Abstract
RecQ DNA helicases are critical for proper maintenance of genomic stability, and mutations in multiple human RecQ genes are linked with genetic disorders characterized by a predisposition to cancer. RecQ proteins are conserved from prokaryotes to humans and in all cases form higher-order complexes with other proteins to efficiently execute their cellular functions. The focus of this review is a conserved complex that is formed between RecQ helicases and type-I topoisomerases. In humans, this complex is referred to as the BLM dissolvasome or BTR complex, and is comprised of the RecQ helicase BLM, topoisomerase IIIα, and the RMI proteins. The BLM dissolvasome functions to resolve linked DNA intermediates without exchange of genetic material, which is critical in somatic cells. We will review the history of this complex and highlight its roles in DNA replication, recombination, and repair. Additionally, we will review recently established interactions between BLM dissolvasome and a second set of genome maintenance factors (the Fanconi anemia proteins) that appear to allow coordinated genome maintenance efforts between the two systems.
Keywords: RecQ helicase, Topoisomerase, Bloom syndrome, Fanconi anemia, DNA repair, Genomic stability
Introduction
Genomic instability is a shared phenotype in multiple hereditary diseases where DNA repair or replication-dependent checkpoint genes are mutated. In these disorders, malfunctions in the normal genome maintenance pathways of cells increase the chance that disease-causing changes will occur that allow for uncontrolled cell proliferation [1–3]. Through the study of one such rare genetic disease, Bloom syndrome (BS), much has been learned about how cells ensure genomic fidelity. The gene mutated in BS encodes a RecQ family DNA helicase, BLM, and the pronounced genomic instability of BS cells reflects the protein’s multiple roles in DNA replication, recombination, and repair [2, 4, 5]. In coordination with a large number of protein partners, BLM aids in cellular responses to replication stress and DNA damage. In this review, we focus on the discovery of a key BLM-containing complex referred to as the “BLM dissolvasome” or “BTR complex” [6] that includes topoisomerase IIIα (Topo IIIα) and the RMI proteins, and on recent advances in understanding its functions. The coordinated action of the BLM helicase and Topo IIIα in this complex is critical for untangling intertwined DNA structures that can arise during DNA replication or repair. Recent insights into interactions between the BLM dissolvasome and FANCM will also be reviewed to provide an overview of the importance of coordinating the functions of these proteins in cells, particularly for the prevention of crossover formation in mitotic cells.
Bloom syndrome
Bloom syndrome is a rare autosomal recessive genetic disorder that was first described in 1954 by dermatologist David Bloom [7]. BS manifestations include proportional dwarfism, sun sensitivity, increased susceptibility to infections and diabetes, and a predisposition to a broad array of cancers. Cancer is a frequent cause of death in BS patients, with the first diagnosis often occurring early in life (mean age is in the mid-20s) [4, 8, 9]. Cells derived from BS patients display a remarkable increase in chromosomal damage as evidenced by DNA rearrangements, gaps, and breaks [8, 10]. The major distinguishing genomic instability feature of somatic BS cells, which is used in diagnosis, is a tenfold elevated frequency of sister-chromatid exchanges (SCEs) [4, 8, 11]. One proposed mechanism that would lead to formation of these crossovers is the asymmetric resolution of Holliday junction (HJ) intermediates formed during homologous recombination (HR) reactions. The abnormal genomic instability behavior of BS cells provided an early clue that the gene mutated in the syndrome (BLM) encoded a protein that had important roles in suppressing chromosomal crossovers and maintaining genomic stability [11].
BLM is a member of the RecQ family of DNA helicases
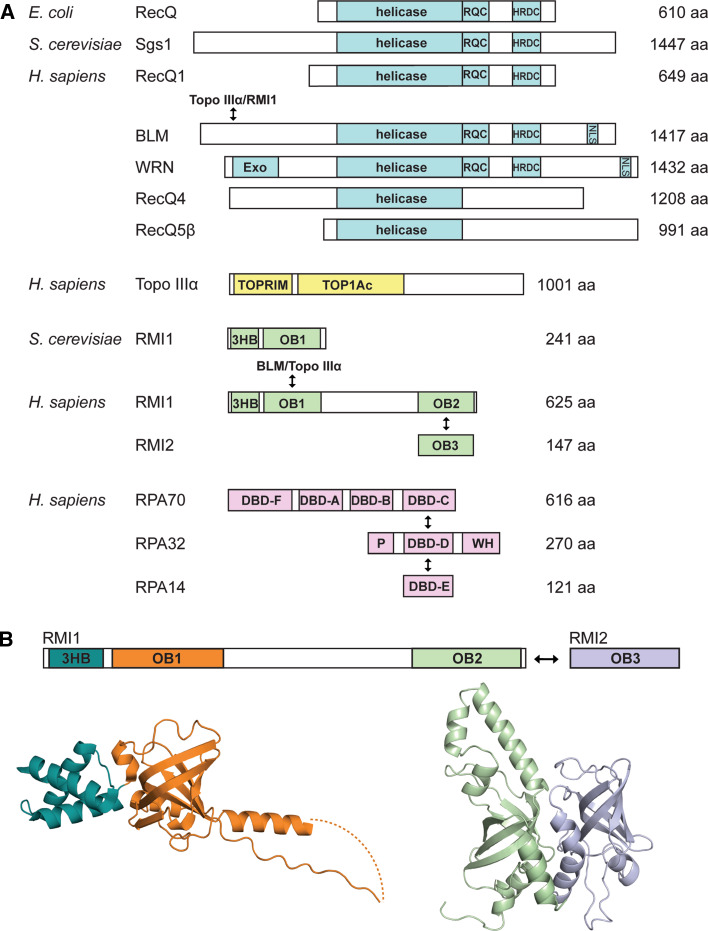
In the mid-1990s, the BLM gene was mapped to chromosome 15 and positional cloning techniques identified its gene product (BLM) as a member of the RecQ family of DNA helicases [12, 13]. Many of the mutations in BLM that give rise to BS are frameshift or nonsense mutations that lead to premature translation termination of BLM, resulting in a truncated protein. Since the nuclear localization signal (NLS) of BLM is encoded in its C-terminus, these truncation products are expected to fail to reach the nucleus (see the Bloom Syndrome Registry, http://med.cornell.edu/bsr/genetics/ and [14]) (Fig. 1a). In addition, there are 13 known BS-causing missense mutations, 7 in the helicase domain and 6 in the “RecQ C-terminal” (RQC) domain [13–17]. Through biochemical and structural studies, these mutations have been shown to confer loss of catalytic activity or are expected to hinder protein folding or stability [17–20].
Fig. 1.
Properties of the RecQ helicases and related proteins. a Domain architectures of RecQs, Topo IIIα, RMI1, RMI2, and RPA. Known interactions are depicted with arrows. RPA is shown because it shares sequence and structural similarities with the RMI subcomplex: RMI1 OB1 resembles RPA 70 DBD-F, RMI1 OB2 resembles RPA70 DBD-C, and RMI2 OB3 resembles RPA32 DBD-D. RQC RecQ-conserved, HRDC helicase and RNaseD C-terminal, NLS nuclear localization signal, Exo exonuclease, TOPRIM topoisomerase-primase, TOP1Ac topoisomerase IA conserved, 3HB three helix bundle (also DUF1767), OB OB-fold, DBD DNA binding domain (also OB-folds), P phosphorylated region, WH winged helix, aa amino acids. b Structures of RMI1 and RMI2. The structures are indicated by the colors in the domains above. On the left is a ribbon diagram of the crystal structure of the N-terminus of RMI1 (residues 2–204, excluding residues 109–119, and this region is connected with a dashed line) [120]. On the right is a ribbon diagram of the crystal structure RMI1 OB2 (residues 475–625), and RMI2 (residues 17–147) [120, 121]. The diagrams were rendered using PyMol [220]
The discovery of the BLM gene helped to explain the molecular basis for BS, since the RecQ helicase in Escherichia coli was already known to play a role in bacterial genomic integrity [21]. At the time that BLM was discovered, ATP-dependent DNA helicase activity had been demonstrated for E. coli RecQ and roles for the protein in the RecF recombination pathway had been established [22]. This background knowledge proved valuable for quickly ascertaining the functions of BLM and other eukaryotic RecQ DNA helicases. Bacteria and yeast typically code for a single RecQ protein, whereas humans have five: RecQ1, BLM, WRN, RecQ4, and RecQ5 (Fig. 1). In addition to BS, two other recessive genetic diseases are caused by mutations in the WRN and RecQ4 genes [Werner’s syndrome (WS) [23] and Rothmund–Thomson syndrome (RTS) [24], respectively]. Two more diseases, RAPADILINO syndrome and Baller–Gerold syndrome, both share some clinical characteristics with RTS and have also been linked to a subset of mutations of RECQ4 [25, 26]. As with BS, WS and RTS are characterized by increased genomic instability and cancer predisposition, although the breadth of cancers observed in WS and RTS is narrower than that of BS, and the characteristic increase in SCEs of BS cells is not seen in cell lines derived from WS and RTS patients.
