Abstract
Much of the cancer-causing effects of ultraviolet radiation from the sun have been linked to the formation of dimerized DNA bases. These dimeric DNA photoproducts include the cyclobutane pyrimidine dimers (CPDs) and the pyrimidine(6–4)pyrimidone photoproducts [(6–4)PPs). CPDs are highly mutagenic and are produced in substantial quantities by UVB radiation. These dimers can form between any two adjacent pyrimidines and can involve thymine, cytosine, or 5-methylcytosine. Very recently, a sixth DNA base, 5-hydroxymethylcytosine (5hmC) has been identified and characterized as a normal component of mammalian DNA. Here, we investigated the formation of CPDs at different DNA sequences containing 5hmC following irradiation with UVA, UVB, or UVC light sources. We show that the formation of CPDs at dipyrimidines containing 5hmC occurs at different DNA sequences but is not enhanced relative to cytosine or 5-methylcytosines at the same sequence positions. In fact, in some sequence contexts, CPDs containing 5hmC are formed at very low levels. Nonetheless, CPD formation at 5hmC pyrimidines is expected to be biologically relevant since three types of human skin-derived cells, fibroblasts, keratinocytes and melanocytes, all contain detectable levels of this modified base.
Introduction
The DNA damaging properties of ultraviolet (UV) radiation have been known for a long time. Of biological relevance for human exposure are the UVA (320–400 nm) and the UVB (280–320 nm) components of sunlight that reach the earth’s surface. While UVA is weakly mutagenic and produces both oxidative DNA damage products, mostly at guanines, and low levels of cis-syn cyclobutane pyrimidine dimers (CPDs) (1–5), UVB is considered a more powerful mutagen due to its ability to effectively produce CPDs and to a lesser extent (6–4)PPs (3, 6). The earth’s atmospheric layer removes most of the radiation with wavelengths less than 300 nm making UVB-induced photoproducts the type of DNA damage with the highest biological relevance. CPDs are much more mutagenic in mammalian cells than (6–4)PPs (7), which is at least in part due to rapid repair of (6–4) photoproducts (8). The mutagenicity of CPDs and their relevance to human skin cancer is best explained by their long persistence in skin, which allows time for deamination of cytosine or 5-methylcytosine to occur when they are part of a CPD (9). When the deaminated CPDs are bypassed by DNA polymerases, for example by the DNA damage-tolerant DNA polymerase eta, a mutagenic event occurs that is predominantly due to deamination rather than is caused a polymerase error (10–12). Indeed, the most common mutation observed after irradiation of DNA or cells with UVB is the transition mutation C to T at dipyrimidine sequences (7, 13, 14). Such mutations are also observed as the by far most common event in sunlight-associated skin cancers. Initially, these types of mutations have been described in the p53 gene in nonmelanoma skin cancers (15, 16). However, they are seen genome-wide in studies of melanoma genomes clearly implicating UVB irradiation in melanoma (17, 18).
5-methylcytosine (5mC) is a DNA base that chiefly occurs at 5’-CpG dinucleotide sequences in mammalian genomes. This base is produced in a postreplicative manner by DNA methyltransferases that methylate the cytosine ring at position 5. 5mC is particularly prone to formation of CPDs, both upon irradiation of cells with sunlight (19) or, to a lesser extent, with UVB (20). This phenomenon is most likely due to the longer wavelength absorbance of 5mC relative to C, which makes the trinucleotide sequences 5’-TmCG and 5’-CmCG some of the most remarkable targets for CPD formation and UVB mutagenesis (7, 14).
Although 5hmC has been long known to occur as a normal DNA base in certain bacteriophages (21), its presence in mammalian cells has only recently been confirmed (22, 23). 5hmC is produced by enzymatic oxidation of 5mC by one of the three members of the TET (ten–eleven translocation) family of proteins (23, 24). TET proteins are iron- and alpha-ketoglutarate-dependent dioxygenases that seem to be encoded in all vertebrate genomes. The functional role of 5hmC in mammalian DNA is still unclear. This base is produced from 5mC during epigenetic reprogramming of the paternal genome after fertilization (25). It is most abundant in brain tissue but lower levels of 5hmC than in brain are present in every cell type analyzed (26, 27). One potential function of 5hmC could be the reversal of repressive effects of 5mC on gene activity. Since 5hmC is a pyrimidine base, its potential involvement in pyrimidine dimer formation is of relevance. Early studies had indicated that pyrimidine dimers containing 5hmC can be detected in the DNA of UV-irradiated bacteriophage T4 (28, 29). However, since 5hmC in mammalian DNA has long been viewed as a DNA damage product itself, no further studies have been reported to our knowledge. Using synthetic oligonucleotides containing 5hmC, we have analyzed the propensity of this modified base towards CPD formation following irradiation with different UV light sources and in different sequence contexts.
Experimental Procedures
Oligonucleotides
All site-specifically modified oligonucleotides were synthesized at the W.M. Keck oligonucleotide synthesis facility at Yale University. Sixty-four-mer oligonucleotides (sequence 5’-CCTCACCATCTCAACCAATATTACGCGTATATCCGGTATTTTCGAATTGAGGGAGAAGTGGTGA) contained C, 5mC or 5hmC at the four underlined 5’-CG sequences. Forty-five- mer oligonucleotides (sequence 5’ – CATAGCATGTGAATAGGTACAATXGGTATGTGATAGAACTACTGA or 5’ – CATAGCATGTGAATAGGTACAACXGGTATGTGATAGAACTACTGA) contained C, 5mC or 5hmC (“X”) at the single underlined 5’-CG sequence. Opposite strand oligonucleotides containing the modified bases at the equivalent CpG positions were also synthesized and used for annealing to form double-stranded DNA.
