Abstract
Chemical glycobiology is emerging as one of the most uniquely powerful sub-disciplines of chemical biology. The previous scarcity of chemical strategies and the unparalleled structural diversity have created a uniquely fertile ground that is both rich in challenges and potentially very profound in implications. Glycans (oligosaccharides, polysaccharides, and glycoconjugates) are everywhere in biological systems and yet remain disproportionately neglected – reviews highlighting this ‘Cinderella status’ abound. Yet, the last two decades have witnessed tremendous progress, notably in chemical and chemoenzymatic synthesis, ‘sequencing’ and arraying, metabolic engineering and imaging. These vital steps serve to highlight not only the great potential but just how much more remains to be done. The vast chemical and functional space of glycans remains to be truly explored. Top-down full-scale glycomic and glycoproteomic studies coupled with hypothesis-driven, bottom-up innovative chemical strategies will be required to properly realize the potential impact of glycoscience on human health, energy, and economy. In this review, we cherry-pick far-sighted advances and use these to identify possible challenges, opportunities and avenues in chemical glycobiology.
Introduction
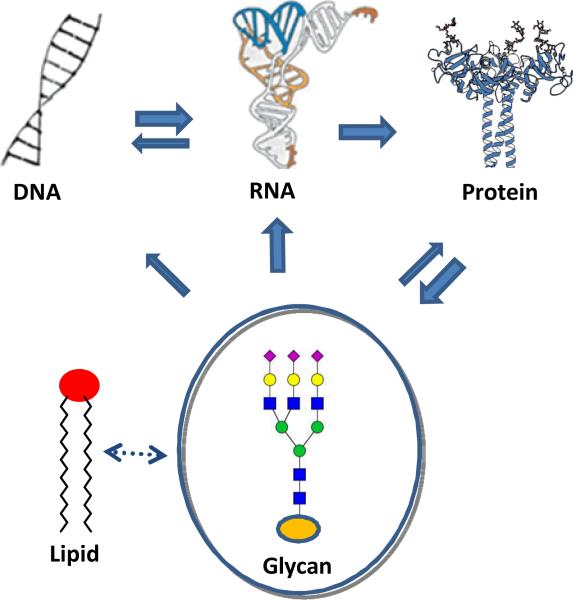
25 years on from its first concise definition [1], the field of Glycobiology has continued to successfully highlight the myriad of complex processes in which carbohydrates play a vital role. It becomes clear that glycans, in the form of oligosaccharides, polysaccharides, glycoproteins, glycolipids, proteoglycans and other glycoconjugates can be key players in a number of important biological recognition processes: intracellular trafficking, cell adhesion, development, cancer progression, host-pathogen interaction, and immune response, to name a few [2-6]. They may now be reasonably added to the ‘central dogma molecules’ (nucleic acids, proteins) as playing a distinct role in information transfer [7] – they are an additional ‘language of life’ (Fig. 1), whose roles are constantly emerging [8]. Yet decoding this language has barely started. In comparison with proteins and nucleic acids that contain linear sequences of building blocks, glycans frequently form branching structures and the inter-residue linkages (glycosidic) can exist in different configurations (anomeric isomers). The biosynthetic assembly of glycans is also more complex – it is not template-driven, as is largely the case for proteins and nucleic acids – and involves complex sugar chain processing and trimming under the action of a series of competitive enzymes along secretory pathways. As a result, glycans are generally more diverse in structure and are often present as heterogeneous mixtures that are difficult to separate for detailed structure-activity relationship studies. However, this structural complexity, which has often been blamed for the slow development of hypothesis testing within the field, also offers powerful opportunities in the design of molecular experiments that will unpick the mechanism of this Biology. In this way Chemistry offers one unique, and as yet largely unrealized, strategy for dissecting this complexity where strictly biological approaches may fail.
Fig. 1.
A modified central dogma of biological information flow
The increasing importance and realization of Chemical Biology as a field have seen some successful analyses of its intellectual thrust. We use here a concept of the study of Biological questions and problems through molecular insight. Current moves towards the cellular and organismal are vital but at the same time can create an imbalance in skills and a lack of molecular knowledge. This creates a current opportunity for the Chemical Biologist. The aesthetic of synthesis is a self-consistent goal that does not need to justify itself in disciplines. And without genuine biological questions, the synthesis of structural permutations (‘countless numbers of grapes’ [Bacon]) is a strategy that is unlikely to work. This is particularly pertinent to Glycoscience, where permutations are too vast [9]. Chemical Biologists, therefore, have an essential and powerful role in posing the ‘right’ molecular questions in Biology. In Chemical Glycobiology, the exciting depth and breadth of the challenges that we face precludes devotion to only one discipline or sub-discipline. The recently released US National Academy of Sciences report, entitled “Transforming the glycosciences: a roadmap for the future”, offers one blueprint for future studies. It too emphasizes the essence of a multi-disciplinary approach (http://www.nap.edu/catalog.php?record_id=13446). This will forge superbly exciting ‘joined-up’ strategies and here we guess at how these might develop. With technical and conceptual development, the time is now ripe to bring glycoscience to the mainstream of research.
Chemical Glycobiology, as broadly defined, explores chemical approaches and concepts for deciphering and understanding the function of entire glycomes and related partners. Remarkable progress has been achieved within the last two decades, and this has been discussed in a number of excellent recent reviews [10-16]. While we should be proud of this, we must be also fully aware of the great structural and functional space that remains unexplored. The purpose of this speculative review is therefore not to recapitulate and list the many studies and reviews that comprehensively cover what has been tried in the past, but instead to speculate on challenges that remain.
The generalities of sugar synthesis and glycosylation
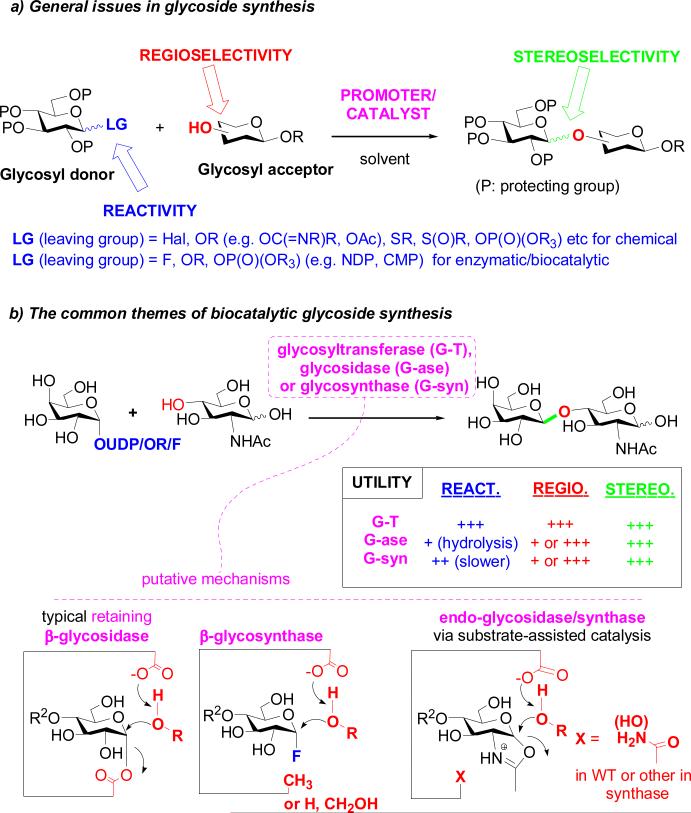
Synthesis of glycans and related chemical probes is essential if we are to precisely decipher structure, biosynthesis, metabolism and function in biological systems. The relative importance of synthetic glycans in glycobiology is exacerbated since homogeneous glycans (often found as part of glycoconjugates) are typically difficult to obtain from natural sources because of their structural micro-heterogeneity and, in many cases, their natural scarcity. Such synthetic glycans are also starting to play essential roles in developing carbohydrate-based therapeutics, such as vaccines [17]. However, the issues involved in the construction of complex oligosaccharides and glycoconjugates, including branching structures, anomeric stereochemistry, regiochemistry (even in linear oligomers), and coupling efficiency, pose many synthetic challenges. The complexity and diversity of oligosaccharides and glycoconjugates demand the use of a range of the most efficient synthetic methods with sufficient flexibility and breadth of use, regardless of whether these are chemical, enzymatic, or chemoenzymatic.
