Abstract
Drosophila larval locomotion is a splendid model system in developmental and physiological neuroscience, by virtue of the genetic accessibility of the underlying neuronal components in the circuits1-6. Application of optogenetics7,8 in the larval neural circuit allows us to manipulate neuronal activity in spatially and temporally patterned ways9-13. Typically, specimens are broadly illuminated with a mercury lamp or LED, so specificity of the target neurons is controlled by binary gene expression systems such as the Gal4-UAS system14,15. In this work, to improve the spatial resolution to "sub-genetic resolution", we locally illuminated a subset of neurons in the ventral nerve cord using lasers implemented in a conventional confocal microscope. While monitoring the motion of the body wall of the semi-intact larvae, we interactively activated or inhibited neural activity with channelrhodopsin16,17 or halorhodopsin18-20, respectively. By spatially and temporally restricted illumination of the neural tissue, we can manipulate the activity of specific neurons in the circuit at a specific phase of behavior. This method is useful for studying the relationship between the activities of a local neural assembly in the ventral nerve cord and the spatiotemporal pattern of motor output.
Keywords: Neuroscience, Issue 77, Molecular Biology, Neurobiology, Developmental Biology, Bioengineering, Cellular Biology, Motor Neurons, Neurosciences, Drosophila, Optogenetics, Channelrhodopsin-2, Halorhodopsin, laser, confocal microscopy, animal model
Introduction
Forward peristaltic locomotion in Drosophila larvae occurs by the propagation of muscular contraction from posterior to anterior segments. This motion is realized by sequential activation of motor neurons within the ventral nerve cord along the longitudinal body axis. To examine the circuitry behind this patterned propagating activity, local perturbation of neural activity could be an informative approach. Although pharmacological assay can control specific substance, such as neurotransmitters, in neural tissue, the effect of pharmacological drugs can be similar among the almost all segments because the larval ventral nerve cord is repetitive structure composed of similar neuromeres along the longitudinal axis. Alternatively, one could express optogenetic or temperature-sensitive probe molecules in a small number of segments. However, such specific gal4 lines are very difficult to generate. Here we present a method for manipulating neurons in a few segments using laser illumination. Taking advantage of the spatially confined illumination in confocal microscopy, one can perturb the activity of neurons in a local area. Combined with the expression pattern of the probe by subsets of neuron-specific gal4 lines, this laser illumination method greatly improves spatial resolution for perturbation. And because this method requires only a conventional confocal microscope, the purchase of additional laboratory equipment is typically not necessary.
Using this technique, we examined the role of motor neuronal activity during the propagation of motor output13. By blocking the activities of motor neurons within a few segments during peristaltic motion, we were able to test whether the activity of motor neurons themselves is required for the propagation of the motor output signal. Local and transient blockade of motor neurons arrested the propagation of peristaltic motion. After the blockade was removed, propagation appeared at the segment where the wave had been arrested. This phenomenon suggests that without motor neuronal activation, the propagative wave cannot progress along the ventral nerve cord any further, and thus the activation of motor neurons is necessary for peristaltic motion. We have previously reported detailed data and discussions about this phenomenon in Inada et al. (2011) 13. Here, we describe the method to perturb neural activity interactively while monitoring the motion of the dissected larvae. Using various gal4 lines and series of spatiotemporal patterns for activity manipulation, one can investigate circuitry logic through perturbation response properties of the motor circuit.
Protocol
1. Larvae Preparation
Maintain fly lines of OK6-Gal4, UAS-ChR2 or OK6-Gal4, UAS-NpHR2 in plastic vials containing standard fly food.
Spread yeast paste containing all-trans retinal (ATR) at appropriate concentrations (1 mM for ChR2, 10 mM for NpHR2 ("NpHR" for short) on an apple juice agar plate.
Pick up 2nd or 3rd instar larvae from the vials and put them on the ATR-containing plate.
Rear them at 25 °C in the dark for an appropriate period of time (1 day for ChR2, 2 days for NpHR.)
2. Microscope Setup
Attach a CCD camera (XCD-V60, Sony) to a conventional confocal microscope (in our case, FV1000, Olympus) with a C-mount attachment (magnification 0.35x).
If there is a shutter along the light path from the objective lens to the CCD camera, which closes during laser scanning, remove it carefully. As this step depends on the microscope setup, contact the microscope manufacturer for technical support, if necessary.
3. Dissection
Rinse the ATR-fed larvae with water to remove residual food from the body.
Put the larva on a sylgard coated dish, dorsal side up (the dorsal side has two tracheal tubes running longitudinally, one either side of the dorsal midline). The thickness of the sylgard is about 5mm.