UV irradiation
A Sellas Sunlight System (Medizinische Geräte GmbH; Gevelsberg, Germany) with an average fluence rate of 60 mW/cm2 was used for UVA irradiation. This UVA source exclusively emits long-wave UVA (UVA1: 340–400 nm). The UVB source consisted of three fluorescent light tubes (Philips TL 20 W/12R) and has a peak spectral emission at 312 nm. The UVB dose was determined with a UVX radiometer (Ultraviolet Products; Upland, CA). The UVC irradiation was carried out with five 254 nm UV light bulbs from a distance of 20 cm (Stratalinker UV Crosslinker 2400; Stratagene; La Jolla, CA).
Mapping of CPDs at modified cytosine-containing oligonucleotides
3’-end labeling of oligonucleotides was performed with terminal deoxynucleotidyl transferase (Invitrogen; Carlsbad, CA) and biotin-16-ddUTP (Roche; Indianapolis, IN) under the following conditions. Three hundred nanograms of each sense strand oligonucleotide was incubated at 37°C for 30 min with 15 units of deoxynucleotidyl transferase in 50 µl reaction mixture containing 2 µM biotin-16-ddUTP, 0.1 M potassium cacodylate (pH 7.2), 2 mM CoCl2, and 0.2 mM DTT. After this incubation, we added 1 µl of 0.5 M EDTA (pH 8.0) and 50 µl of chloroform:isoamyl alcohol (24:1). The mixture was centrifuged at 13,000×g for 3 min, and then the upper phase was recovered. The 3’-labeled sense strand oligonucleotide was annealed to the non-labeled anti-sense strand oligonucleotide. The 3’-biotin-labeled double-stranded oligonucleotide was irradiated on Parafilm placed directly on ice to avoid heat production. The UV doses were 10,000, 20,000 or 40,000 J/m2 for UVB, 1,000 and 2,500 J/m2 for UVC and 72 J/cm2 for UVA. After irradiation, the oligonucleotide was treated with T4 endonuclease V (Trevigen; Gaithersburg, MD) as follows: The enzyme digestion was performed in 1x REC™ Buffer 11 (Trevigen; 25 mM sodium phosphate pH 6.8, 1 mM EDTA, 100 mM NaCl, 1 mM DTT, 0.01% Triton X-100), 0.1 mg/ml BSA with 20 units of T4 endonuclease V in 20 µl reaction volume for 3 hours at 37°C. After T4 endonuclease V treatment, the sample was added to 94% formamide loading dye (94% formamide, 2 mM EDTA (pH 8.0), 0.05% xylene cyanole, 0.05% bromophenol blue), and then heated at 95°C for 2 min before separation on a 10% polyacrylamide gel containing 7 M urea. The gel was blotted onto a charged nylon-based membrane (Perkin Elmer; Waltham, MA) using an electrotransfer device (Trans blot SD; Bio-Rad; Hercules, CA). The signal was detected by using the Lightshift Chemiluminescent EMSA Kit (Thermo Scientific; Kalamazoo, MI) following the manufacturer’s instructions.
Detection of CPDs containing 5hmC by anti-CPD antibodies
The 45-mer oligonucleotide top-strand containing the TX sequence was irradiated with UVC at different doses (250, 500 and 1,000 J/m2). 250 ng of oligonucleotide was used for blotting. After irradiation, the DNA was spotted onto charged nylon membranes (Perkin Elmer). The membrane was placed on a filter paper presoaked in 0.4 N NaOH for 20 min at room temperature. Subsequently, the membrane was blocked with 0.15 % PBS-T (PBS plus 0.15% Tween 20) containing 5 % non-fat milk at 4°C overnight. After washing three times with PBS-T, the membrane was incubated with anti-CPD antibody (Kamiya Biomedical Company; Seattle, WA; dilution 1:2000) and horseradish peroxidase-conjugated anti-mouse IgG (eBioscience; San Diego, CA; dilution 1:5000) in PBS-T containing 5 % non-fat milk for 2 hours at room temperature. The signal on the membrane was detected by using the ECL-Plus system (GE Healthcare; Pittsburgh, PA). The membrane was exposed to X-ray film, and then the relative intensity of the signals was determined using a Quantity One image analyzer (Bio-Rad Laboratories).
Cell culture and DNA isolation
Normal human foreskin fibroblasts were grown in DMEM (Invitrogen) containing 10% fetal bovine serum. Normal human keratinocytes (Clonetics; San Diego) were grown in keratinocyte medium (All serum free; Lonza; Allendale, NJ). Normal human melanocytes (Lonza) were grown in Melanocyte Medium (All serum free, Lonza). The cells were trypsinized and collected, and then DNA was isolated with Quick gDNA-Miniprep Kit (Zymo Research; Irvine, CA).
Immuno-dot-blot assay to detect 5hmC
0.5 µg of DNA in TE buffer (pH 7.5) was denatured by heating for 5 min at 95°C and was then immediately placed on ice. The denatured DNA was blotted onto a charged nylon-based membrane (PerkinElmer) using the Bio-Dot Microfiltration System (Bio-Rad). The membrane was processed as described above. After washing three times with PBS-T, the membrane was incubated with rabbit polyclonal anti-5hmC antibody (Active Motif; Carlsbad, CA; dilution 1:7000) in PBS-T containing 5 % non-fat milk for 2 hours at room temperature. The membrane was washed with PBS-T, and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad; 1:25000) in PBS-T containing 5 % non-fat milk for 1 hour at room temperature. Detection of the signal was achieved as described above.