The general aspects of chemical glycosylation and major glycosylation methods have been well reviewed and discussed elsewhere [13,18-21] but key aspects are worth re-iterating. Carbohydrate chemistry often takes selective protection and deprotection methods to their most comprehensive and elegant heights to achieve regio-selectivity in glycosylation [13,18-22]. This lack of ‘atom efficiency’, of course, begs strategic questions but nonetheless it has proven a successful reliable approach especially when coupled with novel glycoside-forming strategies. These include manipulations of stereoelectronic effects, neighboring/remote protecting group participation, and conformational constraints – all usefully explored to control stereoselectivity at the anomeric center. In addition, there is a continuous effort to develop glycosyl donors with novel leaving groups and new promoters/catalysts in order to enhance glycosylation efficiency, and to tune reactivity often with useful knock on consequences for the better control of stereochemistry (Fig. 2). The progress in chemical synthesis has been remarkable, as exemplified by the successful total chemical synthesis of complex heparin oligosaccharide-based drugs [23], highly complex carbohydrate-containing natural products [24], and even homogeneous natural glycoproteins [25-28]. From those looking-in from outside the field, such effort spent on the creation of what is in essence just a mixed acetal or ketal linkage may seem a stunning self-indulgence for the carbohydrate community to still be so heavily involved in more than 100 years after the first realization of a real glycosylation reaction. Yet, we must be clear, there is no general synthetic method available that can cover all of the diverse structures of glycans and glycoconjugates and, in this era of oversimplified chemical strategy (“Click” and “automated synthesis”), it is pertinent to realize that each glycan target may pose unique challenges of reactivity and selectivity. More ‘programmable’ strategies remain one option to create more general approaches, whilst on the other hand ‘tricks’ in regioselectivity and stereoselectivity remain attractive as bespoke solutions to target structures. Even today, there remains an argument on what active intermediates may be involved in fundamental glycosylation reactions and how the reactivity of the active species contributes to the control of stereo-selectivity [29]. Innovations in achieving near perfect stereocontrol in glycosylation remain as urgently sought-after goals [30] and the mechanistic ambiguities of reactions at acetals ensures that there is still rich chemistry to be explored in glycosidic bond formation.
Fig. 2.
Chemical and enzymatic synthesis of glycosides and oligosaccharides. a) General issues in glycoside synthesis; b) the common themes of biocatalytic glycoside construction.
Enzymatic and chemoenzymatic glycoside synthesis and manipulation
The need to simultaneously control reactivity, regio-selectivity, and stereo-selectivity, some might argue, has created a double-bind for carbohydrate synthesis: such complexity requires the greatest use of the available tools (e.g., extensive protecting group usage, arsenals of donors) and yet it is increasingly and strikingly atom inefficient. In addition, much of the role of the protecting group in glycan synthesis is to provide the ‘grease’ to solubilize the substrate in a manner suitable for our traditional organic solvents. In contrast, nature has devised many thousands of biocatalysts (enzymes) to achieve often perfect control of stereo- and regio-selectivity and usually high efficiency in transformations for glycan assembly in aqueous solution (arguably the ideal solvent for glycans), without the need of protecting groups. Thus, there seems no reason why we chemical biologists should not embrace such special and efficient tools in our synthetic business and we would argue that synthetic carbohydrate scientists have been forward-thinking in their adoption of biocatalysis. In particular, combined processes (incorporating enzymatic transformations as key steps in a chemical synthesis, the so-called chemoenzymatic approach), can rapidly simplify synthetic schemes [31].
In theory, all types of enzymes involved in glycan biosynthesis, modification, and metabolism can be borrowed for synthesis. The opportunities for catalysis are huge since the numbers of enzymes involved in glycan assembly and metabolism far exceed those involved in the transformations of any other type of biomolecule. Glycosyltransferases are the natural enzymes responsible for constructing glycosidic bonds. Their high selectivity in glycosidic bond formation and tolerance of certain modifications on the donor and acceptor substrates make glycosyltransferases unique biocatalyst for synthesizing natural and selectively modified glycans [32]. Sugar nucleotide regeneration systems have been used now for some to by-pass the stoichiometric use of the expensive sugar nucleotides but this creation of ‘catalytic donors’ has not gained the prominence that it perhaps should in the synthetic community. The perceived intolerance of some enzymes can, in fact, sometimes be ‘forced’ in vitro using concentrations above the KM; this has actually allowed broad use of quite different donors in many cases. Moreover, mutational studies have led to mutant enzymes that demonstrate broader substrate tolerance, capable of accepting modified and altered substrates [31,33]. Furthermore, enzymatic synthesis should not be limited to purely in vitro enzymatic transformations. For example, functional transfer of glycan biosynthetic genes or even genes cluster into bacterium Escherichia coli (E. coli) has enabled a large-scale in situ production of heparin-like oligosaccharides, sialyloligosaccharides, ganglioside oligosaccharides and human milk oligosaccharides [34,35]. Thus, E. coli, biotechnology's “work horse”, as well as other promising host systems (yeast, insect cells, and plant cells) could well be further explored as factories for producing diverse glycans on large scale.
Glycosidases, the natural function of which is to hydrolyze glycosidic bonds in glycan metabolism, have strong potential in transglycosylation by virtue of their ability to manipulate bond formation at the anomeric center [36-38]. One of the most striking advance in this area is the invention of glycosynthases [39,40]: logically-designed glycosidase mutants that are devoid of product hydrolysis activity (a clear limitation in glycosidase-associated synthesis) but that are able to take activated donor substrates (e.g., glycosyl fluorides or sugar oxazolines) for transglycosylation [36-38]. Since the report of the first glycosynthase in 1998 [39,40], various glycosynthases have been constructed from glycosidases belonging to more than a dozen of glycoside hydrolase families. Yet, this reflects only a small fraction of many thousands of existing glycosidases; often the principles of glycosynthase design do not translate. One reason for this could be that observed reaction rates, in some cases, are simply below the threshold for detection. Even a little perturbation at the catalytic site caused by mutation could result in a large impact on the reaction rates. It may also be that our assumptions regarding modes of such catalysis may need refining; more detailed mechanistic studies are undoubtedly required to unveil why some enzymes have more potent transglycosylation activities than others (Fig. 2b).
In addition to the enzymes exploited in glycosidic bond formation, a number of glycan-modifying enzymes, such as sulfotransferases, acetyltransferases, oxidases, and epimerases, to name a few, also bear high value for post-glycosylation modifications of oligosaccharides and glycoconjugates [31,41]. A very large space of enzyme capacity for synthesis remains to be explored. Two notable examples showcase the power of the chemoenzymatic approach. One is the recently reported, in vitro chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins (heparin hepta-saccharides) with well-defined sequence and sulfation patterns, which was fulfilled by a combined use of glycosyltransferases, sulfotransferases, and epimerase, starting from simple disaccharides [42]. The synthesis took only 10 to 12 steps and resulted in a 45% overall yield. This was achieved on the basis of an in depth understanding of the biosynthetic pathways, together with the efficient cloning and expression of those enzymes involved [42]. In comparison, the pure chemical synthesis of the related anticoagulant drug, Arixtra (a heparin pentasaccharide), a landmark in target chemical oligosaccharide synthesis, took more than 50 steps leading to a low (1%) overall yield [23]. The other example is the ab initio glycan remodeling of glycoproteins via endoglycosidase-catalyzed deglycosylation and block oligosaccharide transfer [43,44]. This highly convergent approach enables a quick assembly of well-defined glycoforms, such as the homogeneously Fc-glycosylated IgG antibodies, for structural and functional studies.
While often specificity is an advantage in enzymatic transformations, the great diversity in the types of glycosidic linkages also potentially means that large numbers of enzymes must be collected to cover this diversity. However, the once popular notion of “one linkage - one enzyme” might not be quite accurate, as some enzymes such as glycosyltransferases, particularly those from bacteria, can be, in fact, quite promiscuous in substrate recognition, enabling “glycorandomization” to diversify the glycosylation patterns of complex natural products [45,46]. Moreover, directed evolution, coupled with mechanistic studies and protein engineering, provides a powerful method to improve catalytic efficiency and plasticity of enzymes [33,47-51]. It is expected that studies along these lines will significantly expand the repertoire of enzymes useful for synthesis. In our opinion, the chemoenzymatic approach holds great promise, particularly in constructing those targets that are very large and contain complex carbohydrates, such as proteoglycans, glyco(sphingo)lipids, and glycoproteins.
