure 5.
a) Scatter plot of SD obtained with the PPWM algorithm as a function of SD obtained with the PPWM algorithm. The dotted line represents the unity line. b) Scatter plot of SD as a function of median R2* values for both PPWM and TPWM algorithms.
Intra- and inter-observer Variability
The results of the intra- and inter-observer variability analysis for mean and median R2* values obtained with both the algorithms are indicated in Table 2. Variability was not statistically different for the two methods. The ICC was excellent.
Table 2.
Intra- and inter-observer variability data.
| Traditional pixelwise mapping | ||||
|---|---|---|---|---|
| Intra-observer | Inter-observer | |||
| Median R2* | Mean R2* | Median R2* | Mean R2* | |
| ICC | 1.000 | 1.000 | 0.999 | 0.998 |
| CoV (%) | 0.70 | 1.96 | 2.28 | 4.15 |
| Bland-Altman limits (Hz) | −9.3 to 10.2 | −24.5 to 28.5 | −31.4 to 32.2 | −50.6 to 61.3 |
| Bland-Altman bias (Hz) | 0.4 | 2.0 | 0.4 | 5.3 |
| Pseudo pixelwise mapping | ||||
| Intra-observer | Inter-observer | |||
| Median R2* | Mean R2* | Median R2* | Mean R2* | |
| ICC | 1.000 | 0.999 | 0.999 | 0.997 |
| CoV (%) | 1.10 | 2.48 | 2.26 | 4.74 |
| Bland-Altman limits (Hz) | −16.7 to 12.6 | −35.9 to 28 | −32.6 to 29.6 | −67.4 to 55.9 |
| Bland-Altman bias (Hz) | −2.0 | −3.9 | −1.5 | −5.8 |
Accuracy of the algorithms
Reference median R2* values were 633.2 ± 466.7 (range: 25.3 – 1700.6 Hz). For noisy magnitude images, median R2* values were 631.9 ± 464.8 Hz (range: 24.9 – 1702.7 Hz) when using the TPWM and 632.1 ± 467.5 Hz (range: 24.3 – 1701.2 Hz) when using the PPWM. Table 3 shows the accuracy of both algorithms for quantifying median R2* values (compared with the reference standard). The PPWM had twofold lower coefficient of variation and Bland-Altman bias/range as well as fourfold lower mean squared error.
Table 3.
Accuracy of the two algorithms for calculating median R2* values, considering as reference values the median R2* values obtained from noiseless magnitude images.
| TPWM | PPWM | |
|---|---|---|
| CoV (%) | 16.13 | 7.86 |
| MSE (Hz) | 208.28 | 49.49 |
| Bland-Altman limits (Hz) | −29.8 to 27.3 | −14.9 to 12.6 |
| Bland-Altman bias (Hz) | −1.3 | −1.2 |
On a pixel-by-pixel basis, the absolute values of the bias averaged were not significantly different (12.1 ± 17.2 Hz for TPWM vs 11.5 ± 13.5 Hz, PPWM P=0.775). However, the TPWM showed significantly increased lower limits of agreement (−284.9 ± 287.1 Hz vs −270.9 ± 271.0 Hz, P=0.008), higher limits of agreement (268.9 ± 297.8 Hz vs 259.1 ± 270.5 Hz, P=0.028) and ranges (553.7 ± 553.9 Hz vs 530.0 ± 510.4 Hz, P<0.0001).
DISCUSSION
LIC assessment is critical for the diagnosis and management of iron overload patients. MRI R2* has been proven to accurately quantify hepatic iron across a clinically relevant range of LIC [14]. R2* estimates are obtained by fitting an appropriate decay model to the MRI signal at various echo times. The monoexponential function plus a constant has been demonstrated to be an optimal model for large regions of interest [15]. The constant corrects for the signal offset due to the noise and the presence of iron-poor regions (i.e. blood) [14, 16]. The use of this conventional model, however, is computationally-intensive, requiring iterative nonlinear fitting, an excessively timeconsuming process if pixel-wise mapping is desired. To reduce processing time some laboratories compromise by examining small regions of interest in vessel poor areas [17]. While this can be clinically acceptable, this practice can generate misleading data as illustrated in Figure 1.
The PPWM presented in this report represents a hybrid between these two approaches. The entire liver in the single slice acquired is used, reducing inter-observer error, but the liver slice is then subdivided into multiple ROIs containing tissue of similar relaxivity based upon a rapid linear least squares estimate. The rapid linear least squares fit is not sufficient alone for accurate R2* quantization because the natural logarithm overemphasizes fitting contribution from the weaker signal even for a constant variance noise [18]. Moreover, it has been shown that the single exponential model introduces a large error for T2* values lower than 5 ms [15]. In fact, at high liver iron concentrations, the “tail” of the decay curve reflects long T2 species such as blood volume and bile as well as rectified noise bias. One accepted method for dealing with this effect is to truncate the fit. The value of 2 ms was chosen because it provided at least a 2-point estimator of R2* across the multicenter cohort. Although a crude approximation to R2*, but this rapid estimate worked sufficiently well to rank order pixels with respect to iron load.
