Abstract
Originally identified as a mediator of DNA damage response (DDR), checkpoint kinase 1 (Chk1) has a broader role in checkpoint activation in DDR and normal cell cycle regulation. Chk1 activation involves phosphorylation at conserved sites. However, recent work has identified a splice variant of Chk1, which may regulate Chk1 in both DDR and normal cell cycle via molecular interaction. Upon activation, Chk1 phosphorylates a variety of substrate proteins, resulting in the activation of DNA damage checkpoints, cell cycle arrest, DNA repair, and/or cell death. Chk1 and its related signaling may be an effective therapeutic target in diseases such as cancer.
Keywords: Chk1, Cell cycle, DNA damage, Checkpoint
Introduction
Checkpoint kinase 1 (Chk1) was initially identified in fission yeast as a serine/threonine protein kinase that is essential for DNA damage-induced cell cycle arrest [1]. Chk1 homologs were subsequently identified in other species such as Drosophila, Xenopus, mouse, and human [2–4]. Functional studies in these model systems showed that Chk1 phosphorylates the key regulators of cyclin-dependent kinase 1 (CDK1) during DNA damage, resulting in CDK1 inactivation and blockade of G2/M transition. More recent work has established important roles of Chk1 not only in DNA damage response (DDR) but also in unperturbed cell cycle. In the normal cell cycle, Chk1 mediates the checkpoints in S and M phases as well as G2/M transition. In this review, we discuss Chk1 regulation, its role in DDR and unperturbed cell cycle, and the possibility of targeting Chk1 for cancer therapy.
Cell cycle and DNA damage response
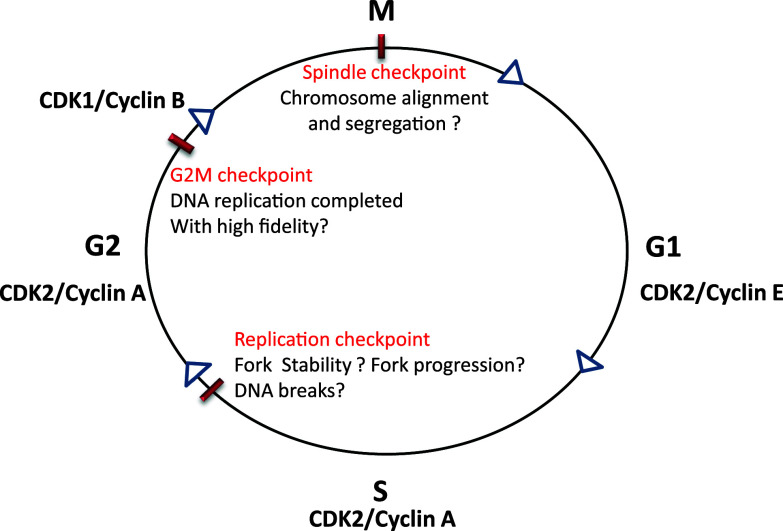
The cell cycle consists of a series of highly ordered, sequential phases that lead to cell division [5]. A notable feature of cell cycle progression is that cells do not enter the next phase until the previous phase is completed. This feature, known as cell cycle checkpoints, provides an important surveillance mechanism for faithful replication and division of the cells [5] (Fig. 1). Progression from one phase of cell cycle to the next is governed by Cdks, their cyclin partners, protein kinases, and phosphatases [6]. Specifically, the cell cycle is driven by temporal activation of Cdks, which depends on the association of cyclin partners, phosphorylation by specific protein kinases, and dephosphorylation by Cdc25 family phosphatases. For example, CDK1, the critical CDK for G2/M phase transition, is activated by cyclin B binding, phosphorylation at serine-161, and dephosphorylation of the inhibitory tyrosine-14 and -15 sites by Cdc25.
Fig. 1.
Cell cycle checkpoints. The cell cycle consists of G1, S, G2, and M phases, which are driven by various cyclin/CDK complexes. Progression from one phase to the next phase in the cell cycle is monitored by different checkpoints. S phase is regulated by replication checkpoint that monitors the initiation of replication, replication fork stability, fork progression, and DNA lesions. G2/M checkpoint monitors the completion of DNA replication with high fidelity. Spindle checkpoint makes sure that chromosomes are aligned and segregated for even distribution into two daughter cells
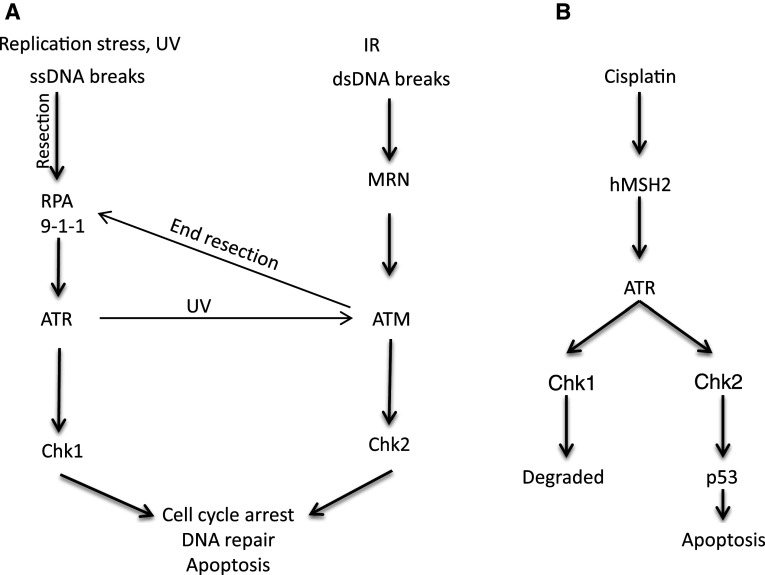
DNA damage response is a network of signal transduction pathways activated in a cell that leads to checkpoint activation, regulate cell cycle transitions, DNA repair and apoptosis in response to DNA damage [7, 8]. DNA damage is sensed by several molecular complexes or pathways, the most notable of which include ATM and ATR that activate DNA damage checkpoint (Fig. 2). In general, double-strand DNA breaks are sensed by MRE11-Rad 50-Nbs1 (MRN) complex, which localizes to the DNA damage site and recruits ATM [9, 10]. ATM usually exists as homo-dimers that are inactive; however, upon localization to DNA damage sites, ATM is autophosphorylated to become monomers leading to its activation [11]. Activated ATM phosphorylates downstream targets including Chk2, H2AX, and MDC1 etc. [12]. Different from ATM, ATR is mainly activated by single-stand DNA generated as a result of replication stress, UV induced DNA damage, nuclease activity consequent to double-strand breaks, enzymatic and helicase remodelling following ICL [13]. The single-strand DNA is recognized and bound by replication protein A (RPA), resulting in the recruitment of ATR through its interacting partner protein called ATRIP [7, 14, 15]. Independently, RAD9-RAD1-HUS1 (9-1-1) complex binds to the ssDNA and dsDNA junction through Rad17 [16]. The 9-1-1 complex in turn helps to recruit topoisomerase binding protein 1 (TOPBP1), an activator of ATR, resulting in ATR activation [17, 18]. In addition to this canonical pathway, recent work has revealed a novel pathway of ATR activation involving mismatch repair proteins [19] (Fig. 2). Upon activation, ATR can phosphorylate downstream targets including Chk1. ATR-mediated Chk1 phosphorylation also requires another intermediate protein called claspin, which binds and stabilizes activated Chk1. As a result, Chk1 phosphorylation was abolished in the absence of claspin in Xenopus egg extracts [20]. Although ATM and ATR are generally known to respond to double- and single-strand DNA damage respectively, their signaling pathways are not mutually exclusive. For example, double-strand breaks activate ATM, which along with MRE11 nuclease activity, generates single-strand DNA leading to consequent ATR activation [7, 21]. ATR activated in response to UV-induced DNA damage activate ATM by phosphorylating ATM at its autophosphorylation site S1981 [22]. In addition, the downstream signaling molecules are not exclusively responsive to ATM or ATR. For example, in cisplatin-induced DNA cross-linking and replication stress, both Chk1 and Chk2 are activated in an ATR-dependent manner. While Chk1 is degraded after initial activation, Chk2 phosphorylates p53 leading to its stabilization and activation to induce apoptosis [23].