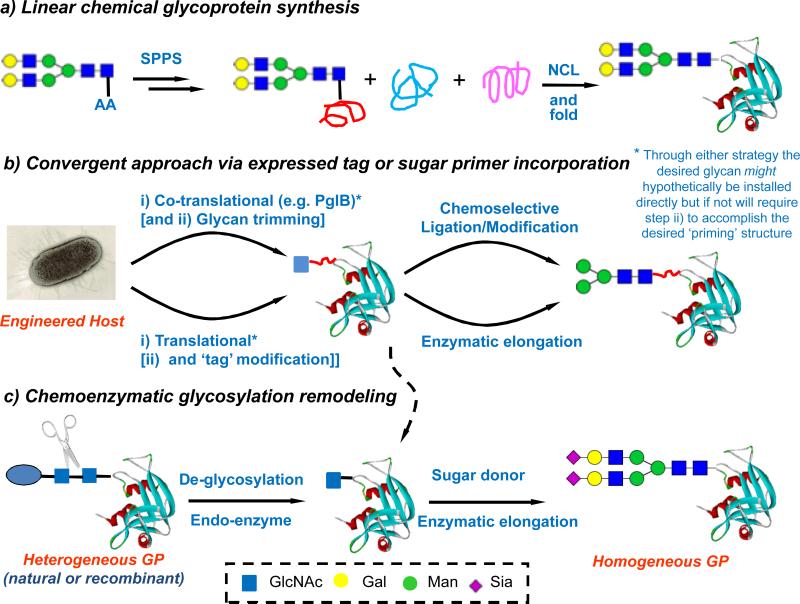
The construction of homogeneous glycoproteins poses a special challenge (Fig. 3). One question to be asked is: Can we develop routine protocols to selectively alter the nature of glycans at specific sites in a multiply glycosylated glycoprotein, in analogy to site-directed mutagenesis in recombinant technology? Theoretically, total and semi-chemical synthesis, via native chemical ligation (NCL) and expressed protein ligation, offers a possibility to install different glycans at different sites in the context of a polypeptide (Fig. 3a) [25-28]. However, in reality, chemical technologies for total glycoprotein synthesis are far from maturation. The exploitation of various native chemical ligation strategies has significantly improved synthetic quality. Nevertheless, the choice of ligation strategy, the design of ligation components, and the efficiency of tandem ligations should all rightly be considered on a case-by-case basis for individual targets. Convergence and efficiency of ligations are a key to success and the installation of glycan prior to protein backbone assembly necessarily places a more linear emphasis on NCL routes.
Fig. 3.
Strategies for glycoprotein synthesis. a) Linear chemical glycoprotein synthesis; b) Convergent approach via expressed tag or sugar primer incorporation; c) Chemoenzymatic glycosylation remodeling
In this sense, enzymatic glycan remodeling of natural and recombinant glycoproteins (Fig. 3c) offers an exciting opportunity to convert heterogeneous glycoproteins to homogeneous glycoforms in a global and convergent glycosylation remodeling manner [52], as it combines some advantages of recombinant DNA technology for protein synthesis and the selectivity of in vitro enzymatic manipulation for attaching desired glycans. However, despite its power this method can struggle to control the site for convergent glycan attachment; methods are urgently needed to achieve differential site-selective glycan remodeling. One, yet to be demonstrated, solution would be novel endoglycosidases that can distinguish the nature of glycans for de-glycosylation and/or the location (site) of the monosaccharide primer needed for en bloc transfer of glycans. Co-translational glycosylation to determine the site selectivity provides another broad strategy (Fig. 3b). In this regard, one of the ‘killer apps’ in the form of stop codon suppression by a glyco-amino acid has not yet been clearly demonstrated in a useable form [53-55], but hope remains. The bacterial PglB oligosaccharyltransferase system has been nicely exploited in vivo to create valuable substrates using pathway recapitulation and repositioning of the appropriate DNXS/T consensus motif [56]. In particular, the relaxed specificity of the PglB toward glycan structures has enabled the transfer of certain large bacterial oligosaccharides and polysaccharides (such as O-antigens) to proteins in a site-specific manner to form linkage-defined polysaccharide-protein conjugates, which points to a promising new avenue to making bacterial glycoconjugate vaccines in E. coli [57,58]. Another way to determine the site of glycosylation is to do this chemically by attaching a primer glycan through chemical site-selective protein chemistry. In this sense, the ‘tag-and-modify strategy’ has successfully allowed conversion of recombinant proteins into homogeneously glycosylated proteins, particularly when combined with the in vitro enzymatic sugar chain elongation [59,60]. Although these systems are tolerated by endoglycosidases to allow transglycosylation, one goal that remains to be addressed is how to introduce the tags with a fully native linkage found in natural glycoproteins.
Automated and programmable synthesis
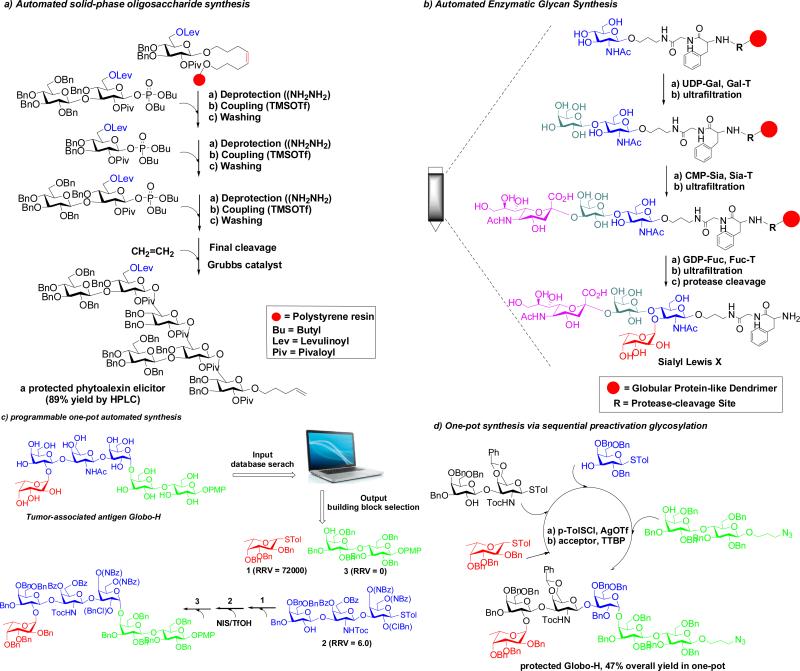
Conventional oligosaccharide synthesis often involves tedious time-consuming manipulations of a similar reaction type – protection and deprotection steps to define the regioselectivity and sometimes intensive chromatographic steps to purify the desired anomer(s) after each glycosylation. Automated synthesis is therefore a sensible and logical strategy to streamline glycan assembly by minimizing the intermediate purification steps and by minimizing the manual nature of any repetition [61]. Major strategies that hold promise for automated and programmable oligosaccharide synthesis are summarized in Fig. 4. Solid-phase synthesis seems an appropriate adjunct and, indeed, di- or tri-saccharides were targets first addressed in the 1970s soon after the success of automated polymer-supported synthesis of polypeptides [62]. However, it wasn't until 2001 that an automated oligosaccharide synthesis strategy (using glycosyl phosphates on a solid support) captured general attention (Fig. 4a) [63]. This remarkable demonstrated success showed that an automated synthesis of complex oligosaccharides was conceptually and technologically feasible. Some have correctly highlighted that this disclosure did not provide a general solution but its power was to throw down a gauntlet to other synthetic methods – there is no doubt it has prompted a dramatic development in synthetic sophistication of both automated and non-automated methods.
Fig. 4.
Strategies for automated oligosaccharide synthesis
Nonetheless, many exciting associated goals remain. Recent studies have started to effectively address initial drawbacks faced in this and other automated oligosaccharide syntheses, exemplified by the synthesis of increasingly more complex targets [64-66]. Nevertheless, a number of problems still remain when diverse glycan structures are concerned. In addition to the issues associated with choices of linkers and solid-support, and the accessibility of selectively protected monosaccharide building blocks, the greatest problems surround control of anomeric stereochemistry and the unpredictability of coupling efficiency in each glycosylation steps. Thus, more robust glycosylation method with perfect control of stereo-selectivity (particularly for the 1,2-cis glycosidic linkages) with excellent associated coupling yields (>99%) are welcome in this regard [30] and more are urgently needed, if compounded inefficiencies are not to continue to prove strategically unwieldy. Although more rarely considered, enzymatic transformations also hold great potential for automation; in a recent report, an automated enzymatic synthesis of sialyl Lewis X was fulfilled that applies sequential enzymatic glycosylations on a globular protein-like dendrimer as the solid support [67] albeit in a relatively low overall yield (mainly due to the low recovery efficiency of the polymer) (Fig. 4b). In another study, a digital microfluidic technology was interestingly employed for enzymatic modifications of heparin sulfate chains immobilized on manipulable magnetic nanoparticles [68]. As more-and-more enzymes become available, biocatalytic automated platforms may allow high-throughput access of diverse naturally occurring glycans and a range of non-natural derivatives as well. Such concepts deserve further exploration in order to develop artificial systems that might, one day, effectively mimic the natural Golgi apparatus for oligosaccharide assembly and modification. Interestingly both groups cited here viewed their very different technological approaches as possible ‘Golgi mimics’.