Results and Discussion
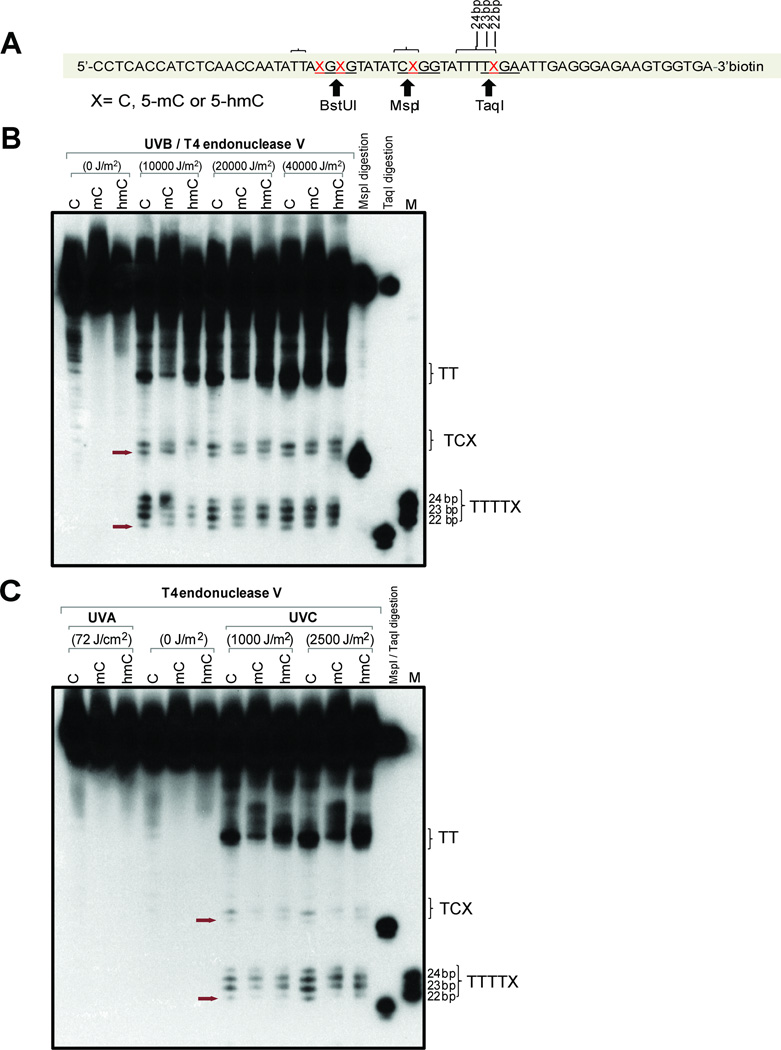
Cytosine and its modified derivatives, 5mC and 5hmC, may form CPDs when part of a dipyrimidne sequence (Fig. 1). We examined the susceptibility of 5hmC in different sequence contexts towards CPD formation. Synthetic oligonucleotides were prepared that contain cytosine, 5-methylcytosine, or 5-hydroxymethylcytosine at specific sequence positions within the CpG dinucleotide context. We first used 64-mer oligonucleotides that contain four modified cytosines on each DNA strand (Fig. 2A). Two of the modified bases were in a dipyrimidine sequence context and were located near MspI and TaqI restriction sites. These oligonucleotides were either left unirradiated (0 J/m2) or were irradiated with different doses of UVB (Fig. 2B) or UVA or UVC (Fig. 2C). After irradiation, the oligonucleotides were incubated with T4 endonuclease V, which specifically recognizes and cleaves DNA at CPDs including CPDs formed from 5hmC (28, 30). After cleavage, the oligonucleotides were separated on 10% denaturing polyacrylamide gels. Using size standards and digestion of the oligonucleotides with MspI or TaqI, we were able to localize the CPDs that involve a 5mC or 5hmC base (Fig. 2B; arrows) and provide semi-quantitative data for their relative intensities. The sequence near the MspI site (5’-TCXGG, where X corresponds to C, 5mC, or 5hmC and the restriction site is underlined) revealed two CPD bands with the lower one on the gel corresponding to the modified cytosine. The gels show that bands with similar intensity were produced at C, 5mC and 5hmC, respectively. Somewhat surprisingly, CPD formation in this sequence context was not enhanced at 5mC bases. The sequence near the TaqI site (5’-TTTTXGA) can theoretically produce four CPDs. This is what we observed; the lowest band corresponds to the CPD at the modified cytosine (Fig. 2B). Also here, CPD formation occurs at C, 5mC and 5hmC and is only slightly different between unmodified and modified cytosines.
