Abstract
During the establishment of an infection, bacterial pathogens encounter oxidative stress resulting in the production of DNA lesions. Majority of these lesions are repaired by base excision repair (BER) pathway. Amongst these, abasic sites are the most frequent lesions in DNA. Class II apurinic/apyrimidinic (AP) endonucleases play a major role in BER of damaged DNA comprising of abasic sites. Mycobacterium tuberculosis, a deadly pathogen, resides in the human macrophages and is continually subjected to oxidative assaults. We have characterized for the first time two AP endonucleases namely Endonuclease IV (End) and Exonuclease III (XthA) that perform distinct functions in M.tuberculosis. We demonstrate that M.tuberculosis End is a typical AP endonuclease while XthA is predominantly a 3′→5′ exonuclease. The AP endonuclease activity of End and XthA was stimulated by Mg2+ and Ca2+ and displayed a preferential recognition for abasic site paired opposite to a cytosine residue in DNA. Moreover, End exhibited metal ion independent 3′→5′ exonuclease activity while in the case of XthA this activity was metal ion dependent. We demonstrate that End is not only a more efficient AP endonuclease than XthA but it also represents the major AP endonuclease activity in M.tuberculosis and plays a crucial role in defense against oxidative stress.
Introduction
Cellular DNA is subjected to continuous assaults by a wide variety of intimidating endogenous and exogenous agents. Amongst these, the predominant damage is caused by reactive oxygen species (ROS) that are formed as by-products of oxidative metabolism in organisms with aerobic respiration. Exogenous agents such as chemical carcinogens and ionizing radiation also generate ROS. These oxygen radicals mostly produce non-bulky DNA lesions that are substrates for the base excision repair (BER) pathway [1], [2]. This pathway is highly conserved from bacteria to humans [3]. Apurinic and apyrimidinic (AP) sites (also known as abasic sites), are the most common DNA lesions that arise during the spontaneous loss of normal or damaged bases or by the action of DNA glycosylases that release modified or mismatched bases from DNA [4], [5], [6]. The AP sites threaten genetic stability because they block replication and are mutagenic [7], [8]. Thus, the removal of AP sites in DNA by a Class II AP endonuclease is a crucial step in BER. AP endonuclease enzymes initiate the repair of abasic sites in DNA by cleavage of the DNA backbone immediately 5′ of an AP residue. A single-nucleotide gap is then created in the DNA by the action of 5′ deoxyribose phosphodiesterase. The base excision repair is completed by resynthesis of the DNA by DNA polymerase followed by the action of DNA ligase [9].
The Class II AP endonucleases have been classified into two families, the exonuclease III (ExoIII) and endonuclease IV (EndoIV) families, based on their homology to the two Escherichia coli enzymes. In E.coli the major AP endonuclease is ExoIII (Ec-ExoIII) representing 90% of the cellular AP endonuclease activity, while EndoIV (Ec-EndoIV) accounts for 10% of the total activity [10]. The E.coli AP endonucleases also exhibit additional 3′ phosphatase and 3′ phosphodiesterase activities in common. These activities are responsible for removing a multitude of blocking groups, including 3′ phosphate and 3′ phosphoglycolate, that are present at single-stranded breaks in DNA, induced by oxidative agents [11], [12]. In addition, Ec-EndoIV and Ec-ExoIII exhibit 3′→5′ exonuclease activity. Saccharomyces cerevisiae also possesses two AP endonucleases, the Apn1 and Apn2 proteins that represent the EndoIV and the ExoIII family, respectively [13]. However, the major AP endonuclease in this organism is Apn1, that exhibits a strong AP endonuclease activity in yeast cells while the Apn2 protein, is a weak AP endonuclease that exhibits strong 3′→5′ exonuclease and 3′ phosphodiesterase activities [13], [14], [15], [16]. Contrary to the above discovery in budding yeast, in the fission yeast Schizosaccharomyces pombe, Apn2 provides the major AP endonuclease activity while the Apn1 serves only as a back up activity [17]. The human AP endonucleases, Ape1 and Ape2, are both members of the ExoIII family where Ape1 is the major human AP endonuclease, while EndoIV homologs are not known in humans [18]. Amidst this vast heterogeneity observed amongst different species with respect to the function(s) of AP endonucleases, in the current study, we were interested in deciphering the role of AP endonucleases in mycobacteria.
Mycobacterium tuberculosis, is a highly successful pathogen, killing approximately 1.4 million infected people annually [19]. In the host, this bacterium resides in macrophage in an environment rich in reactive oxygen and nitrogen intermediates capable of damaging its genome [20], [21]. To maintain its genome integrity, the bacterium must possess robust DNA repair machinery. Further, the GC rich (∼66%) genome of this pathogen renders it much more susceptible to cytosine deamination (generating uracil) and guanine oxidation [predominantly generating 8-oxoguanine (8-oxoG)] than other intracellular bacteria. This has led to special interest in the BER pathways that repair uracil [22] and 8-oxoG [23] in mycobacteria and it is thought that BER may play a central role in maintaining the integrity of DNA in this bacterium in the absence of any recognized homologs of mismatch repair [6], [24]. However, in spite of the predicted importance of base excision repair to this pathogen [6], [25], studies in BER have been primarily restricted to biochemical and functional characterization of DNA glysosylases such as uracil DNA glycosylase (Ung,UdgB) [22], [26], formamidopyrimidine DNA glycosylase (MutM) [27], [28], [29] and adenine DNA glycosylase (MutY) [23]. The AP sites that result from the action of DNA glycosylases are cytotoxic and mutagenic. In fact, the inability to repair these AP sites is detrimental to the survival in the host as seen in various pathogens such as Salmonella, Neisseria, Toxoplasma etc. [30], [31], [32]. However, to the best of our knowledge no studies have focused on the role and characterization of AP endonucleases in M.tuberculosis. An understanding about the role of these enzymes in M.tuberculosis would shed light on the repair processes that this pathogen employs to counter the host defense mechanism(s) and help evolve strategies to combat TB.
The sequencing of M.tuberculosis genome revealed the presence of Ec-EndoIV and Ec-ExoIII homologs namely Endonuclease IV (End) and Exonuclease III (XthA), that are encoded by the genes end (Rv0670) and xthA (Rv0427c), respectively [33]. In the current study, for the first time, biochemical and functional characterization of these proteins has been carried out in M.tuberculosis.
Materials and Methods
Bacterial Strains and Growth Conditions
E.coli XL-1Blue (Stratagene) and BL21 (λDE3) (Novagen) strains were employed for cloning and protein expression, respectively. E.coli was propagated in Luria-Bertani (LB) media. Mycobacterial strains were grown on Middlebrook 7H11 (MB7H11) agar supplemented with 10% oleic-acid albumin dextrose catalase (OADC) or in MB7H9 medium supplemented with 10% ADC, 0.2% glycerol, and 0.05% Tween 80 at 37°C with shaking at 200 rpm. Recombineering method [34] was employed to generate mutants of end (MtbΔend), xthA (MtbΔxthA) and both the genes together (MtbΔendΔxthA) in M.tuberculosis H37Rv. end and xthA mutants were complemented extra-chromosomally on a plasmid to generate complements MtbΔendComp and MtbΔxthAComp, respectively. The strains were confirmed by immunoblotting (Puri, R.V. et. al, unpublished data). Ampicillin was used at a concentration of 50 µg/ml for E.coli. Kanamycin and chloramphenicol were used at concentrations of 25 µg/ml and 30 µg/ml, respectively. Hygromycin was used at a concentration of 50 µg/ml for mycobacteria or at 150 µg/ml for E.coli.
Expression of end and xthA and Purification of the Encoded Proteins
The AP endonuclease genes end (Rv0670) and xthA (Rv0427c) were PCR amplified by using M.tuberculosis H37Rv genomic DNA as the template by using primers (Table S1) designed from the published genome sequence. The primers were designed such that the expressed proteins comprise of a streptavidin tag at their C-terminus. PCR amplified end and xthA were separately cloned into the pet-21c(+) expression vector (Novagen) by using NheI and HindIII restriction sites to generate pet-21c-end and pet-21c-xthA constructs, respectively. The constructs were confirmed by DNA sequencing.
E.coli BL21 (λDE3) cells were separately transformed with pet-21c-end and pet-21c-xthA . Cells were grown to an A600 nm of 0.8 in LB media containing 50 µg/ml of ampicillin and synthesis of proteins was induced by the addition of 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) followed by incubation at 18°C for 18 hours with constant shaking at 200 rpm. Cells were resuspended in 20 mM Tris-HCl (pH 8.0), containing 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 mM β-mercaptoethanol (buffer A) followed by sonication; lysates were clarified by centrifugation. These lysates were subjected to Strep-tactin® Superflow® affinity chromatography columns (IBA) pre-equilibrated with buffer A for further purification. The column was washed with two column volumes of the same buffer to remove unbound proteins. Proteins were eluted with 2.5 mM desthiobiotin in buffer A and analysed on SDS-PAGE. The fractions containing proteins (End and XthA) were pooled separately and dialyzed against 20 mM Tris-HCl (pH 8.0). Further purification of End was carried out by anion exchange chromatography by using Resource Q (GE Healthcare) pre-equilibrated with 20 mM Tris-HCl (pH 8.0), on an AKTA FPLC purifier (GE Healthcare). End was eluted from the Resource Q column by using a linear gradient of 0 to 0.5 M NaCl in the same buffer; protein eluted at 100 mM NaCl and was dialyzed against 20 mM Tris-HCl (pH 8.0) prior to storage. XthA was purified by using gel filtration chromatography by using Superdex 200 (GE Healthcare) column, pre-equilibrated with 20 mM Tris-HCl (pH 8.0). The purity of each protein was analyzed by SDS–PAGE on a 12.5% polyacrylamide gel. Purified proteins were collected and stored as aliquots at −80°C till further use.
DNA Substrates
Oligonucleotides (Table 1 and Table S1) were purchased from Sigma except the oligonucleotide containing a central synthetic (tetrahydrofuranyl, THF) AP site that was purchased from Glen research.