In parallel to solid-phase synthesis, there are several solution-based, one-pot, multi-step synthetic strategies that hold promise for automation. These include: programmable one-pot multi-step oligosaccharide synthesis based on relative reactivity of selected glycosyl donors (Fig. 4c) [18,69], one-pot orthogonal activation strategy [70], and one-pot pre-activation glycosylation strategy (Fig. 4d) [71-73]. Again, here, control is the name of the game – the success of these strategies for any given complex synthetic target relies on how one can efficiently fine tune the many factors involved, even the stoichiometry and mode-of-use of the reactants and activators, as well as their identity. As for all the strategies highlighted in this section, these methods serve to illustrate that far from there being one general solution, the answers will continue to also lie in robust and rapid access to rapidly optimized bespoke syntheses.
Glycan (and associated) arrays
Glycan (micro)arrays are emerging as a popular and important tool in deciphering glycan functions [14,16,74]. Microarray technology typically consists of immobilization on a surface (e.g., microplates / glass slides) and subsequent probing of interactions between the printed analyte and (glycan) binding partners (lectins, antibodies, enzymes, etc.) either in an isolated form or present in a biological context (sera, cell surface, or cell lysates). This format can offer several important advantages for glycomic studies, including high-throughput screening, sensitive and quantitative analysis, and the need for reduced amounts of glycans. A major application of glycan microarrays has been to detect and quantify specific interactions between glycans and diverse glycan binding proteins in mammalian, microbial and plant systems [16]. This has probed changes in affinity and specificity of glycan-protein interactions aimed at identifying specific biomarkers for the early diagnosis of diseases. Screening the sera from patients or vaccinated individuals in a glycan microarray format also allows quick assessment of immune responses, facilitating biomarker discovery and vaccine development [75,76]. In combination with sensitive mass spectrometric analysis, glycan microarray format has also been successfully used for probing enzyme substrate specificity [77] and for discovering new glycosyltransferases [78].
Despite these significant advances, the technology is still far from reaching its full potential. Perhaps the most significant limitation of current glycan microarray format is a lack of sufficient number and structural diversity of glycans. For example, the most “comprehensive” glycan microarrays so far available from the Consortium of Functional Glycomics (CFG) (http://www.functionalglycomics.org) contain only 600 or so glycans. This covers only a very small fraction of basic natural glycan structures, which are estimated to be in the tens of thousands. Accelerated synthesis, including development of more efficient chemical and chemoenzymatic methods (likely employed for ‘on array’ synthesis), is urgently needed to fill the gap. But synthesis is not the only avenue to expanding the chemical space of glycans. In fact, a number of natural glycans can be adequately prepared directly from natural sources of glycoproteins, glycolipids, and glycosaminoglycans: over 200 natural glycans from several rich sources such as human milk, chicken ovalbumin, egg yolks, and glycosaminoglycans can be readily conjugated with a bifunctional fluorescent linker to facilitate their tagging, separation and printing in a microarray format [79,80]. Here, the fluorescent tag introduced in the glycans can also serve as a means for quantifying the immobilization density and efficiency – these particular glycan arrays were successfully used for identifying the natural ligands for galectins [80]. It should be noted that, in this pursuit, complete separation may not even be necessarily required for initial screening; once hits are narrowed to subgroups, they can be identified by further separation and characterization. Thus, development of more robust methods for isolation, characterization, and tagging of glycans from diverse natural sources should be a high priority for rapidly expanding the chemical space of glycans and as a way of accessing glycomes in a number of useful guises.
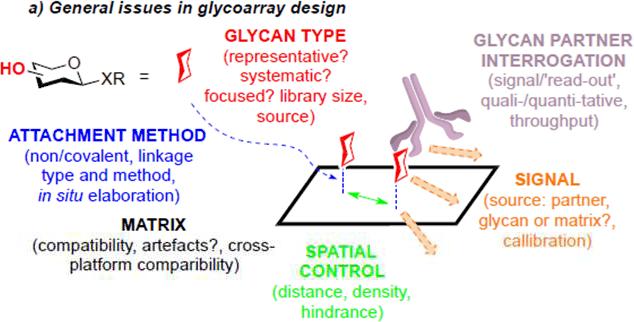
Another major limitation of current glycan array technology is the lack of adequate control of glycan presentations (Fig. 5). Firstly, different array platforms clearly result in differences in the density and orientation of glycan presentations; these may dramatically change the affinity (avidity) and even specificity in glycan-protein interactions. Continuing cross-comparison among different array platforms is necessary in order to evaluate such factors in array fabrication [81]. Secondly, the presentations of glycans printed on microplates or glass slides could be quite different from those of glycans present in the context of glycoproteins and glycolipids. Synthesis of neoglycoproteins and neoglycolipids for array presentation may partially address this issue [82-84]. However, future generations of glycan microarrays should routinely include arrays of natural glycoproteins (varied glycoforms also) and glycolipids in order to understand the precise contributions of the lipid and protein portions to glycan-protein recognitions. This will be particularly important when micro- and macro-multivalency is involved (likely with variable mechanisms). Thirdly, future generations of glycan microarrays should also consider the dynamic nature of glycans present on cell surface. Fluidic glycan arrays offer an intriguing avenue to mimicking the mobility of glycoconjugates (rafts) imbedded in a lipid bilayer [85]. With efforts in the development of new approaches and tools, glycan microarray technology is expected to continue to play key roles in accelerating the decoding of the glycome. Nonetheless, we should not lose track of the vital importance of also delineating quantitative mechanistic information from glycan-protein interactions where we can – striking a balance between rapid, qualitative and semi-quantitative methods on one hand, with a need to understand the subtleties of modes of binding, and how these influence selectivity and kinetics on the other hand will prove vital. Moreover, whilst we perhaps naturally gravitate to arrays that contain glycans (since they loosely mimic glycocalyx) we should also recognize that arraying of carbohydrate-binding proteins as analytical tools for glycoconjugate solutes can also prove powerful [86].
Fig. 5.
General issues in glycan array design
Glycan sequencing
Decoding the sequence and structure of glycans is of immediate importance in elucidating their biological functions and associated structure-activity relationships. Whilst proteins and nucleic acids can be routinely sequenced using well-established and robust methods, the sequencing of glycans is more challenging. In contrast to the well-defined unmodified building blocks for proteins (20 amino acids) and DNA (4 basic nucleotides) and their single backbone linkages (amide and phosphodiester, respectively), the possible natural monosaccharide building blocks for glycans far exceed the number for amino acids or nucleotides. Moreover, these monosaccharide building blocks display more diversity in their context of glycans (D- / L-, furanosyl / pyranosyl rings, regioisomerism of linkage, linear / branching, α- or β- anomers) when forming glycosidic bonds. These structures can be further complicated by diverse post-glycosylation modifications such as sulfation, acetylation, deoxygenation, amination, and phosphorylation. Moreover, natural glycans in many cases appear in the form of glycoconjugates (glycoproteins, glycolipids, proteoglycans) and are usually present as mixtures associated also with heterogeneity of their context in those platforms.
This diversity and heterogeneity of glycans therefore presents a unique challenge in sequencing and structural determination. Yet the level of difficulty and complexity in structural analysis varies depending on the nature and source of the glycans, which also dictate the choice of analytical strategies. Conventional and longstanding monosaccharide and permethylation analysis can provide useful initial information on monosaccharide composition and site of linkages on monosaccharide building blocks, but it has not typically been able to provide information on linkage types, linkage sequence, and anomeric configuration. MS and NMR are two most important tools for glycan sequencing and structural analysis and both technologies have now begun to take on greater challenges in structural characterization of complex oligosaccharides and glycoconjugates. For example, advances in mass spectrometry (nanoLC-ESI-MS, tandem MS, electron-capture dissociation and electron-transfer dissociation) have made it possible to reliably characterize N- and O-glycosylation of glycoproteins [87]. However, using MS it has been hitherto difficult to characterize regio- and stereo-isomers of glycans. Recent work on MS assignment of the stereochemistry and anomeric configuration of monosaccharides in oligosaccharides has shown promise in this direction [88,89]. MS coupled with specific enzymatic transformations also provides a useful tool for sequencing glycans, yet one current limitation is the availability of sufficiently diverse linkage-specific glycosidases.