Fig. 2.
Chk1 and Chk2 activation in response to DNA damage. a Double-strand DNA (dsDNA) breaks are sensed by the MRN complex, which recruits and activates ATM. MRN-mediated nuclease activity generates SS DNA, which can activate ATR. ATM is also activated in UV-induced DNA damage in ATR-dependent manner. Activated ATM phosphorylates several substrates including Chk2. RPA senses and binds single-strand DNA (ssDNA) lesions, which recruit ATR through ATRIP. 9-1-1 (Rad9-Rad1-Hus1) complex binds to ssDNA and dsDNA junction, which recruits TOPBP1 to activate ATR, which phosphorylates Chk1. b ATR is also activated by DNA damage induced by cisplatin. Cisplatin-induced DNA lesion recruits MSH2 (mismatch repair protein 2), which in turn helps in the recruitment and activation of ATR. Activated ATR phosphorylates both Chk1 and Chk2, however, Chk1 is degraded and Chk2 remains active, which phosphorylates and activates p53 leading to apoptosis
Intertwined relationship between the cell cycle and DDR
DNA damage response and the cell cycle are two intertwined cellular processes. On the one hand, as discussed above, DDR leads to cell cycle arrest by activating checkpoint kinases. On the other hand, an emerging idea is that even the unperturbed cell cycle has a constitutive surveillance mechanism that is related to DDR [24, 25]. This is particularly relevant to DNA replication in S-phase, when single-strand DNA (ssDNA) and DNA breaks may be induced in several ways. During replication, template DNA is unwound at the replication fork by DNA helicases resulting in ssDNA. ssDNA is highly susceptible to damage such as those induced by free radicals. Breaks in ssDNA at the replication fork can be converted to a double-strand break following replication. As a result, both single-strand and double-strand DNA breaks-associated DDR may be activated to slow down the progression of the cell cycle to repair the damage to ensure a faithful DNA replication and genome integrity. In this process, ATR and Chk1 play a critical role in sensing the initial ssDNA breaks to activate DDR. Consistently, deficiency in either ATR or Chk1 leads to embryonic lethality in mice and embryonic stem cells of these models show cell cycle abnormalities, accumulate double-stand breaks and fragile site breaks [26–29]. The intertwined relationship between DDR and cell cycle provides an explanation as to why DDR proteins, such as ATR and Chk1, play such critical roles in the unperturbed cell cycle.
Chk1 in cell cycle regulation and tissue physiology
Checkpoint kinase 1 was originally identified as a gene that can rescue a CDK1/cdc2 mutant in fission yeast [1]. Constitutive overexpression of Chk1 in fission yeast leads to mitotic delay, whereas loss of Chk1 has no effect on cell cycle progression. However, in the absence of Chk1, the yeast cells become more sensitive to UV-induced DNA damage and fail to arrest in G2-phase following DNA damage [1]. In Drosophila, Grapes (Chk1 homolog in Drosophila) regulates syncytial cell division fidelity, mitotic entry, and cyclin A degradation and as a result, it is indispensable for embryogenesis [3, 30]. In addition, Grapes may delay the accumulation of cyclin B1 in the nucleus to prevent nuclear CDK1 activation and premature entry into mitosis in Drosophila embryos [31, 32], supporting a role of Chk1 in cell cycle in early stages of embryogenesis in Drosophila. In Xenopus eggs, addition of exogenous, active Chk1 delays mitosis whereas immunodepletion of Chk1 results in an early entry into mitosis [2]. Unlike yeast, mammalian cells do not show noticeable phenotypes following Chk1 overexpression; however Chk1 knockdown and inhibition leads to reduced proliferation and cell death [33]. In mice, Chk1 knockout leads to embryonic death in utero at E6.5 stage [28, 29]. Chk1 knockout blastocysts as well as embryonic stem cells also fail to proliferate and fail to activate the G2/M checkpoint in response to DNA damage and die shortly in culture [28, 29]. Embryonic death of Chk1-null mice is independent of p53 since p53-deficiency fail to rescue the phenotype [28]. Although viable, Chk1-deficient chicken lymphoma DT40 cells are deficient in G2 checkpoint in response to DNA damage and are defective in both DNA replication and cell cycle [34]. Collectively, these studies indicate that Chk1 is master regulator of cell cycle, cell survival and embryogenesis.
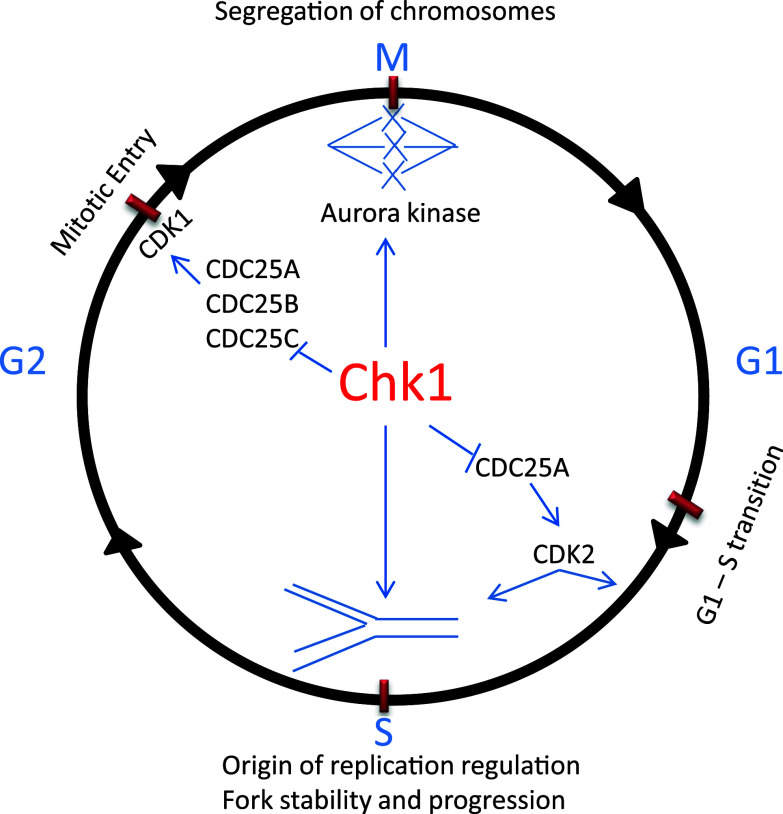
In unperturbed cell cycle, Chk1 regulates DNA replication in S phase, G2/M transition or mitotic entry, and spindle checkpoint in M phase (Fig. 3). In S phase, Chk1 arrests cell cycle for DNA replication mainly by inducing Cdc25A degradation, resulting in the inhibition of CDK2. In U2OS cells, Chk1 phosphorylates Cdc25A at multiple sites and inhibition of Chk1 results in a marked accumulation of Cdc25A, suggesting that phosphorylation by Chk1 targets Cdc25A for degradation [35–37]. Consistently, conditional deletion of Chk1 from mammary cells in mice results in Cdc25A accumulation, which is accompanied by increases in S phase cells, accumulation of DNA damage, premature mitotic entry, and cell death [38]. Knockdown and inhibition of Chk1 lead to increased initiation of DNA synthesis, increased single-strand DNA, aberrant fork structures, accumulation of double-strand breaks, DDR, and increased phosphorylation of ATR targets [33, 39]. These studies suggest that Chk1 may suppress late initiation of replication to prevent DNA damage for normal progression of S phase. However, the ATR/Chk1 pathway can also sense the strand stability and progression and initiate late replication to repair DNA breaks [40]. Collectively, all these observations suggest that Chk1 regulates replication checkpoint and is required for normal S phase progression and cell survival.