Table 1. DNA substrates employed in the study.
| Oligo number | Name of oligomers | Sequence (5′→ 3′) | Length (nt) | Oligomer no. annealed for DS substrate | Name of DS substrate |
| Oligonucleotides for AP endonuclease activity | |||||
| 1 | subSS-19AP | *ggcgaacgagacgagggc(AP)gctggaaagg | 29 | ||
| 2 | subSS-19AP(T) | cctttccagcTgccctcgtctcgttcgcc | 29 | 1and 2 | subAP·T/subDS-AP |
| 3 | subSS-19AP(A) | cctttccagcAgccctcgtctcgttcgcc | 29 | 1and 3 | subAP·A |
| 4 | subSS-19AP(G) | cctttccagcGgccctcgtctcgttcgcc | 29 | 1and 4 | subAP·G |
| 5 | subSS-19AP(C) | cctttccagcCgccctcgtctcgttcgcc | 29 | 1and 5 | subAP·C |
| Oligonucleotides for 3′→5′ exonuclease activity | |||||
| 6 | subSS-Exo | *gggactctcgaggaatgcg | 19 | ||
| 7 | subSS-Rec | ggcgcattcctcgagagtccc | 21 | 6 and 7 | subDS-REC |
| 8 | subSS-Blunt | cgcattcctcgagagtccc | 19 | 6 and 8 | subDS-BLUNT |
| 9 | subSS-Nick1 | cagctaatggctagcggc | 18 | ||
| 10 | subSS-Nick2 | gccgctagccattagctgcgcattcctcgagagtccc | 37 | 6, 9 and 10 | subDS-NICK |
SS is single-strand; DS is double-strand; nt is nucleotides; AP is the abasic site; asteriks (*) denotes the γ-32P radioactively labelled terminus.
The substrates employed in the enzymatic assays are mentioned in Table 1. 5′ end labeling of the oligonucleotides was performed in a 50 µl reaction at 37°C for 30 min by using T4 polynucleotide kinase (New England Biolabs) and [γ-32P] ATP (3000 Ci/mmol) (BRIT, India) in the kinase buffer supplied by the manufacturer. Free nucleotides were then removed from the reaction mix by using QIAquick® Nucleotide Removal Kit (QIAGEN) and the oligo was eluted in nuclease-free water (QIAGEN). The 5′ end-labeled oligonucleotides were annealed in equimolar quantities to their appropriate complementary oligonucleotides in a buffer containing 50 mM NaCl and 10 mM Tris-HCl (pH 8.0) by heating to 95°C for 5 min followed by cooling to room temperature as described previously [35]. The single-stranded oligonucleotide substrate bearing the synthetic AP site, tetrahydrofuranyl (AP) was designated as subSS-19AP. The double-stranded form of the substrate, was generated by annealing subSS-19AP with complementary strand subSS-19AP(T), subSS-19AP(A), subSS-19AP(G) or subSS-19AP(C) where either thymine, adenine, guanine or cytosine was paired opposite to the AP site; the resulting duplex oligonucleotides were referred to as subAP·T (A, G, C), respectively. Routinely, subAP·T was used as substrate for AP endonuclease assays and designated as subDS-AP. To perform 3′→5′ exonuclease assays, subSS-Exo was employed as the single-stranded substrate. To generate duplex substrates comprising of a 3′ recessed or blunt terminus, subSS-Exo was separately annealed to subSS-Rec or subSS-Blunt to generate subDS-REC or subDS-BLUNT, respectively. A duplex substrate comprising of a nick (subDS-NICK) was also generated by annealing the oligonucleotides subSS-Nick1 and subSS-Nick2 with subSS-Exo.
Enzymatic Assays
The AP endonuclease assays were performed by using 800 fmol of the 5′ end-labeled substrate and indicated amount of purified protein (either End or XthA) in a standard buffer comprising of 10 mM Tris-HCl (pH 8.0) and 100 µg/ml bovine serum albumin (BSA) in a total volume of 10 µl. For the exonuclease activity of both proteins, 20 fmol of the 5′ end-labeled substrate was employed in standard buffer (same as above) in a volume of 10 µl. To evaluate the exonuclease activity of XthA, the standard buffer was supplemented with 10 mM MgCl2. All reactions were carried out for 15 min at 37°C unless stated otherwise. All assays were carried out in duplicates.
The reactions were stopped by transferring tubes to ice and adding an equal volume of loading solution (98% formamide, 10 mM EDTA, 0.5% xylene cyanole and 0.5% bromphenol blue) [36]. The products of the reactions were resolved by denaturing gel electrophoresis (20% polyacrylamide, 8 M urea). The gels were dried and visualized by using a phosphorimager (model FLA-9000, FUJIFILM Corporation, Minato-ku, Tokyo, Japan) and quantification of the results was carried out by using the Multi Gauge V3.0 software (FUJIFILM Corporation, Minato-ku, Tokyo, Japan). To eliminate the error due to unequal loading, if any, the concentration of the reaction products were calculated from the ratio of the intensity of bands of products to the intensity of the total radioactivity in each lane. Graphs were generated by using Prism 5 software (Version 5.01; GraphPad Software Inc., CA, USA).
AP Endonuclease Assay by Employing Mycobacterial Cell-free Protein Extracts
The cultures of parental and mutant (lacking either End or XthA or both) mycobacterial strains were grown to an A600 nm of 0.8 in MB7H9 medium at 37°C in an orbital shaker at 200 rpm. 50 ml culture was harvested by centrifugation, washed once with equal volume of phosphate buffered saline (PBS), pH 7.4. Cells were resuspended in 750 µl of above buffer containing 1 mM PMSF and 1.6 g of 0.1 mm zirconium beads were added. Cells were lysed by bead beating at 5000 vibrations per minute for 8 times with 30 second pulses. Lysates were centrifuged at 12000 rpm for 20 min at 4°C. The cell-free protein extract was filtered through 0.2 µm filter followed by sonication for 2 min. The amount of protein in the lysate was quantified by using Bradford’s method. The lysate was used for AP endonuclease assay in a 10 µl reaction volume by employing 20 fmol of 5′ end-labeled substrate (subDS-AP), 1 µg of M.tuberculosis lysates, 100 µg/ml BSA, 2 mM MgCl2 in 10 mM Tris-HCl (pH 8.0). Reactions were carried out at 37°C for 30 min.
Disk Diffusion Assay
The parental and mutant strains (lacking either End or XthA or both) were exposed to oxidative stress inducing agent cumene hydroperoxide (CHP) by employing disk diffusion assay as described previously [37]. Briefly, all the mycobacterial strains were grown to an A600 nm of 0.8 in MB7H9 supplemented medium. The cells were washed once with equal volume of PBS (pH 7.4) followed by dilution to an A600 nm of 0.5 and 200 µl (2×106 cells) was plated on MB7H11 agar supplemented with 10% OADC. The plates were allowed to dry following which a 5 mm sterile disk was placed on the plate. CHP was added at 5 mM and 10 mM concentrations on to the disk (in a 10 µl volume). DMSO was used as the solvent control. After incubation for 2 weeks at 37°C, the growth inhibition zone was measured.
Results
Purification and AP Endonuclease Activity of End and XthA
The cloning of end and xthA in pet-21c(+) vector was confirmed by restriction digestion (data not shown). The constructs were verified for the absence of errors by sequencing of plasmid inserts. End (26.8 kDa) and XthA (32.1 kDa) were purified as described in materials and methods. Analysis of the purified fractions by SDS-PAGE and Coomassie Blue staining showed that the End and XthA proteins were >95% pure (Fig. 1). These proteins were assayed for AP endonuclease activity.
Figure 1. Analysis of expression of End and XthA by SDS - PAGE using a 12.5% gel.
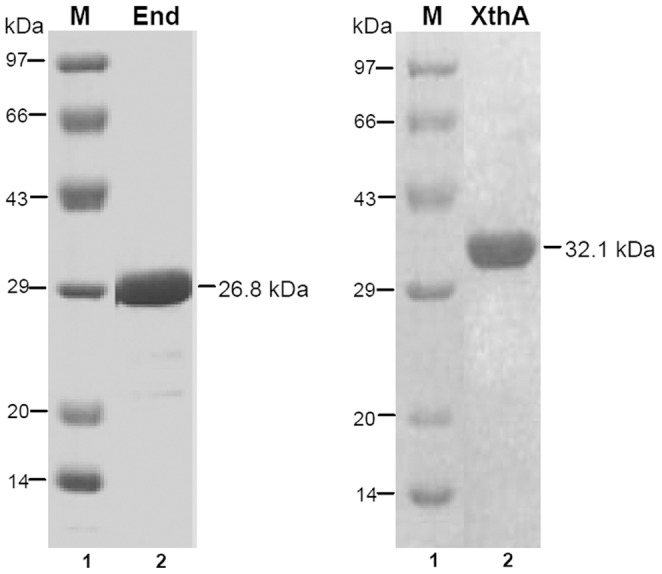
Purified proteins End (26.8 kDa) and XthA (32.1 kDa) were analyzed on a 12.5% SDS PAGE. Lane 1, protein molecular weight marker; lane 2, 2.5 µg of purified protein.
The natural substrate for AP endonuclease enzyme is an AP site within a double-stranded DNA. Initially, to determine whether the M.tuberculosis AP endonucleases were active at the AP site, a 5′ end-labeled double-stranded 29-nt substrate (subDS-AP,100 fmol), in which one strand contains a central synthetic AP site, tetrahydrofuranyl (THF), at position 19 was incubated for 30 minutes separately with 10 fmol of either purified End or XthA as described in materials and methods. Class II AP endonuclease would characteristically result in cleavage at the AP site in this substrate, generating two primary cleavage products: an end-labeled 18-nt fragment and an unlabelled 11-nt fragment. Incubation of the 29-nt duplex DNA substrate with the M.tuberculosis AP endonucleases generated an 18-nt labeled oligomer indicating that End and XthA catalyze the hydrolysis of 5′phosphodiester bond upstream to an abasic site. Significant AP endonuclease activity was observed for each of these proteins (Fig. 2A, lanes 2 and 3). End and XthA cleaved 92% and 71% of the substrate, respectively, after 30 minutes of incubation. However, when these proteins were tested for AP endonuclease activity against the single-strand DNA substrate (subSS-19AP), the activity was observed only for End (Fig. 2B, lane 3) but not for XthA (Fig. 2B, lane 2). Hence, End endonucleolytically cleaves both single-strand and double-strand AP substrate, while XthA acts only on double-strand AP substrate. In addition, it was observed that the AP endonuclease activity of the End enzyme was ∼2 fold higher for double-stranded DNA in comparison to single-stranded DNA as quantified by the level of cleavage at the AP site in the substrates. The high degree of purity of both the proteins strongly suggested that the observed AP endonuclease activity was intrinsic to the M.tuberculosis AP endonucleases.