Linear oligomers allow some simplification; for glycosaminoglycans (GAGs), recently, a top-down Fourier transform mass spectrometric sequencing technology was employed for sequencing bikunin, one of the simplest proteoglycans. Surprisingly, the GAG chain of proteoglycan bikunin was shown to have a “homogeneous”, defined sequence for activity [90]. ‘Top-down’ strategy preserves whole sequences that conventional bottom-up strategy might miss. This example also emphasizes the importance of the choice of method – for samples consisting of mixtures of numerous different glycosaminoglycans, such a top-down strategy might prove tedious and difficult.
NMR has been particularly useful for polysaccharide sequencing through the application of various homo- and hetero-nuclear coupling techniques, which allows reliable assignment of the ring forms of monosaccharides, anomeric configurations, linkage types, and monosaccharide sequential connections [91]. In addition, it can also provide detailed three-dimensional structures of glycans in the context of glycoconjugates [92]. However, NMR usually requires a relatively large quantity of samples, and the sensitivity of natural isotope abundance is an issue. Thus, development of NMR techniques with greater sensitivity for polysaccharides and glycoconjugates of natural isotope abundance is a high priority, whilst at the same time new methods that allow easy isotope labeling and enrichment of glycans with specific isotopes (13C, 15N, and/or 18O, by chemical, enzymatic, or biosynthetic means) would prove powerful.
Novel approaches to glycan functions
For chemical biologists, function should be a major goal. One criticism leveled at Chemical Glycobiology is that this aspect of our field is in its infancy as compared with methods for building and characterizing glycans. Key methods may offer incisive options as follows (Fig. 6).
Fig. 6.
Some chemical biology approaches to glycan functions
Glyco-chemical genetics
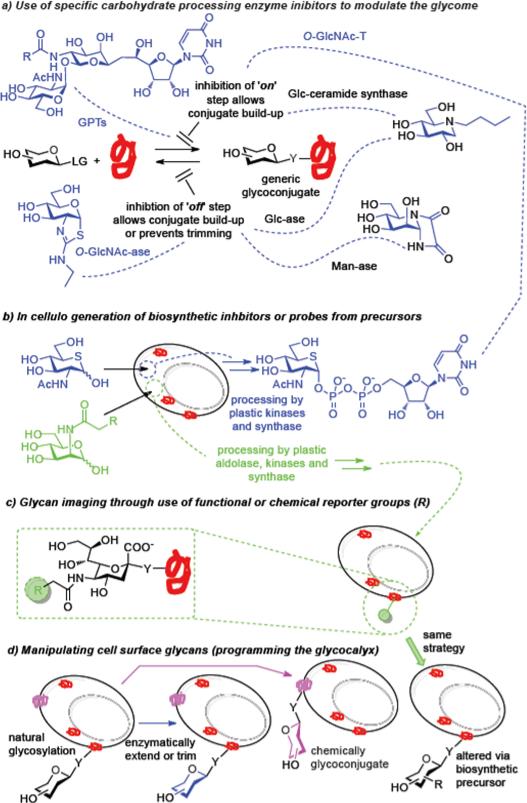
An important approach in Chemical Biology is to explore specific exogenous (small molecule) ligands to probe gene-product functions in a cellular or organismal context [93,94]. In this so-called ‘Chemical Genetics’ approach, small molecule ligands that bind directly to proteins with high affinity and specificity are used to perturb protein functions in cellular processes, resembling effects of genetic mutations. By analogy, novel small molecule probes that can specifically perturb the functions of glycans should be particularly useful in functional glycomic studies. In fact, glyco-chemical genetics is not new to our field, and many specific (natural or synthetic) inhibitors of glycan processing enzymes, including glycosidases and glycosyltransferases, have been discovered and implemented for manipulating functions in cells or in vivo [95]. Tunicamycin, a uridine analog isolated from Streptomyces that blocks the early steps of N-glycan assembly thus deleting N-glycosylation globally, has played a key role in elucidating the functions of protein N-glycosylation in protein folding, intracellular trafficking, and cellular communications. Inhibitors against polypeptide N-acetylgalactosaminyltransferases (ppGalNAcTs), a class of enzymes responsible for the attachment of the first sugar in mucin O-glycosylation have been discovered by screening a synthetic uridine-based small molecule library [96]; these inhibitors were shown to abrogate mucin-type O-linked glycosylation and to induce apoptosis in cultured cells. Recently, potent inhibitors against the two key enzymes (OGA and OGT) responsible for a dynamic O-GlcNAc glycosylation [97] were discovered [98,99]. As demonstrated in a mouse model of Alzheimer's disease, direct use of a sugar thiazoline-based O-GlcNAcase (OGA) inhibitor led to an increase of O-GlcNAc glycosylation level, resulting in the slowing of neurodegeneration without apparent adverse effects [100]. Precursors/prodrugs may also prove powerful; the acetylated 5-thio analog of GlcNAc could be taken into the biosynthetic pathway of UDP-GlcNAc in cells, leading to the in situ formation of UDP-5SGlcNAc, which inhibited the OGT (O-GlcNAc transferase) activity [99] that uses UDP-GlcNAc as a donor. Similarly, it was more recently demonstrated that selectively fluorinated sialic acids and L-fucose analogues could be taken up and transformed into respective sugar nucleotides that can also act as substrate-based inhibitors of enzymes that process them inside the cell [101]. Their application resulted in global down-regulation of sialylation and fucosylation with clear consequent changes to glycome. The resulting functional consequence in myeloid cells was a loss of selectin binding and impaired leukocyte rolling [101] due to a reduced display of the associated sialylated and fucosylated glycoconjugates that act as selectin ligands.
In addition to global inhibition of glycosylation, individual glycan functions can be also dissected through the use of more specific inhibitors or molecular probes. These include the use of chemical primers serving as competing acceptor substrates for glycosyltransferases, the use of inhibitors against specific glycan-processing enzymes such as distinct α-mannosidase inhibitors, and the metabolic incorporation of sugar chain terminators to generate truncated glycoforms [11]. We know that structure-based design can lead to the discovery of potent inhibitors, as powerfully and now archetypally exemplified by the class of influenza sialidases – these were successfully developed into two widely used anti-influenza drugs, Relenza (zanamivir) and Tamiflu (oseltamivir) [102,103]. These inhibitors were designed to mimic the transition state of a sialoside during enzymatic hydrolysis, with the installation of a guanidinium or ammonium group at the C-4 position for ion pairing with the positively charged residues in the binding pocket, thus enhancing the affinity and specificity for viral sialidases over human enzymes. We are therefore aware as a field that mechanism can prove uniquely important in designs that generate potent small molecules as probes and drugs. More recently, novel mechanism-based influenza sialidase inhibitors were designed based on the structures of Relenza and Tamiflu in which fluorine atoms were introduced at the C-2 and C-3 positions [104]. These new inhibitors react specifically with influenza sialidases and inactivate the viral enzymes via the formation of a stable, covalently linked inhibitor-enzyme intermediate. Notably, these mechanism-based inhibitors demonstrate broad-spectrum antiviral activity against those drug-resistant influenza strains, making them attractive for further drug development.
As well as targeting catalytic function, synthetic glycan-based ligands are emerging as highly valuable tools to probe specific glycan-protein interactions in cellular processes. An example is the discovery of multivalent sialylated glycan ligands for CD22, a sialic acid–binding lectin from the Siglec family that attenuates B cell receptor signaling, for suppressing B cell activation [105,106]. It was found that exposure of B cells to sialylated antigens led to inhibition of key steps in B cell receptor signaling and induced B cell tolerance. These studies showcase the value of the glycochemical genetic approach to dissecting cellular functions that would be otherwise difficult to unveil by traditional genetic methods. The power of chemistry therefore allows us to consider the creation of even quite large hybrid molecules in pure form to act as such perturbing probes or molecules. A so-called glycodendrinanoparticle (with a valency of up to 1620) was constructed in homogeneous form recently through the use of a nested multivalency in both glycodendrimer attachments and the self-assembly of many proteins into a virus-like particle [107]. The resulting constructs proved to be powerful binders of dendritic cell (DC) lectin DC-SIGN and picomolar inhibitors showing large multivalency effects in models of DC infection by Ebola virus.
