Abstract
Our previous study showed that selenamide reagents such as ebselen and N-(phenylseleno) phthalimide (NPSP) can be used for selective and rapid derivatization of protein/peptide thiols in high conversion yield. This paper reports the systematic investigation of MS/MS dissociation behaviors of selenamide-derivatized peptide ions upon collision induced dissociation (CID) and electron transfer dissociation (ETD). In the positive ion mode, derivatized peptide ions exhibit tag-dependent CID dissociation pathways. For instance, ebselen-derivatized peptide ions preferentially undergo Se–S bond cleavage upon CID to produce a characteristic fragment ion, the protonated ebselen (m/z 276), which allows selective identification of thiol peptides from protein digest as well as selective detection of thiol proteins from protein mixture using precursor ion scan (PIS). In contrast, NPSP-derivatized peptide ions retain their phenylselenenyl tags during CID, which is useful in sequencing peptides and locating cysteine residues. In the negative ion CID mode, both types of tags are preferentially lost via the Se–S cleavage, analogous to the S–S bond cleavage during CID of disulfide-containing peptide anions. In consideration of the convenience in preparing selenamide-derivatized peptides and the similarity of Se–S of the tag to the S–S bond, we also examined ETD of the derivatized peptide ions to probe the mechanism for electron-based ion dissociation. Interestingly, facile cleavage of Se–S bond occurs to the peptide ions carrying either protons or alkali metal ions, while backbone cleavage to form c/z ions is severely inhibited. These results are in agreement with the Utah-Washington mechanism proposed for depicting electron-based ion dissociation processes.
Keywords: CID, ETD, Peptide ions, Selenamide, Thiol derivatization, Selective detection
Introduction
Biological thiols such as glutathione and thiol proteins are critical physiologic components found in animal tissues and are involved in a plethora of important cellular functions [1]. Due to the inherent sensitivity and chemical specificity (e.g., molecular weight and structural information), mass spectrometry (MS) [2, 3] has attracted significant attention in the characterization of biological thiols. A number of elegant MS studies of thiols and disulfides of proteins/peptides based on the novel ion chemistry have been reported [4-9]. For MS analysis of thiol proteins/peptides, the derivatization of thiol groups with a suitable chemical reagent is often necessary for increasing thiol stability and improving detection selectivity or for enrichment and purification purposes. Furthermore, tandem MS analysis (MS/MS) can benefit from the modification of thiols. For instance, N,N-dimethyl-2-chloro-ethylamine [10] was used to increase the charge state of cysteine-containing toxin ions to meet the requirement for performing electron-transfer dissociation (ETD) [11]. In addition, it has been shown that thiol derivatization using (3-acrylamidopropyl)-trimethyl ammonium chloride significantly improves the percent fragmentation and sequence coverage for peptide ions upon ETD [12]. Remarkably, quinone-modified cysteine was shown to facilitate site-selective cleavage by photodissociation [13].
Traditional thiol labeling strategies are mainly based on nucleophilic substitution (e.g., using heptafluorobutyl chloroformate [14] and iodoacetamide [15]), Michael-addition with unsaturated C=C bonds (e.g., using acrylate [16] or maleimide derivatives [17]), or thiol exchange reaction (e.g., using the Ellman’s reagent [18, 19]). However, these protocols have some limitations such as low reaction selectivity, long reaction time, or low conversion yield. Recently we developed a new methodology to label biological thiols using selenamide reagents for MS analysis, which involves the cleavage of Se–N bond by thiol to form a new Se–S bond [20]. Our data show that the reaction is highly selective, rapid, reversible and efficient. For instance, among 20 natural amino acids, only cysteine is reactive towards Se–N containing selenamide reagents such as ebselen or N-(phenylseleno)phthalimide (NPSP), and the reaction occurs in seconds. In the case of protein β-lactoglobulin A containing one free cysteine residue, the derivatization can be completed in 30 s with 100% conversion yield. By adding reductant of dithiothreitol (DTT), peptides derivatized by selenamide reagents can be recovered. In order to better implement this new thiol derivatization reaction for protein/peptide analysis by mass spectrometry, it is therefore necessary to well understand the tandem MS dissociation behavior of derivatized protein/peptide ions, which is the motivation and focus of this study. On the other hand, due to the significance of electron capture dissociation (ECD) [21] and ETD [11] in top-down proteomics research, their mechanisms arouse much attention and are still under debate [21-23]. One of the approaches for mechanism elucidation is to introduce various tags with different electron or proton affinities to model peptides and then test the effects of the tags on the dissociation [24-27]. Since the preparation of selenamide-derivatized proteins/peptides is convenient as mentioned above and that the formed Se-S bond is structurally similar to the S–S bond, we also investigated ETD of the derivatized peptide ions in effort to provide some new insight to the electron-based ion dissociation mechanism.