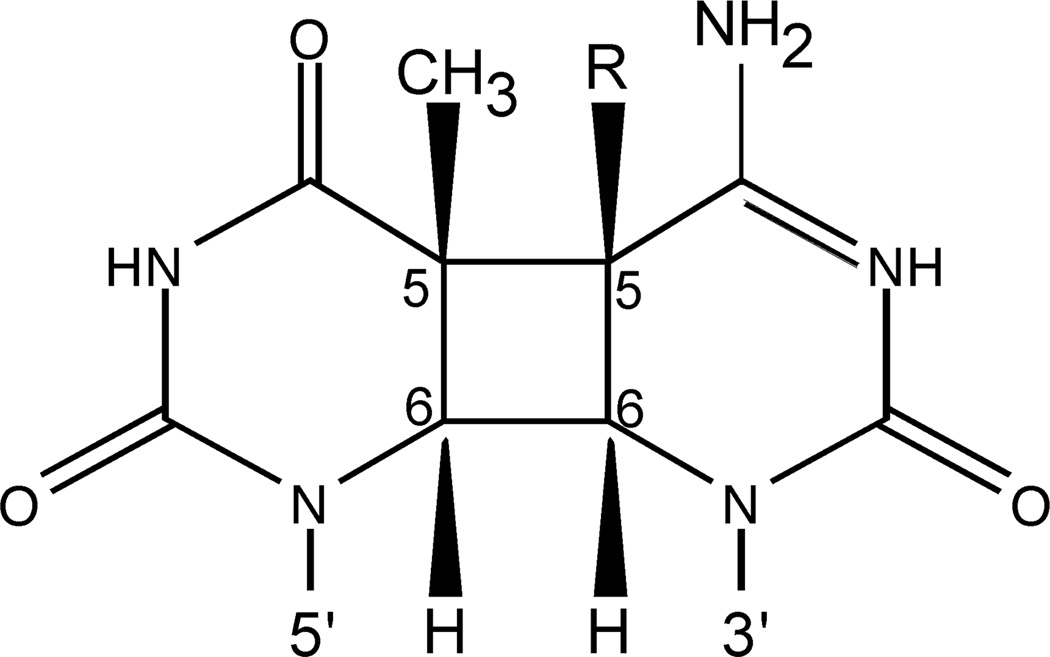
Figure 1. CPD containing cytosine or modified cytosine.
A cis-syn cyclobutane pyrimidine dimer forming at a 5’TC sequence is shown. R represents H for cytosine, CH3 for 5-methylcytosine, or CH2OH for 5-hydroxymethylcytosine.
Figure 2. Formation of CPDs in 64-mer oligonucleotides containing cytosine, 5-methylcytosine or 5-hydroxymethylcytosine.
A. Sequence of the 64-mers containing C, 5mC, or 5hmC at positions X. The oligonucleotides were labeled with biotin at the 3’ end. Positions of restriction endonuclease cleavage sites are indicted. The brackets indicate sites of CPD formation as shown in panels B and C.
B. The oligonucleotides were irradiated with different doses of UVB, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The positions of CPDs containing the modified cytosine base are indicated by arrows. The position marked ‘TT’ on the right side of the gel indicates CPD formation at the 5’TT sequence of 5’TTAXGXG, which includes the BstUI site (underlined) as shown in panel A.
C. The oligonucleotides were irradiated with 72 J/cm2 of UVA or two different doses of UVC, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The position of CPDs containing the modified cytosine base is indicated by arrows.
Interestingly, the cytosine modifications affected CPD formation at a 5’TT sequence that was 2–3 bases distant from a 5mC or 5hmC position (Fig. 2A, position labeled ‘TT.” In this sequence context, the 5mC-containing sequence was characterized by lower levels of CPD formation, and the oligonucleotide migrated more slowly in the gel. Presence of 5hmC, however, further enhanced CPD formation at the adjacent 5’TT sequence relative to 5mC and C.
We next analyzed CPD formation after UVA and UVC irradiation. It has been observed that CPDs can form upon irradiation of DNA with UVA (2, 4). However, using 72 J/cm2 of UVA, we could not detect CPDs with any of the unmodified or modified oligonucleotides (Fig. 2C). Presumably, this dose is too low to induce detectable CPDs. Irradiation with UVC did produce CPDs at all four possible dipyrimidine positions. However, the position involving the modified cytosine produced relatively low levels of these dimers (Fig. 2C).
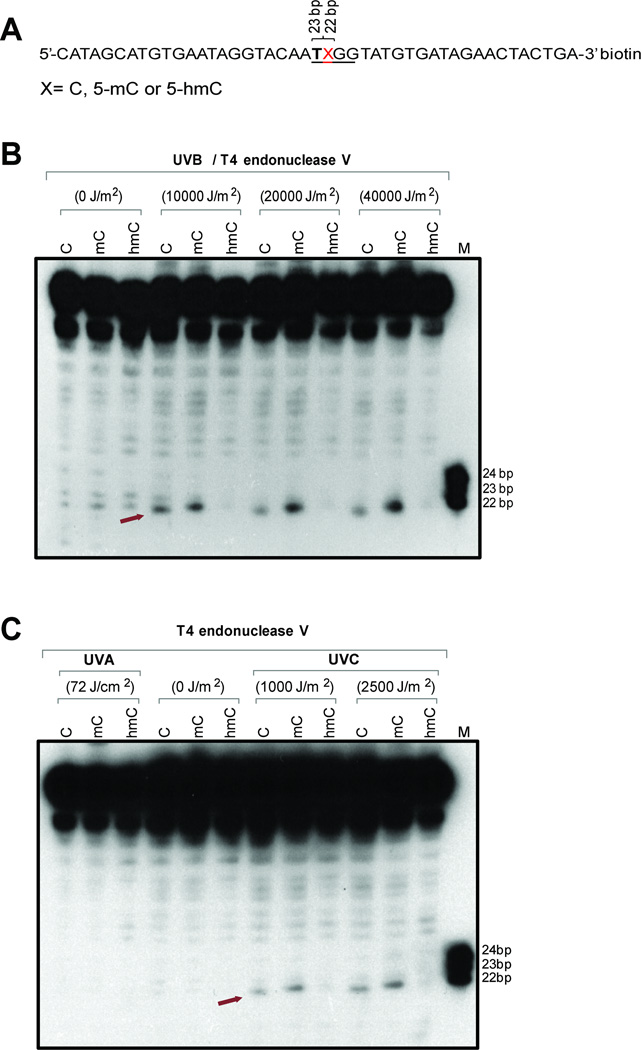
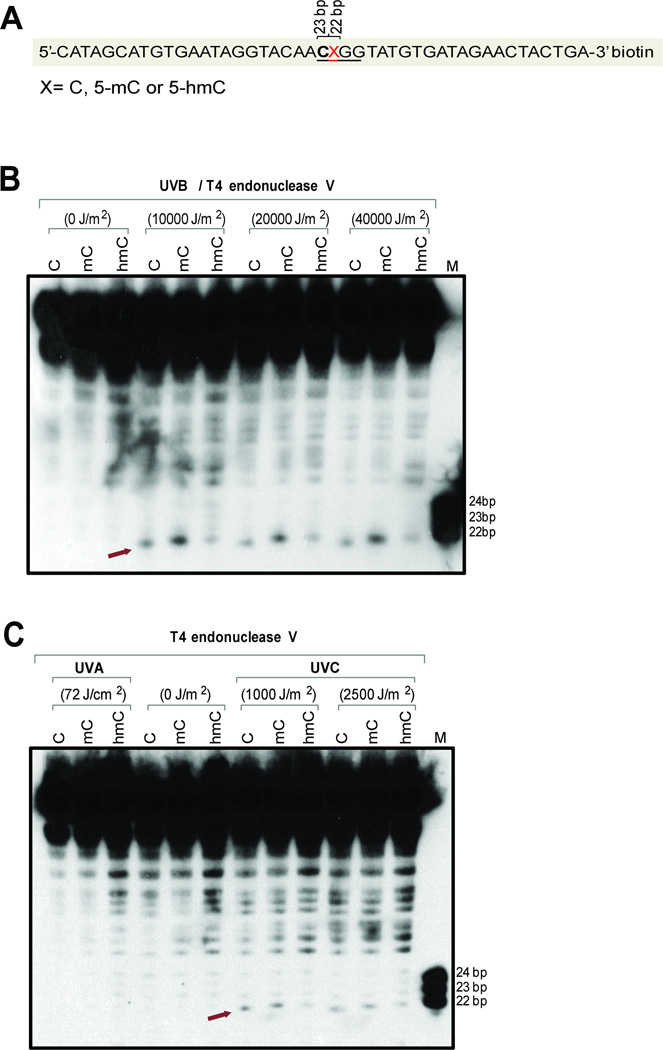