Fig. 3.
Cell cycle checkpoints regulated by Chk1. Chk1 helps maintain genomic integrity by regulating DNA replication, G2/M, and spindle checkpoints. Chk1 monitors DNA replication, slows down the replication to favor fork stability, prevent stalling of forks and DNA breaks. Chk1 prevents premature entry into mitosis before DNA replication is completed with high fidelity by inhibiting Cdc25 family phosphatases. Chk1 also regulates segregation of chromosomes through activation of aurora kinase to prevent genomic instability
Mitotic entry is promoted by the activation of the cyclin B-CDK1 complex, which also involves the regulation by Chk1 [41]. Briefly, in late G2-phase, Chk1 accumulates at centrosome to phosphorylate Cdc25B (the phosphatase and activator of CDK1), resulting in the inhibition of Cdc25B and consequent suppression of CDK1. This regulation is critical to the prevention of premature mitotic entry in unperturbed cell cycle [41].
Finally, Chk1 has a critical role in spindle checkpoint in M-phase of the cell cycle. In support of this conclusion, Chk1-deficient chicken lymphoma DT 40 cells are viable, but they have increased levels of chromosome mis-segregation and genomic instability and fail to arrest in mitosis when treated with Taxol, a microtubule-stabilizing agent [42]. Similarly, Chk1 haploinsufficient primary mammary epithelial cells also exhibit misaligned chromosomes, chromosome missegregation, and enhanced binucleation [43]. Checkpoint kinase 1 knockdown U2OS cells undergo aberrant mitosis and fail to activate spindle checkpoint in response to Nocodazole treatment [37]. These studies suggest defective spindle checkpoint in the absence of Chk1. Mechanistically, Chk1 regulates spindle checkpoint through the activation and localization of Aurora B kinase, a regulator of spindle checkpoint that recruits BubR1 and MAD1 to the kinetochores [42, 43] to prevent APC/C activation and delay anaphase onset [44]. Chk1 deficiency leads to mislocalization of Aurora B, BubR1 and MAD1 proteins, resulting in spindle checkpoint failure [42, 43] (Fig. 3).
Chk1 in DNA damage response
Originally identified in yeast, Chk1 is now recognized as an important mediator and signal transducer in DDR in other eukaryotes including mammals. In response to DNA damage, Chk1 is rapidly phosphorylated at serine-317 and serine-345 by ATR and becomes highly activated, resulting in the activation of DNA damage checkpoints [28, 45, 46]. Activated Chk1 phosphorylates several downstream targets to bring about cell cycle arrest, activate DNA repair pathways, and induce apoptosis when DNA damage is severe [8, 47–49].
Chk1-induced cell cycle arrest in DDR
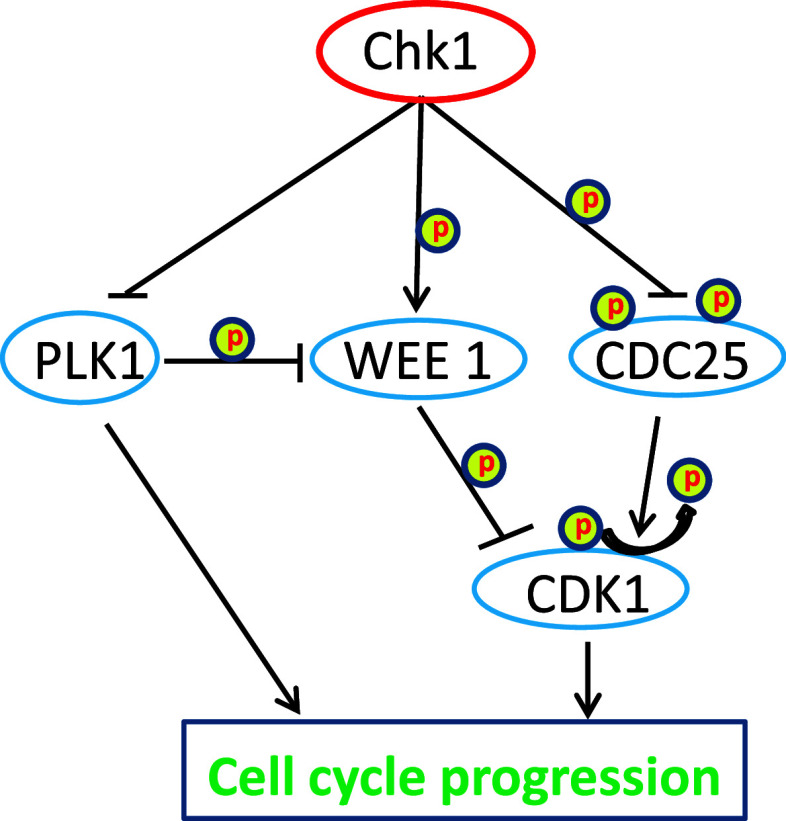
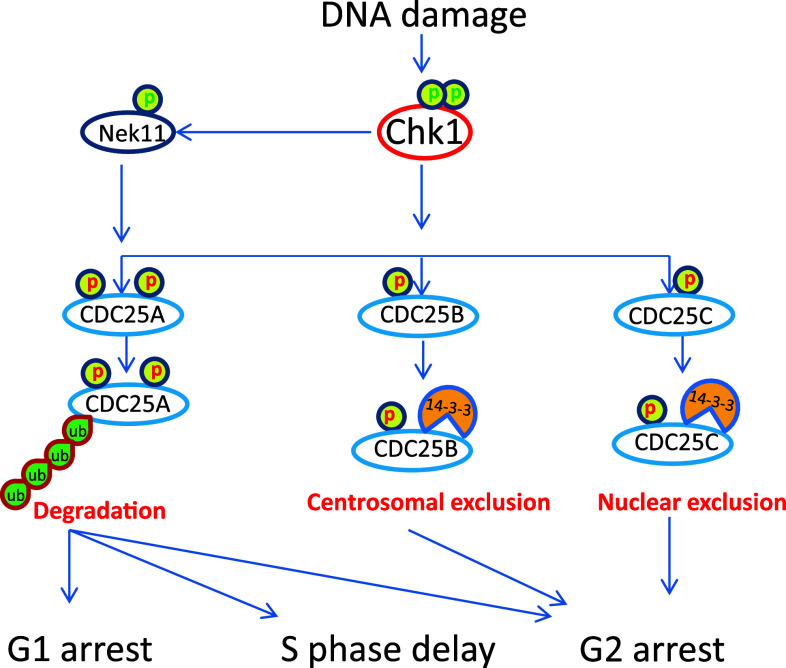
Checkpoint kinase 1 induces cell cycle arrest in DDR mainly by phosphorylating Cdc25 family phosphatases, WEE1 kinase and controlling PLK1 (Fig. 4). The Cdc25 family is dual-specificity phosphatases that can dephosphorylate Cdks in their ATP-binding loop, resulting in the activation of Cdks and cell cycle progression. In mammalian cells, there are three (A, B and C) isoforms of Cdc25, all of which can be phosphorylated by Chk1 in DDR. Interestingly, upon phosphorylation Cdc25A, B and C may employ distinct mechanisms to arrest the cell cycle [50, 51] (Fig. 5). For Cdc25A, phosphorylation by Chk1 targets it for proteasomal degradation, which leads to the inhibition of Cdk1 and Cdk2 resulting in cell cycle arrest at G1/S transition, S phase, and G2/M transition [52, 53]. Interestingly, besides direct phosphorylation, Chk1 can also activate Nek11, which in turn phosphorylates Cdc25A to induce cell cycle arrest during DDR [54]. For Cdc25C, a main regulation seems to be its cellular localization. Specifically, phosphorylation of Cdc25C by Chk1 promotes its binding to 14-3-3 proteins leading to the sequestration of Cdc25C from Cdk1, Cdk1 inactivation and G2 arrest [4]. Cdc25B regulation by Chk1 seems to occur at centrosomes, where Chk1 phosphorylates Cdc25B leading to its sequestration from centrosome and inhibition of centrosomal Cdk1 [41].