In this study, we chose several free cysteine-containing peptides as examples and systemically investigated their ion dissociation pathways after derivatization by selenamide reagents such as ebselen and NPSP, under CID (in both positive and negative ion modes) and ETD conditions. For the ebselen tag, it appears to be easily lost upon CID, probably due to the presence of an adjacent amide group in the tag, consistent with previous observation [20]. The phenylselenenyl tag from NPSP derivatization can survive in the CID process, leading to the formation of a series of b/y ions from backbone cleavage of peptides in the positive ion mode. Interestingly, both tags can be removed upon CID in the negative ion mode, which is similar to the known CID behavior of disulfide bonds [28]. During ETD of derivatized peptide ions, selenium tag loss is dominant while the backbone cleavage is suppressed. Similar phenomenon was also observed for alkali metal-containing peptide ions, which can be explained by the Utah-Washington mechanism proposed for ECD dissociation [22, 23].
Experimental
Chemicals
Peptides HCKFWW (MW 905.4 Da), NRCSQGSCWN (MW 1153.4 Da), bovine pancreas insulin (MW 5733.5 Da), α-lactalbumin from bovine milk (type III, calcium depleted, ≥85%, MW 14178 Da), β-lactoglobulin A from bovine milk (MW 18369 Da), lysozyme from chicken egg white (MW 14300 Da), pepsin from porcine gastric mucosa (MW~35 KDa), NPSP, and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ebselen was purchased from Calbiochem (Cincinnati, OH, USA). HPLC-grade methanol from GFS Chemicals (Columbus, OH, USA) and acetonitrile from Sigma-Aldrich (St. Louis, MO, USA) were used and acetic acid was purchased from Fisher Scientific (Pittsburgh, PA, USA). The deionized water used for sample preparation was obtained using a Nanopure Diamond Barnstead purification system (Barnstead International, Dubuque, IA, USA).
Preparation of Selenamide-Derivatized Peptides
Peptide thiols were derivatized by reaction with 5 ~10 time excess amount of ebselen or NPSP in either methanol/water (1:1 by volume) containing 1% acetic acid or acetonitrile/ water (1:1 by volume) containing 1% acetic acid at room temperature.
The derivatized insulin chain B was obtained via reacting the reduced insulin with the selenamide reagents. First TCEP in 20 mM ammonium bicarbonate aqueous solution was added to insulin in acetonitrile/water (1:1 by volume) containing 0.1% trifluoroacetic acid in the molar ratio of 1:50 (protein/TCEP) for 3.5 h at room temperature. Then Millipore-ZipTip Pipette Tips were used to remove TCEP and ammonium bicarbonate via desalting. After that, excess amount of ebselen (ebselen:protein=6:1) or NPSP (NPSP: protein=11:1) were added to derivatize the reduced insulin.
Derivatization of Insulin Peptic Digest and Protein Mixture
Digestion of insulin was performed by incubating insulin and pepsin at a molar ratio of 50:1 in water containing 1% acetic acid at 37 °C for 11 h. Then TCEP was added to the peptic digested insulin in the molar ratio of 1:20 (protein: TCEP) for several days at room temperature. After that, ebselen (ebselen:protein=20:1) was added to derivatize the insulin digest.