To study CPD formation at modified cytosines in a different sequence context and including contexts with only a single dipyrimidine sequence, we placed the modified cytosines into 45-mer oligonucleotides (Fig. 3, Fig. 4). The modified base was in the middle of these oligonucleotides and was in the dinucleotide 5’-TX (Fig. 3A) or 5’-CX (Fig. 4A) sequence context. Using UVB irradiation (Fig. 3B and Fig. 4B), we observed formation of CPDs at cytosines and enhanced levels at 5-methylcytosines in both the 5’-TX and 5’-CX sequence contexts. However, placement of 5hmC into these sequences led to a reduction of CPD formation. In the 5’-TX context, the 5hmC-containing CPD was barely detectable (Fig. 3B), whereas it was still visible in the 5’-CX sequence context. With UVA, there was no detectable formation of CPDs (Fig. 3C, Fig. 4C). Irradiation with UVC produced similar levels of CPDs at the three cytosine derivatives at 5’-CX (Fig. 4C) but we observed diminished levels of CPDs in the 5’-TX sequence context when 5hmC was the modified base (Fig. 3C).
Figure 3. Formation of CPDs in 45-mer oligonucleotides containing cytosine, 5-methylcytosine or 5-hydroxymethylcytosine within a 5’-TX sequence.
A. Sequence of the 45-mers containing C, 5mC, or 5hmC at position X. The oligonucleotides were labeled with biotin at the 3’ end.
B. The oligonucleotides were irradiated with different doses of UVB, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The position of CPDs containing the modified cytosine base is indicated by an arrow.
C. The oligonucleotides were irradiated with 72 J/cm2 of UVA or two different doses of UVC, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The position of CPDs containing the modified cytosine base is indicated by an arrow.
Figure 4. Formation of CPDs in 45-mer oligonucleotides containing cytosine, 5-methylcytosine or 5-hydroxymethylcytosine within a 5’-CX sequence.
A. Sequence of the 45-mers containing C, 5mC, or 5hmC at position X. The oligonucleotides were labeled with biotin at the 3’ end.
B. The oligonucleotides were irradiated with different doses of UVB, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The position of CPDs containing the modified cytosine base is indicated by an arrow.
C. The oligonucleotides were irradiated with 72 J/cm2 of UVA or two different doses of UVC, cleaved with T4 endonuclease V and then separated on a 10% denaturing polyacrylamide gel. The position of CPDs containing the modified cytosine base is indicated by an arrow.