Fig. 4.
Chk1 regulation of cell cycle progression. Chk1 regulates progression of cell cycle by inhibiting Cdc25 family phosphatases and polo-like kinase 1(PLK1), and activating WEE 1 kinase. Cdc25 is a phosphatase and activator of CDK1, whereas WEE1 inhibits CDK1 by phosphorylation. PLK1 can activate CDK1 by inhibiting WEE1. PLK1 can also directly promote cell cycle progression
Fig. 5.
Chk1-mediated cell cycle arrest in response to DNA damage. Chk1 phosphorylates Cdc25 family resulting in their inhibition. Chk1 can also activate Nek11, which further phosphorylates Cdc25A on multiple sites to target it for degradation. Cdc25A is crucial for G1-S transition, S phase progression, and mitotic entry; as a result, Cdc25A inhibition leads to G1 arrest, slows down replication, and G2 arrest. Chk1 phosphorylates Cdc25B to promote its binding to 14-3-3 protein and sequestration from centrosome. Chk1 also phosphorylates Cdc25C to induce its binding to 14-3-3 protein and nuclear exclusion. Cdc25B and Cdc25C inhibition mainly causes G2 arrest
In addition to Cdc25 phosphotases, Chk1 phosphorylates WEE1, the protein kinase that is responsible for the inhibitory phosphorylation of Cdk1 (Fig. 4). As a result, in response to DNA damage, the phosphorylation and activation of WEE1 by Chk1 leads to the inhibition of Cdk1 and cell cycle arrest at G2 phase [55, 56]. Chk1 is also a negative regulator of PLK1 (polo like kinase 1), a mitotic kinase involved in centrosome maturation, spindle formation, and cytokinesis [57]. PLK1 phosphorylates WEE1 and targets it for degradation, leading to Cdk1 activation and mitotic entry (Fig. 4). In addition, recent work suggests that, in response to DNA damage, Chk1 dissociates from the chromatin leading to decreased phosphorylation and acetylation of histone H3, which may contribute to DNA damage-associated transcriptional repression [58]. Interestingly, the repressed genes include cyclin B and Cdk1, although the role of the transcriptional repression in cell cycle arrest remains unclear.
Chk1-mediated DNA repair during DDR
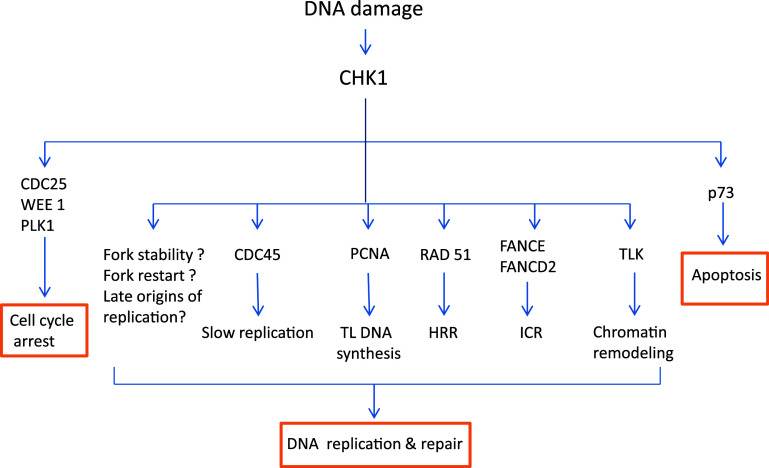
As discussed above, Chk1 induces checkpoint activation resulting in cell cycle arrest. In addition, in S-phase, Chk1 may slow down DNA replication by blocking the initiation factor CDC45 in a Cdk2-independent manner [59]. Together, these mechanisms provide time for DNA repair to maintain genomic integrity in response to DNA damage. Notably, emerging evidence from the past few years has suggested a more direct role of Chk1 in DNA repair (Fig. 6).
Fig. 6.
Chk1-mediated DNA repair. Activated Chk1 phosphorylates several substrates to mediate DNA repair and apoptosis. Chk1 slows down DNA replication directly by affecting CDC45 loading on replication complex to allow time for DNA repair. Chk1 promotes monoubiquitination of PCNA, which recruits translesion polymerases to help translesion DNA synthesis. Chk1 phosphorylates TLK-tousled like kinase to promote chromatin assembly. Chk1 phosphorylates Rad51 to target it to double-strand breaks to promote HRR-homologous recombination and repair. Chk1 phosphorylates FANCE and monoubiquitinates FANCD2, which help resolve ICR-interstrand crosslink DNA lesion. Chk1 also regulates replication fork stability, fork restart, and late origins of replication, the mechanistic details of which are currently unclear. In addition to DNA repair, Chk1 may also induce cell cycle arrest and apoptosis by regulating different substrates
In this regard, Chk1 induces post-translational modification and activation of several important DNA repair factors. (1) Proliferating cell nuclear antigen (PCNA). One of the mechanisms of Chk1-mediated DNA repair is by inducing monoubiquitination of PCNA [60]. Upon ubiquitination, PCNA can activate translesion synthesis, a process that allows the replication machinery to pass or tolerate DNA lesions for replication. Specifically, at DNA damage sites, monoubiquitinated PCNA recruits specific translesion synthesis DNA polymerases to replace classical polymerases [61, 62], which can bypass or repair the damaged sites (lesion) for DNA replication [63]. (2) FANCE. In the repair of DNA cross-links, Chk1 phosphorylates the FANCE subunit of the Fanconi anemia (FA) core complex on two conserved sites serine-346 and serine-S374, resulting in its co-localization with FANCD2 at nuclear foci or DNA damage sites [44, 64]. Together with BRCA1 and 2, FANCE and ubiquitinated FANCD2 help resolve interstrand DNA crosslinks [44]. (3) Rad51. Checkpoint kinase 1 participates in homologous recombination repair by regulating Rad51. Knockdown or inhibition of Chk1 leads to accumulation of double-strand breaks and increased cell death in response to double-strand breaks induced by hydroxyurea and camptothecin. Mechanistically, Chk1 phosphorylates Rad51 at serine-309 and recruits it to DNA repair foci to promote homologous recombination repair and cell survival [65]. (4) TLK. In response to double-strand DNA breaks, Chk1 can also phosphorylate the serine-695 site of TLK (tousled-like kinase), an evolutionarily conserved serine/threonine kinase that is involved in chromatin assembly, DNA replication, and repair. Interestingly, the phosphorylation of TLK by Chk1 depends on ATM [66]. Finally, it is noteworthy that Chk1 has also been suggested to stabilize replication forks and/or restart stalled replication forks; however, the underlying mechanisms remain unclear.