Figure 2. End displays stronger AP endonuclease activity in comparison to XthA.
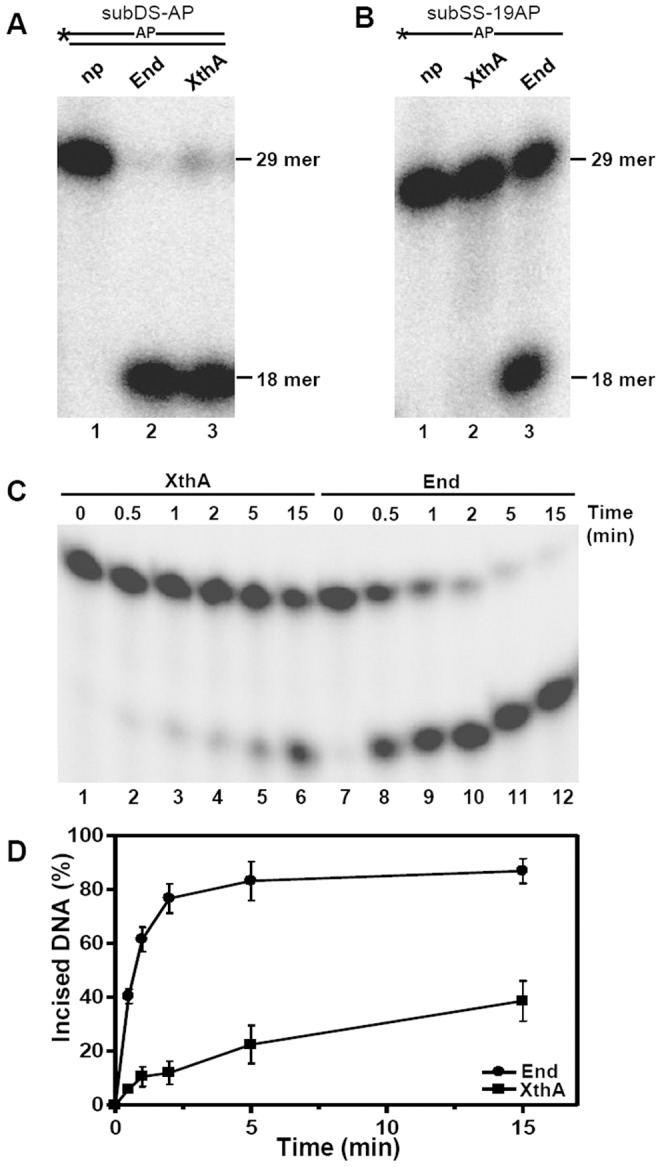
Schematic diagram of oligonucleotide substrates in which asterisk (*) and AP denote the radioactively labeled γ-32P terminus and the abasic site in the substrate, respectively. 5′ end-labeled substrate (200 fmol) was incubated with 5 fmol of either End or XthA at 37°C. (A) Lanes 1–3, subDS-AP was incubated without protein (np), with End or with XthA, respectively, for 30 min. (B) Lanes 1–3, subSS-AP was incubated without protein, with XthA or with End, respectively, for 30 min. (C) Time dependence of End and XthA on AP endonuclease activity. The reaction was carried out for different intervals of time (0–15 min). The products were analyzed by denaturing PAGE on 20% polyacrylamide gels containing 8 M Urea, and the DNA bands were visualized by autoradiography. The position of 29 mer substrate and 18 mer incision product is indicated. (D) Graphical representation of results in C. The data in the graph represent mean (±standard errors) of two independent experiments carried out in duplicates.
Examination of activity profiles for these enzymes revealed that the kinetics for both enzymes were significantly dissimilar, with End exhibiting a much faster rate of conversion of the substrate (subDS-AP) to the product in comparison to XthA (Fig. 2C and Fig. 2D). 40% of the substrate was cleaved by End within 30 sec of incubation, while XthA exhibited an incision approximating only to 5%. After 5 min of incubation, the incision of the substrate by End and XthA increased to ∼80% and 20%, respectively. Hence, AP endonuclease activity of End was observed to be 3–4 fold higher than XthA. Moreover, the reaction employing End proceeded to near completion within 15 min whereas, XthA exhibited only 38% incision activity of the substrate in the same time substantiating the higher efficiency of End in comparison to XthA. We calculated the kinetic parameters for these enzymes by employing substrate subDS-AP. The Vmax and Kcat values of the End were estimated to be 41.31±2.44 pmoles/min/µg and 0.0193 sec−1, respectively, while for XthA these were estimated to be 10.39±0.86 pmoles/min/µg and 0.0055 sec−1. End and XthA displayed classical Michaelis-Menten kinetics for the substrate subDS-AP with Km values of 6.91±1.24 nM and 7.52±2.35 nM, respectively. Moreover, the specificity constant (Kcat/Km) for End (2.79 µM−1sec−1) was four fold higher than for XthA (0.74 µM−1sec−1). From the above results we hypothesize that M.tuberculosis End, which displays a higher efficiency in comparison to XthA may be playing a more crucial role in the repair of damaged DNA comprising of AP sites.
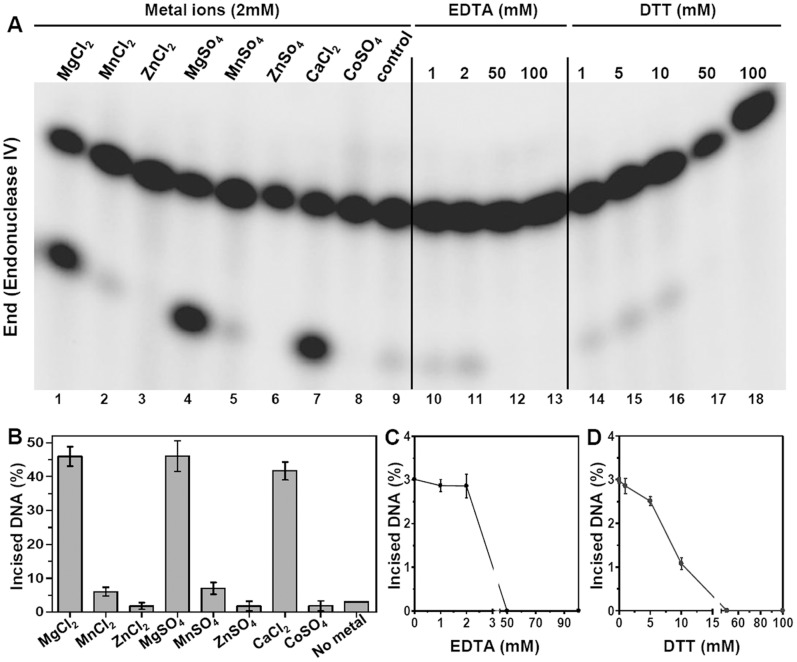
Effect of Metal Ions and Reducing Conditions on the AP Endonuclease Activity of M.tuberculosis End and XthA
To evaluate the effect of different metal ions on the AP endonuclease activity of End and XthA, 2 fmol of each protein was separately incubated with 800 fmol of 5′ end-labeled AP substrate (subDS-AP) and reactions were carried out in the presence and absence of various metal ions. End cleaved ∼3% of the AP sites in the substrate in the absence of metal ions, whereas in the presence of MgCl2, MnCl2, ZnCl2, MgSO4, MnSO4, ZnSO4, CaCl2 and CoSO4, the cleavage was ∼46, 6, 3, 46, 7, 0.7, 41 and 0.7%, respectively (Fig. 3A, lanes 1–9 and Fig. 3B). Hence, it was apparent that the AP endonuclease activity of End was maximally stimulated in the presence of Mg2+ (Fig. 3A, lanes 1 and 4) and Ca2+ (Fig. 3A, lane 7), both leading to an approximately 15 fold increase in AP endonuclease activity in comparison to the reaction without addition of metal (Fig. 3A, lane 9). End displayed a slight (∼2-fold) increase in its AP endonuclease activity on the substrate when incubated with Mn2+ (Fig. 3A, lanes 2 and 5) while no change in activity was observed in the presence of Zn2+ or Co2+ (Fig. 3A, lane 3, 6 and 8) in comparison to the activity without addition of any metal.
Figure 3. Endonuclease IV is stimulated by the addition of Magnesium and Calcium ions.
The 5′ end-labeled substrate subDS-AP (800 fmol) was incubated with 2 fmol of End with various metal ions at a concentration of 2 mM (lanes 1–8), with EDTA (lanes 10–13) and with DTT (lanes 14–18). Concentrations (mM) of EDTA and DTT are indicated on the figures. Substrate incubated with the enzyme without the addition of metal ion, EDTA or DTT was used as control (lane 9). Graphical representation of lanes 1–9, 10–13 and 14–18 of figure A is shown in B, C and D, respectively. The data in the graphs represent mean (±standard errors) of two independent experiments carried out in duplicates. The reaction was carried out for 15 min at 37°C and the products were analyzed on 20% polyacrylamide gels containing 8 M Urea and the DNA bands were visualized by autoradiography.