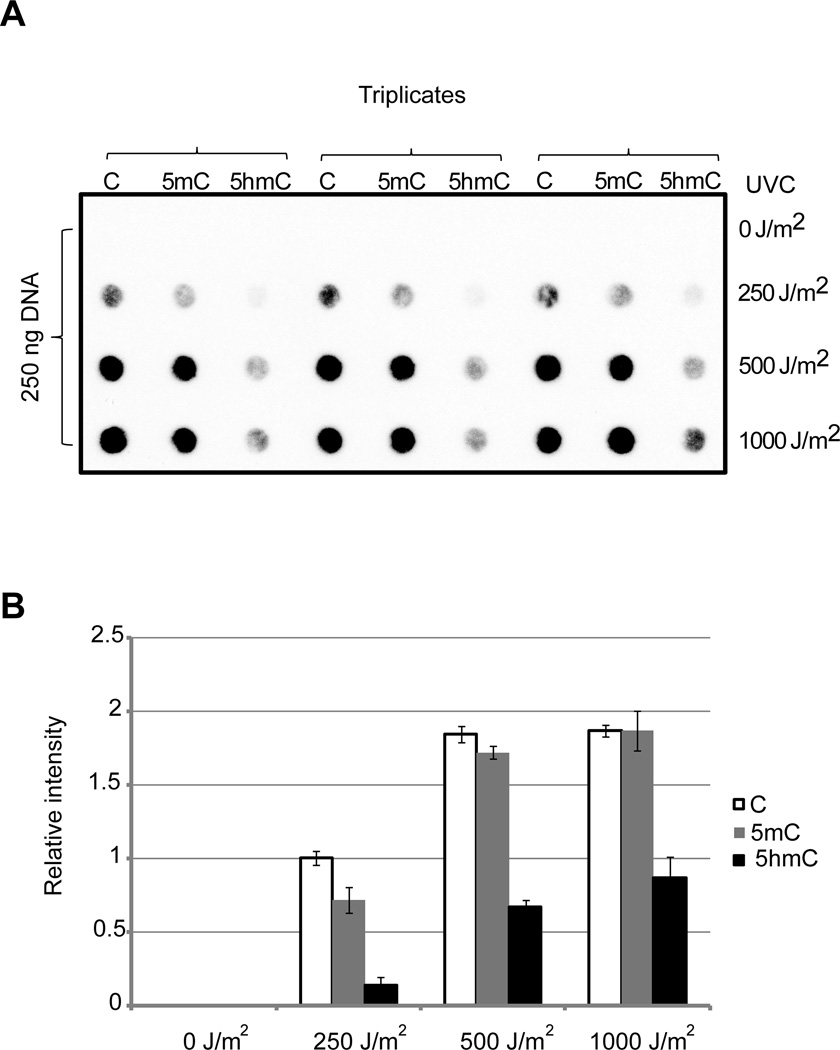
To verify this data by an independent approach, we probed the presence of CPDs in oligonucleotides with an anti-CPD antibody (Fig. 5), which is based on the assumption that the antibody recognizes CPDs with all cytosine modifications. This data also shows a strong reduction of CPDs when 5hmC was the dimerized base in the UVC-irradiated 5’-TX 45-mer sequence (Fig. 5) and is consistent with the T4 endonuclease cleavage data (Fig. 3C). This oligonucleotide also contains two cytosine-containing 5’-CT sequences, which most likely explain the residual dimer formation detected by the antibody in the 5hmC-containing oligonucleotides.
Figure 5. Detection of CPDs by immuno-dot blot.
The 45-mer oligonucleotides containing the 5’-TX sequence (see Fig. 3A) was irradiated with UVC at the indicated doses, applied to a nylon membrane and the CPD signals were detected with anti-CPD antibody.
A.Original triplicate experiments are shown.
B. Semi-quantitative analysis was conducted by image analysis (+/− S.D.).
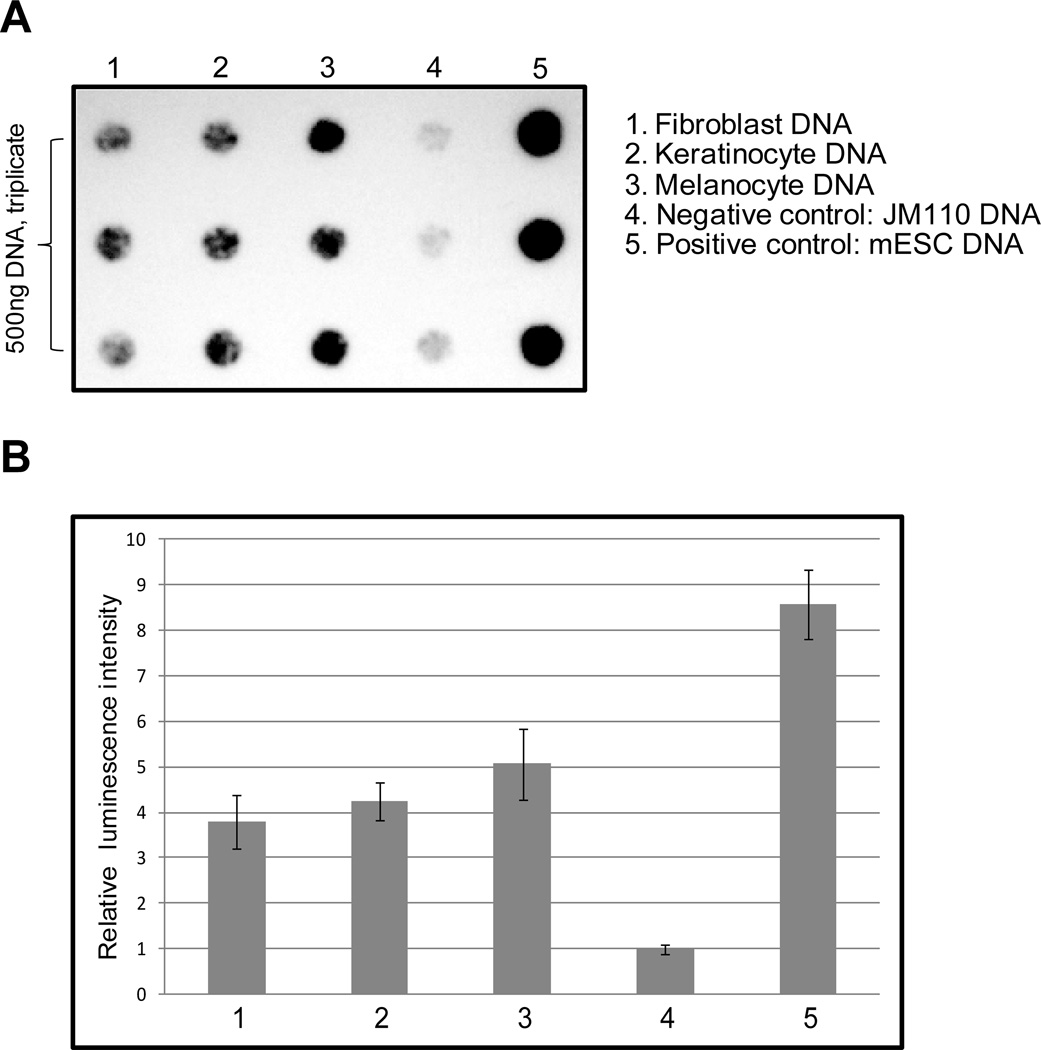
The combined data with the different sequence contexts indicate that 5hmC is susceptible to CPD formation but the exact levels of CPDs depend on the sequence context. There was clearly no enhancement of CPDs at 5hmC positions relative to C or 5mC. Nonetheless, its presence would create sites in genomic DNA susceptible to dimer formation. Therefore, we investigated if human skin-derived cells do actually contain 5hmC bases. A sensitive assay for detection of 5hmC in the genome is based on a specific antibody raised against this modified base. This antibody does not cross-react with DNA containing only unmodified cytosines or DNA containing 5mC (25, 31). We used this antibody to determine levels of 5hmC in DNA isolated from human fibroblasts, keratinocytes and melanocytes (Fig. 6). As a negative control, we used E. coli strain JM110 genomic DNA, which lacks 5mC, and as a positive control, we used DNA from mouse embryonic stem cells. The immuno-dot-blot data show that all three skin-derived cell types contain substantial levels of 5hmC (Fig. 6). The highest levels were found in melanocytes. Compared to mouse ES cells, 5hmC levels in human skin cells were 2–3 times lower. Since the level of 5hmC in mouse ES cells is known to be 0.1 % of all cytosines as determined previously by quantitative LC/MS/MS analysis (32), we can estimate the levels of this base to be approximately 0.03 to 0.05% of cytosine in melanocytes.