Chk1-induced cell death in DDR
There are few reports implicating Chk1 in apoptosis or cell death. Chk1 phosphorylates p73 at serine-47, resulting in a drastic increase in p73 expression and transcription activity in response to DNA damage. Although p73 is a p53-related protein, it induces apoptosis independently of p53 [67, 68]. In contrast, there are instances where Chk1 inhibits apoptosis through ATM/ATR-caspase2 and RPA-caspase3 pathway [69, 70]. Therefore, it remains unclear whether (and to what extent) Chk1 contributes to apoptosis and its regulation.
Regulation of Chk1
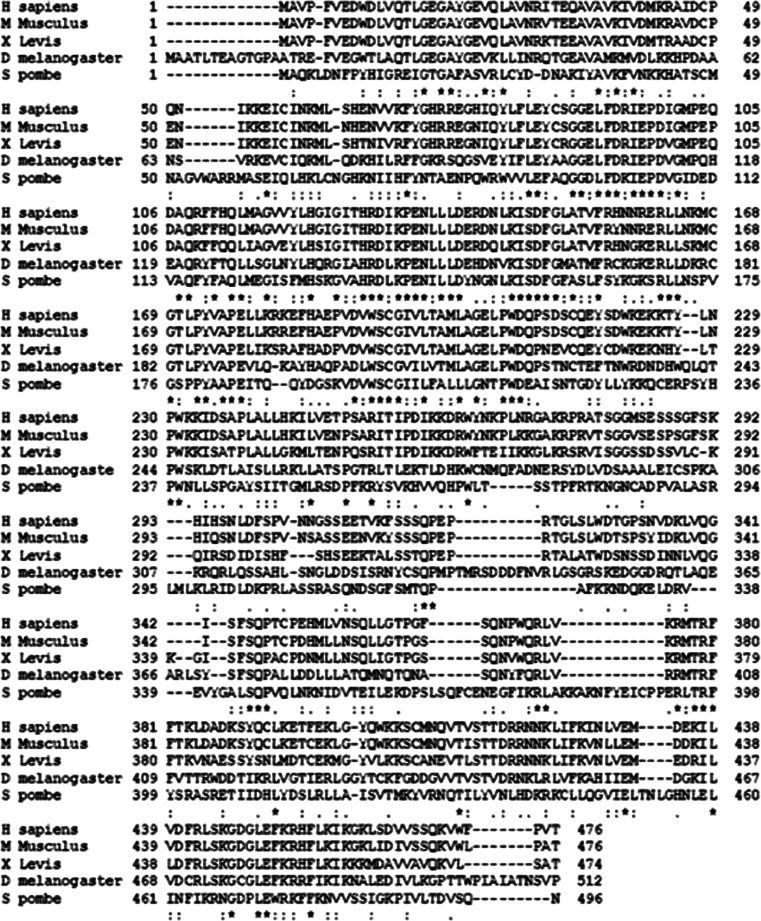
At the sequence level, Chk1 is highly conserved from yeast to humans [52] (Fig. 7). Structurally, Chk1 has a conserved N-terminal kinase domain, a linker region, a regulatory SQ/TQ domain, and a C-terminal domain with unclear functions [52, 71]. It is noteworthy that Chk1 and Chk2 are not homologous in sequence, although they both have checkpoint function and share some phosphorylation substrate proteins.
Fig. 7.
CHk1 sequence alignment between human, mice, fruit fly, Xenopus and yeast. Chk1 sequences of human, mice, Drosophila, Xenopus, and yeast were analyzed using T-coffee, a Web server for multiple sequence alignment tool. The N-terminal kinase domain and regulatory SQ/TQ domain of Chk1 are highly conserved, while the C-terminal domain is less conserved. Asterisk indicates highly conserved sequence, Colon indicates conserved substitution, Dot indicate semi conserved substitutions
It is well recognized that Chk1 is activated upon phosphorylation at two conserved sites, serine-317 and serine-345 [28, 46, 72]. Although phosphorylation at these two sites is indispensable for the function of Chk1, it remains elusive as to how exactly the phosphorylation activates Chk1 [45]. The study of crystallographic structure revealed that in the absence of phosphorylation, the Chk1 kinase domain assumes an open, active conformation [73]. Indeed, the kinase domain has significantly higher kinase activity when compared to full-length Chk1. These observations indicate that Chk1 kinase domain has a constitutively active conformation that does not rely on phosphorylation for activation. One model of Chk1 activation proposes that normally the C-terminal domain interacts with the kinase domain to mask the active site and the phosphorylation at serine-317 and serine-345 dissociates these two domains leading to Chk1 activation. This model is supported by several lines of evidence. First, C-terminal truncation mutants show higher catalytic activity than intact Chk1 [73]. Second, in binding assays, ectopically expressed C-terminal domain of rat Chk1 binds to the N-terminal kinase domain [74]. In addition, in Xenopus egg extracts, ectopically expressed C-terminal domain can interact with N-terminal Chk1 domain and inhibit its activity but not full-length Chk1 [71]. The inhibitory effect of C-terminal Chk1 domain on N-terminal kinase domain can be overcome by treatment with aphidicolin and phosphomimetic mutations of ATR [71], suggesting that the C-terminal domain indeed inhibits kinase activity. Together, these observations suggest that the C-terminal domain is inhibitory to the kinase domain in Chk1 and, upon phosphorylation, this inhibition is disabled. While this “intramolecular inhibition” model is supported by several lines of evidence, it has been challenged recently. For example, several C-terminal truncation mutants lead to loss of function in Chk1 [45, 75], suggesting that removal of the C-terminal domain is not sufficient for Chk1 activation and Chk1 regulation may be more complex than “intramolecular inhibition”. In addition to serine-317 and serine-345, Chk1 has also been reported to have autophosphorylation at serine-296, which plays a role in G2 checkpoint [76]. Interestingly, the autophosphorylation is consequent to phosphorylation at serine-317 and serine-345 through intramolecular mechanisms. It is noteworthy that serine-296 phosphorylation does not affect the kinase activity of Chk1 [77].