Insert an insect pin (Austerlitz Insect pins, Φ0.10mm, stainless) into the tail between the tracheal tubes with forceps (#5 Inox, FST by Dumont, Switzerland). The pins, about 10mm long, should be bent or cut to be short enough (~2mm long) to avoid hitting and damaging the surface of the objective lens. Then put the second insect pin into the head of the larva near the mouth hook, black claw-like structure at the anterior end.
Add Ca2+-free normal saline (NaCl 140 mM, KCl 2 mM, MgCl2 6 mM, HEPES-NaOH 5 mM, Sucrose 36 mM (pH7.1)) to keep the larva moist.
Make a small incision near the tail with micro scissors (MB-50-7, Napox, Japan).
From the incision, make a longitudinal cut along the dorsal midline toward the head. Be careful not to damage the ventral nerve cord (VNC) and axons.
Make a small incision at the head laterally.
Place 4 pins at each corner of the dissected bodywall. The body wall should be stretched enough to visualize a segmental pattern of the body wall, but not too much to hinder peristaltic motion.
Remove the internal organs except for the brain and the VNC and rinse the sample with Ca2+-free normal saline.
Adjust the orientation of the ventral nerve cord to be bilaterally symmetric by changing the position and orientation of insect pins on body wall. To fix the position of the ventral nerve cord, insert a pin through tissue between the brain and the mouth hook down to the sylgard.
Replace the buffer with 2 mM Ca2+ Ringer solution (NaCl 130 mM, KCl 5mM, MgCl2 2mM, CaCl2 2 mM, HEPES-NaOH 5 mM, Sucrose 36 mM (pH7.3)).
4. Imaging with Laser Illumination
Attach a 4x dry objective lens (UPlanSApo 4x, NA 0.16, Olympus, Japan) in the confocal microscope and set the larva preparation on the stage.
Obtain a transmission image of the dissected preparation with a red laser (633nm) to locate the ventral nerve cord by confocal microscope. For this scanning, be sure not to use a 488nm or 559nm laser, which induces unwanted stimulation of ChR2 or NpHR, respectively.
Define the region of interest (ROI) on the transmission image. Zoomed mode could be useful for detailed determination of the ROI.
Change the filter set of the microscope to illuminate with 488nm or 559nm light.
Illuminate the preparation with a halogen lamp to visualize the body wall. The preparation can be monitored with a CCD camera attached to the confocal microscope.
Record the motion of the body wall and switch the laser beam on and off while monitoring the motion.
Representative Results
Local and transient activation of motor neurons with ChR2.
We expressed ChR2 in motor neurons. The axons of motor neurons emerging from each VNC segment project to a corresponding segment of the body wall. When we stimulated one or two segments of the VNC with a blue laser, muscles in the corresponding segments exhibited contractions (Figure 1).
Local and transient inhibition of motor neurons with NpHR.
We expressed NpHR in motor neurons. When we stimulated a few (1~2) segments of the VNC with a yellow laser during the peristaltic muscle contractions, the propagating wave was arrested at the corresponding segments of the body wall (Figure 2). Then the wave restarted from the arrested segments when we switched off the laser illumination13.
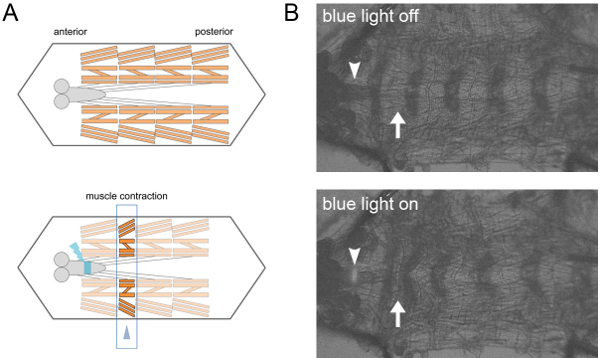 Figure 1. Local activation of motor neurons with ChR2. A. Scheme of the local activation of filleted preparation. By blue-light illumination of a few segments of the ventral nerve cord expressing ChR2 in motor neurons, muscles in the corresponding segments are contracted (arrow head). B. Transmission images of a dissected larva. Local illumination of the ventral nerve cord (arrow heads) induces segmental contraction of body wall muscles (arrows).
Figure 1. Local activation of motor neurons with ChR2. A. Scheme of the local activation of filleted preparation. By blue-light illumination of a few segments of the ventral nerve cord expressing ChR2 in motor neurons, muscles in the corresponding segments are contracted (arrow head). B. Transmission images of a dissected larva. Local illumination of the ventral nerve cord (arrow heads) induces segmental contraction of body wall muscles (arrows).