As is the case for all RecQ family members, BLM is a superfamily 2 (SF2) helicase that uses the energy derived from ATP hydrolysis to unwind DNA [13, 27]. In addition to a conserved helicase domain that provides the motor function for DNA unwinding, BLM and most other identified RecQs also contain RQC and the “helicase and RNaseD C-terminal” (HRDC) domains [28] (Fig. 1a). The RQC domain is comprised of a Zn2+-binding scaffold and a winged-helix (WH) subdomain, and in some cases this domain has been shown to be important for structure-specific binding of RecQ proteins to replication fork, HJ, or G-quartet DNA structures [20, 29–33]. The HRDC domain is an independent globular domain that can confer further substrate specificity to RecQ helicases and can potentially enhance DNA unwinding processivity [34–37]. Interestingly, although nonsense and frame-shift insertion/deletion mutations that cause BS span the entire coding-region of the BLM gene, missense mutations are restricted to the helicase and RQC domains, indicating that DNA binding and unwinding functions are critical for BLM cellular functions [14]. In addition to these conserved domains, many eukaryotic RecQ helicases contain additional N- and C-terminal elements that encode for additional enzymatic functions (e.g., the exonuclease domain of WRN; Fig. 1a), provide binding sites for heterologous proteins, receive post-translational modifications, and/or facilitate proper subcellular localization [5, 27].
Biochemical properties of BLM
The BLM helicase is a 1,417-residue ATP-dependent 3′–5′ DNA helicase [4, 38]. The 3′–5′ directionality indicates that the preferred substrate for BLM unwinding contains a 3′ single-stranded (ss) extension to which the helicase is thought to bind and translocate along; this directionality is conserved among RecQ helicase family members. Although BLM is capable of unwinding simple double-stranded (ds) DNA substrates, it preferentially acts on more elaborate DNA structures such as G-quartet, D-loop, and HJ DNAs [32, 39–43]. When combined with the high level of chromosomal abnormalities observed in BS cell lines and the known roles for the prototypical RecQ in E. coli, these preferences suggested early on that the BLM helicase could have an important role in resolving unusual DNA structures that can arise in cells through processes such as HR.
BLM has been shown to form a large number of functionally important homooligomeric and heterooligomeric complexes. Full-length BLM can form homohexameric structures in vitro, and an isolated domain comprised of the N-terminal 431 residues of BLM can self-associate, suggesting that this region aids in BLM oligomerization [44, 45]. More recent results have suggested that BLM oligomers can dissociate into monomers upon ATP hydrolysis, and the helicase may function as a monomer during DNA unwinding [46]. However, this does not rule out the possibility that higher-order oligomers are required to bind and/or unwind more complex DNA structures such as G-quartet and HJ DNA. Interestingly, the N-terminal region of another human RecQ, RecQ1, is required for both oligomerization and HJ unwinding [42]. Furthermore, RecQ1 has been shown to form higher-order oligomers upon binding DNA, but these dissociate to monomers or dimers upon ATP-binding, suggesting that RecQ1 may unwind DNA as monomers or lower-order oligomers [47]. The N-terminal exonuclease domain of WRN also appears to be responsible for hexamer formation, and this isolated exonuclease domain acts catalytically as a hexamer [48] (Fig. 1a). Oligomerization of full-length WRN is stimulated by DNA binding in a similar manner to RecQ1 [49, 50]. For BLM, constructs lacking the N-terminal oligomerization domain are able to unwind HJ DNA, and instead it appears that the HRDC domain is critical for HJ unwinding [37, 52] (Fig. 1a). More research will be required in order to elucidate the roles of oligomerization for the BLM helicase for DNA binding and unwinding in vivo.
By itself, BLM is a poorly processive helicase and is only able to unwind duplexes of less than ~100 bp [53]. However, the addition of the eukaryotic single-stranded DNA-binding protein [replication protein A (RPA)] strongly stimulates BLM DNA unwinding of longer duplexes, whereas the E. coli single-stranded DNA-binding protein (SSB) fails to stimulate BLM unwinding [53]. Since BLM also interacts with RPA, it is thought that BLM/RPA complex formation is important for increasing the processivity of BLM DNA unwinding [53]. BLM has also been shown to possess ssDNA annealing activity; however, RPA inhibits this activity. Since RPA or other DNA-binding proteins are bound to exposed ssDNA, the cellular relevance of this activity is unclear [54]. Similar interactions and/or stimulation by SSBs have been observed in several other RecQ family members including E. coli RecQ/SSB, Saccharomyces cerevisiae Sgs1/RPA, human and Caenorhabditis elegans WRN/RPA, human RecQ1/RPA, and human RecQ5/RPA [51, 55–71]. In addition to its interactions with RPA, numerous studies have shown that BLM can interact with a large number of protein partners that have roles in DNA replication, recombination, and repair, including topoisomerase 3α (Topo IIIα), RMI1, Topo IIα, PICH, RAD51, EXO1, DNA2, FANCJ, RIF1, ATM, ATR, FEN1, DNA polymerase δ, MLH1, p53, WRN, and TRF2 [72–92]. The focus of this review is the complex formed by BLM, Topo IIIα, RMI1, and RMI2, and therefore many of these other important interactions will not be discussed further (see reviews[4, 5]).
RecQ complex formation with Topo III
Physical and functional interactions with topoisomerases have been found to be critical for the genome maintenance activities of many RecQ proteins. Topoisomerases function to relieve the torsional stress that arises from DNA supercoiling by transiently breaking the DNA backbone in either one (type-I) or both (type-II) strands and then passing intact DNA through the opening before resealing the break [93]. Although RecQ proteins have been found to associate with both type-I and type-II topoisomerases, the partner that we focus on in this review is Topo III, a type-I topoisomerase. Linkages between RecQ proteins and Topo IIIs have been apparent since the discovery of the S. cerevisiae RecQ protein (Sgs1) as a suppressor of the slow-growth phenotype of cells lacking Topo III [94]. The N-terminus of Sgs1 interacts with Topo III, and this interaction appears to be the sole function of the first 100 amino acids of Sgs1 [94–97]. The discovery of this interaction in yeast prompted investigations in other organisms; for example, E. coli RecQ was shown to functionally interact with Topo III as well to stimulate DNA catenation/decatenation and the resolution of converging DNA replication forks [98–100]. In humans, the N-terminus of BLM is responsible for interaction with Topo IIIα and both proteins localize to promyelocytic leukemia protein (PML) nuclear bodies [72, 101, 102]. Furthermore, a study examining the effects of deleting the Topo IIIα-interaction domain from BLM (residues 1–133) revealed an arrangement in which interaction with BLM recruits Topo IIIα to PML bodies and showed that this interaction is important in suppressing SCEs [102]. BLM appears to recruit Topo IIIα to DNA and to stimulate its decatenase activity [102–104]. These functional RecQ/Topo III interactions appear to be conserved from bacteria to humans, and, in all cases, the RecQ helicase stimulates the enzymatic activity of the topoisomerase [94, 98, 103, 104].
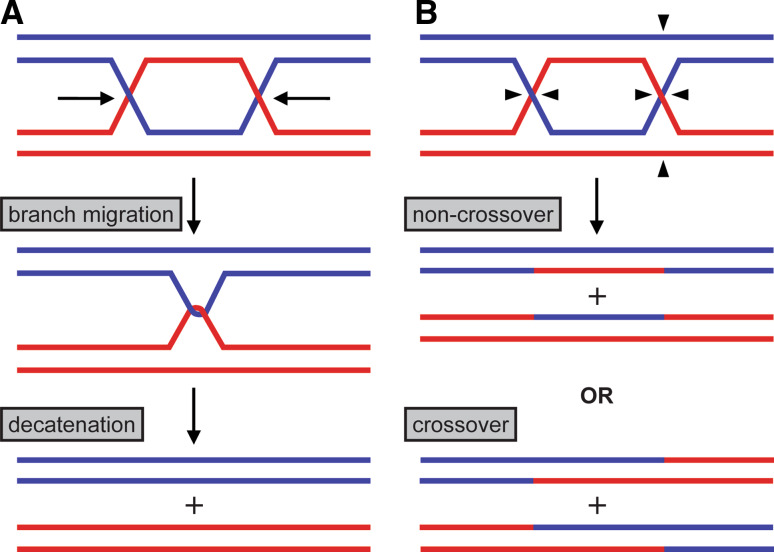
Although the cellular importance of RecQ/Topo III interactions had been established by the late 1990s, the precise substrates on which the BLM/Topo IIIα complex operated (especially with respect to the elevated SCEs observed in BS cells) remained unresolved for some time. An important breakthrough on this question was made by the Hickson group when they considered the types of substrate that might require both BLM and Topo IIIα [105]. Since there are no torsional constraints in simple HJ substrate that would mimic DNA in vivo, they sought out a constrained HJ-based substrate—a double Holliday junction (dHJ). It was discovered that BLM and Topo IIIα worked together to resolve this substrate in a way that prevented exchange of flanking DNA, a process termed “dissolution” [105] (Fig. 2a). This discovery also provided a satisfying explanation for the high levels of SCEs in BS cells. dHJ structures can arise in HR-dependent double-strand break (DSB) repair [106], and can be acted upon by either the BLM/Topo IIIα dissolution pathway or by HJ resolvases, which cleave symmetrically or asymmetrically to form a mixture of crossover and non-crossover products [107, 108] (Fig. 2b). In BLM-deficient BS cells, resolvase-mediated cellular processes would be the dominant pathways for dealing with dHJ structures, which would lead to the exchange of large pieces of homologous chromosomes. In the process of dissolution, BLM is proposed to catalyze branch migration to merge the two HJs into a hemicatenated structure upon which Topo IIIα acts to create exclusively non-crossover products [6] (Fig. 2a). This exclusivity is thought to be important to avoid a “loss of heterozygosity” (LOH) that can occur when the chromatids are separated into daughter cells, which might be tied to the increased cancer predisposition seen in BS patients [109, 110]. While this mechanism provides an explanation for the increase in SCEs observed in BS cells, BLM and the BLM dissolvasome have multiple roles in cells beyond dHJ dissolution, and their roles in other pathways also prevent crossover formation (see “BLM and the BLM dissolvasome in DSB repair”).