Interestingly, a recent study by Walker and colleagues indicates that Chk1 may be subjected to a new mechanism of regulation, called “de-repression”. In this model, Chk1 is antagonized by an inhibitory or repressing factor(s), the dissociation of which leads to Chk1 activation. In support of this possibility, Chk1 immunoprecipitated from untreated cells demonstrates significantly higher kinase activity after being washed with a more stringent buffer. Notably, Chk1 immunoprecipitated from DNA damaged (by aphidicolin) cells also shows significantly higher kinase activity when washed with the stringent buffer, although the increase is somewhat smaller than that of untreated cells. In addition, stringent buffer washes also increase the kinase activity of Chk1 (S345A) mutant from transfected cells [78], suggesting that phosphorylation at serine-345 is not obligatory for Chk1 kinase activity per se, although it plays a role in Chk1 activation in cells. Furthermore, Chk1 can be activated in a claspin-dependent manner in response to genotoxic stress independently of ATM/ATR and associated phosphorylation [79]. Together, these studies support the possibility of additional regulatory mechanisms for Chk1, in addition to phosphorylation. The “de-repression” model is intriguing and tantalizing, but the identity of the “repressing” factor(s) of Chk1 is not known.
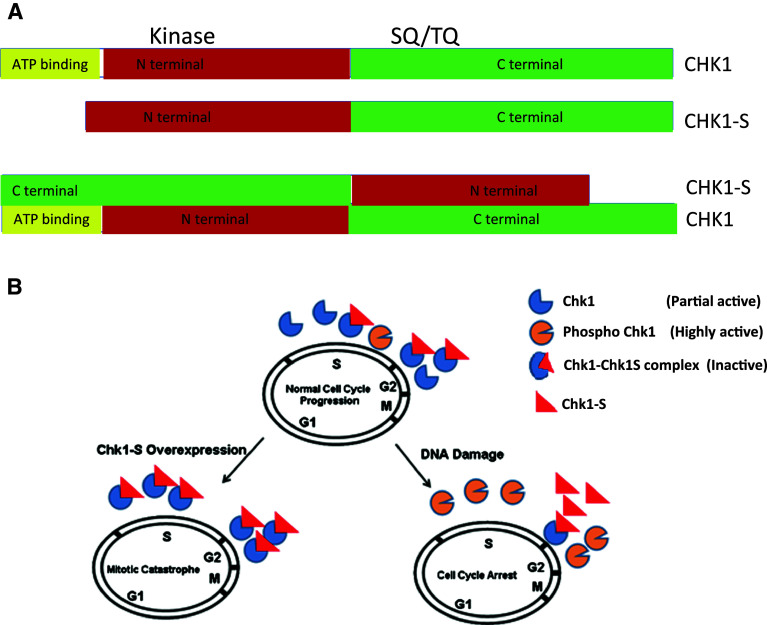
Our latest work has identified a splice variant of Chk1, which may act as an endogenous repressor of Chk1 [80]. This variant is spliced in a way that the exon 3 of Chk1 is deleted, leading to the production of an N-terminally truncated protein, which we call Chk1-short or Chk1-S. Due to the truncation, Chk1-S lacks part of the kinase domain including the ATP-binding (Fig. 8a) and, as expected, does not possess protein kinase activity. Importantly, a series of in vitro and in vivo tests showed that Chk1-S directly interacts with Chk1 at its N-terminal domain and inhibits Chk1 kinase activity. In addition, we confirmed the finding by Walker and colleagues that stringent buffer washes can increase the activity of Chk1 immunoprecipitated from cells. Moreover, we showed that the addition of exogenous Chk1-S can reverse the effect of stringent buffer washing. Together, these results suggest that Chk1-S may be one of the key endogenous repressing factors of Chk1. In an unperturbed cell cycle, Chk1-S expression increases significantly in late G2 and early M phases coinciding with the decrease in Chk1 activity for mitotic entry. Overexpression of Chk1-S leads to premature mitotic entry followed by cell death in the form of mitotic catastrophe. These observations support an important role of Chk1-S in the regulation of normal cell cycles by binding and antagonizing Chk1. In response to DNA damage, the interaction between Chk1 and Chk1-S is lost in an ATR-dependent manner (Fig. 8b). Remarkably, mutation of Chk1 at serine-317 and serine-345 prevents the dissociation of Chk1 from Chk1-S during DNA damage, indicating that the dissociation of Chk1 from Chk1-S in DDR depends on Chk1 phosphorylation. Based on these observations, we propose that Chk1-S is a key regulator of Chk1 in both normal cell cycles and DDR. Mechanistically, Chk1-S binds to Chk1 to act as a repressor to block Chk1 activity. In normal cell cycles, Chk1-S is drastically increased at late G2 phase to repress Chk1 to promote mitotic entry. In DDR, ATR-mediated Chk1 phosphorylation at serine-317 and serine-345 leads to the dissociation of Chk1-S from Chk1, resulting in Chk1 activation and cell cycle arrest (Fig. 8b).
Fig. 8.
Physical and functional interactions between Chk1 and Chk1-S. a Schematic representation of Chk1 and Chk1S domains and their interaction. Checkpoint kinase 1 has ATP binding domain, N-terminal kinase domain, SQ/TQ domain, and C-terminal domain. Chk1S lacks ATP binding domain. C-terminal domain of Chk1 interacts with N-terminal domain of CHK1-S. b Regulation of Chk1 by Chk1-S. Chk1-S interacts with Chk1 to inhibit its kinase activity. In unperturbed cell cycles, Chk1-S is expressed in late S to G2 phase to inhibit Chk1 and promote mitotic entry. In DNA damage, Chk1 is phosphorylated to prevent Chk1-S binding and inhibition, resulting in Chk1 activation. When Chk1-S is overexpressed, it antagonizes Chk1, leading to premature mitotic entry and consequent cell death via mitotic catastrophe. Figure 8b is adopted from Pabla et al. PNAS 109, 197-202, 2012
In addition to the above-described regulatory mechanisms, Chk1 is also known to be subjected by proteasomal degradation. Interestingly, proteasomal degradation of Chk1 may depend on its phosphorylation status. In camptothecin-induced DNA damage, Chk1 phosphorylation at serine-345 targets the protein for proteosomal degradation [81]. It was later shown that the phosphorylation exposes a dragon-like region at the C-terminus, which is recognized by the F-box protein called Fbx6, leading to Chk1 ubiquitination and proteasomal degradation and termination of the checkpoint [82]. Consistently, there is an inverse correlation between Chk1 and Fbx6 expression in cultured cancer cells and breast cancer tissues. In addition, overexpression of Fbx 6 leads to degradation of chk1 and increased cellular sensitivity to camptothecin [82].
Chk1 substrates
Despite the multiple roles of Chk1 in normal cell cycles and DDR, only a few phosphorylation substrates of Chk1 have been identified thus far. As a result, identification of functional substrates of Chk1 is currently a subject of intense investigation. The consensus sequence of Chk1-specific substrates has been suggested by analyzing the peptide derived from Xenopus. Cdc25C containing serine-287, a typical Chk1-phosphorylation site. Indeed, the consensus sequence has been useful for predicting Chk1 substrates including PDS1 (an anaphase inhibitor) and Wee1 [83]. Nonetheless, the consensus sequence is insufficient to explain other Chk1 substrates. Some of the Chk1 substrates may be context-dependent and do not rely on the consensus sequence for phosphorylation. In this regard, a substrate may be phosphorylated by Chk1 through interaction with adaptor proteins. A recent study has conducted a global phospho-proteomic screen using analogue-sensitive Chk1 and suggested 171 proteins as its potential phosphorylation substrates. Moreover, KAP1, a protein phosphorylated in response to DNA damage, has been validated to be a Chk1 substrate in both in vitro and in vivo studies and KAP1 phosphorylation can be used as a read out for Chk1 activation [47]. However, most of the substrates implied in this study remain to be validated. It is expected that new and important functional substrates of Chk1 will be discovered in the next few years to provide new insights into the multiple functions of Chk1 in the cell cycle and DDR.