In our “hybrid” approach, the choice of bin-size creates a trade-off between noisesuppression and smoothness of the resulting map and its histogram. Note that each bin contains a constant number of points but a range of R2* values that varies according to the underlying distribution, i.e. the bin width adapts to the histogram shape. Many “optimal” binning algorithms have been proposed for histograms of known density functions. However, liver R2 and R2* histograms are often at least biphasic, so we chose to set the bin size equal to the square-root of the number of pixels. This empiric algorithm scales signal to noise ratio (SNR) improvements (#pixels/bin) and resolution (#bins) in a balanced manner and was robust across region of interest sizes that we tested (50 to 50,000 pixels). Our choice of histogram bin size cannot be claimed to be optimal in any mathematical sense, but it is a common initial choice for spreadsheets and statistical programs. Our algorithm produce histograms that were more “peaky” than the pixel-wise representation; this reflects the reduction in data dimensionality, similar to parametric power spectral analysis. Higher order iteration could be used to better refine histogram estimation, but this would require considerably more computation. Our goal was more modest, since we simply wanted to gain speed and robustness through signal averaging without overly blurring the spatial distribution of iron overload. Our approach preserves anatomic boundaries unlike low pass filtering and generates tissue relaxivity maps that correspond well with traditional pixelwise maps.
Concordance between the PPWM and TPWM methods was excellent. No systematic difference was observed in median R2* value, the value we use for LIC estimation, and CoV of only 2.4%. Intra-observer and inter-observer errors were nearly identical and exceeding low, comparable to prior work [15]. Small systematic differences were observed in the right tail of the R2* distribution (see Figure 2) resulting in slightly lower mean R2* values and moderately lower SD of R2* values for the PPWM. We believe that these differences represent fitting errors by the traditional pixelwise method resulting from either poor SNR at high iron concentrations and partial volume-effects near vessels. This hypothesis is supported by the concave-upward relationship between SD and median R2* value for traditional pixelwise estimation observed in Figure 5b. The PPWM maintains a linear scaling of SD with median R2* because binning improves SNR more than five-fold, suppressing spuriously high R2* estimates.
The qualitative impression presented in Figure 2 was born out in our test images where an absolute standard existed. We showed that the deviation of median R2* estimates from the reference values was lower when using the PPWM algorithm (Table 3). Moreover, the agreement is better also on a pixelwise basis. These differences are unlikely to be important if the clinical target is solely an LIC estimate, given that both methods produced statistically identical LIC results and superb reproducibility. However, if spatial variation in iron distribution proves to contain information regarding disease progression, the PPWM is visually and quantitatively superior.
In conclusion, a fast algorithm for approximate liver R2* maps was presented and validated: its reproducibility was comparable to that of the traditional pixelwise mapping and its accuracy was superior. The algorithm combines the statistical advantages of pixel binning but the use of noncontiguous pixels minimizes blurring. Although the fast algorithm for R2* mapping was originally developed to quantify hepatic iron, it is now used to estimate iron concentrations in other organs, notably the heart where R2* values are generally < 50 Hz). When mean R2* is low, R2* histograms are generally monotonic and signal to noise is high, so both TPWM and PPWM methods work indistinguishably well; the only difference is computational speed. Moreover, this two-stage approach to generating relaxivity maps may be useful for other applications in which R2* mapping provides visualization of the R2* properties in a quantitative fashion. These include functional imaging dependent on blood oxygenation [19], quantitative assessment of brain iron in patients with Friedreich ataxia or relapsing–remitting multiple sclerosis [20] and detection and tracking of super paramagnetic iron oxides [21].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gordeuk VR, Bacon BR, Brittenham GM. Iron overload: causes and consequences. Annu Rev Nutr. 1987;7:485–508. doi: 10.1146/annurev.nu.07.070187.002413. [DOI] [PubMed] [Google Scholar]