Fig. 2.
Pathways for the resolution of dHJ structures. a Proposed pathway for dHJ dissolution. In this pathway, BLM migrates the two HJs towards each other, then Topo IIIα decatenates the structure in the second step, producing exclusively non-crossover products. b Pathway for HJ resolvases. In this pathway, each HJ is cleaved by an HJ resolvase and an equal mixture of non-crossover and crossover products result, depending on if the HJs are cleaved symmetrically or asymmetrically, as depicted by the arrows
BLM interactions with RMI1 and RMI2
Although the BLM/Topo IIIα connection provided an important link for understanding the origins of SCEs in BS cell lines, two additional proteins were subsequently found to be important for stabilizing the complex in HeLa cell nuclear extracts—RMI1 and RMI2 (RMI stands for RecQ-mediated genome instability) (Fig. 1). Together, the BLM/Topo IIIα/RMI1/RMI2 complex is referred to as the “BLM dissolvasome” or “BTR complex.”
RMI1 was identified as a 625-residue protein that co-purified at near-stoichiometric levels with human BLM and Topo IIIα [73] (Fig. 1). The N-terminus of RMI1 directly interacts with BLM and Topo IIIα and stimulates dHJ dissolution greater than tenfold in vitro [111–113]. This is accomplished by stimulation of Topo IIIα, since a variant of RMI1, K166A, which is able to bind BLM but not Topo IIIα, cannot stimulate dHJ dissolution [104, 113]. RMI1 contains N- and C-terminal oligonucleotide/oligosaccharide binding (OB) domains, which led to speculation that it may bind DNA and stimulate Topo IIIα through recruitment effects [73, 114] (Fig. 1). However, RMI1 has only very weak DNA binding affinity in its C-terminal OB domain, which is dispensable for dHJ stimulation in vitro [73, 111, 113, 115]. Instead, RMI1 appears to be important to the stability of the BLM/Topo IIIα/RMI1 complex, since depletion of RMI1 significantly increases SCE levels and affects cellular protein levels of Topo IIIα and, to a lesser extent, BLM [73]. Since RMI1 is able to stimulate Topo IIIα activity in vitro, it might also stabilize a conformation of Topo IIIα with increased catalytic activity [111, 112]. In S. cerevisiae, a homolog of the human RMI1 (Rmi1) was identified as a smaller, 241-residue protein that appears to be most similar to the N-terminus of human RMI1 in that it forms a complex with Sgs1 and Topo III and stimulates Topo III activity [116–118].
RMI2 was subsequently discovered as a binding partner to RMI1 in human cells, and the two together comprise the RMI subcomplex. RMI2 is a 147-residue protein comprised entirely of an OB fold that interacts with the C-terminal OB fold of RMI1 [115, 119] (Fig. 1). The RMI subcomplex is formed through interactions between these OB folds in a manner similar to that of RPA70/RPA32; however, the RMI subcomplex lacks the ability to bind DNA and there is no conservation with the DNA-binding residues found in the RPA complex [115, 120, 121] (Fig. 1). In HeLa cells, depletion of either RMI1 or RMI2 by siRNA significantly reduces the protein levels of the other, indicating their interdependence [115, 119]. Furthermore, their knockdown leads to a strong decrease in Topo IIIα protein levels and a more modest decrease in BLM protein levels [115, 119]. Therefore, it appears that RMI1, RMI2, and Topo IIIα are more highly interdependent in vivo (perhaps forming a stable complex), whereas BLM may be a dissociable component. In addition, studies in hyper-recombinant chicken DT40 cells reveal that knockdown of RMI2 leads to an increase in SCE levels, and when both BLM and RMI2 are knocked down the levels are the same as BLM alone, indicating that RMI2 works with BLM to prevent crossovers [73, 115] (Table 1). Mutations that destabilize the RMI1/RMI2 interface also lead to increased levels of SCEs [121]. In response to DNA damaging agents and replication inhibitors, RMI1 and RMI2 both co-localize with BLM to nuclear foci associated with DNA damage [115]. Additionally, when either RMI1 or RMI2 is depleted, BLM localizes to significantly fewer nuclear foci [73, 115]. These studies confirm the importance of the RMI subcomplex and highlight the central importance of the BLM dissolvasome in DNA repair.
Table 1.
A summary of studies examining cellular SCE
| Protein disrupted or mutated | WT SCE | Mean SCE | Fold increase | Cell line | Significance |
|---|---|---|---|---|---|
| BLM [11] | 6.9 | 89.0 | 12.9 | BS patient cells | Initial SCE report in BS cells |
| Topo IIIα [221] | – | – | – | Mouse embryonic | Embryonic lethal |
| Topo IIIα [222] | – | – | – | DT40 | Knockout is lethal |
| BLM [223] | 2.1 | 26.4 | 12.6 | DT40 | |
| RAD54 [223] | 2.1 | 1.7 | 0.8 | DT40 | RAD54 is a DSB repair protein |
| BLM/RAD54 [223] | 2.1 | 8.2 | 3.9 | DT40 | Many SCEs come from DSB repair |
| BLM [73] | ~0.2a | ~0.5a | 2–3 | HeLa siRNAb | |
| RMI1 [73] | ~0.2a | ~0.5a | 2-3 | HeLa siRNAb | BLM and RMI1: same pathway |
| BLM [115] | 2.1 | 19.0 | 9.0 | DT40 | |
| RMI2 [115] | 2.1 | 11.4/9.5 | 5.4/4.5 | DT40 | Two different experiments |
| BLM/RMI2 [115] | 2.1 | 18.1 | 8.6 | DT40 | BLM and RMI2: same pathway |
| RMI2 Lys121Ala [115] | 2.1 | 8.2 | 3.9 | DT40 | Disrupts RMI/FANCM interface |
| RMI2 [121] | 1.85 | 11.3 | 6.1 | DT40 | |
| RMI2 Lys121Ala [121] | 1.85 | 10.2 | 5.5 | DT40 | Disrupts RMI/FANCM interface |
| RMI2 Lys121Glu [121] | 1.85 | 10.9 | 5.9 | DT40 | Also weakens RMI1/RMI2 (in Co-IPs) |
| RMI2 Asp141Lys [121] | 1.85 | 15.3 | 8.3 | DT40 | Disrupts RMI1/RMI2 interface |
| RMI2 Lys121Glu/Asp141Lys [121] | 1.85 | 11.6 | 6.3 | DT40 | Disrupts both interfaces |
| FANCC [173] | 1.4 | 5.1 | 3.6 | DT40 | |
| FANCC [175] | 1.8 | 4.1 | 2.3 | DT40 | |
| FANCJ [175] | 1.8 | 5.2 | 2.9 | DT40 | |
| FANCM [172] | 1.8 | 9.0 | 5.0 | DT40 | SCEs higher than other FA disruptions |
| FANCC [172] | 1.8 | 6.0 | 3.3 | DT40 | |
| FANCM/FANCC [172] | 1.8 | 14.9 | 8.3 | DT40 | SCEs are additive: two pathways |
| FANCM D203A [172] | 1.8 | 8.9 | 4.9 | DT40 | FANCM catalytic activity required |
| BLM [172] | 1.8 | 14.0 | 7.8 | DT40 | |
| BLM/FANCM D203A [172] | 1.8 | 15.2 | 8.4 | DT40 | BLM and FANCM: same pathway |
| FANCJ [79] | 0.31 | 0.59 | 1.9 | EUFA30-Fc | Also no SCE increase in FA-C cells |
| FANCJ [79] | 0.21 | 0.36 | 1.7 | PSNF5c/FANCJ siRNA | |
| BLM [79] | 0.21 | 1.38 | 6.6 | PSNG13c | “WT” data is from above PSNF5 cells |
| BLM/FANCJ [79] | 0.21 | 1.21 | 5.8 | PSNG13c/FANCJ siRNA | BLM and FANCJ: same pathway |
| FANCM [156] | ~6a | ~12.5a | 2-3 | 293 siRNAb with MMC | |
| FANCM FF >AA [156] | ~6a | ~12.5a | 2-3 | 293 siRNAb with MMC | Disrupts RMI/FANCM interface [183] |
| FANCM [183] | 2.0 | 17.7 | 8.9 | DT40 | |
| FANCM F1232A [183] | 2.0 | 14.2 | 7.1 | DT40 | Disrupts RMI/FANCM interface [183] |
| RIF1 [80] | 1.7 | 1.7 | 1.0 | DT40 | Not involved in SCE suppression |