The unique advantage of the glycochemical genetic approach using molecular ligands or inhibitors as probes is to perturb functions of a cellular process in a dynamic, reversible and tunable manner. This complements well the traditional genetic approaches that make “permanent” alternations of functions. However, the success of glycochemical genetic approach relies on the discovery of high-quality and specific carbohydrate-based or noncarbohydrate-based probes. Future studies should be directed not only to systematic, high-throughput screening of small-molecule compounds targeting specific sites or stages of glycan functions but also to the rational design of high-affinity ligands or inhibitors specific for glycan-recognition receptors and enzymes; and the design of novel probes that are suitable for in vivo (not just in vitro) perturbation of glycan functions.
Glycan imaging
The ability to directly detect the changes of glycans in biological systems offers an exciting opportunity to decipher their roles and functions in normal physiological and disease states. While lectins and glycan-specific antibodies have been widely used for many years for detecting cell-surface glycans and for glycan visualization on tissue sections, they are not widely suitable for all in vivo imaging (e.g., due to cytotoxicity and/or membrane impermeability) and the truly characterized selectivities of many ‘anti-sugar’ antibodies can be unclear or even absent. One emerging technology for glycan imaging is the metabolic engineering of glycans with so-called bioorthogonal chemical reporters [108,109]. In this approach, a specific chemical reporter is incorporated into the glycan via biosynthetic incorporation of a sugar precursor modified with the chemical reporter. The reporter group is then visualized by covalent reaction with an imaging probe that can specifically detect and react with the chemical reporter. The currently most common chemical reporters are azide or alkyne groups, since they are small and relatively biologically inert and they have the potential to react specifically under physiological conditions. For example, an azide reporter has been detected by reactions with a biotin-tagged phosphine via Staudinger reaction or with a fluorescence-tagged alkyne via copper-catalyzed azide-alkyne cycloaddition reaction [110]. To avoid perceived cytotoxicity of the copper catalyst in living system, strain-promoted azide-alkyne cycloadditions has also been devised that use cyclooctyne derivatives as reactants, enabling reaction without copper mediation [111-114]. Early focus was dominated by imaging of cell surface sialylated glycans using azide- or alkyne-tagged ManNAc variants (ManNAz or ManNAl) as precursors. Since then the metabolic glycan imaging approach has been expanded to the imaging of other types of glycans including mucin-type O-glycans (using azide-tagged GalNAc as the precursor), O-GlcNAc glycoproteins (using azide-tagged GlcNAc as the precursor), and fucosylated glycans (using 6-azido-fucose or alkynyl fucose as the precursor). In addition, the imaging technology has also expanded from its use in cultured cell systems to living organisms including zebrafish and mouse [110].
Glycan imaging provides an important tool for deciphering the localization and trafficking of glycans in cellular processes. Moreover, noninvasive glycan imaging in living animals offers a unique opportunity to detect, monitor, and quantify the dynamic changes of glycans associated with development and disease states, which may unveil novel biomarkers for diagnosis and important targets for drug discovery. However, a potential limitation of the metabolic glycan imaging is the lack of cell type or tissue specificity, as the injected sugar precursors would be incorporated into all the shared glycan biosynthetic pathways. A recently reported cell-specific metabolic labeling strategy that uses ligand-targeted liposomes to encapsulate and selectively deliver azidosugar precursor to target cells provides a promising solution to this circumvent this problem [115]. Future studies should be directed to the exploration of novel biosynthetic pathways to cover additional types of glycans in the glycome, and to understanding their limitations, when sugar precursors are biosynthetically used for more than one purpose [116]; new unnatural sugar precursors can be envisaged that possess better biocompatibility and favorable pharmacokinetic properties; and new orthogonal chemical reporters are needed that permit simultaneous imaging of different types of glycans in real time.
Selective cell-surface glycan remodeling
Diverse subtypes of glycans are present on cell surfaces. Site-specific remodeling of this glycocalyx provides an attractive approach to altering and understanding cell surface functions as well gaining novel ones. One notable example is the ex vivo glycan engineering of CD44 on human mesenchymal stem cells (hMSCs) that enabled redirection of cell migration to bone [117]. Thus, the native CD44 glycoform on MSCs that lacks the key α-1,3-fucosyl residue was converted into a hematopoietic cell E-selectin/L-selectin ligand via enzymatic α-1,3-fucosylation under mild physiological conditions; this conferred potent E-selectin binding and tropism to bone, without drastic effects on cell viability or multipotency. This and a subsequent study [118] unveils a great potential to program cellular trafficking via chemical engineering of glycans on a distinct membrane glycoprotein for directing cellular migration.
Another example is the development of universal red blood cells through enzymatic removal of blood group ABO antigens on the cell surface [119]. While this idea had been considered previously, a practical approach to remodeling the ABO antigens had not been fulfilled because of the lack of enzymes to efficiently remove the GalNAc and galactose residues present in the group A and group B antigens, respectively, without affecting red blood cells’ integrity. The discovery of two specific bacterial glycosidases, an α-GalNAcase and an α-galactosidase, provided new hope; these are efficient and specific for cleavage of the A and B immunodominant monosaccharides under the mild conditions suitable for maintaining the integrity and functions of red blood cells [120]. This study suggests a practical approach to transforming group A, B, and AB red blood cells to the universal group O type for transfusion. The study also emphasized the importance of discovering new, more efficient methods for glycan-specific cell-surface engineering for basic and clinical applications. Whilst many of these methods have relied to date on alterations using naturally-derived enzyme activities other approaches can be envisaged. The directed evolution of novel glycose oxidases offers tantalizing opportunities in this regard [51] . Catalytic chemical methods may also be considered; the Pd-catalyzed remodeling of cell surfaces through direct C–C bond formation [121] has recently been applied to the remodeling of bacterial glycocalyx leading to a ‘chemical differentiation’ that alters cellular interactions as determined by appropriate, different chemical glycosylating agents [122].
Conclusion
As a final general comment, we suggest that in this burgeoning discipline we may need to be more clear-minded than in many other fields. Whilst an unnecessary (and often false) opposition is sometimes created between “blue skies” research and applied science there is, nonetheless, a clear and useful distinction between the creation of knowledge (science) and its use (technology). This is particularly pertinent to Glycoscience since one tempting way that we might consider solving some problems that we face is through the use of ‘bigger-and-better’ and ‘more-and-more’. In the field of arrays there is an argument that once we have a big enough array with enough sugars then we will be able to ‘probe’ every interaction. In the field of ‘glycoproteomics’ there is an argument that if we perform large enough, comprehensive ‘top-down’ proteomic studies then we will see sufficient examples to get a better handle on the patterns that are relevant to Glycobiology.
Both of us would like to finish by highlighting that whilst such ‘powerful’ resource-driven analyses will create great quantities of very useful data, the strength of many academic institutions is best found in asking the right questions and creating the experiment to answer them. The size of the many challenges in our field means that this may end up being even more vital for our community than for others; it may be that Chemical Glycobiology is one of the first to be tested by the temptations of large amounts of correlative data that lack causal links. Sometimes we decide as a community that where we currently do not yet understand or see the direct causal links that we wish for, this might simply be because we haven't yet looked widely enough or deeply enough. This argument to ‘do more’ is one approach. Yet another approach, perhaps, is to pause and think about new mechanisms and roles for sugars in Biology? There is the temptation for us to imbue sugars with ‘meaning’ that might not be there. Why does nature preserve such a diversity and complexity of glycan after so many years of evolution? Is the heterogeneity of glycoforms a requirement for functions or is it simply the results of a less perfect control of the biosynthetic endeavors? Why might an additional GlcNAc on a protein have function cf an additional hydroxyl? Although we know that hydroxylation and GlcNAcylation have specific functional effects it may also be that sometimes they provide simply a register or ‘read-out’ of a background organismal state. There will be a long list of questions and we have just begun to decode the function and dynamics of the glycome of any given organism. It becomes clear that Chemical Biology, which provides unique molecular tools and approaches, will continue to play increasingly important roles in deciphering the diverse functions of glycans in biological systems.