Figure 6. Detection of 5hmC in human fibroblasts, keratinocytes and melanocytes.
A. The levels of 5hmC were determined by dot blot analysis using 500 ng of DNA in triplicates. E. coli JM110 DNA was used as a negative (background) control. As a positive control, we used DNA from mouse ES cells (mESC).
B. Using densitometric scanning, the levels of 5hmC were determined semi-quantitatively. Data are from triplicates and show the mean +/− S.D. The numbers correspond to the samples in panel A.
Such levels are substantial enough so that a biological role of 5hmC in CPD formation in human skin can be expected. There are several questions that arise from these findings and warrant further study. Once formed, CPDs containing 5hmC would need to be repaired by the cell to preempt mutagenic events occurring during DNA replication. Repair of such a modified dimer may be different from repair of dimers containing C or 5mC. Another interesting question is how such dimers may undergo deamination. We know that CPDs containing 5mC are more or less prone towards deamination depending on the sequence than dimers containing cytosine (9, 33–35), and the deamination process may also be DNA sequence-dependent (34) and dependent on nucleosome association of the sequence (33). Deamination rates will eventually be reflected in mutagenic properties of a dimer; the more easily it deaminates, the more mutagenic it is expected to be owing to ‘correct’ bypass of deaminated CPDs by polymerases (11, 12). Other parameters that could influence CPD formation at sequences containing 5hmC include the possible interaction of 5hmC-containing DNA with specific proteins. As of today, several proteins, including UHFR1 (36), MBD3 (37), MECP2 (38) and others (39) have been implicated as potential 5hmC-binding proteins. Several other methyl-CpG binding proteins, including MBD1, MBD2 and MBD4, do not show any appreciable binding to 5hmCcontaining DNA (31). DNA-protein complexes tend to modulate CPD formation at the binding sites and often a strong enhancement or reduction of dimer formation has been observed (40, 41). The discovery of a “new” DNA pyrimidine in mammalian DNA is intriguing and further studies will provide important insights into how these specialized pyrimidines play a role in the photochemistry and photobiology of DNA following UVB radiation.
Acknowledgment
We thank Steven Bates for culturing of cells. This work was supported by a grant from the National Institutes of Health (ES006070) awarded to G.P.P.