aMean SCE was not reported, inferred from data
bExperiments using siRNA are known to not completely mimic the SCE levels observed in BS patients
c EUFA30-F FA-J fibroblasts, PSNF5 BLM+/+, PSNG13 BLM−/−
BLM and the BLM dissolvasome in DSB repair
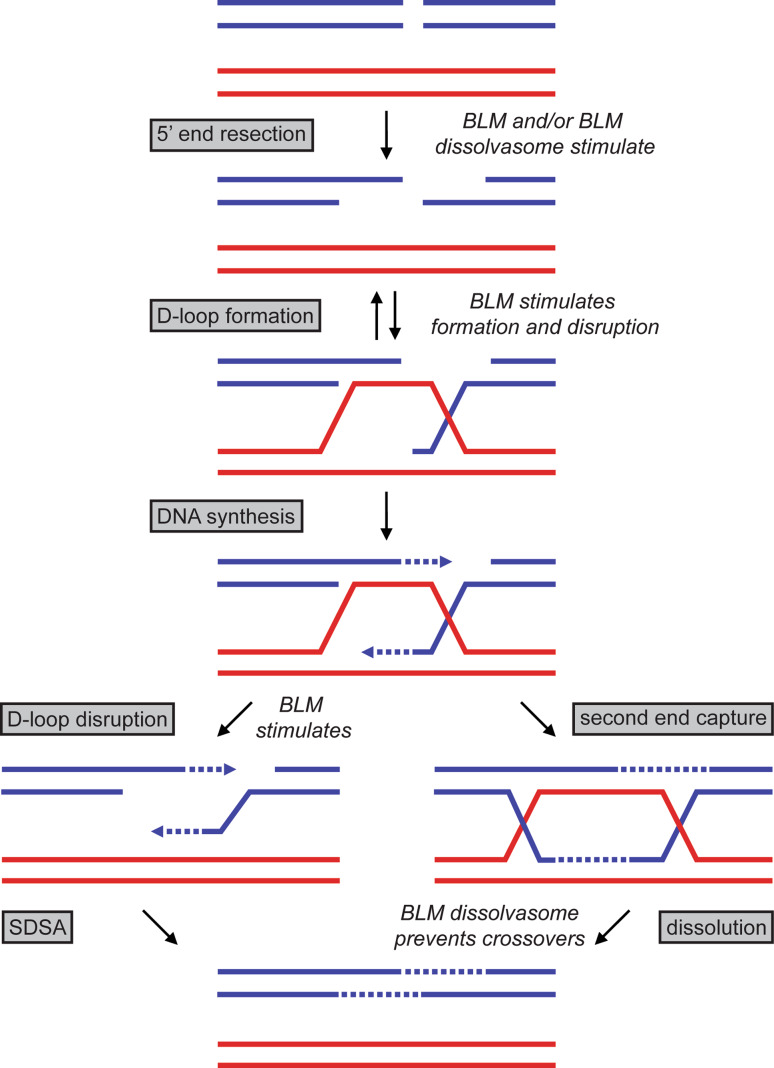
Numerous studies have identified both pro- and anti-recombinogenic activities for BLM in DSB repair, even though the high levels of crossovers that form in its absence suggest a primarily anti-recombinogenic role. In fact, BLM may shuttle DSB repair intermediates towards a “pro-recombination” pathway in order to ensure they are resolved via dissolution, and therefore without crossover [9]. Evidence supports roles for BLM in multiple steps in HR-dependent DSB repair, including 5′ end resection, RAD51 filament and D-loop formation, and, as described earlier, in resolving dHJ structures (Fig. 3). These roles are reviewed in the remainder of this section.
Fig. 3.
Roles for the BLM dissolvasome in HR-dependent DSB repair. Shown here is the pathway for DSB repair, showing the different steps where BLM and/or the BLM dissolvasome may act or stimulate. The pathways shown here are resolved to non-crossover products, either via synthesis-dependent strand annealing (SDSA) (left) or dHJ dissolution (right); however, the dHJ substrate may also be resolved via HJ resolvases as shown in Fig. 2. New DNA synthesis is depicted with a dashed line
Multiple laboratories have shown that BLM is able to stimulate nuclease activity in 5′ end resection [9, 77, 78, 122] (Fig. 3). In humans, two pathways for end resection have been discovered, one involving the nuclease DNA2 and one involving exonuclease 1 (EXO1), and in both these pathways BLM physically interacts with and stimulates the nuclease [77, 78]. While both RecQ1 and WRN are able to simulate other EXO1 catalytic activities, only BLM has been shown to specifically stimulate the end resection activity of EXO1 and DNA2 in vitro [77, 78, 123, 124]. In Xenopus laevis cell-free extracts, WRN was shown to function with DNA2 in end resection, but more research is required to determine whether this represents a conserved role for WRN [125]. The most extensive studies have been performed in the yeast system, where Sgs1 has been shown to stimulate Dna2 [126, 127]. This is relevant in vivo, since, when exo1 and sgs1 are simultaneously deleted in yeast (thus disrupting both pathways for end resection), cells become sensitive to a wide range of DNA-damaging agents [122]. By monitoring end resection in cells, it was further shown that Sgs1, Topo IIIα, and Rmi1 are all involved in the same resection pathway [127]. In addition, the Sgs1/Dna2 pathway has been reconstituted in vitro, and end resection was stimulated by the addition of Rmi1/Topo IIIα [128, 129]. In this context, Rmi1/Topo IIIα are together able to stimulate the helicase activity of Sgs1 and are thought to aid Sgs1 DNA binding; however, this stimulation does not appear to require the catalytic activity of Topo IIIα [128, 129]. Further research will be required to determine the importance of this stimulation in vivo and whether similar biochemical attributes are conserved in humans. Nonetheless, the role of BLM in 5′ end resection is pro-recombinogenic since it aids in initiation of HR. This activity may serve to shuttle DSBs away from non-homologous end joining (NHEJ) pathways, which are more error prone and have been observed at higher levels in BS cells [130]. NHEJ is active during the entire cell cycle; however, it cannot be initiated from the free 3′ ssDNA end that results from end resection, and in this way BLM’s role in end resection may be one of the factors that helps determine DSB repair pathway choice [131]. Since BLM is cell cycle regulated, this would help ensure that the HR-dependent DSB repair is the primary pathway when an available sister chromatid exists to act as a template [132–134]. Moreover, cells in S- and G2-phase appear to predominantly use HR to repair DSBs, and to complete this repair via dissolution to prevent exchange of genetic material [9].
In the second step of HR, the RAD51 recombinase forms a helical filament on the free 3′ DNA end. A homology search for the DNA in the RAD51/ssDNA complex produces a D-loop structure as a result of invasion of the ssDNA into a homologous sister chromatid or chromosome [135] (Fig. 3). In this step, BLM interacts with RAD51 and is able to migrate and unwind D-loops, which disrupts nascent pairing in the first steps of HR [43, 136, 137]. However, this activity appears to depend on the status of RAD51: BLM stimulates strand exchange of active ATP-bound RAD51 filaments, but dismantles inactive ADP-bound filaments [137–139]. This may indicate that BLM surveys nascent D-loops to assure they are appropriate for HR. This activity could be related to a function of the prototypical E. coli RecQ in preventing illegitimate recombination between homologous but non-identical DNA sequences [140]. Finally, some of the dismantled filaments may be repaired via single-strand annealing (SSA), which relies on flanking DNA repeat sequences commonly found in mammalian DNA [135].
Following D-loop formation and DNA synthesis, BLM has additional anti-recombinogenic rolls. First, in synthesis-dependent strand annealing (SDSA), the resulting D-loop product is dismantled and the broken chromosome is then able to re-anneal for repair in a non-crossover fashion (Fig. 3). BLM may play a role in unwinding this D-loop [43, 136, 137]. Second, in the HR-dependent pathway, the D-loop remains intact and second end capture recruits the second strand to form a dHJ substrate (Fig. 3). This intermediate is the proposed substrate for dissolution via the BLM dissolvasome, as described above [6, 105] (Fig. 2a). However, the dHJ substrate could also be resolved through cleavage by HJ resolvases, which leads to an equal distribution of non-crossover and crossover products (Fig. 2b). Overall, it appears that BLM and potentially the entire BLM dissolvasome have multiple roles in HR, and while these roles may be categorized as pro- and anti-recombinogenic, it is likely that BLM activity is focused on ensuring HR fidelity and the prevention of crossovers.
The role of the BLM dissolvasome in replication restart
Early observations with BS cell lines revealed a replication deficiency and hinted that BLM might help repair stalled or damaged replication forks. Cells from BS patients are slower to progress through S-phase and accumulate abnormal replication intermediates [141–143]. Additionally, BLM levels are regulated in a cell cycle-dependent manner, accumulating in S-phase and persisting through G2/M before diminishing in G1 [132–134]. Cells from BS patients are hypersensitive to replication stalling with hydroxyurea (HU), and BLM localizes to these stalled forks [82, 144].