Ebselen (50 μM) was reacted with β-lactoglobulin A (10 μM) in the presence of α-lactalbumin, lysozyme, and insulin (10 μM for each) in methanol/water (1:1 by volume) containing 1 % acetic acid for 30 s.
Mass Spectrometry
A LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA, USA) with an electrospray source was used for collecting CID and ETD data of derivatized peptide ions. Peptide samples were directly ionized by ESI with the injection flow rate of 3–5 μL/min. The spray voltage was 4–4.5 kV. The automatic gain control (AGC) value was set at 1×106 (Orbitrap) and 3×104 (linear ion trap) with maximum injection time of 500 ms (Orbitrap) and 200 ms (linear ion trap). The high resolution product-ion spectra were acquired at high mass resolving power (60 000 for ions of m/z 400). The LTQ Orbitrap was externally calibrated using a standard calibration mixture of caffeine, MRFA, and Ultramark 1621. CID experiments were conducted in the linear ion trap with helium as the collision gas. The normalized collision energy used was 5%–35% with the activation time of 30 ms. The isolation width was ±2 Da. All ETD experiments were conducted with fluoranthene radical anion as the electron-transfer reagent in the linear ion trap. The ETD reaction time was 60–120 ms. The precursor ion scan (PIS) was performed using a hybrid triple quadrupole/linear ion trap mass spectrometer (Q-trap 2000; Applied Biosystems/MDS SCIEX, Concord, Canada) equipped with an electrospray source. Samples were injected for ESI at the flow rate of 5 μL/min. The spray voltage was +5 kV. Curtain gas was 20. Collision energy (CE) used was 50–80 eV. PIS was used to screen the ebselen tag-containing precursor ions based on the formation of the characteristic fragment ion of m/z 276 (i.e., the protonated ebselen) and N2 was used as the collision gas.
Results and Discussion
Various biological thiols, such as peptide HCKFWW containing one free cysteine, NRCSQGSCWN having two free cysteines, and insulin chain B carrying two cysteines, were chosen for examination in this study. Following derivatization by ebselen or NPSP, the samples were directly ionized by ESI, and it was found that the number of tags in the main derivatization products agrees with the number of the free cysteines of the peptide substrates reacted (shown in Figure S-1, Supporting Information). Then, the derivatized peptide ions were mass-selected for tandem MS analysis.
CID MS/MS of the Positive Ions of Selenamide-Derivatized Peptides
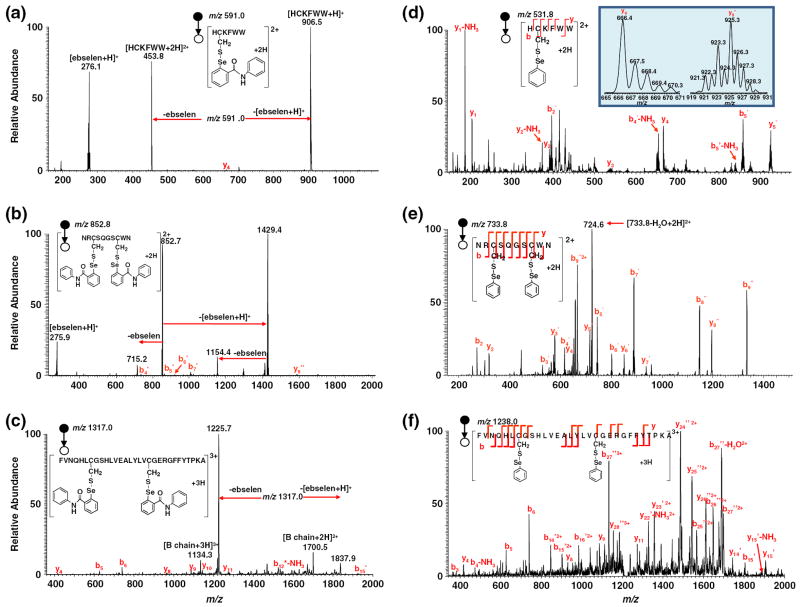
Positive ion CID MS/MS of ebselen-derivatized peptide ions was first carried out. Figures 1a–c show the CID MS/MS mass spectra of doubly charged [HC*KFWW+2 H]2+ (m/z 591.0; see its structure in Figure 1a; the asterisk “*” denotes one ebselen tag on the peptide cysteine group and such a denotation is applicable throughout the text), doubly charged [NRC*SQGSC*WN+2 H]2+ (m/z 852.8; structure shown in Figure 1b), and triply charged [FVNQHLC*GSHLVEALYLVC*GERGFFYTPKA+3 H]3+ (m/z 1317.0; structure shown in Figure 1c), respectively. Upon CID, facile Se–S bond cleavage occurs to m/z 591.0 (Figure 1a), leading to the formation of singly and doubly charged ions of HCKFWW (m/z 906.5 