Acknowledgements
This work was supported in part by the US National Institutes of Health (NIH grants R01 GM080374 and R01 GM096973 to LXW) and the Bill and Melinda Gates Foundation, Cancer Research UK and the UK research councils (EPSRC, BBSRC, MRC, to BGD). BGD is Royal Society Wolfson Merit Award recipient.
References
- 1.Rademacher TW, Parekh RB, Dwek RA. Annu. Rev. Biochem. 1988;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 2.Dwek RA. Chem. Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 4.Hart GW, Copeland RJ. Cell. 2010;143:672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marth JD, Grewal PK. Nat. Rev. Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jefferis R. Nat. Rev. Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 7.Crick F. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laine RA. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 10.Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 11.Prescher JA, Bertozzi CR. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Seeberger PH. Nat. Chem. Biol. 2009;5:368–372. doi: 10.1038/nchembio0609-368. [DOI] [PubMed] [Google Scholar]
- 13.Boltje TJ, Buskas T, Boons GJ. Nat. Chem. 2009;1:611–622. doi: 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiessling LL, Splain RA. Annu. Rev. Biochem. 2010;79:619–653. doi: 10.1146/annurev.biochem.77.070606.100917. [DOI] [PubMed] [Google Scholar]
- 15.Wu CY, Wong CH. Chem. Commun. (Camb) 2011;47:6201–6207. doi: 10.1039/c0cc04359a. [DOI] [PubMed] [Google Scholar]
- 16.Rillahan CD, Paulson JC. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verez-Bencomo V, Fernandez-Santana V, Hardy E, Toledo ME, Rodriguez MC, Heynngnezz L, Rodriguez A, Baly A, Herrera L, Izquierdo M, Villar A, Valdes Y, Cosme K, Deler ML, Montane M, Garcia E, Ramos A, Aguilar A, Medina E, Torano G, Sosa I, Hernandez I, Martinez R, Muzachio A, Carmenates A, Costa L, Cardoso F, Campa C, Diaz M, Roy R. Science. 2004;305:522–525. doi: 10.1126/science.1095209. [DOI] [PubMed] [Google Scholar]
- 18.Sears P, Wong CH. Science. 2001;291:2344–2350. doi: 10.1126/science.1058899. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaou KC, Mitchell HJ. Angew. Chem. Int. Ed. 2001;40:1576–1624. [PubMed] [Google Scholar]
- 20.Boons GJ, Demchenko AV. Chem. Rev. 2000;100:4539–4566. doi: 10.1021/cr990313g. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, Schmidt RR. Angew. Chem. Int. Ed. 2009;48:1900–1934. doi: 10.1002/anie.200802036. [DOI] [PubMed] [Google Scholar]
- 22.Wang CC, Lee JC, Luo SY, Kulkarni SS, Huang YW, Lee CC, Chang KL, Hung SC. Nature. 2007;446:896–899. doi: 10.1038/nature05730. [DOI] [PubMed] [Google Scholar]
- 23.Petitou M, van Boeckel CA. Angew. Chem. Int. Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 24.Galonic DP, Gin DY. Nature. 2007;446:1000–1007. doi: 10.1038/nature05813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Y, Chen J, Wan Q, Wilson RM, Danishefsky SJ. Biopolymers. 2010;94:373–384. doi: 10.1002/bip.21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne RJ, Wong CH. Chem. Commun. (Camb) 2010;46:21–43. doi: 10.1039/b913845e. [DOI] [PubMed] [Google Scholar]
- 27.Piontek C, Varon Silva D, Heinlein C, Pohner C, Mezzato S, Ring P, Martin A, Schmid FX, Unverzagt C. Angew. Chem. Int. Ed. 2009;48:1941–1945. doi: 10.1002/anie.200804735. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto I, Tezuka K, Fukae K, Ishii K, Taduru K, Maeda M, Ouchi M, Yoshida K, Nambu Y, Igarashi J, Hayashi N, Tsuji T, Kajihara Y. J. Am. Chem. Soc. 2012;134:5428–5431. doi: 10.1021/ja2109079. [DOI] [PubMed] [Google Scholar]
- 29.Huang M, Retailleau P, Bohe L, Crich D. J. Am. Chem. Soc. 2012;134:14746–14749. doi: 10.1021/ja307266n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boltje TJ, Kim JH, Park J, Boons GJ. Nat. Chem. 2010;2:552–557. doi: 10.1038/nchem.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmaltz RM, Hanson SR, Wong CH. Chem. Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 32.Palcic MM. Curr. Opin. Chem. Biol. 2011;15:226–233. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 34.Barreteau H, Richard E, Drouillard S, Samain E, Priem B. Carbohydr. Res. 2012;360:19–24. doi: 10.1016/j.carres.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Fierfort N, Samain E. J. Biotechnol. 2008;134:261–265. doi: 10.1016/j.jbiotec.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Wang LX, Huang W. Curr. Opin. Chem. Biol. 2009;13:592–600. doi: 10.1016/j.cbpa.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perugino G, Trincone A, Rossi M, Moracci M. Trends Biotechnol. 2004;22:31–37. doi: 10.1016/j.tibtech.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Hancock SM, Vaughan MD, Withers SG. Curr. Opin. Chem. Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 39.MacKenzie LF, Wang Q, Warren RAJ, Withers SG. J. Am. Chem. Soc. 1998;120:5583–5584. [Google Scholar]
- 40.Malet C, Planas A. FEBS Lett. 1998;440:208–212. doi: 10.1016/s0014-5793(98)01448-3. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Chen X. Org. Biomol. Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J. Science. 2011;334:498–501. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodfellow JJ, Baruah K, Yamamoto K, Bonomelli C, Krishna B, Harvey DJ, Crispin M, Scanlan CN, Davis BG. J. Am. Chem. Soc. 2012;134:8030–8033. doi: 10.1021/ja301334b. [DOI] [PubMed] [Google Scholar]
- 44.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. J. Am. Chem. Soc. 2012;134:12308–12318. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 46.Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat. Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- 47.Shaikh FA, Withers SG. Biochem. Cell Biol. 2008;86:169–177. doi: 10.1139/O07-149. [DOI] [PubMed] [Google Scholar]
- 48.Williams GJ, Goff RD, Zhang C, Thorson JS. Chem. Biol. 2008;15:393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams GJ, Zhang C, Thorson JS. Nat. Chem. Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 50.Hancock SM, Rich JR, Caines ME, Strynadka NC, Withers SG. Nat. Chem. Biol. 2009;5:508–514. doi: 10.1038/nchembio.191. [DOI] [PubMed] [Google Scholar]
- 51.Rannes JB, Ioannou A, Willies SC, Grogan G, Behrens C, Flitsch SL, Turner NJ. J. Am. Chem. Soc. 2011;133:8436–8439. doi: 10.1021/ja2018477. [DOI] [PubMed] [Google Scholar]
- 52.Wang LX, Lomino JV. ACS Chem. Biol. 2012;7:110–122. doi: 10.1021/cb200429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. Science. 2009;326:1187. doi: 10.1126/science.326.5957.1187-a. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. Science. 2004;303:371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt MJ, Summerer D. ChemBioChem. 2012;13:1553–1557. doi: 10.1002/cbic.201200321. [DOI] [PubMed] [Google Scholar]
- 56.Schwarz F, Huang W, Li C, Schulz BL, Lizak C, Palumbo A, Numao S, Neri D, Aebi M, Wang LX. Nat. Chem. Biol. 2010;6:264–266. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. Proc. Natl. Acad. Sci. USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wetter M, Kowarik M, Steffen M, Carranza P, Corradin G, Wacker M. Glycoconj. J. 2012 doi: 10.1007/s10719-012-9451-9. [DOI] [PubMed] [Google Scholar]
- 59.Chalker JM, Bernardes GJ, Davis BG. Acc. Chem. Res. 2011;44:730–741. doi: 10.1021/ar200056q. [DOI] [PubMed] [Google Scholar]
- 60.Fernández-González M, Boutureira O, Bernardes GJL, Chalker JM, Young MA, Errey JC, Davis BG. Chem. Sci. 2010;1:709–715. [Google Scholar]
- 61.Seeberger PH. Carbohydr. Res. 2008;343:1889–1896. doi: 10.1016/j.carres.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 62.Schuerch C, Frechet JM. J. Am. Chem. Soc. 1971;93:492–496. [Google Scholar]
- 63.Plante OJ, Palmacci ER, Seeberger PH. Science. 2001;291:1523–1527. doi: 10.1126/science.1057324. [DOI] [PubMed] [Google Scholar]