Multiple studies point to pivotal roles for BLM in the repair of stalled DNA replication forks [145, 146]. DNA replication processes can stall due to blockage of the replication fork by encounters with physical barriers, such as proteins that are tightly bound to the DNA template or impassable damage to the template. For example, replication through a nick in parental DNA creates a DSB when the replicative helicase proceeds through the nicked region. In these instances, the BLM-dependent DSB repair pathways described above would be triggered to drive replication restart (for review, see [147]). In addition, because BLM is a helicase that specifically acts upon DNA substrates that resemble replication and recombination substrates, a separate possible role for BLM is to produce DNA structures that are competent for replication restart. For example, a lesion on the leading strand that prevents progression of the replication fork could lead to replication fork regression and HJ formation (creating an intermediate referred to as a “chicken foot” structure), whose formation could be driven by BLM branch migration [148–150]. This structure can promote lesion bypass by supplying an intact template for the nascent leading strand (template switching) [145]. Following lesion bypass, BLM may also act to reverse the regressed fork via branch migration so that replication can proceed past the lesion, or, instead, replication may restart through an HR-mediated process that would proceed through a dHJ structure that the BLM dissolvasome could act on [149].
More recently, single-molecule DNA fiber and molecular combing techniques have allowed detailed interrogation of the roles of BLM in mammalian DNA replication [151]. These studies have shown that BLM is required for efficient recovery of replication forks blocked by aphidicolin or HU [152], and that BS cells exhibit reduced fork velocity and more incidences of fork pausing [153]. Moreover, the BLM dissolvasome has been implicated in fork progression, as a recent study has shown that depletion of RMI1 leads to a reduction in replication fork rate and a failure to recover from replication fork arrest [154]. Interestingly, these defects in replication fork rate can be suppressed when BLM is also depleted, which indicates that RMI1 and potentially the rest of the BLM dissolvasome act downstream of BLM [154]. Finally, the protein RIF1 has been shown to interact with the BLM dissolvasome to promote replication fork restart; however, RIF1 does not play a role in SCE suppression [80] (Table 1). RIF1 can bind directly to replication fork and HJ DNA and it may coordinate fork regression with BLM [80]. Therefore, the BLM dissolvasome may have roles in replication restart that are not related to its function in suppressing SCEs. In summary, a growing body of literature supports roles for the BLM dissolvasome at replication forks to help maintain integrity through DSB repair and efficient replication restart.
Link with Fanconi anemia protein FANCM
The discovery that the BLM dissolvasome was linked to proteins involved in the autosomal recessive disease Fanconi anemia (FA) made an important connection in genome maintenance that further solidified the importance of the BLM dissolvasome in DNA replication restart [155, 156]. As is the case for BS cells, FA cell lines have significantly heightened levels of chromosomal instability, although they are exemplified by radial chromosomes and a hypersensitivity to agents that induce DNA interstrand crosslinks (ICLs). Mutations in at least 15 genes [FANCA, B, C, D1 (BRCA2), D2, E, F, G, I, J, L, M, N, O, and P] give rise to this highly heterogeneous disease, which includes symptoms of developmental abnormalities, progressive bone marrow failure, and a high occurrence of cancer [157–160]. Prior to the discovery that key proteins in FA and BS directly interacted with one another, roles for the FA proteins in repair of DNA damage at ICL sites that have caused replication fork failure were known [161]. In this response, a core complex of eight FA proteins (FANCA, B, C, E, F, G, L, and M) recognizes the DNA damage and catalyzes monoubiquitination of the FANCD2/FANCI heterodimer. Ubiquitinated FANCD2/FANCI in turn recruits downstream FA proteins such as FANCJ to facilitate removal of the ICL, which is followed by translesion synthesis, nucleotide excision repair, and HR to fully repair the DNA [160–162]. Further details on the FA pathway can be found in a recent review by Kim and D’Andrea [160].
The initial finding that linked the BLM dissolvasome to FA was identification of an ICL-induced super-complex called BRAFT (BLM, RPA, FA, and Topo IIIα) that included components of both the BLM dissolvasome and FA proteins [155, 163]. A subsequent study revealed that BLM and FA proteins co-localized in response to treatment with DNA crosslinkers or replication arrest and could be co-immunoprecipitated [164]. Of the FA proteins, FANCM is the most conserved, with orthologs in archaebacteria (Hef) and yeast (MPH1 in S. cerevisiae, and Fml1 in Schizosaccharomyces pombe) [165–168]. FANCM is a large, 2,048-residue subunit of the FA core complex that contains an N-terminal DEAH-box helicase domain and a degenerate C-terminal ERCC4-like endonuclease domain linked together by a highly dynamic linker element that has only short regions of conserved sequence [156, 165]. FANCM is thought to act as a sensor to detect blocked replication forks in coordination with partner proteins FAAP24 and MHF [169–171] (Fig. 4a). This complex is then thought to recruit other FA proteins in order to repair the DNA damage, and, subsequently, FANCM may act catalytically to remodel the replication fork in order for replication to proceed [158, 169, 170]. FANCM-deficient cells are also characterized by a large increase in SCEs, which has also been described for other FA proteins such as FANCC and FANCJ, though to a lesser degree than that observed with FANCM-depleted cells [172–175] (Table 1). This striking phenotypic parallel between FANCM- and BLM-deficient cells foreshadowed an interaction between FANCM and the BLM dissolvasome.
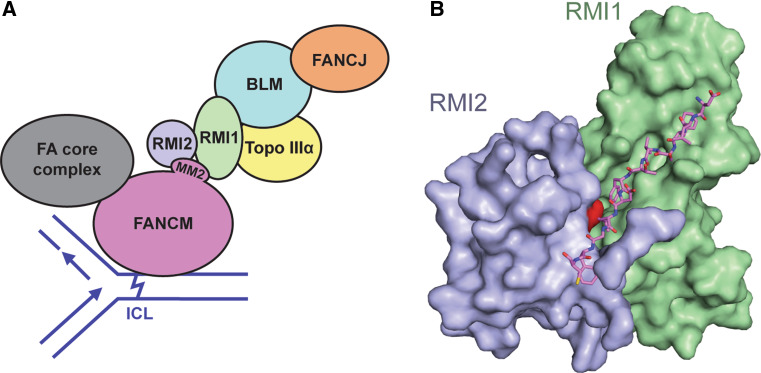
Fig. 4.
BLM dissolvasome and FA protein interactions described in this review. a A potential model for the protein interactions between the BLM dissolvasome, FANCM, and FANCJ. FANCM is shown at a site of DNA damage, depicted as an ICL, that may be encountered during replication and impede fork progression. In this model, FANCM then recruits the BLM dissolvasome through the MM2 interaction with the RMI subcomplex. FANCM is also part of the FA core complex and can localize the rest of the core complex to an ICL. FANCJ may also be recruited to sites of DNA damage via its interaction with BLM. b The overall structure of the RMI core complex with bound MM2 peptide from FANCM. MM2 residues 1,226–1,237 are shown in purple, and RMI2 Lys121 is highlighted in red [183]
FANCM appears to have additional roles beyond recruitment of the FA core complex to ICL-damaged DNA [172, 176, 177]. FANCM might also aid in general coordination of replication restart, and only initiate the FA pathway under specific circumstances such as ICLs. While FANCM has not demonstrated helicase activity, as a translocase FANCM can act on DNA substrates that mimic replication forks and HJs, and has been implicated in replication fork remodeling [165, 176, 178, 179]. Molecular combing experiments have shown that the translocase activity of FANCM is important for the restart of stalled replication forks [180, 181]. Moreover, these stalled replication forks have been shown to be prone to collapse, which generates DSBs that are subsequently repaired through HR [182]. At these sites, the coordinated activity of the BLM dissolvasome and FANCM may be most evident.
A direct interaction between a conserved 34-residue motif in the middle of FANCM (called MM2, for FANCM Motif 2) and the C-terminal OB-fold of RMI1 was first demonstrated in 2009 [156] (Fig. 4). A recent crystal structure of a proteolytically stable RMI1/RMI2 fragment bound to the MM2 peptide has revealed that the full interface is comprised of both the C-terminal OB-fold of RMI1 and the full-length RMI2 protein [183] (Fig. 4b). In this complex, the side chains of conserved hydrophobic residues from FANCM MM2 dock into hydrophobic pockets on the RMI1/RMI2 surface, forming a stable tripartite complex [183]. A FANCM mutation in two of the docking residues from MM2 (FANCM FF >AA) leads to an increase in SCEs upon treatment with mitomycin C (MMC), which induces ICLs [156] (Table 1). Moreover, mutation of a key residue on the RMI surface (Lys121 of RMI2) leads to an increase of SCEs in chicken DT40 cells that is similar to levels seen when FANCM or members of the BLM dissolvasome are depleted [73, 115, 121, 169, 172, 183] (Table 1; Fig. 4b). These data indicate that coordination between FANCM and the BLM dissolvasome is critical in preventing crossover formation. Beyond their roles in SCE suppression, the BLM dissolvasome, FANCM, and FANCJ (see below) all appear to be important in replication fork progression, remodeling, and integrity. Further research will be required to delineate their specific roles and interdependence.