Chk1 in cancer
Checkpoint kinase 1 mutations or loss are very rare in cancer. A logical explanation is that Chk1 plays an essential role in cell cycle regulation, cell proliferation and survival [28, 29, 80], and as a result, cells with defective Chk1 are eliminated during tumorigenesis. Nonetheless, partial ablation of Chk1 (heterozygous) favors tumor formation in WNT1 oncogenic mice [28]. In addition, loss-of-function Chk1 mutations have been reported in stomach, endometrial, and colorectal cancers [84–87]. Mutations have been mapped to the microsatellite instability region in the coding region of Chk1 with a stretch of nine adenine nucleotides [86]. Frame-shift mutations lead to the formation of a truncated protein in colorectal and endometrial cancers lacking part of the kinase domain and the C-terminal domain [86]. Intriguingly, in these cancers is that the second allele is normal, resulting in the expression of functional Chk1 albeit at a lower level than that of normal tissues [88]. Monoallelic mutations leading to partial loss of expression are also observed in ATR in endometrial and stomach cancers [85]. These studies support the haploinsufficient tumor model in which monoallelic mutations in several genes of the same pathway favor tumorigenesis [88]. In mice, conditional deletion of Chk1 in mammary epithelial cells fails to produce tumors; instead it leads to cell death [38]. However, the mice haploinsufficient for both Chk1 and p53 develop tumors in mammary glands [89]. Similarly, complete loss of Chk1 in skin suppresses chemically induced carcinogenesis, whereas partial (haploinsufficient) deletion of Chk1 promotes benign malignant tumor progression [90]. Triple-negative (Estrogen-/progesterone-/HER2-) breast cancers express a very high level of Chk1 and have poor clinical outcomes, suggesting that Chk1 favors cell proliferation [91]. Together, these studies indicate that Chk1 is essential for cell survival and its haploinsufficiency may promote cancer, especially in the presence of mutations of other relevant genes. Of note, Chk1-S, the splice variant and endogenous inhibitor of Chk1, is expressed at significantly higher levels in cancer tissues than normal tissues and notably, Chk1-S expression correlates with the degree of malignancy in ovarian and testicular tumors [80]. Interestingly, Chk1 expression is also higher in cancer. It is suggested that relatively high levels of Chk1 and Chk1-S may favor cell proliferation in cancer.
Targeting Chk1 for cancer therapy
DNA damaging agents are the most commonly used drugs for cancer therapy. By damaging DNA, these chemotherapy drugs induce cell cycle arrest to prevent cell proliferation and trigger cell death in cancers. Many of these agents induce G1 arrest in a p53-dependent manner and G2 arrest in a Chk1-dependent manner. G1 arrest is defective in many cancers since more than 50 % of cancers are defective in p53; under these situations, G2 arrest becomes a main pathway for cancer cell survival. Hence, Chk1-dependent G2 checkpoint is a major target for chemotherapy [92–95]. In this regard, Chk1 inhibition or knockdown increases the sensitivity of p53-deficient cancer cells to DNA damage agents [33, 96]. However, it has also been reported in certain cell lines that Chk1 is essential for G2 checkpoint irrespective of the p53 status and hence Chk1 inhibition may not preferantially kill p53-deficient cells [97]. It has also been shown that Chk1 inhibition increases the sensitivity of cancer cells to anti-mitotic agents [98, 99]. In addition, Chk1 is a client of HSP90 and inhibition of HSP90 leads to the loss of Chk1 resulting in the sensitization of cancer cells to Gemcitabine, an S phase chemotherapeutic agent [100]. Chk1 and Wee1 kinase inhibitors in combination have more than additive effect on different cancer cell line proliferation and human xenograft models [101]. Therefore, Chk1 inhibitors may target multiple cell cycle phases to override the checkpoint responses during cancer therapy, leading to increased therapeutic efficacy. Currently, Chk1 inhibitors are being developed and tested alone or in combination with DNA damaging agents in clinical trials for cancer therapy [8, 36]. However, so far, none of these have passed phase III clinical trials. Obviously, the specificity and potency of Chk1 inhibitors are critical to their clinical use. In addition, their side effects have to be carefully evaluated because Chk1 has an important role in cell cycle regulation and its inhibition may adversely affect cell viability and function in normal tissues.
Concluding remarks
Checkpoint kinase 1 plays an essential role in cell cycle regulation and DNA damage response. In unperturbed cell cycle, Chk1 regulates G1/S transition, S phase, mitotic entry, and mitosis. In DDR, Chk1 is an important signal transducer and the trigger of G2 checkpoint activation. The role of Chk1 in unperturbed cell cycle and tissue physiology is only beginning to be understood. Chk1 regulates S phase progression by controlling DNA replication, fork stability and late origins of replication the mechanistic details of which are yet to be identified. The role of Chk1 in the development and pathogenesis in different tissues are yet to be investigated. In addition, although Chk1 regulates several checkpoints, only a few Chk1 substrate proteins have been identified. The discovery of Chk1-S as an important regulator of Chk1 in both unperturbed cell cycle and DDR has opened new areas of investigation. Finally, Chk1 may be an effective therapeutic target in diseases. In this regard, Chk1 inhibitors, when used together with other therapeutic agents, may significantly enhance the chemotherapy efficacy in cancer treatment.