- 2.Borgna-Pignatti C, Rugolotto S, De Stefano P, et al. Survival and complications in patients with thalassemia major treated with transfusion and deferoxamine. Haematologica. 2004;89(10):1187–1193. [PubMed] [Google Scholar]
- 3.Shander A, Cappellini MD, Goodnough LT. Iron overload and toxicity: the hidden risk of multiple blood transfusions. Vox Sang. 2009;97(3):185–197. doi: 10.1111/j.1423-0410.2009.01207.x. [DOI] [PubMed] [Google Scholar]
- 4.Angelucci E, Brittenham GM, McLaren CE, et al. Hepatic iron concentration and total body iron stores in thalassemia major. N Engl J Med. 2000;343(5):327–331. doi: 10.1056/NEJM200008033430503. [DOI] [PubMed] [Google Scholar]
- 5.Ambu R, Crisponi G, Sciot R, et al. Uneven hepatic iron and phosphorus distribution in beta-thalassemia. J Hepatol. 1995;23(5):544–549. doi: 10.1016/0168-8278(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 6.Emond MJ, Bronner MP, Carlson TH, Lin M, Labbe RF, Kowdley KV. Quantitative study of the variability of hepatic iron concentrations. Clin Chem. 1999;45(3):340–346. [PubMed] [Google Scholar]
- 7.Villeneuve JP, Bilodeau M, Lepage R, Cote J, Lefebvre M. Variability in hepatic iron concentration measurement from needle-biopsy specimens. J Hepatol. 1996;25(2):172–177. doi: 10.1016/s0168-8278(96)80070-5. [DOI] [PubMed] [Google Scholar]
- 8.Anderson LJ, Holden S, Davis B, et al. Cardiovascular T2-star (T2*) magnetic resonance for the early diagnosis of myocardial iron overload. Eur Heart J. 2001;22(23):2171–2179. doi: 10.1053/euhj.2001.2822. [DOI] [PubMed] [Google Scholar]
- 9.Pepe A, Lombardi M, Positano V, et al. Evaluation of the efficacy of oral deferiprone in beta-thalassemia major by multislice multiecho T2*. Eur J Haematol. 2006;76(3):183–192. doi: 10.1111/j.1600-0609.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 10.Au WY, Lam WW, Chu W, et al. A T2* magnetic resonance imaging study of pancreatic iron overload in thalassemia major. Haematologica. 2008;93(1):116–119. doi: 10.3324/haematol.11768. [DOI] [PubMed] [Google Scholar]
- 11.Noetzli LJ, Coates TD, Wood JC. Pancreatic iron loading in chronically transfused sickle cell disease is lower than in thalassaemia major. Br J Haematol. 2011;152(2):229–233. doi: 10.1111/j.1365-2141.2010.08476.x. [DOI] [PubMed] [Google Scholar]
- 12.Noetzli LJ, Panigrahy A, Mittelman SD, et al. Pituitary iron and volume predict hypogonadism in transfusional iron overload. Am J Hematol. 2012;87(2):167–171. doi: 10.1002/ajh.22247. [DOI] [PubMed] [Google Scholar]
- 13.Ghugre NR, Wood JC. Relaxivity-iron calibration in hepatic iron overload: probing underlying biophysical mechanisms using a Monte Carlo model. Magn Reson Med. 2011;65(3):837–847. doi: 10.1002/mrm.22657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood JC, Enriquez C, Ghugre N, et al. MRI R2 and R2* mapping accurately estimates hepatic iron concentration in transfusion-dependent thalassemia and sickle cell disease patients. Blood. 2005;106(4):1460–1465. doi: 10.1182/blood-2004-10-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Positano V, Salani B, Pepe A, et al. Improved T2* assessment in liver iron overload by magnetic resonance imaging. Magn Reson Imaging. 2009;27(2):188–197. doi: 10.1016/j.mri.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 16.St Pierre TG, Clark PR, Chua-anusorn W, et al. Noninvasive measurement and imaging of liver iron concentrations using proton magnetic resonance. Blood. 2005;105(2):855–861. doi: 10.1182/blood-2004-01-0177. [DOI] [PubMed] [Google Scholar]
- 17.Gianesin B, Zefiro D, Musso M, et al. Measurement of liver iron overload: Noninvasive calibration of MRI-R2) * by magnetic iron detector susceptometer. Magn Reson Med. 2012;67(6):1782–1786. doi: 10.1002/mrm.23148. [DOI] [PubMed] [Google Scholar]
- 18.Bonny JM, Zanca M, Boire JY, Veyre A. T2 maximum likelihood estimation from multiple spin-echo magnitude images. Magn Reson Med. 1996;36(2):287–293. doi: 10.1002/mrm.1910360216. [DOI] [PubMed] [Google Scholar]
- 19.Ziyeh S, Rick J, Reinhard M, Hetzel A, Mader I, Speck O. Blood oxygen level-dependent MRI of cerebral CO2 reactivity in severe carotid stenosis and occlusion. Stroke. 2005;36(4):751–756. doi: 10.1161/01.STR.0000157593.03470.3d. [DOI] [PubMed] [Google Scholar]
- 20.Khalil M, Enzinger C, Langkammer C, et al. Quantitative assessment of brain iron by R2)* relaxometry in patients with clinically isolated syndrome and relapsing-remitting multiple sclerosis. Mult Scler. 2009;15(9):1048–1054. doi: 10.1177/1352458509106609. [DOI] [PubMed] [Google Scholar]
- 21.Lee N, Kim H, Choi SH, et al. Magnetosome-like ferrimagnetic iron oxide nanocubes for highly sensitive MRI of single cells and transplanted pancreatic islets. Proc Natl Acad Sci U S A. 2011;108(7):2662–2667. doi: 10.1073/pnas.1016409108. [DOI] [PMC free article] [PubMed] [Google Scholar]