BLM/FANCJ interaction
In addition to the RMI/FANCM interaction, interactions between other FA- and BS-associated proteins have been reported. Notably, a functional and physical interaction has been described between BLM and FANCJ, a helicase that acts downstream of the FA core complex [79] (Fig. 4a). As with BS- and FANCM-deficient cells, SCEs are elevated in FANCJ-deficient cells (although only ~two- to threefold for FANCJ; see Table 1), and this elevation is synergistic with BLM [79, 175]. FANCJ is an ATP-dependent SF2 helicase that acts in the 5′–3′ direction and is also sensitive to replication stress, and therefore it may cooperate with BLM replication restart [79, 184] (for recent discussion on this interaction, see [185]). In addition, both BLM and FANCJ are able to unwind G-quartet DNA, and both may be important in replication to remove these secondary structure elements from DNA so that replication can proceed [39, 186, 187]. A recent study in DT40 cells implied a genetic interaction between FANCJ, BLM, and WRN that may allow coordination of replication through G-quartet DNA [188].
BLM and FA proteins both act at the replication-dependent checkpoint
In parallel with their activities in replication fork stability and restart, roles for BLM and multiple FA proteins have been established in the replication-dependent or S-phase cell cycle checkpoint. This checkpoint triggers cell cycle arrest when replicative processes are blocked due to DNA damage, which leads to the activation of ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) kinases [189]. ATM and ATR activation initiates a signaling cascade that stalls the cell cycle, providing the time needed to recover from replicative stress. While the two pathways are interdependent, ATR generally signals replication-dependent damage through checkpoint kinase 1 (CHK1), whereas ATM signals in response to DSBs via checkpoint kinase 2 (CHK2) [190, 191].
Within this checkpoint mechanism, BLM, FANCD2, and FANCI are targets of both the ATR/CHK1 and the ATM/CHK2 pathways within the pathway, whereas FANCA and FANCE appear to be acted upon preferentially by the ATR/CHK1 pathway [81, 82, 192–199]. BLM is phosphorylated at T99 and T122, and BS cells expressing an unphosphorylatable BLM variant are unable to recover following treatment with HU, and instead enter an extended arrest at the G2/M checkpoint [81, 82]. Furthermore, this recovery is dependent on the helicase activity of BLM [152]. Taken together, these data indicate a critical role for BLM at the replication-dependent checkpoint, and suggest that, after replication has stalled, BLM is phosphorylated, which signals its localization to replication forks in order to aid in faithful restart. BLM may act without the rest of the BLM dissolvasome in this role, as phosphorylation on T99 by ATM or ATR causes BLM to dissociate from Topo IIIα, and presumably also from the RMI subcomplex [192].
FANCM and its binding partner FAAP24 have been implicated as upstream effectors in activation of the ATR pathway, which is independent from the rest of the FA core complex [179–181]. However, a more recent study has suggested that FANCM and FAAP24 have some independent functions, and that FAAP24 alone is responsible for checkpoint activation [200]. In a similar fashion, FANCJ appears to play a role in ATR activation [201]. These insights have led to a model in which FANCM/FAAP24 (or FAAP24 alone) and FANCJ act as sensors of replication stress in cells to initiate activation of ATR/CHK1 and the replication-dependent checkpoint. The activation of this checkpoint then leads to phosphorylation of other FA proteins and BLM to aid in DNA repair and replication restart [158, 181]. Further research will be required to tease apart the roles of each at different stages and how they vary depending on the type of genetic insult.
The BLM dissolvasome and FA proteins are both found associated with ultra-fine anaphase bridges
DNA staining of sister chromatids as they separate during mitosis has revealed the presence of anaphase DNA bridge structures that link the two chromatids. More recently, a subclass of anaphase bridges, termed “ultra-fine anaphase bridges” (UFBs), have been discovered [202–204]. These UFBs are more thread-like and cannot be visualized with conventional DNA dyes; instead, they are localized with immuno-staining of proteins found on the UFBs. Several proteins, including members of the BLM dissolvasome and the PLK1-interacting checkpoint helicase (PICH), are localized to UFBs, and BLM may be specifically recruited by its interaction with PICH [75, 202, 203]. Localization of Topo IIIα and RMI1 to UFBs is dependent on BLM, and therefore it appears that BLM recruits the rest of the BLM dissolvasome to UFBs [203]. The majority of UFBs arise at centromeres, where it has been proposed that the BLM dissolvasome and PICH might cooperate to facilitate the association of Topo IIα, a type-II topoisomerase, to drive centromere disjunction [75, 203, 205–207]. To further support this hypothesis, BLM and Topo IIα have been shown to interact both in vitro and in vivo [74, 208]. The majority of these bridges appear to resolve as mitosis progresses without the formation of DSB intermediates, and may be normal structures that assist in centromeric cohesion [204].
A less common class of UFBs that are exclusively found at common fragile sites on chromosomes are proposed to result from incomplete DNA replication that is induced by replication stalling agents [209, 210]. Normally, under replication stress, additional replication initiation events will occur so that the DNA can be fully duplicated, but recent studies indicate that, at fragile sites, there is a lack of these extra initiation events, and so mitosis may begin before these regions have completed replication, resulting in a UFB [211, 212]. Apart from their spatial differences, the other distinguishing feature of this class of UFBs is the localization of FA proteins FANCD2 and FANCI at their termini, which is consistent with their presence being related to replication stress [209, 210]. FANCD2/I are proposed to be recruited to sites of incomplete replication in the late S- and G2-phases, which then become sites of UFBs that are coated by BLM and PICH in mitosis. FANCM has also been localized to UFBs in a BLM-dependent manner, though FANCM is observed on UFBs later in mitosis after BLM has already dissociated from UFBs [213]. It has therefore been hypothesized that the BLM dissolvasome can recruit FANCM to UFBs in a hand-off mechanism; however, another possibility is that BLM alters the structure of the DNA in the bridge to make it accessible for FANCM later in mitosis [204, 213].
Most of these FA-dependent bridges disappear as mitosis progresses, although some persist, and presumably the cell divides before replication is completed at these sites [209, 210]. One hypothesized fate of UFBs is to resolve before replication is complete and form symmetrical DNA lesions in daughter cells that are marked as sites of DNA damage with 53BP1 [214]. These sites become sequestered and protected by 53BP1 nuclear bodies until they can be repaired properly in the next S-phase [204, 214, 215]. Alternatively, a more detrimental result of unresolved UFBs is chromosomal breakage, which leads to the formation of micronuclei. These products are observed at higher frequency when BLM or FA proteins are depleted in cells [10, 203, 209, 210, 213].
Concluding remarks
From the initial studies of the relatively simple E. coli RecQ protein to the more complex BLM dissolvasome and its many interactions, it has become clear that RecQ proteins and protein complexes have evolved central roles in maintaining genomic stability in all organisms. In all cases, these proteins do not act alone, but instead function as components in direct and indirect networks that integrate diverse enzymatic activities and cellular regulatory systems. In this review, we have focused on BLM and its interactions with Topo IIIα, RMI1, and RMI2, which comprise the BLM dissolvasome in humans. Overall, BLM appears to coordinate its functions with these and other proteins in order to maintain genetic stability in somatic cells. These functions include roles in replication restart, HR-dependent DSB repair, the replication-dependent checkpoint, and at UFBs. In many of these pathways, a role for BLM has only recently been described, and future studies will be required to understand the precise role for BLM and/or other human RecQ helicases in vivo.
We have also explored the role of the interaction between the BLM dissolvasome and FA proteins, specifically FANCM and FANCJ. It appears that BLM and FA proteins work together in many pathways, and we have described one potential model for the localization of these proteins at ICLs (Fig. 4a). The BLM dissolvasome and FA proteins coordinate their roles in dHJ dissolution, both respond to stalled replication forks, are involved in the replication-dependent checkpoint, and are found at UFBs. Another interesting collaboration may also exist in DSB repair, as FA proteins have been speculated to be involved in repair pathway choice, especially if the damage is an ICL. Studies in C. elegans, chicken DT40, and mammalian cells have provided evidence that the FA pathway helps to suppress NHEJ and promote repair via HR [216, 217]. A recent study showing that DNA2 and FANCD2 can be copurified provides further evidence that the FA pathway is involved in end resection [218]. However, the network of direct protein–protein interactions required to support this function remains to be defined.
With the stark effects of mutations in three of the five human RecQ genes, the central nature for RecQ proteins is well appreciated. Future research will continue to define the biochemical and cellular roles for RecQ proteins, leading to a better understanding of the molecular basis by which these critical enzymes function and the cellular consequences that result from their dysfunction. These studies may also pave the way for the development of new chemotherapeutics and allow for better treatment of patients with BS and FA. As an example, one recent study identified a small molecule that competitively inhibits BLM and induces an increase in SCEs [219]. This compound will be useful to more specifically target BLM inactivation in further studies and as a potential chemotherapeutic. Other compounds may similarly be discovered that inhibit specific proteins or interactions and could be used to sensitize cancer cells to DNA-damaging agents in cancer treatment.
Acknowledgments
We apologize to all authors whose work we could not cite due to space limitations. Work in our laboratory was funded by a grant from the National Institutes of Health (GM068061) and K.A.M. was supported in part by a National Institutes of Health training grant in Molecular Biosciences (GM07215).