and 453.8) via the loss of one protonated ebselen and one neutral ebselen, respectively. Indeed, the protonated ebselen (m/z 276.1) was also observed in Figure 1a. This process might be driven by the re-formation of the five-membered ring of ebselen as assisted via the nucleophilic attack of selenium by the adjacent amide nitrogen in the ebselen tag. The result is consistent with our previous observation in the case of ebselen-derivatized glutathione, a tripeptide with one cysteine residue [20]. As illustrated in Scheme 1, the cleavage of Se–S bond during CID takes place in two pathways: one is to lose neutral ebselen (pathway a, eq 1) and the other is to lose the protonated ebselen (pathway b, eq 1). Also, in comparison to the Se–S bond dissociation, the backbone cleavage to b/y ions is negligible in this case and only a tiny peak of y4 ion was seen. The facile Se–S bond cleavage appears to be a general trend and was also observed during CID of the other two peptides containing two ebselen tags (Figures 1b and c). In Figure 1b, two major fragment ions of m/z 1429.4 and 715.2 were observed, corresponding to the singly and doubly charged ions of NRCSQGSCWN with one ebselen tag, respectively, arising from the losses of one protonated ebselen and one neutral ebselen from [NRC*SQGSC*WN+2 H]2+ (m/z 852.8). The m/z 1429.4 was also further examined by MS/MS/MS, which shows that the ebselen tag is either on its Cys-3 or on the Cys-8 residue (shown in Figure S-2, Supporting Information). It indicates that there is no regioselectivity for the dissociation of its precursor ion of m/z 852.8 with regard to the ebselen tag loss. Another fragment ion of m/z 1154.4 (Figure 1b), the protonated NRCSQGSCWN originating from m/z 1429.4 dissociation by further loss of the ebselen tag, was detected along with the protonated ebselen and b/y fragment ions such as b4*, b5*, b6*, b7*, and y9** with low abundances resulting from backbone cleavage (again, the asterisk indicates one ebselen tag and the double asterisks indicate two ebselen tags). In the case of insulin chain B, m/z 1837.9 and 1225.7 representing the doubly and triply charged ions of chain B with one ebselen tag were produced upon CID, again, by losses of one protonated ebselen and one neutral ebselen from the precursor ion of m/z 1317.0, respectively. Also, the doubly and triply charged ions of the free chain B were seen at m/z 1700.5 and 1134.3, respectively, as a result of secondary dissociation of m/z 1837.9 and 1225.7 by further loss of the remaining ebselen tag. These results clearly suggest that ebselen-derivatized peptide cations preferentially undergo Se–S bond cleavage during CID via losses of both neutral and protonated ebselen (the exception of no protonated ebselen observed in Figure 1c is probably due to the low mass cut-off in the linear ion trap of the LTQ-Orbitrap used). Both the formation of the characteristic fragment ion of m/z 276 and the facile loss of neutral ebselen from the CID of derivatized thiol peptide ions would be useful to identify thiol-containing peptides from mixtures such as protein digests, using either PIS or neutral loss scan (NLS), and the former application is demonstrated below.