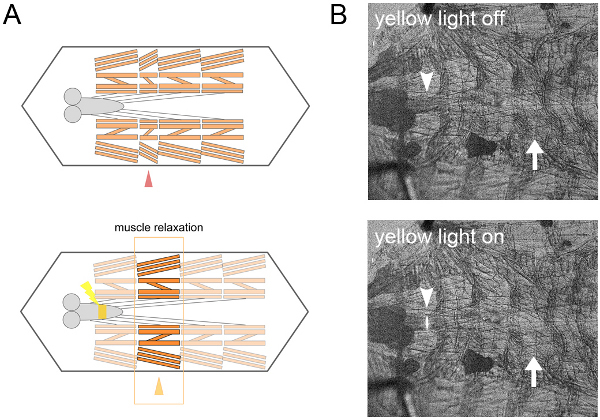 Figure 2. Local inhibition of motor neurons with NpHR. A. Scheme of the local inhibition of filleted preparation. By yellow-light illumination of a few segments of the ventral nerve cord expressing NpHR in motor neurons, muscles contraction during spontaneously-occurred peristaltic motion (arrow head in the top) are relaxed (arrow head in the bottom.) B. Transmission images of a dissected larva. Local illumination of the ventral nerve cord (arrow heads) relaxed the spontaneously-contracted body wall segments (arrows).
Figure 2. Local inhibition of motor neurons with NpHR. A. Scheme of the local inhibition of filleted preparation. By yellow-light illumination of a few segments of the ventral nerve cord expressing NpHR in motor neurons, muscles contraction during spontaneously-occurred peristaltic motion (arrow head in the top) are relaxed (arrow head in the bottom.) B. Transmission images of a dissected larva. Local illumination of the ventral nerve cord (arrow heads) relaxed the spontaneously-contracted body wall segments (arrows).
Discussion
Temporal and local perturbation of neural activity is an invaluable technique for analyzing the network dynamics of neural circuits. In this protocol, we present a method to manipulate neural activity by optogenetics using a laser. The laser's high directionality allows more localized optogenetic stimulation than wide field stimulation using mercury or Xenon lamp. Though laser illuminations have already been applied to optogenetics in previous studies, special setups such as glass fiber, micromanipulator, and laser source, are required in most previous studies. In this protocol, we use a conventional confocal microscopy for local illumination. Since the confocal microscope system is widely used, this method will open the opportunity to apply higher resolution optogenetics in many labs.
The protocol has two critical points: larval dissection and laser power. Firstly, if the brain, the ventral nerve cord or a motor nerve is damaged during dissection, the larvae will exhibit less or none spontaneous peristaltic motion. Precision is therefore essential, especially when making an incision on the dorsal side with spring scissors and removing internal organs with forceps. Details about the dissection have been reported previously21. Secondly, if sufficient stimulation is to be achieved, a laser power of about 0.1~1 mW/ mm2 for ChR2 or 1~10 mW/mm2 for NpHR is required. We evaluated the laser power as total light power below an objective lens divided by an estimated area of illumination. We measured the total light power using a power meter (mobiken, Sanwa M.I. Technos, Japan). We estimated the area of illumination as circular constant times the square of the wave length of the laser, which roughly gives illuminated area in diffraction limit. Effective illumination power on the sample can be adjusted not only by the power of the laser output, but also by changing the scanning speed and number of repetitions. We scanned laser with 20-100 μsec/pixel for 63 times. In addition, the expression level of optogenetics protein and the amount of ATR are also critical for stimulation efficiency. Accordingly, if larvae showed no optogenetic response, the following points should be checked and corrected: 1) laser power (by optimizing microscope system), 2) expression level of optogenetics protein (by checking genotype and rearing temperature), and 3) amount of ATR (by adjusting concentration of ATR and feeding period). NpHR requires higher concentration of ATR than ChR2 does by unknown reasons 13. ChR2 works well in larvae reared in food containing lesser ATR (e.g. 0.1 mM) than described here (1 mM). As feeding larvae to 1 mM ATR is sufficient for making ChR2 photo-reactive, we recommend this concentration for the first trial in ChR2 experiments. The concentration of ATR can subsequently be titrated down from 1 mM. As to exposure duration for ATR, we recommend the duration described here for robust optogenetic control. We often failed to observe optogenetic response when feeding larvae to ATR for shorter duration.
In this protocol, laser illumination and image acquisition are operated by a separate computer. So, clear detection of spatiotemporal pattern of laser illumination by CCD camera is critical for data analysis. If laser spot or line is dim on CCD image, you should optimize the power of halogen lamp and gain value of the CCD camera to visualize the laser illumination while keeping body wall visible.