- 64.Seeberger PH. Chem. Soc. Rev. 2008;37:19–28. doi: 10.1039/b511197h. [DOI] [PubMed] [Google Scholar]
- 65.Jaipuri FA, Pohl NL. Org. Biomol. Chem. 2008;6:2686–2691. doi: 10.1039/b803451f. [DOI] [PubMed] [Google Scholar]
- 66.Hsu CH, Hung SC, Wu CY, Wong CH. Angew. Chem. Int. Ed. 2011;50:11872–11923. doi: 10.1002/anie.201100125. [DOI] [PubMed] [Google Scholar]
- 67.Matsushita T, Nagashima I, Fumoto M, Ohta T, Yamada K, Shimizu H, Hinou H, Naruchi K, Ito T, Kondo H, Nishimura S. J. Am. Chem. Soc. 2010;132:16651–16656. doi: 10.1021/ja106955j. [DOI] [PubMed] [Google Scholar]
- 68.Martin JG, Gupta M, Xu Y, Akella S, Liu J, Dordick JS, Linhardt RJ. J. Am. Chem. Soc. 2009;131:11041–11048. doi: 10.1021/ja903038d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mong TK, Lee HK, Duron SG, Wong CH. Proc. Natl. Acad. Sci. USA. 2003;100:797–802. doi: 10.1073/pnas.0337590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaeothip S, Demchenko AV. Carbohydr. Res. 2011;346:1371–1388. doi: 10.1016/j.carres.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang X, Huang L, Wang H, Ye XS. Angew. Chem. Int. Ed. 2004;43:5221–5224. doi: 10.1002/anie.200460176. [DOI] [PubMed] [Google Scholar]
- 72.Sun B, Srinivasan B, Huang X. Chem. Eur. J. 2008;14:7072–7081. doi: 10.1002/chem.200800757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Xu Y, Yang B, Tiruchinapally G, Sun B, Liu R, Dulaney S, Liu J, Huang X. Chem. Eur. J. 2010;16:8365–8375. doi: 10.1002/chem.201000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oyelaran O, Gildersleeve JC. Curr. Opin. Chem. Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang PH, Wu CY, Greenberg WA, Wong CH. Curr. Opin. Chem. Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oyelaran O, Gildersleeve JC. Expert Rev. Vaccines. 2007;6:957–969. doi: 10.1586/14760584.6.6.957. [DOI] [PubMed] [Google Scholar]
- 77.Laurent N, Haddoub R, Voglmeir J, Flitsch SL. Methods Mol. Biol. 2012;808:269–284. doi: 10.1007/978-1-61779-373-8_19. [DOI] [PubMed] [Google Scholar]
- 78.Ban L, Pettit N, Li L, Stuparu AD, Cai L, Chen W, Guan W, Han W, Wang PG, Mrksich M. Nat. Chem. Biol. 2012;8:769–773. doi: 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat. Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 80.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Chem. Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Padler-Karavani V, Song X, Yu H, Hurtado-Ziola N, Huang S, Muthana S, Chokhawala HA, Cheng J, Verhagen A, Langereis MA, Kleene R, Schachner M, de Groot RJ, Lasanajak Y, Matsuda H, Schwab R, Chen X, Smith DF, Cummings RD, Varki A. J. Biol. Chem. 2012;287:22593–22608. doi: 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oyelaran O, Li Q, Farnsworth D, Gildersleeve JC. J. Proteome Res. 2009;8:3529–3538. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Godula K, Rabuka D, Nam KT, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:4973–4976. doi: 10.1002/anie.200805756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feizi T, Chai W. Nat. Rev. Mol. Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 85.Zhu XY, Holtz B, Wang Y, Wang LX, Orndorff PE, Guo A. J. Am. Chem. Soc. 2009;131:13646–13650. doi: 10.1021/ja902783n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Propheter DC, Hsu KL, Mahal LK. Methods Mol. Biol. 2011;723:67–77. doi: 10.1007/978-1-61779-043-0_6. [DOI] [PubMed] [Google Scholar]
- 87.North SJ, Hitchen PG, Haslam SM, Dell A. Curr. Opin. Struct. Biol. 2009;19:498–506. doi: 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Konda C, Bendiak B, Xia Y. J. Am. Soc. Mass Spec. 2012;23:347–358. doi: 10.1007/s13361-011-0287-5. [DOI] [PubMed] [Google Scholar]
- 89.Fang TT, Bendiak B. J. Am. Chem. Soc. 2007;129:9721–9736. doi: 10.1021/ja0717313. [DOI] [PubMed] [Google Scholar]
- 90.Ly M, Leach FE, 3rd, Laremore TN, Toida T, Amster IJ, Linhardt RJ. Nat. Chem. Biol. 2011;7:827–833. doi: 10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin-Pastor M, Canales-Mayordomo A, Jimenez-Barbero J. J. Biomol. NMR. 2003;26:345–353. doi: 10.1023/a:1024096807537. [DOI] [PubMed] [Google Scholar]
- 92.Barb AW, Prestegard JH. Nat. Chem. Biol. 2011;7:147–153. doi: 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stockwell BR. Nat. Rev. Genetics. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schreiber SL. Nat. Chem. Biol. 2005;1:64–66. doi: 10.1038/nchembio0705-64. [DOI] [PubMed] [Google Scholar]
- 95.Gloster TM, Vocadlo DJ. Nat. Chem. Biol. 2012;8:683–694. doi: 10.1038/nchembio.1029. [DOI] [PubMed] [Google Scholar]
- 96.Hang HC, Yu C, Ten Hagen KG, Tian E, Winans KA, Tabak LA, Bertozzi CR. Chem. Biol. 2004;11:337–345. doi: 10.1016/j.chembiol.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 97.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, Vocadlo DJ. Nat. Chem. Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 99.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Nat. Chem. Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Nat. Chem. Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 101.Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Nat. Chem. Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 103.von Itzstein M. Nat. Rev. Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 104.Kim JH, Resende R, Wennekes T, Chen HM, Bance N, Buchini S, Watts AG, Pilling P, Streltsov VA, Petric M, Liggins R, Barrett S, McKimm-Breschkin JL, Niikura M, Withers SG. Science. 2013 doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 105.Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, Kiessling LL. Proc. Natl. Acad. Sci. USA. 2009;106:2500–2505. doi: 10.1073/pnas.0807207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, Kubitz M, Bovin N, Paulson JC, Nemazee D. J. Exp. Med. 2010;207:173–187. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ribeiro-Viana R, Sanchez-Navarro M, Luczkowiak J, Koeppe JR, Delgado R, Rojo J, Davis BG. Nat. Commun. 2012;3:1303. doi: 10.1038/ncomms2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 109.Laughlin ST, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sletten EM, Bertozzi CR. Acc. Chem. Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 113.Sanders BC, Friscourt F, Ledin PA, Mbua NE, Arumugam S, Guo J, Boltje TJ, Popik VV, Boons GJ. J. Am. Chem. Soc. 2011;133:949–957. doi: 10.1021/ja1081519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Poloukhtine AA, Mbua NE, Wolfert MA, Boons GJ, Popik VV. J. Am. Chem. Soc. 2009;131:15769–15776. doi: 10.1021/ja9054096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xie R, Hong S, Feng L, Rong J, Chen X. J. Am. Chem. Soc. 2012;134:9914–9917. doi: 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- 116.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2011 doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, Wohlgemuth R. Nat. Med. 2008;14:181–187. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- 118.Thankamony SP, Sackstein R. Proc. Natl. Acad. Sci. USA. 2011;108:2258–2263. doi: 10.1073/pnas.1018064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Daniels G, Withers SG. Nat. Biotechnol. 2007;25:427–428. doi: 10.1038/nbt0407-427. [DOI] [PubMed] [Google Scholar]
- 120.Liu QP, Sulzenbacher G, Yuan H, Bennett EP, Pietz G, Saunders K, Spence J, Nudelman E, Levery SB, White T, Neveu JM, Lane WS, Bourne Y, Olsson ML, Henrissat B, Clausen H. Nat. Biotechnol. 2007;25:454–464. doi: 10.1038/nbt1298. [DOI] [PubMed] [Google Scholar]
- 121.Spicer CD, Triemer T, Davis BG. J. Am. Chem. Soc. 2012;134:800–803. doi: 10.1021/ja209352s. [DOI] [PubMed] [Google Scholar]
- 122.Spicer CD, Davis BG. Chem. Commun. (Camb) 2013;49:2747–2749. doi: 10.1039/c3cc38824g. [DOI] [PubMed] [Google Scholar]