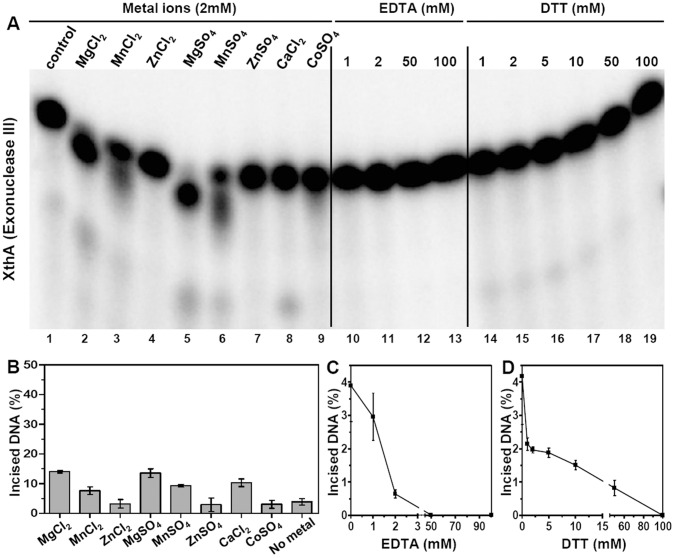
Similarly, the effect of different metal ions on the AP endonuclease activity of XthA was examined (Fig. 4A, lanes 1–9 and Fig. 4B). In the absence of any added metal, XthA cleaved ∼4% of the AP sites in the subDS-19AP substrate. In the presence of MgCl2, MnCl2, ZnCl2, MgSO4, MnSO4, ZnSO4, CaCl2 and CoSO4, the cleavage was ∼14, 8, 3, 14, 9, 4, 10 and 4%, respectively. Similar to the results observed for End, the AP endonuclease activity of XthA was also maximal in the presence of Mg2+ (Fig. 4A, lanes 2 and 5). Though, the maximal activity achieved by XthA, in the presence of Mg2+ under the standard reaction conditions was observed to be ∼3.5 fold higher than the activity achieved without any metal ion (Fig. 4A, lane 1), it corresponded to only 14% of the cleaved product, as against 46% product observed in the case of End on addition of this metal. The activity of XthA observed in the presence of Mn2+ (Fig. 4A, lanes 3 and 6) or Ca2+ (Fig. 4A, lane 8) was comparable resulting in ∼2 fold increase in AP endonuclease activity while addition of Zn2+ or Co2+ resulted in no significant enhancement of AP endonuclease activity (Fig. 4A, lanes 4, 7 and 9). Interestingly, it was observed during this study that in the presence of Magnesium and Manganese, XthA cleaved the 29-nt substrate to not only 18-nt primary cleavage product, but to products that appeared ∼1–2 nucleotides smaller than the 29-nt substrate as well, indicating the possibility of XthA exhibiting an 3′→5′ exonuclease activity in addition to the AP endonuclease activity. For this reason, it appeared that these metals are important for both AP endonuclease and exonuclease activity of XthA.
Figure 4. Exonuclease III is stimulated by the addition of Magnesium and Calcium ions.
The 5′ end-labeled substrate subDS-AP (800 fmol) was incubated with 2 fmol of XthA with various metal ions at a concentration of 2 mM (lanes 2–9), with EDTA (lanes 10–13) and with DTT (lanes 14–19). Concentrations (mM) of EDTA and DTT are indicated on the figures. Substrate incubated with the enzyme without the addition of metal ion, EDTA or DTT was used as control (lane 1). Graphical representation of lanes 1–9, 10–13 and 14–19 of figure A is shown in B, C and D, respectively. The data in the graphs represent mean (±standard errors) of two independent experiments carried out in duplicates. The reaction was carried out for 15 min at 37°C and the products were analyzed on 20% polyacrylamide gels containing 8 M Urea and the DNA bands were visualized by autoradiography.
The AP endonuclease activity of both the proteins was tested for their sensitivity to the metal chelator, EDTA (Fig. 3A and 4A, lanes 10–13). End was tolerant to EDTA upto a concentration of 2 mM (Fig. 3C) while XthA was highly sensitive to increasing concentrations of EDTA and a significant reduction in AP endonuclease activity was observed (Fig. 4C). The activity was abolished completely at 50 mM EDTA in case of both the proteins. This result again emphasized the importance of metal ions in the AP endonuclease activity of both these mycobacterial proteins.
The effect of the reducing agent DTT on the AP endonuclease activity of End (Fig. 3A, lanes 14–18 and Fig. 3D) and XthA (Fig. 4A, lanes 14–19 and Fig. 4D) was investigated. Addition of increasing concentrations of DTT to the reaction mixture reduced the AP endonuclease activity of both the enzymes. The complete inhibition of AP endonuclease activity of End and XthA was seen at DTT concentrations of 50 mM and 100 mM, respectively. These results indicate that the AP endonucleases may be more active under oxidative rather than reductive environments.
Base Pair Specificity of End and XthA during AP Endonuclease Activity
To examine the influence of the base opposite to an AP site on the incision activity, we assayed the M.tuberculosis AP endonucleases. The base pair specificity of End and XthA was investigated by using 2 fmol of enzyme and 800 fmol of the duplex oligonucleotides (subAP·T, subAP·A, subAP·G or subAP·C) containing each of the four naturally occurring bases opposite the AP site. The incision activity was measured for each enzyme on the DNA substrates as relative activity of the enzyme when acting upon the four different duplexes (Fig. 5). The repair of AP residues exhibited a marked preference for the opposite base. End (Fig. 5A) and XthA (Fig. 5B) both strongly preferred an AP site paired opposite to cytosine (AP·C), exhibiting a 5 fold higher incision efficiency in comparison to AP·T mismatch. AP·T was incised least efficiently by either enzyme. AP·A and AP·G mismatches were incised with quite similar efficiency in DNA substrates by individual enzymes. These results demonstrate that the incision activity of M.tuberculosis AP endonucleases is influenced by the base that is paired opposite to the AP site, displaying a marked preference for Cytosine residue.
Figure 5. Preferential recognition of cytosine opposite the abasic site by End and XthA.
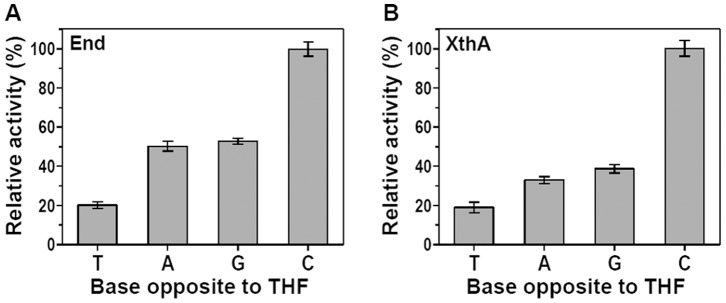
Each of the four 5′ end-labeled substrates subAP·T/A/G/C (800 fmol) were separately incubated with 2 fmol of either End (A) or XthA (B) for 25 min at 37°C. The AP endonuclease activity was measured under standard assay conditions and plotted as relative activities when acting on different duplex substrates. The data represents mean (±standard errors) of two independent experiments carried out in duplicates.
3′→5′ Exonuclease Activity of M.tuberculosis AP Endonucleases
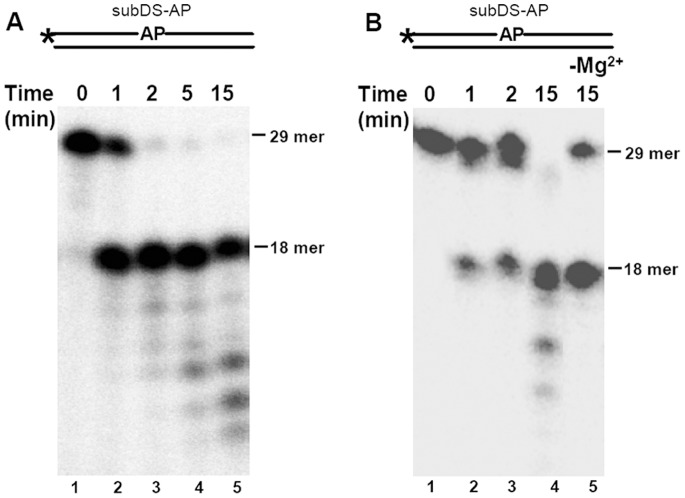
We next evaluated the exonuclease activity of the M.tuberculosis AP endonucleases by using subDS-AP as the substrate. However, a higher amount of enzyme to substrate ratio was used for the characterization to facilitate examination of the effect of enzyme on the activity and the extent to which the enzyme progressed on the substrate. 20 fmol of the substrate was incubated with 10 fmol of either End (Fig. 6A) or XthA (Fig. 6B) for different time intervals. Under these reaction conditions, besides the endonucleolytic cleavage at the AP site, a significant 3′→5′ exonuclease activity of the substrate was observed in the case of each enzyme. The exonuclease activity was observed after 2 min and 15 min of incubation of the substrate with End and XthA, respectively. Moreover, the exonuclease activity of End did not require any additional metal ion (data not shown). In contrast, XthA was highly dependent on metal ion for its AP endonuclease and 3′→5′ exonuclease activities. The AP endonuclease activity of XthA reduced by 29% in the absence of metal ions (Fig. 6B, lane 5) in comparison to its activity in the presence of metal ion (Fig. 6B, lane 4). More importantly, a complete abrogation of the exonuclease activity was observed in the absence ofmetal ions. Hence, XthA is dependent on the metal ions for complete manifestation of both the activities.
Figure 6. 3′→5′ exonuclease activity of End and XthA on AP substrate.
Schematic diagram of oligonucleotide substrates in which asterisk (*) and AP denote the radioactively labeled γ-32P terminus and the abasic site in the substrate, respectively. 5′ end-labeled substrate subDS-AP (20 fmol) was incubated with 10 fmol of either End (A) or XthA (B) for variable time at 37°C. (A) Lanes 1–5, incubation of substrate with End in standard buffer for 0, 1, 2, 5 and 15 min, respectively. (B) Lanes 1–4, incubation of substrate with XthA in standard buffer containing 10 mM MgCl2 for 0, 1, 2 and 15 min, respectively. Lane 5, XthA was incubated with the substrate for 15 min in standard buffer without Magnesium. The position of 29 mer substrate and 18 mer incision product is indicated.