Spatiotemporal illumination shown in this protocol provides information on larval motor circuits; however, the method has some limitations. As to the "spatial" aspect, location of the illumination is recorded by low magnification CCD image used for videotaping body wall movements. Accordingly, illumination area can be assigned at segment level, but not at cellular level. As to the "temporal" aspect, time lag between switching on the laser scan by confocal microscope and illumination by laser depends on the confocal microscope system and scanning condition. Consequently, some testing by trial and error is required to adjust illumination timing. Since the timing of illumination can be determined in CCD image movies using adequate software like ImageJ, the critical factor in temporal resolution is frame rate of CCD camera imaging. We typically set the frame rate for 7.5-15 frames per second.
Advances in optogenetic tools provide us a chance to modify this protocol. We used 3rd instar larvae possessing OK6-Gal4 and UAS-ChR2[H134R] or UAS-NpHR2. Other optogenetic tools instead of ChR2[H134R] or NpHR2 such as ChR2[T159C/E123T], NpHR3 or Arch can boost the efficiency of the activity control22. In addition, using other gal4 lines or analyzing in other developmental stages can provide more information about the motor circuits.
In this protocol, motor activity was monitored by transmission images of the bodywall contraction with a CCD camera. The activity of motor neurons with calcium-sensitive fluorescence molecules (e.g. GCaMP) can also be directly monitored while manipulating neural activity with ChR2 or NpHR. In this case, blue light illumination in a broad area and a highly sensitive CCD camera (e.g. EMCCD camera) are used instead of a halogen lamp and the normal CCD camera previously described herein. To improve spatial resolution of stimulation while monitoring bodywall movement, using an additional objective lens may be a more advanced method: illumination of the ventral nerve cord by a high magnification objective lens (e.g. 40x) from above the sample and monitoring bodywall by low magnification lens (e.g. 4x) below the sample. This dual lens system may allow us to stimulate nervous system in higher spatial resolution.
The protocol shown here can be applied to other neural circuits, provided optogenetic tools can be expressed in the nervous system and preparation in which sufficient light can be delivered into the neural tissue can be established.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas "Mesoscopic Neurocircuitry" (No. 22115002) and "Comprehensive Brain Science Network" (No. 221S0003) of The Ministry of Education, Culture, Sports, Science, and Technology, Japan to A.N. and Grant-in-Aid for Young Scientists (B) (No. 21700344) of Japan Society for the Promotion of Science (JSPS) to H.K.
References
- Cattaert D, Birman S. Blockade of the central generator of locomotor rhythm by noncompetitive NMDA receptor antagonists in Drosophila larvae. J. Neurobiol. 2001;48:58–73. doi: 10.1002/neu.1042. [DOI] [PubMed] [Google Scholar]
- Fox LE, Soll DR, Wu CF. Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J. Neurosci. 2006;26:1486–1498. doi: 10.1523/JNEUROSCI.4749-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LE, Song W, Looger LL, Jan LY, Jan YN. The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron. 2010;67:373–380. doi: 10.1016/j.neuron.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Marin A, Louis M. Active sensation during orientation behavior in the Drosophila larva: more sense than. Curr. Opin. Neurobiol. 2011. [DOI] [PubMed]
- Gershow M, et al. Controlling airborne cues to study small animal navigation. Nat. Methods. 2012. [DOI] [PMC free article] [PubMed]
- Suster ML, Bate M. Embryonic assembly of a central pattern generator without sensory input. Nature. 2002;416:174–178. doi: 10.1038/416174a. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, Kevrekidis IG. Optical imaging and control of genetically designated neurons in functioning circuits. Annu. Rev. Neurosci. 2005;28:533–563. doi: 10.1146/annurev.neuro.28.051804.101610. [DOI] [PubMed] [Google Scholar]
- Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Hwang RY, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv. Physiol. Educ. 2011;35:82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada K, Kohsaka H, Takasu E, Matsunaga T, Nose A. Optical dissection of neural circuits responsible for Drosophila larval locomotion with halorhodopsin. PLoS One. 2011;6:e29019. doi: 10.1371/journal.pone.0029019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 1073;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E, Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975;257:46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Mochizuki Y, Kamo N, Kobatake Y. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem. Biophys. Res. Commun. 1985;127:99–105. doi: 10.1016/s0006-291x(85)80131-5. [DOI] [PubMed] [Google Scholar]
- Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Brent JR, Werner KM, McCabe BD. Drosophila larval NMJ dissection. J. Vis. Exp. 2009. p. e1107. [DOI] [PMC free article] [PubMed]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]