Abbreviations
- Chk1
Checkpoint kinase 1
- Chk2
Checkpoint kinase 2
- Chk1
S-checkpoint kinase 1-short
- DDR
DNA damage response
- ATM
Ataxia telangiectasia mutated
- ATR
ATM and Rad3 related
- Cdc25
Cell division cycle 25
- CDK
Cyclin-dependent kinases
References
- 1.Walworth N, Davey S, Beach D. Fission yeast Chk1-protein kinase links the rad checkpoint pathway to Cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fogarty P, Kalpin RF, Sullivan W. The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle 13. Development. 1994;120:2131–2142. doi: 10.1242/dev.120.8.2131. [DOI] [PubMed] [Google Scholar]
- 4.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14–3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 5.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 6.Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100:71–78. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- 7.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 11.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 12.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 13.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 15.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 16.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9–1-1 for DNA damage: the Rad9-Hus1-Rad1 (9–1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9–1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 19.Pabla N, Ma Z, McIlhatton MA, Fishel R, Dong Z. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J Biol Chem. 2011;286:10411–10418. doi: 10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 21.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 22.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–6583. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 24.Enders GH. Expanded roles for Chk1 in genome maintenance. J Biol Chem. 2008;283:17749–17752. doi: 10.1074/jbc.R800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 27.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 29.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 30.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 31.Purdy A, Uyetake L, Cordeiro MG, Su TT. Regulation of mitosis in response to damaged or incompletely replicated DNA require different levels of Grapes (Drosophila Chk1) J Cell Sci. 2005;118:3305–3315. doi: 10.1242/jcs.02454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Royou A, McCusker D, Kellogg DR, Sullivan W. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J Cell Biol. 2008;183:63–75. doi: 10.1083/jcb.200801153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zachos G. Chk1-defcient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO Rep. 2003;22(3):713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 36.Carrassa L, Damia G. Unleashing Chk1 in cancer therapy. Cell Cycle. 2011;10:2121–2128. doi: 10.4161/cc.10.13.16398. [DOI] [PubMed] [Google Scholar]
- 37.Carrassa L, Sanchez Y, Erba E, Damia G. U2OS cells lacking Chk1 undergo aberrant mitosis and fail to activate the spindle checkpoint. J Cell Mol Med. 2009;13:1565–1576. doi: 10.1111/j.1582-4934.2008.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 39.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maya-Mendoza A. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO Rep. 2007;26(11):2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 42.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, Gillespie DA. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci USA. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guervilly JH, Mace-Aime G, Rosselli F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum Mol Genet. 2008;17:679–689. doi: 10.1093/hmg/ddm340. [DOI] [PubMed] [Google Scholar]
- 45.Tapia-Alveal C, Calonge TM, O’Connell MJ. Regulation of chk1. Cell Div. 2009;4:8. doi: 10.1186/1747-1028-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Poon RY. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front Biosci. 2008;13:5016–5029. doi: 10.2741/3060. [DOI] [PubMed] [Google Scholar]
- 50.Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003;4:671–677. doi: 10.1038/sj.embor.embor887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–3396. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 53.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 54.Melixetian M, Klein DK, Sorensen CS, Helin K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11:1247–1253. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- 55.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14–3-3 proteins. Mol Biol Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci USA. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 60.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 62.Freudenthal BD, Gakhar L, Ramaswamy S, Washington MT. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat Struct Mol Biol. 2010;17:479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 66.Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J, Hansen K. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J. 2003;22:1676–1687. doi: 10.1093/emboj/cdg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez S, Prives C, Cordon-Cardo C. p73 alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–8171. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, Kanki JP, Green DR, D’Andrea AA, Look AT. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katsuragi Y, Sagata N. Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell. 2004;15:1680–1689. doi: 10.1091/mbc.E03-12-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Capasso H, Palermo C, Wan S, Rao H, John UP, O’Connell MJ, Walworth NC. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–4564. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- 73.Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, Kan CC, O’Connor PM. The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell. 2000;100:681–692. doi: 10.1016/s0092-8674(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 74.Shann YJ, Hsu MT. Cloning and characterization of liver-specific isoform of Chk1 gene from rat. J Biol Chem. 2001;276:48863–48870. doi: 10.1074/jbc.M108253200. [DOI] [PubMed] [Google Scholar]
- 75.Kosoy A, O’Connell MJ. Regulation of Chk1 by its C-terminal domain. Mol Biol Cell. 2008;19:4546–4553. doi: 10.1091/mbc.E08-04-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okita N, Minato S, Ohmi E, Tanuma S, Higami Y. DNA damage-induced CHK1 autophosphorylation at Ser296 is regulated by an intramolecular mechanism. FEBS Lett. 2012;586:3974–3979. doi: 10.1016/j.febslet.2012.09.048. [DOI] [PubMed] [Google Scholar]
- 77.Kasahara K, Goto H, Enomoto M, Tomono Y, Kiyono T, Inagaki M. 14–3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–2812. doi: 10.1038/emboj.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene. 2009;28:2314–2323. doi: 10.1038/onc.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez-Bravo V, Guaita-Esteruelas S, Florensa R, Bachs O, Agell N. Chk1- and claspin-dependent but ATR/ATM- and Rad17-independent DNA replication checkpoint response in HeLa cells. Cancer Res. 2006;66:8672–8679. doi: 10.1158/0008-5472.CAN-05-4443. [DOI] [PubMed] [Google Scholar]
- 80.Pabla N, Bhatt K, Dong Z. Checkpoint kinase 1 (Chk1)-short is a splice variant and endogenous inhibitor of Chk1 that regulates cell cycle and DNA damage checkpoints. Proc Natl Acad Sci. 2011;109:197–202. doi: 10.1073/pnas.1104767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 82.Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, Manning G, Abraham RT, Hunter T. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–453. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hutchins JR, Hughes M, Clarke PR. Substrate specificity determinants of the checkpoint protein kinase Chk1. FEBS Lett. 2000;466:91–95. doi: 10.1016/s0014-5793(99)01763-9. [DOI] [PubMed] [Google Scholar]
- 84.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Gene Chromosome Canc. 1999;26:176–180. [PubMed] [Google Scholar]
- 85.Menoyo A. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61(21):7727–7730. [PubMed] [Google Scholar]
- 86.Codegoni AM, Bertoni F, Colella G, Caspani G, Grassi L, D’Incalci M, Broggini M. Microsatellite instability and frameshift mutations in genes involved in cell cycle progression or apoptosis in ovarian cancer. Oncol Res. 1999;11:297–301. [PubMed] [Google Scholar]
- 87.Vassileva V. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002;62(14):4095–4099. [PubMed] [Google Scholar]
- 88.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Canc Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 89.Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, Lahusen T, Xu X, Lee MH, Liu Q, Elledge SJ, Ried T, Deng CX. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene. 2010;29:4007–4017. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tho LM, Libertini S, Rampling R, Sansom O, Gillespie DA. Chk1 is essential for chemical carcinogen-induced mouse skin tumorigenesis. Oncogene. 2012;31:1366–1375. doi: 10.1038/onc.2011.326. [DOI] [PubMed] [Google Scholar]
- 91.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, Verstuyf A. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res. 2007;67:6574–6581. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 92.Zhou BB, Anderson HJ, Roberge M. Targeting DNA checkpoint kinases in cancer therapy. Cancer Biol Ther. 2003;2:S16–S22. [PubMed] [Google Scholar]
- 93.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 94.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15:360–367. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 95.Tenzer A, Pruschy M. Potentiation of DNA-damage-induced cytotoxicity by G2 checkpoint abrogators. Curr Med Chem Anticancer Agents. 2003;3:35–46. doi: 10.2174/1568011033353533. [DOI] [PubMed] [Google Scholar]
- 96.Koniaras K. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20(51):7453–7463. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 97.Zenvirt S, Kravchenko-Balasha N, Levitzki A. Status of p53 in human cancer cells does not predict efficacy of CHK1 kinase inhibitors combined with chemotherapeutic agents. Oncogene. 2010;29:6149–6159. doi: 10.1038/onc.2010.343. [DOI] [PubMed] [Google Scholar]
- 98.Blagosklonny MV. Sequential activation and inactivation of G2 checkpoints for selective killing of p53-deficient cells by microtubule-active drugs. Oncogene. 2002;21:6249–6254. doi: 10.1038/sj.onc.1205793. [DOI] [PubMed] [Google Scholar]
- 99.Xiao Z, Xue J, Semizarov D, Sowin TJ, Rosenberg SH, Zhang H. Novel indication for cancer therapy: Chk1 inhibition sensitizes tumor cells to antimitotics. Int J Cancer. 2005;115:528–538. doi: 10.1002/ijc.20770. [DOI] [PubMed] [Google Scholar]
- 100.Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278:52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- 101.Guertin AD, Martin MM, Roberts B, Hurd M, Qu X, Miselis NR, Liu Y, Li J, Feldman I, Benita Y, Bloecher A, Toniatti C, Shumway SD. Unique functions of CHK1 and WEE1 underlie synergistic anti-tumor activity upon pharmacologic inhibition. Cancer cell Int. 2012;12:45. doi: 10.1186/1475-2867-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]