References
- 1.Cadetand J, Douki T. Oxidatively generated damage to DNA by UVA radiation in cells and human skin. J Invest Dermatol. 2011;131:1005–1007. doi: 10.1038/jid.2011.51. [DOI] [PubMed] [Google Scholar]
- 2.Douki T, Reynaud-Angelin A, Cadetand J, sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer GP, Youand YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 4.Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetskyand EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastien N, Millau JF, Rouabhia M, Daviesand RJ, Drouin R. The sunscreen agent 2-phenylbenzimidazole-5-sulfonic acid photosensitizes the formation of oxidized guanines in cellulo after UV-A or UV-B exposure. J Invest Dermatol. 2010;130:2463–2471. doi: 10.1038/jid.2010.150. [DOI] [PubMed] [Google Scholar]
- 6.Mitchelland DL, Fernandez AA. Different types of DNA damage play different roles in the etiology of sunlight-induced melanoma. Pigment Cell Melanoma Res. 2011;24:119–124. doi: 10.1111/j.1755-148X.2010.00789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You YH, Lee DH, Yoon JH, Nakajima S, Yasuiand A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 8.Mitchelland DL, Nairn RS. The biology of the (6–4) photoproduct. Photochem. Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 9.Leeand DH, Pfeifer GP. Deamination of 5-methylcytosines within cyclobutane pyrimidine dimers is an important component of UVB mutagenesis. J. Biol. Chem. 2003 doi: 10.1074/jbc.M212696200. in press. [DOI] [PubMed] [Google Scholar]
- 10.Choiand JH, Pfeifer GP. The role of DNA polymerase eta in UV mutational spectra. DNA Repair (Amst) 2005;4:211–220. doi: 10.1016/j.dnarep.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Jiangand N, Taylor J-S. In vivo evidence that UV-induced C-T mutations at dipyrimidine sites could result from the replicative bypass of cis-syn cyclobutane dimers or their deamination products. Biochemistry. 1993;32:472–481. doi: 10.1021/bi00053a011. [DOI] [PubMed] [Google Scholar]
- 12.Vu B, Cannistraro VJ, Sunand L, Taylor JS. DNA synthesis past a 5-methylCcontaining cis-syn-cyclobutane pyrimidine dimer by yeast pol eta is highly nonmutagenic. Biochemistry. 2006;45:9327–9335. doi: 10.1021/bi0602009. [DOI] [PubMed] [Google Scholar]
- 13.Sage E, Lamolet B, Brulay E, Moustacchi E, Chteauneufand A, Drobetsky EA. Mutagenic specificity of solar UV light in nucleotide excision repair-deficient rodent cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:176–180. doi: 10.1073/pnas.93.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Y-H, Liand C, Pfeifer GP. Involvement of 5-methylcytosine in sunlightinduced mutagenesis. J. Mol. Biol. 1999;293:493–503. doi: 10.1006/jmbi.1999.3174. [DOI] [PubMed] [Google Scholar]
- 15.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperinand AJ, Pontén J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumaz N, Drougard C, Sarasinand A, Daya-Grosjean L. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1993;90:10529–10533. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeifer GP. Environmental exposures and mutational patterns of cancer genomes. Genome Med. 2010;2:54. doi: 10.1186/gm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tommasi S, Denissenkoand MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- 20.Drouin R, Therrien JP. UVB-induced cyclobutane pyrimidine dimer frequency correlates with skin cancer mutational hotspots in p53. Photochem Photobiol. 1997;66:719–726. doi: 10.1111/j.1751-1097.1997.tb03213.x. [DOI] [PubMed] [Google Scholar]
- 21.Wyattand GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kriaucionisand S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravindand L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowersand LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010 doi: 10.1038/nature09303. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal K, Jin SG, Pfeiferand GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin SG, Wu X, Liand AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szwagierczak A, Bultmann S, Schmidt CS, Spadaand F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Childs JD, Paterson MC, Smithand BP, Gentner NE. Evidence for a near UVinduced photoproduct of 5-hydroxymethylcytosine in bacteriophage T4 that can be recognized by endonuclease V. Mol Gen Genet. 1978;167:105–112. doi: 10.1007/BF00270326. [DOI] [PubMed] [Google Scholar]
- 29.Childs JD, Ellisonand MJ, Pilon R. Formation of 5-hydroxymethylcytosinecontaining pyrimidine dimers in UV-irradiated bacteriophage T4 DNA. Photochem Photobiol. 1983;37:513–519. doi: 10.1111/j.1751-1097.1983.tb04510.x. [DOI] [PubMed] [Google Scholar]
- 30.Meneghiniand R, Hanawalt P. T4-endonuclease V-sensitive sites in DNA from ultraviolet-irradiated human cells. Biochim Biophys Acta. 1976;425:428–437. doi: 10.1016/0005-2787(76)90007-1. [DOI] [PubMed] [Google Scholar]
- 31.Jin SG, Kadamand S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Luand Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Q, Cannistraroand VJ, Taylor JS. Rotational position of a 5-methylcytosine-containing cyclobutane pyrimidine dimer in a nucleosome greatly affects its deamination rate. J Biol Chem. 2011;286:6329–6335. doi: 10.1074/jbc.M110.183178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu Y, Dammannand R, Pfeifer GP. Sequence and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J. Mol. Biol. 1998;284:297–311. doi: 10.1006/jmbi.1998.2176. [DOI] [PubMed] [Google Scholar]
- 35.Celewicz L, Mayerand M, Shetlar MD. The photochemistry of thymidylyl-(3'– 5')-5-methyl-2'-deoxycytidine in aqueous solution. Photochem Photobiol. 2005;81:404–418. doi: 10.1562/2004-06-15-ra-201.1. [DOI] [PubMed] [Google Scholar]
- 36.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antesand I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Randoand OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellen M, Ayata P, Dewell S, Kriaucionisand S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carelland T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifer GP, Drouin R, Riggsand AD, Holmquist GP. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol. Cell. Biol. 1992;12:1798–1804. doi: 10.1128/mcb.12.4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tornalettiand S, Pfeifer GP. UV light as a footprinting agent: modulation of UV-induced DNA damage by transcription factors bound at the promoters of three human genes. J. Mol. Biol. 1995b;249:714–728. doi: 10.1006/jmbi.1995.0331. [DOI] [PubMed] [Google Scholar]