References
- 1.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8):3230–3239. [PubMed] [Google Scholar]
- 2.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev. 2008;129(7–8):425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374(Pt 3):577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117(3):219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 6.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem Sci. 2007;32(12):538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. Am J Dis Child. 1954;88(6):754–758. [PubMed] [Google Scholar]
- 8.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72(6):393–406. [PubMed] [Google Scholar]
- 9.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9(9):644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 10.Rosin MP, German J. Evidence for chromosome instability in vivo in Bloom syndrome: increased numbers of micronuclei in exfoliated cells. Hum Genet. 1985;71(3):187–191. doi: 10.1007/BF00284570. [DOI] [PubMed] [Google Scholar]
- 11.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71(11):4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDaniel LD, Schultz RA. Elevated sister chromatid exchange phenotype of Bloom syndrome cells is complemented by human chromosome 15. Proc Natl Acad Sci USA. 1992;89(17):7968–7972. doi: 10.1073/pnas.89.17.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 14.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom’s Syndrome Registry. Hum Mutat. 2007;28(8):743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 15.Foucault F, Vaury C, Barakat A, Thibout D, Planchon P, Jaulin C, Praz F, Amor-Gueret M. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum Mol Genet. 1997;6(9):1427–1434. doi: 10.1093/hmg/6.9.1427. [DOI] [PubMed] [Google Scholar]
- 16.Barakat A, Ababou M, Onclercq R, Dutertre S, Chadli E, Hda N, Benslimane A, Amor-Gueret M. Identification of a novel BLM missense mutation (2,706T>C) in a Moroccan patient with Bloom’s syndrome. Hum Mutat. 2000;15(6):584–585. doi: 10.1002/1098-1004(200006)15:6<584::AID-HUMU28>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 17.Guo RB, Rigolet P, Ren H, Zhang B, Zhang XD, Dou SX, Wang PY, Amor-Gueret M, Xi XG. Structural and functional analyses of disease-causing missense mutations in Bloom syndrome protein. Nucleic Acids Res. 2007;35(18):6297–6310. doi: 10.1093/nar/gkm536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neff NF, Ellis NA, Ye TZ, Noonan J, Huang K, Sanz M, Proytcheva M. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of Bloom syndrome cells. Mol Biol Cell. 1999;10(3):665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahr A, De Graeve F, Kedinger C, Chatton B. Point mutations causing Bloom’s syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene. 1998;17(20):2565–2571. doi: 10.1038/sj.onc.1202389. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 2003;22(19):4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195(3):474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama K, Irino N, Nakayama H. The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Mol Gen Genet. 1985;200(2):266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 23.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 24.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat Genet. 1999;22(1):82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 25.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 26.Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, Plon SE, Fourneau C, Kestila M, Gillerot Y, Megarbane A, Verloes A. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller–Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43(2):148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killoran MP, Keck JL. Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006;34(15):4098–4105. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem Sci. 1997;22(11):417–418. doi: 10.1016/S0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 29.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106(4):1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18(2):177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 31.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278(52):52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 32.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol. 2006;358(4):1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 33.Vindigni A, Hickson ID. RecQ helicases: multiple structures for multiple functions? HFSP J. 2009;3(3):153–164. doi: 10.2976/1.3079540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13(8):1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J Biol Chem. 2007;282(4):2717–2728. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- 36.Sato A, Mishima M, Nagai A, Kim SY, Ito Y, Hakoshima T, Jee JG, Kitano K. Solution structure of the HRDC domain of human Bloom syndrome protein BLM. J Biochem. 2010;148(4):517–525. doi: 10.1093/jb/mvq097. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24(14):2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J Biol Chem. 1997;272(49):30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 39.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273(42):27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 40.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30(18):3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29(13):2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283(26):17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 43.van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39(47):14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 44.Karow JK, Newman RH, Freemont PS, Hickson ID. Oligomeric ring structure of the Bloom’s syndrome helicase. Curr Biol. 1999;9(11):597–600. doi: 10.1016/S0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 45.Beresten SF, Stan R, van Brabant AJ, Ye T, Naureckiene S, Ellis NA. Purification of overexpressed hexahistidine-tagged BLM N431 as oligomeric complexes. Protein Expr Purif. 1999;17(2):239–248. doi: 10.1006/prep.1999.1135. [DOI] [PubMed] [Google Scholar]
- 46.Xu YN, Bazeille N, Ding XY, Lu XM, Wang PY, Bugnard E, Grondin V, Dou SX, Xi XG. Multimeric BLM is dissociated upon ATP hydrolysis and functions as monomers in resolving DNA structures. Nucleic Acids Res. 2012;40(19):9802–9814. doi: 10.1093/nar/gks728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5(2):e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, Redinbo MR. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′–5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry. 2002;41(9):2901–2912. doi: 10.1021/bi0157161. [DOI] [PubMed] [Google Scholar]
- 49.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and Holliday junction DNAs as an oligomer. J Biol Chem. 2008;283(36):24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′–5′ exonuclease. Nucleic Acids Res. 2000;28(12):2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harmon FG, Kowalczykowski SC. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J Biol Chem. 2001;276(1):232–243. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 52.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J Mol Biol. 2003;330(1):29–42. doi: 10.1016/S0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 53.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275(31):23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 54.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33(12):3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umezu K, Nakayama H. RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J Mol Biol. 1993;230(4):1145–1150. doi: 10.1006/jmbi.1993.1231. [DOI] [PubMed] [Google Scholar]
- 56.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12(8):1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282(26):19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 58.Lecointe F, Serena C, Velten M, Costes A, McGovern S, Meile JC, Errington J, Ehrlich SD, Noirot P, Polard P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007;26(19):4239–4251. doi: 10.1038/sj.emboj.7601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB’s C terminus. J Mol Biol. 2009;386(3):612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J Biol Chem. 2010;285(11):8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22(16):4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guenole A, van Attikum H, Thoma NH, Gasser SM. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012;31(18):3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia PL, Bradley G, Hayes CJ, Krintel S, Soultanas P, Janscak P. RPA alleviates the inhibitory effect of vinylphosphonate internucleotide linkages on DNA unwinding by BLM and WRN helicases. Nucleic Acids Res. 2004;32(12):3771–3778. doi: 10.1093/nar/gkh709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the replication protein a interaction domain of the Werner and Bloom syndrome helicases. J Biol Chem. 2005;280(33):29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 65.Ahn B, Lee JW, Jung H, Beck G, Bohr VA. Mechanism of Werner DNA helicase: POT1 and RPA stimulates WRN to unwind beyond gaps in the translocating strand. PLoS ONE. 2009;4(3):e4673. doi: 10.1371/journal.pone.0004673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sowd G, Wang H, Pretto D, Chazin WJ, Opresko PL. Replication protein A stimulates the Werner syndrome protein branch migration activity. J Biol Chem. 2009;284(50):34682–34691. doi: 10.1074/jbc.M109.049031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Machwe A, Lozada E, Wold MS, Li GM, Orren DK. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J Biol Chem. 2011;286(5):3497–3508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyun M, Park S, Kim E, Kim DH, Lee SJ, Koo HS, Seo YS, Ahn B. Physical and functional interactions of Caenorhabditis elegans WRN-1 helicase with RPA-1. Biochemistry. 2012;51(7):1336–1345. doi: 10.1021/bi200791p. [DOI] [PubMed] [Google Scholar]
- 69.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32(7):2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui S, Klima R, Ochem A, Arosio D, Falaschi A, Vindigni A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem. 2003;278(3):1424–1432. doi: 10.1074/jbc.M209407200. [DOI] [PubMed] [Google Scholar]
- 71.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23(14):2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275(13):9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 73.Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24(7):1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase IIalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284(22):14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ke Y, Huh JW, Warrington R, Li B, Wu N, Leng M, Zhang J, Ball HL, Yu H. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 2011;30(16):3309–3321. doi: 10.1038/emboj.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276(22):19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 77.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105(44):16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM., Jr Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011;30(4):692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D, 3rd, Meetei AR, Wilson L, Nguyen H, Weng NP, Brill SJ, Li L, Vindigni A, Pommier Y, Seidman M, Wang W. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29(18):3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem. 2002;277(34):30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- 82.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24(3):1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J Biol Chem. 2004;279(11):9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 84.Wang W, Bambara RA. Human Bloom protein stimulates flap endonuclease 1 activity by resolving DNA secondary structure. J Biol Chem. 2005;280(7):5391–5399. doi: 10.1074/jbc.M412359200. [DOI] [PubMed] [Google Scholar]
- 85.Selak N, Bachrati CZ, Shevelev I, Dietschy T, van Loon B, Jacob A, Hubscher U, Hoheisel JD, Hickson ID, Stagljar I. The Bloom’s syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase delta. Nucleic Acids Res. 2008;36(16):5166–5179. doi: 10.1093/nar/gkn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Bebenek K, Kunkel TA, Groden J. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem. 2001;276(32):30031–30035. doi: 10.1074/jbc.M009664200. [DOI] [PubMed] [Google Scholar]
- 87.Pedrazzi G, Perrera C, Blaser H, Kuster P, Marra G, Davies SL, Ryu GH, Freire R, Hickson ID, Jiricny J, Stagljar I. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001;29(21):4378–4386. doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang XW, Tseng A, Ellis NA, Spillare EA, Linke SP, Robles AI, Seker H, Yang Q, Hu P, Beresten S, Bemmels NA, Garfield S, Harris CC. Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem. 2001;276(35):32948–32955. doi: 10.1074/jbc.M103298200. [DOI] [PubMed] [Google Scholar]
- 89.Garkavtsev IV, Kley N, Grigorian IA, Gudkov AV. The Bloom syndrome protein interacts and cooperates with p53 in regulation of transcription and cell growth control. Oncogene. 2001;20(57):8276–8280. doi: 10.1038/sj.onc.1205120. [DOI] [PubMed] [Google Scholar]
- 90.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277(24):22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 91.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11(25):3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 92.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277(43):41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 93.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 94.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type-I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275(35):26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 96.Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem. 2001;276(12):8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bennett RJ, Wang JC. Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc Natl Acad Sci USA. 2001;98(20):11108–11113. doi: 10.1073/pnas.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3(5):611–620. doi: 10.1016/S1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 99.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30(6):779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278(43):42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 101.Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60(5):1162–1167. [PubMed] [Google Scholar]