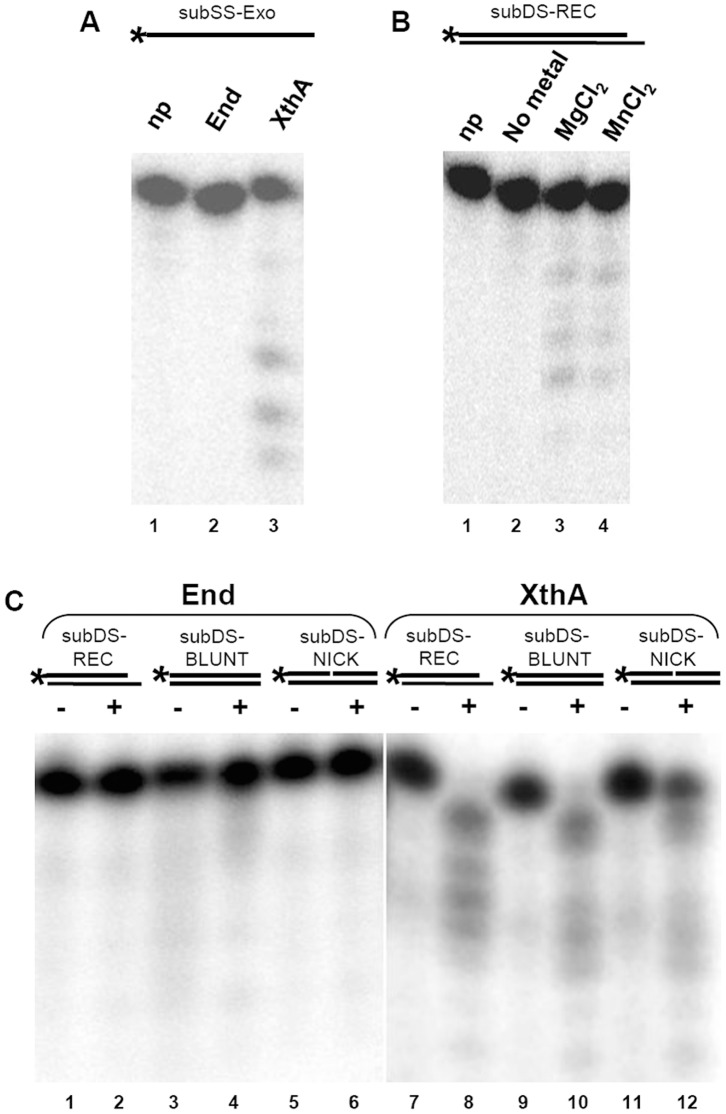
AP endonucleases have been reported to be most active on 3′ recessed DNA substrates in comparison to other substrates for the display of exonuclease activity [38], [39]. Hence, to study the exonuclease activity of mycobacterial AP endonucleases a similar substrate was generated by annealing a 5′ end-labeled 19-nt oligomer (subSS-Exo) with a 21-nt oligomer (subSS-Rec) to generate a double-stranded exonuclease substrate subDS-REC (3′ recessed substrate). End did not display exonuclease activity when incubated with either single-stranded (subSS-Exo, Fig. 7A, lane 2) or double-stranded substrate (subDS-REC, Fig. 7C, lanes 1 and 2). On the other hand, it was observed that incubation of XthA with either a 5′ end-labeled single-strand oligomer, subSS-Exo (Fig. 7A, lane 3), or with a double-strand 3′ recessed oligomer, subDS-REC (Fig. 7B, lanes 3 and 4), resulted in the gradual shortening of the 19 mer template that indicated an exonucleolytic digestion of the DNA from the 3′ end. Examination of exonuclease activity on the subDS-Rec by employing XthA, demonstrated the exonuclease digestion of the substrate both in the presence of Mg2+ or Mn2+ ions which was abolished in the absence of metal ions (Fig. 7B, lane 2). Hence, the exonuclease activity of XthA was strongly dependent upon metal ion. To further determine the DNA substrate specificity, we compared the exonuclease activity of End and XthA for their ability to hydrolyze DNA duplexes by employing substrates with either a blunt (subDS-BLUNT) or a recessed 3′-terminus (subDS-REC) or by using a substrate containing an internal nick (subDS-NICK) in the heteroduplex DNA. End displayed no activity on any of the above substrates (Fig. 7C, lanes 1–6). In contrast, XthA exhibited strong exonuclease activity on a recessed 3′ end containing partial DNA duplex, with 10% uncleaved substrate (Fig. 7C, lane 8) and on a blunt-end containing heteroduplex DNA, with 14% uncleaved substrate (Fig. 7C, lane 10). In both the above cases, more than 85% of the substrate was cleaved further to multiple bands smaller in size than the labeled 19-nt fragment indicating the presence of exonuclease activity. Though, the nicked substrate was cleaved and the band sizes decreased approximately one base at a time, XthA showed weaker activity on a nicked DNA in comparison to recessed and blunt ended substrates (Fig. 7C, lane 12) displaying ∼35% of uncleaved subDS-NICK.
Figure 7. XthA displays a metal ion dependent 3′→5′ exonuclease activity on a large variety of substrates.
Schematic diagram of oligonucleotide substrates (refer Table 1 for details) in which asterisk (*) denotes the radioactively labeled γ-32P terminus. (A) 20 fmol of 5′ end-labeled substrate subSS-Exo was incubated without protein (np,lane 1), with 2 fmol of either End (lane 2) or XthA (lane 3) in a buffer containing 10 mM MgCl2 for 15 min at 37°C. (B) 20 fmol of 5′ end-labeled substrate subDS-REC (3′ recessed substrate) was incubated without protein (lane 1), with 2 fmol of XthA without any metal (lane 2), XthA in a buffer containing 10 mM MgCl2 (lane 3) or XthA in a buffer containing 10 mM MnCl2 (lane 4). (C) 20 fmol of each of the 5′ end-labeled DNA substrates (subDS-REC, subDS-BLUNT and subDS-NICK) were incubated in the absence (–) or presence (+) of 10 fmol of End (lanes 1–6) and XthA (lanes 7–12).
AP Endonuclease Activity in Cell-free Protein Extracts of M.tuberculosis
To understand the specific role(s) of these AP endonucleases in M.tuberculosis, we generated single (MtbΔend and MtbΔxthA) and double (MtbΔendΔxthA) mutants (Puri, R.V. et. al, unpublished data) by employing recombineering method [34]. Cell-free protein extracts of the parental and mutant strains were prepared and examined for AP endonuclease activity (Fig. 8A and Fig. 8B) as described in materials and methods. Incubation of the 29-nt duplex DNA substrate (subDS-AP) with the M.tuberculosis parental cell-free extract generated a 18-nt labeled oligomer (17% incised substrate, Fig. 8A, lane 2) indicating that AP endonucleases catalyze the hydrolysis of the 5′ phosphodiester bond upstream to the abasic site. On incubation of this substrate with MtbΔend, a complete abrogation of AP endonuclease activity was observed (Fig. 8A, lane 3). However, this activity was restored when the end gene was complemented on a plasmid and was in fact, 4 fold higher than the parental strain (76% incised substrate, Fig. 8A, lane 4). This increased activity of MtbΔendComp may be attributed to the higher copy number of the plasmid used for the complementation [40], [41], [42]. The activity was comparable in the parental and MtbΔxthA (20% incised substrate) implying that XthA did not contribute to the AP endonuclease activity in the mycobacterial cells under the experimental conditions. The activity of MtbΔxthAComp was similar to the parental strain (18% incised substrate). These results indicate that under normal physiological conditions, most of the AP endonuclease activity in M.tuberculosis is represented by End.
Figure 8. End is the major AP endonuclease in the mycobacterial cell-free protein extracts.
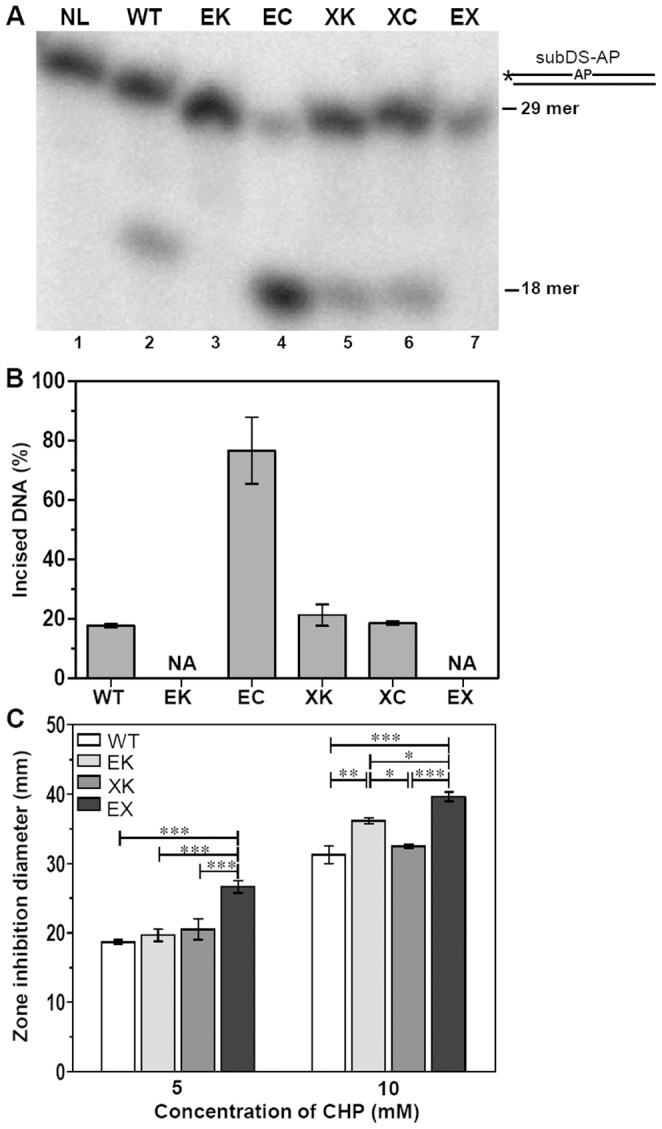
(A) Schematic diagram of oligonucleotide substrate in which asterisk (*) and AP denote the radioactively labeled γ-32P terminus and the abasic site in the substrate, respectively. The 5′ end-labeled substrate subDS-AP (20 fmol) was incubated with cell-free protein extracts (1 µg) of parental and mutant strains of M.tuberculosis for 30 min at 37°C. (B) The graph represents mean (±standard errors) of three independent experiments, of which one is shown in (A). (C) Influence of disruption of single or both the AP endonucleases on the ability of M.tuberculosis to withstand oxidative stress generated by cumene hydroperoxide (CHP). Graph depicts the range of the inhibition zones observed in the presence of various concentrations of cumene hydroperoxide. The values are represented as the mean (± standard errors) of two independent experiments carried out in triplicates. ∗,P<0.05; ∗∗,P<0.01; ∗∗∗,P<0.001 (Two way ANOVA). NL, no lysates; NA, no activity; WT, M.tuberculosis; EK, MtbΔend; EC, MtbΔendComp; XK, MtbΔxthA; XC, MtbΔxthAComp; EX, MtbΔendΔxthA.
Further, to evaluate the role of AP endonucleases in the protection of M.tuberculosis against the DNA damages caused by oxidative radicals, the toxic oxidant cumene hydroperoxide (CHP) was employed. CHP, an alkylperoxide, is a stable free radical source produced inside the cells by metabolism of unsaturated fatty acids and nucleic acids [43]. Figure 8C depicts that the MtbΔendΔxthA double mutant exhibited an enhanced sensitivity to CHP as demonstrated by larger zones of inhibition relative to either parental or the single mutant strains in the disc diffusion assay. At low concentration of CHP (5 mM), both MtbΔend and MtbΔxthA displayed zones of inhibition similar to the parental strain. However, as the level of oxidative damage increased (10 mM CHP), the absence of End (MtbΔend mutant) resulted in a significant increase in the inhibition zone in comparison to the parental strain, while mutation in xthA alone (MtbΔxthA mutant) did not result in any enhancement in the sensitivity to CHP. Our results suggest that in M.tuberculosis, the major role of protecting the DNA against oxidative damage is played by End. These observations are in agreement with our hypothesis that End, the more efficient AP endonuclease, is of higher significance to M.tuberculosis in the repair of damaged DNA by BER pathway.
Discussion
M.tuberculosis encounters the assaults on all its biomolecules, including DNA, that are inflicted upon by the reactive oxygen and nitrogen species produced as a part of the host’s innate immune response. In its struggle to survive in the host, this pathogen must possess efficient DNA repair mechanisms to safeguard its genetic machinery. AP endonucleases are a class of enzymes responsible for the removal of abasic sites in DNA, which are generated in response to spontaneous hydrolysis or modifications of the bases. However, there have been no earlier reports on the biochemical characterization of AP endonucleases from this important pathogen [6]. Based on the sequence analysis of these AP endonuclease genes in clinical strains of M.tuberculosis, it has been demonstrated that end displayed variations in sequence in certain clinical strains and in several other strains a single nucleotide deletion was found in this gene [44]. On the other hand, among the 100 strains evaluated in this study, xthA was amongst the few genes that displayed no sequence variation or deletion indicating thereby that xthA may play a more important role in M.tuberculosis than end [44]. However, the experimental evidence gathered by us in the current study, has made interesting revelations in support of End being the more important AP endonuclease of M.tuberculosis. We have evaluated the enzymatic activities of the annotated AP endonucleases of M.tuberculosis [33], and show that both Endonuclease IV (End) and Exonuclease III (XthA) are multifunctional enzymes. These enzymes exhibit AP endonuclease and 3′→5′ exonuclease activities with differences in their substrate specificities. More importantly, we show that Endonuclease IV is the major AP endonuclease of M.tuberculosis that also plays an important role in protecting the pathogen against oxidative DNA damage.
Previous studies have indicated that sequence of the DNA substrate may affect the excision activity of enzymes [45], [46], [47], [48]. We have utilized well characterized oligonucleotide substrates that have been employed by several groups for routine BER assays [36], [38]. Evaluation of M.tuberculosis AP endonucleases revealed that End is a highly efficient AP endonuclease while XthA displays weaker AP endonuclease activity. This difference in the efficiency of End is attributed to a ∼4 fold higher Kcat value of End in comparison to XthA when acting on a double-stranded DNA containing an AP site. Mycobacterial End cleaved both the double and single-stranded DNA containing an AP site. This finding is similar to the endonuclease IV of Chlamydia pneumoniae (CpEndoIV), an obligate intracellular bacterium that infects humans and is a major cause of pneumonia [49]. Quantification of the level of cleavage at the AP site in the substrates showed that the AP endonuclease activity of the End enzyme exhibited almost 2-fold higher AP endonuclease activity on double-stranded DNA in comparison to single-stranded DNA; a finding also reported for Ec-EndoIV [36]. Though the significance of excision of AP sites from single-stranded DNA substrates has been a matter of debate, a number of phylogenetically diverse DNA glycosylases in BER have shown activity against damaged bases in single-stranded DNA suggesting that BER may at least be initiated in single-stranded DNA [50], [51], [52], [53]. The DNA glycosylase activity directed against a modified base in single-stranded DNA would generate an AP site in the single-stranded DNA. Besides, the incision activity on AP site in single-stranded DNA has also been reported for other AP endonucleases such as CpEndoIV, Ec-EndoIV and human APE1 [36], [49], [54].The above results promote the understanding of mechanism of AP site recognition by endonuclease IV and are suggestive of a probable role of End in the repair of AP site of single-stranded DNA during replication and/or transcription in vivo apart from its major function of AP site removal from the double-stranded DNA. However, the cleavage activity of mycobacterial XthA on the single-stranded DNA containing an AP site was not detected.
In E.coli, the association of metal ions with Ec-EndoIV and Ec-ExoIII is well established [12], [55], [56] which stimulated us to evaluate the effect of metal ions on the AP endonuclease activity of mycobacterial End and XthA. The primary role of the metal ion is to promote a conformational change in the DNA containing abasic site, priming it for enzyme-mediated hydrolysis [57]. Previous studies have demonstrated the importance of metal ions (Mg2+, Ca2+, Co2+, Zn2+ and Mn2+) for the activity of AP endonucleases across various species [55], [56], [57], [58], [59], [60], [61], [62]. The sequence alignment of mycobacterial AP endonucleases End and XthA with their homologues in Endonuclease IV and Exonuclease III family, reveals complete conservation of metal binding sites in End (His56, His96, Glu129, Asp162, His165, His191, Asp204, His206 and Glu233) and XthA (Asn32, Glu57, Asp180, Asn182 and His281) [63], [64]. Our results demonstrate that the activity of AP endonucleases of M.tuberculosis is stimulated in the presence of Mg2+ or Ca2+ and these metals may play an important role in the catalysis of these enzymes. Inhibitory effect of EDTA on the AP endonuclease activity of both the proteins further substantiates that like other members of Endonuclease IV and Exonuclease III family, both the mycobacterial AP endonucleases appear to require transition metals for their activity. In the host macrophages, the pathogen encounters oxidative stress that results in a variety of DNA damaging lesions including AP sites. Moreover, there is experimental evidence for an increase in the number of AP sites in DNA after exposure to oxidative stress implicating the role of AP endonucleases under such conditions [65]. Our results demonstrate that the addition of the reducing agent DTT inhibited the progression of the enzyme on the substrate suggesting that these enzymes may be more active when the cell is under oxidative stress substantiating their in vivo role.
We next analyzed the influence of the base opposite the AP site (THF) on the nicking activity of End and XthA. The preferential recognition of a modified residue paired opposite to one of the four natural bases in duplex DNA is an important feature of DNA repair enzymes [66]. An oligonucleotide harboring an AP site was annealed with complementary strand with different bases opposite AP site prior to treatment with the enzymes. Both End and XthA were five times as active on a THF residue opposite C as compared to T. End and XthA incised AP·A and AP·G mismatches with a similar efficiency. The data obtained with mycobacterial AP endonucleases acting on THF mismatches in the present study, are in agreement with a previous observation employing S.cerevisiae Apn1. Apn1 has been observed to display a 5-fold higher activity on a THF residue opposite C or T than opposite G or A [66]. However, the opposite base had no effect on the AP endonuclease activity of E.coli AP endonucleases [67]. Cytosine under normal physiological conditions base pairs with guanine. The preferential recognition of AP site opposite the cytosine residue is an important characteristic of mycobacterial AP endonucleases, probably indicating the importance of AP endonucleases in recognizing the modifications in the guanine base. 7,8-dihydro-8-oxoguanine (8-oxoG) is one of the most common damage resulting from the oxidation of DNA, and failure to replace it with the correct base results in mutations [68]. Moreover, it has been demonstrated that DNA polymerase(s) from mycobacteria display a preference for the incorporation of G opposite the 8-oxoG as opposed to an A in E. coli, which further enhances the risk of accumulating guanine modifications like 8-oxoG, in their GC rich DNA [29]. Further, several class II AP endonucleases such as yeast Apn1 and human Ape1 have been demonstrated to remove 3′ incorporated 8-oxoG damaged nucleotide [69], [70]. In light of the above, it is not unlikely that the AP endonucleases of M.tuberculosis play a role in the removal of 8-oxoG damaged nucleotide.