- 102.Hu P, Beresten SF, van Brabant AJ, Ye TZ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum Mol Genet. 2001;10(12):1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 103.Wu L, Hickson ID. The Bloom’s syndrome helicase stimulates the activity of human topoisomerase IIIalpha. Nucleic Acids Res. 2002;30(22):4823–4829. doi: 10.1093/nar/gkf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J Biol Chem. 2010;285(28):21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 106.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 107.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456(7220):357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 108.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471(7340):642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26(4):424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 110.LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, Takeda J, Jasin M. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci USA. 2011;108(29):11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103(11):4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281(20):13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 113.Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent Holliday junction processing. J Biol Chem. 2008;283(23):15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murzin AG. Ob(oligonucleotide oligosaccharide binding)-Fold—common structural and functional solution for nonhomologous sequences. EMBO J. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22(20):2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24(11):2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25(11):4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem. 2007;282(39):28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang F, Yang Y, Singh TR, Busygina V, Guo R, Wan K, Wang W, Sung P, Meetei AR, Lei M. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure. 2010;18(9):1159–1170. doi: 10.1016/j.str.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoadley KA, Xu D, Xue Y, Satyshur KA, Wang W, Keck JL. Structure and cellular roles of the RMI core complex from the Bloom syndrome dissolvasome. Structure. 2010;18(9):1149–1158. doi: 10.1016/j.str.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem. 2005;280(30):28085–28094. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- 124.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem. 2003;278(26):23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 125.Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′–3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008;36(19):6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467(7311):112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae . Nature. 2010;467(7311):108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gaymes TJ, North PS, Brady N, Hickson ID, Mufti GJ, Rassool FV. Increased error-prone non homologous DNA end-joining–a proposed mechanism of chromosomal instability in Bloom’s syndrome. Oncogene. 2002;21(16):2525–2533. doi: 10.1038/sj.onc.1205331. [DOI] [PubMed] [Google Scholar]
- 131.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 132.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 2000;19(23):2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 133.Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91(1–4):217–223. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- 134.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153(2):367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 136.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34(8):2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21(23):3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284(39):26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kikuchi K, Abdel-Aziz HI, Taniguchi Y, Yamazoe M, Takeda S, Hirota K. Bloom DNA helicase facilitates homologous recombination between diverged homologous sequences. J Biol Chem. 2009;284(39):26360–26367. doi: 10.1074/jbc.M109.029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli . Proc Natl Acad Sci USA. 1997;94(8):3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hand R, German J. A retarded rate of DNA chain growth in Bloom’s syndrome. Proc Natl Acad Sci USA. 1975;72(2):758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ockey CH, Saffhill R. Delayed DNA maturation, a possible cause of the elevated sister-chromatid exchange in Bloom’s syndrome. Carcinogenesis. 1986;7(1):53–57. doi: 10.1093/carcin/7.1.53. [DOI] [PubMed] [Google Scholar]
- 143.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom’s syndrome. Cancer Res. 1990;50(11):3141–3145. [PubMed] [Google Scholar]
- 144.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22(5):1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6(7):936–944. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 146.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 147.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443(1):13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 148.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA. 2000;97(12):6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281(32):22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 150.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45(47):13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 151.Tuduri S, Tourriere H, Pasero P. Defining replication origin efficiency using DNA fiber assays. Chromosome Res. 2010;18(1):91–102. doi: 10.1007/s10577-009-9098-y. [DOI] [PubMed] [Google Scholar]
- 152.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14(7):677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 153.Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol Cancer Res. 2007;5(7):713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 154.Yang J, O’Donnell L, Durocher D, Brown GW. RMI1 promotes DNA replication fork progression and recovery from replication fork stress. Mol Cell Biol. 2012;32(15):3054–3064. doi: 10.1128/MCB.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23(10):3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s syndrome and Fanconi anemia. Mol Cell. 2009;36(6):943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 157.Moldovan GL, D’Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24(16):1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123(7):1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 160.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18(4):500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 164.Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23(15):3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37(9):958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Komori K, Fujikane R, Shinagawa H, Ishino Y. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet Syst. 2002;77(4):227–241. doi: 10.1266/ggs.77.227. [DOI] [PubMed] [Google Scholar]
- 167.Scheller J, Schurer A, Rudolph C, Hettwer S, Kramer W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics. 2000;155(3):1069–1081. doi: 10.1093/genetics/155.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32(1):118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, Lin T, Schuster B, Decaillet C, Stasiak A, Stasiak AZ, Stone S, Hoatlin ME, Schindler D, Woodcock CL, Joenje H, Sen R, de Winter JP, Li L, Seidman MM, Whitby MC, Myung K, Constantinou A, Wang W. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37(6):865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, Sung P, Meetei AR. MHF1–MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37(6):879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, el Laghmani H, Joenje H, McDonald N, de Winter JP, Wang W, West SC. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25(3):331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 172.Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37(13):4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15(4):607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 174.Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde JM, Enomoto T, Takeda S, Thompson LH, Takata M. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24(2):418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37(9):953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 176.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet. 2008;17(11):1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, Wahengbam K, Steltenpool J, Andreassen PR, Williams DA, Joenje H, de Winter JP, Meetei AR. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114(1):174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA. 2008;105(42):16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32(3):313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 180.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29(4):795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29(4):806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21(9):2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- 183.Hoadley KA, Xue Y, Ling C, Takata M, Wang W, Keck JL. Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc Natl Acad Sci USA. 2012;109(12):4437–4442. doi: 10.1073/pnas.1117279109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci USA. 2004;101(8):2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Suhasini AN, Brosh RM., Jr Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012;28(1):7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28(12):4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283(52):36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40(4):1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 190.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.O’Driscoll M, Jeggo PA. The role of double-strand break repair—insights from human genetics. Nat Rev Genet. 2006;7(1):45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 192.Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y. Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol. 2005;25(20):8925–8937. doi: 10.1128/MCB.25.20.8925-8937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19(52):5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 194.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D’Andrea AD. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109(4):459–472. doi: 10.1016/S0092-8674(02)00747-X. [DOI] [PubMed] [Google Scholar]
- 195.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 2004;23(5):1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, Tashiro S, Elledge SJ, Takata M. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15(11):1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Collins NB, Wilson JB, Bush T, Thomashevski A, Roberts KJ, Jones NJ, Kupfer GM. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009;113(10):2181–2190. doi: 10.1182/blood-2008-05-154294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27(8):3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y, Leung JW, Jiang Y, Lowery MG, Do H, Vasquez KM, Chen J, Wang W, Li L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell. 2013;49(5):997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37(3):438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128(1):101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 203.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26(14):3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol. 2011;22(8):906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 205.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117(2):123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rouzeau S, Cordelieres FP, Buhagiar-Labarchede G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Gueret M. Bloom’s syndrome and PICH helicases cooperate with topoisomerase IIalpha in centromere disjunction before anaphase. PLoS ONE. 2012;7(4):e33905. doi: 10.1371/journal.pone.0033905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J Cell Sci. 2007;120(Pt 22):3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Russell B, Bhattacharyya S, Keirsey J, Sandy A, Grierson P, Perchiniak E, Kavecansky J, Acharya S, Groden J. Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIalpha. Cancer Res. 2011;71(2):561–571. doi: 10.1158/0008-5472.CAN-10-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 210.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11(6):761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 211.Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470(7332):120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 212.Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43(1):122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 213.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D’Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120(11):3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13(3):243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 215.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193(1):97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39(1):25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 217.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 218.Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11(21):3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nguyen GH, Dexheimer TS, Rosenthal AS, Chu WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M, Savitsky P, Abu M, McHugh PJ, Bohr VA, Harris CC, Jadhav A, Gileadi O, Maloney DJ, Simeonov A, Hickson ID. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol. 2013;20(1):55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Delano WL. The PyMol molecular graphics system. San Carolos: DeLano Scientific; 2002. [Google Scholar]
- 221.Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci USA. 1998;95(3):1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Seki M, Nakagawa T, Seki T, Kato G, Tada S, Takahashi Y, Yoshimura A, Kobayashi T, Aoki A, Otsuki M, Habermann FA, Tanabe H, Ishii Y, Enomoto T. Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol. 2006;26(16):6299–6307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19(13):3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]