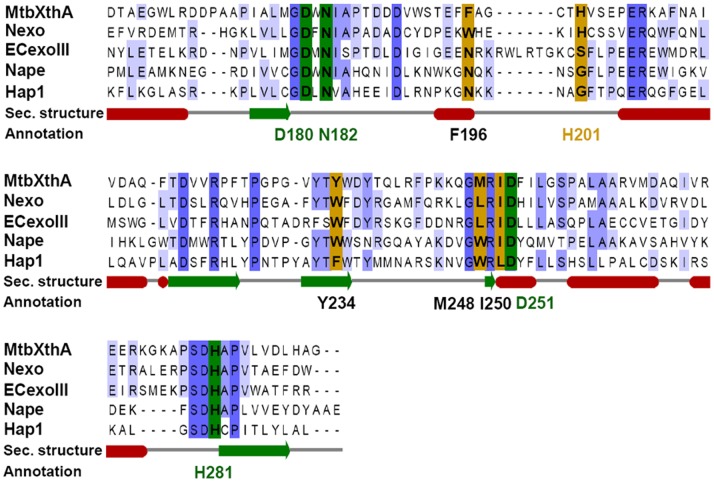
We observed that XthA is a less efficient enzyme than End for its AP endonuclease activity (Fig. 2C and Fig. 2D). Also, incubation of XthA with metal ions enhanced the AP endonuclease activity of XthA, but to lesser extent than that observed in the case of End (Fig. 3 and Fig. 4). Overall, we observed a lower AP endonuclease activity of XthA when compared to End. Sequence comparison of M.tuberculosis XthA, with other well-characterized exonuclease III family AP endonucleases from other species, revealed interesting observations. XthA possesses all the active site residues necessary for AP endonuclease activity (i.e., Asp180, Asn182, Asp251 and His281; Fig. 9). However, XthA comprises of a histidine residue (His201) at the abasic ribose binding pocket, instead of a small amino acid like Glycine or Serine generally observed in functional AP endonucleases (Gly231 in human AP endonuclease, HAP1; Gly170 in Neisseria meningitides AP endonuclease, Nape and Ser178 in E.coli AP endonuclease, ExoIII) [31]. The equivalent residue in Nexo (an exonuclease of N.meningitides) is also a histidine (His167). Similar to our observations on XthA in this study, Nexo has also been demonstrated to be a very weak AP endonuclease [31]. Moreover, Carpenter et al. demonstrated that modification of His167 to Glycine or Serine resulted in a substantial increase in the AP endonuclease activity of Nexo, signifying the critical role of this histidine as a structural determinant for the AP endonuclease activity [31]. We hypothesize that the presence of histidine side chain at the abasic ribose binding site is involved in the impairment of the binding of abasic ribose to the enzyme which would prevent the consequent cleavage of the substrate, in turn leading to a low AP endonuclease activity of XthA. Additionally, in M.leprae, the pathogen causing leprosy, xthA has been converted to a pseudogene [6], further substantiating a less vital role of XthA in pathogenic mycobacteria.
Figure 9. Sequence alignments of XthA with other AP endonuclease homologs.
Alignment of the C-terminal 127 residues of MtbXthA (Rv0427c), with non-functional AP endonuclease paralogue of N.meningitidis (Nexo) and functional AP endonucleases of E.coli (ECExoIII), N.meningitidis (Nape) and Homo sapiens (Hap1) indicates the plausible role of histidine (H201) for low AP endonuclease activity. The residues involved in abasic ribose binding and catalysis are indicated in brown and green color, respectively. Absolutely conserved residues in the alignment are colored in dark blue and light blue for partial identity. Secondary structure elements are shown in red for α-helices and green for β-strands. CLUSTALW was employed for the initial multiple sequence alignment of proteins [71]. This was followed by using the programme JalView [72] for the prediction of conserved residues and secondary structure.
In the ExoIII/Ape1/Apn2 family of proteins, Ec-ExoIII and human Ape1 have been extensively studied, and both of these enzymes have been shown to possess AP endonuclease and 3′→5′ exonuclease activities [38]. Our study demonstrates that XthA, the less active AP endonuclease of mycobacteria, displayed an efficient metal ion dependent 3′→5′ exonuclease activity on a large variety of substrates which includes: duplex DNA with 3′ recessed or blunt termini, duplex DNA with an internal nick or an AP site, as well as a single-stranded DNA, emphasizing the primary role of this protein as an exonuclease. On the other hand, M.tuberculosis End was observed to be competent to digest exonucleolytically, specifically a duplex DNA that comprised of a damaged AP site and not any other substrate employed in this study. Although the function of the 3′→5′ exonuclease activity of Exonuclease III family is unknown and has been considered redundant, the discovery of substantial exonuclease activity associated with Endonuclease IV homologs, in E.coli [36] and M.tuberculosis argues in favor of a significant functional importance for this activity in vivo.
The in vitro observations using purified enzymes and defined oligonucleotide substrates cannot be simply extrapolated to an in vivo situation without taking into account the complex cellular network. In E.coli crude cell extracts, Ec-ExoIII accounts for 90% of total AP endonuclease activity [10]. The measurement of AP endonuclease activity in the mycobacterial cell-free extracts of the parental and mutant strains of end or/and xthA, demonstrate End as the major apurinic endonuclease under normal growth conditions. However, the activity of XthA in these cultures was only conspicuous by its absence. To evaluate if the activity of XthA might be induced under DNA damaging conditions, logarithmically growing cultures of M.tuberculosis were subjected to treatment with DNA damaging agents such as methylmethane sulfonate (MMS), hydrogen peroxide (H2O2) or mitomycin C (MMC) at various concentrations (1– 5 mM MMS, 0.1– 1 mM MMC and 1 mM-10 mM H2O2) for 24 hours followed by determination of XthA activity in the cell free extract. Apart from M.tuberculosis, MtbΔend was also employed in these studies so that even slight induction in the activity of XthA could be measured. However, inspite of exposure to these DNA damaging agents, we did not find any measurable activity of XthA even at the highest concentration of these DNA damaging agents used under our experimental conditions (data not shown). Based on the above results, End seems to represents the major AP endonuclease in M.tuberculosis. However, we cannot rule out the possibility that during the infection of the host by this pathogen there could be situations that could lead to induction of either End or XthA.
To investigate the role of these AP endonucleases in repairing the DNA damage and thereby protecting M.tuberculosis, we employed disc diffusion assay. The response of M.tuberculosis to DNA damage resulting from the exposure of reactive oxygen intermediates in mutant strains lacking one or both the AP endonucleases was measured by growth inhibition zone around a paper disc impregnated with CHP. At 5 mM and 10 mM concentrations of CHP, a significantly enhanced sensitivity was observed in the case of MtbΔendΔxthA strain in comparison to the parental strain. More importantly, the absence of End in M.tuberculosis (MtbΔend) resulted in an enhanced sensitivity in comparison to the parental and MtbΔxthA strains at 10 mM concentration of CHP. These results suggest that in the M.tuberculosis BER pathway, the removal of damaged DNA resulting from oxidative stress is primarily carried out by End. XthA that possesses only a weak AP endonuclease activity with a predominant 3′→5′ exonuclease activity plays a less significant role in the repair of such damage. More importantly, the absence of Endonuclease IV homolog in humans and the contribution of End as the major AP endonuclease in M.tuberculosis, opens up a new arena of research targeting this crucial enzyme for developing anti-tubercular molecules.
In summary, the present study provides the first evidence for the presence of active AP endonucleases in M.tuberculosis and for distinct substrate preferences of these AP enzymes. We demonstrate that End is not only a more efficient AP endonuclease enzyme than XthA but it also represents the major AP endonuclease activity in M.tuberculosis and plays a crucial role in defense against oxidative stress in comparison to XthA. In addition, while End possesses a metal ion independent exonuclease activity; XthA is a metal ion dependent enzyme which predominantly acts as a 3′→5′ exonuclease possessing weak AP endonuclease activity. The AP endonuclease activity of both the M.tuberculosis AP endonucleases is stimulated by Mg2+ and Ca2+ and displays a preferential recognition for abasic site paired opposite to a cytosine residue in DNA. In our future studies, we aim to elucidate the biological implications of AP endonuclease activity in the pathogenesis of M.tuberculosis by employing single and double mutant strains of end and xthA in animal models.
Supporting Information
(PDF)
Acknowledgments
We acknowledge the Central Instrumentation Facility, UDSC for the use of Phosphorimager for all the radioactivity based assays; P. Vineel Reddy (UDSC) is acknowledged for critical proof reading of the manuscript; Amit Kumar is acknowledged for helping with the use of the programme JalView. Technical assistance provided by Sandeep Kumar and Priti Singh is highly acknowledged.
Funding Statement
This work was supported by a research grant from the Department of Biotechnology, Ministry of Science and Technology, Government of India. The Council of Scientific and Industrial Research, Government of India, is acknowledged for Junior/Senior research fellowships to RVP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Seeberg E, Eide L, Bjoras M (1995) The base excision repair pathway. Trends Biochem Sci 20: 391–397. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T (2001) Keynote: past, present, and future aspects of base excision repair. Prog Nucleic Acid Res Mol Biol 68: xvii-xxx. [DOI] [PubMed]
- 3. Ide H, Kotera M (2004) Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol Pharm Bull 27: 480–485. [DOI] [PubMed] [Google Scholar]
- 4. Krokan HE, Standal R, Slupphaug G (1997) DNA glycosylases in the base excision repair of DNA. Biochem J 325: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacobs AL, Schar P (2012) DNA glycosylases: in DNA repair and beyond. Chromosoma 121: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurthkoti K, Varshney U (2011) Base excision and nucleotide excision repair pathways in mycobacteria. Tuberculosis (Edinb) 91: 533–543. [DOI] [PubMed] [Google Scholar]
- 7. Loeb LA (1985) Apurinic sites as mutagenic intermediates. Cell 40: 483–484. [DOI] [PubMed] [Google Scholar]
- 8. Loeb LA, Preston BD (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet 20: 201–230. [DOI] [PubMed] [Google Scholar]
- 9. Dianov G, Lindahl T (1994) Reconstitution of the DNA base excision-repair pathway. Curr Biol 4: 1069–1076. [DOI] [PubMed] [Google Scholar]
- 10. Ljungquist S, Lindahl T, Howard-Flanders P (1976) Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol 126: 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 374: 381–386. [DOI] [PubMed] [Google Scholar]
- 12. Doetsch PW, Cunningham RP (1990) The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res 236: 173–201. [DOI] [PubMed] [Google Scholar]
- 13. Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, et al. (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev 12: 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Popoff SC, Spira AI, Johnson AW, Demple B (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci U S A 87: 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Unk I, Haracska L, Johnson RE, Prakash S, Prakash L (2000) Apurinic endonuclease activity of yeast Apn2 protein. J Biol Chem 275: 22427–22434. [DOI] [PubMed] [Google Scholar]
- 16. Unk I, Haracska L, Gomes XV, Burgers PM, Prakash L, et al. (2002) Stimulation of 3′–>5′ exonuclease and 3′-phosphodiesterase activities of yeast apn2 by proliferating cell nuclear antigen. Mol Cell Biol 22: 6480–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ribar B, Izumi T, Mitra S (2004) The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe . Nucleic Acids Res 32: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parsons JL, Dianova, II, Dianov GL (2004) APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res 32: 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO (2012) Tuberculosis Global Facts 2011/2012. Available : http://www.who.int/tb/publications/2011/factsheet_tb_2011.pdf Accessed 2013 June 5.
- 20. Ferrari CK, Souto PC, Franca EL, Honorio-Franca AC (2011) Oxidative and nitrosative stress on phagocytes' function: from effective defense to immunity evasion mechanisms. Arch Immunol Ther Exp (Warsz) 59: 441–448. [DOI] [PubMed] [Google Scholar]
- 21. Soldati T, Neyrolles O (2012) Mycobacteria and the Intraphagosomal Environment: Take It With a Pinch of Salt(s)! Traffic. 13: 1042–1052. [DOI] [PubMed] [Google Scholar]
- 22. Kumar P, Bharti SK, Varshney U (2011) Uracil excision repair in Mycobacterium tuberculosis cell-free extracts. Tuberculosis (Edinb) 91: 212–218. [DOI] [PubMed] [Google Scholar]
- 23. Kurthkoti K, Srinath T, Kumar P, Malshetty VS, Sang PB, et al. (2010) A distinct physiological role of MutY in mutation prevention in mycobacteria. Microbiology 156: 88–93. [DOI] [PubMed] [Google Scholar]
- 24. Springer B, Sander P, Sedlacek L, Hardt WD, Mizrahi V, et al. (2004) Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol Microbiol 53: 1601–1609. [DOI] [PubMed] [Google Scholar]
- 25. Mizrahi V, Andersen SJ (1998) DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol 29: 1331–1339. [DOI] [PubMed] [Google Scholar]
- 26. Srinath T, Bharti SK, Varshney U (2007) Substrate specificities and functional characterization of a thermo-tolerant uracil DNA glycosylase (UdgB) from Mycobacterium tuberculosis . DNA Repair (Amst) 6: 1517–1528. [DOI] [PubMed] [Google Scholar]
- 27. Olsen I, Balasingham SV, Davidsen T, Debebe E, Rodland EA, et al. (2009) Characterization of the major formamidopyrimidine-DNA glycosylase homolog in Mycobacterium tuberculosis and its linkage to variable tandem repeats. FEMS Immunol Med Microbiol 56: 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Y, Bandaru V, Jaruga P, Zhao X, Burrows CJ, et al. (2010) The oxidative DNA glycosylases of Mycobacterium tuberculosis exhibit different substrate preferences from their Escherichia coli counterparts. DNA Repair (Amst) 9: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jain R, Kumar P, Varshney U (2007) A distinct role of formamidopyrimidine DNA glycosylase (MutM) in down-regulation of accumulation of G, C mutations and protection against oxidative stress in mycobacteria. DNA Repair (Amst) 6: 1774–1785. [DOI] [PubMed] [Google Scholar]
- 30. Suvarnapunya AE, Lagasse HA, Stein MA (2003) The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol Microbiol 48: 549–559. [DOI] [PubMed] [Google Scholar]
- 31. Carpenter EP, Corbett A, Thomson H, Adacha J, Jensen K, et al. (2007) AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. EMBO J 26: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Onyango DO, Naguleswaran A, Delaplane S, Reed A, Kelley MR, et al. (2011) Base excision repair apurinic/apyrimidinic endonucleases in apicomplexan parasite Toxoplasma gondii . DNA Repair (Amst) 10: 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 34. van Kessel JC, Hatfull GF (2007) Recombineering in Mycobacterium tuberculosis . Nat Methods 4: 147–152. [DOI] [PubMed] [Google Scholar]
- 35. Castillo-Acosta VM, Ruiz-Perez LM, Yang W, Gonzalez-Pacanowska D, Vidal AE (2009) Identification of a residue critical for the excision of 3′-blocking ends in apurinic/apyrimidinic endonucleases of the Xth family. Nucleic Acids Res 37: 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kerins SM, Collins R, McCarthy TV (2003) Characterization of an endonuclease IV 3′–5′ exonuclease activity. J Biol Chem 278: 3048–3054. [DOI] [PubMed] [Google Scholar]
- 37. Reddy PV, Puri RV, Khera A, Tyagi AK (2012) Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. J Bacteriol 194: 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Unk I, Haracska L, Prakash S, Prakash L (2001) 3′-phosphodiesterase and 3′→ 5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Molecular and cellular biology 21: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burkovics P, Szukacsov V, Unk I, Haracska L (2006) Human Ape2 protein has a 3′–5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res 34: 2508–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Das Gupta SK, Bashyam MD, Tyagi AK (1993) Cloning and assessment of mycobacterial promoters by using a plasmid shuttle vector. J Bacteriol 175: 5186–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bourn WR, Jansen Y, Stutz H, Warren RM, Williamson AL, et al. (2007) Creation and characterisation of a high-copy-number version of the pAL5000 mycobacterial replicon. Tuberculosis (Edinb) 87: 481–488. [DOI] [PubMed] [Google Scholar]
- 42. Uhlin BE, Nordstrom K (1977) R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid 1: 1–7. [DOI] [PubMed] [Google Scholar]
- 43. Antelmann H, Engelmann S, Schmid R, Hecker M (1996) General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol 178: 6571–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, et al. (2008) Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis . PLoS One 3: e1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson DM 3rd, Takeshita M, Grollman AP, Demple B (1995) Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem 270: 16002–16007. [DOI] [PubMed] [Google Scholar]
- 46. Chou KM, Cheng YC (2002) An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature 415: 655–659. [DOI] [PubMed] [Google Scholar]
- 47. Hadi MZ, Ginalski K, Nguyen LH, Wilson DM 3rd (2002) Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol 316: 853–866. [DOI] [PubMed] [Google Scholar]
- 48. Lebedeva NA, Khodyreva SN, Favre A, Lavrik OI (2003) AP endonuclease 1 has no biologically significant 3(')–>5(')-exonuclease activity. Biochem Biophys Res Commun 300: 182–187. [DOI] [PubMed] [Google Scholar]
- 49. Liu X, Liu J (2005) Chlamydia pneumoniae AP endonuclease IV could cleave AP sites of double- and single-stranded DNA. Biochim Biophys Acta 1753: 217–225. [DOI] [PubMed] [Google Scholar]
- 50. Dou H, Mitra S, Hazra TK (2003) Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem 278: 49679–49684. [DOI] [PubMed] [Google Scholar]
- 51. Hardeland U, Bentele M, Jiricny J, Schar P (2003) The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res 31: 2261–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ishchenko AA, Bulychev NV, Maksakova GA, Johnson F, Nevinsky GA (1999) Single-stranded oligodeoxyribonucleotides are substrates of Fpg protein from Escherichia coli . IUBMB Life 48: 613–618. [DOI] [PubMed] [Google Scholar]
- 53. Boorstein RJ, Cummings A Jr, Marenstein DR, Chan MK, Ma Y, et al. (2001) Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem 276: 41991–41997. [DOI] [PubMed] [Google Scholar]
- 54. Marenstein DR, Wilson DM 3rd, Teebor GW (2004) Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair (Amst) 3: 527–533. [DOI] [PubMed] [Google Scholar]
- 55. Levin JD, Shapiro R, Demple B (1991) Metalloenzymes in DNA repair. Escherichia coli endonuclease IV and Saccharomyces cerevisiae Apn1. J Biol Chem 266: 22893–22898. [PubMed] [Google Scholar]
- 56. Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98: 397–408. [DOI] [PubMed] [Google Scholar]
- 57. Erzberger JP, Wilson DM 3rd (1999) The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J Mol Biol 290: 447–457. [DOI] [PubMed] [Google Scholar]
- 58. Tomanicek SJ, Hughes RC, Ng JD, Coates L (2010) Structure of the endonuclease IV homologue from Thermotoga maritima in the presence of active-site divalent metal ions. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thielmann HW, Hess U (1981) Apurinic endonuclease from Saccharomyces cerevisiae . Biochem J 195: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guyer RB, Deering RA (1985) Apurinic/apyrimidinic-specific endonuclease activities from Dictyostelium discoideum . Biochim Biophys Acta 824: 304–312. [DOI] [PubMed] [Google Scholar]
- 61. Johnson AW, Demple B (1988) Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: substrate specificity and kinetics. J Biol Chem 263: 18017–18022. [PubMed] [Google Scholar]
- 62. Kow YW (1989) Mechanism of action of Escherichia coli exonuclease III. Biochemistry 28: 3280–3287. [DOI] [PubMed] [Google Scholar]
- 63. Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DM 3rd, et al (2001) Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J Mol Biol 307: 1023–1034. [DOI] [PubMed] [Google Scholar]
- 64. Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Bjoras M, et al. (2008) DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol 15: 515–522. [DOI] [PubMed] [Google Scholar]
- 65. Chastain PD 2nd, Nakamura J, Rao S, Chu H, Ibrahim JG, et al (2010) Abasic sites preferentially form at regions undergoing DNA replication. FASEB J 24: 3674–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ishchenko AA, Sanz G, Privezentzev CV, Maksimenko AV, Saparbaev M (2003) Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res 31: 6344–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takeuchi M, Lillis R, Demple B, Takeshita M (1994) Interactions of Escherichia coli endonuclease IV and exonuclease III with abasic sites in DNA. J Biol Chem 269: 21907–21914. [PubMed] [Google Scholar]
- 68. Lindahl T (1982) DNA repair enzymes. Annu Rev Biochem 51: 61–87. [DOI] [PubMed] [Google Scholar]
- 69. Ishchenko AA, Yang X, Ramotar D, Saparbaev M (2005) The 3′–>5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae . Mol Cell Biol 25: 6380–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parsons JL, Dianova, II, Dianov GL (2005) APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res 33: 2204–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)