Figure 1.
Positive ion CID MS/MS mass spectra of (a) [HC*KFWW+2 H]2+ (m/z 591.0); (b) [NRC*SQGSC*WN+2 H]2+ (m/z 852.8); (c) [FVNQHLC*GSHLVEALYLVC*GERGFFYTPKA+3 H]3+ (m/z 1317.0); (d) [HC’KFWW+2 H]2+ (m/z 531.8); (e) [NRC’SQGSC’WN+2 H]2+ (m/z 733.8); (f) [FVNQHLC’GSHLVEALYLVC’GERGFFYTPKA+3 H]3+ (m/z 1238.0)
Scheme 1.
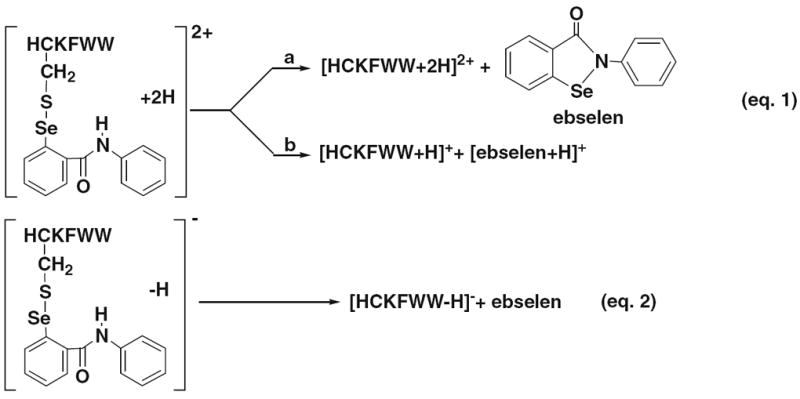
CID MS/MS processes for [HC*KFWW+2 H]2+ (eq 1) and [HC*KFWW – H]− (eq 2)
As a demonstration, insulin (its sequence is shown in the inset of Figure 2b) was digested by pepsin and then reduced by TCEP. The reduced protein digest mixture was derivatized via the selective reaction with ebselen and then subject to ESI-MS analysis. As illustrated in Figure 2a, the ESI-MS spectrum shows that a series of ebselen-derivatized thiol peptide ions [FVNQHLC*GSHL+3 H]3+ (m/z 510.8), [HLC*GSHL +2 H]2+ (m/z 521.4), [LVC*GERGF+2 H]2+ (m/z 579.2), [QCCASVC*SL+2 H]2+ (m/z 596.0) (the exact position of the ebselen tag in this case is uncertain), [LVC*GERGFF+2 H]2+ (m/z 652.0), [YLVC*GERGF+ 2 H]2+ (m/z 659.8), [YC*N+H]+ (m/z 674.3), [VC*SL+H]+ (m/z 696.1), [QCC*ASVC*SL+2 H]2+ (m/z 733.5) (the exact positions of the two ebselen tags are not certain in this case), [FVNQHLC*GSHL+2 H]2+ (m/z 765.6), [LVC*GERGFFYT+ 2 H]2+ (m/z 783.1), [NYC*N+ H]+ (m/z 788.2), [ASVC*SL + H]+ (m/z 854.0), and [LVC*GERGFF+H]+ (m/z 1303.2) were observed (listed in the Figure 2a inset). In addition, [FVNQHLCGSHL + 2 H]2+, [LVCGERGF + H]+, [QCCASVCSL+H]+, [LVCGERGFF+H]+ were seen at m/z 627.8, 880.5, 914.3, and 1027.1, respectively, probably resulting from the incomplete derivatization reaction by ebselen owing to the presence of the excess amount of reductant TCEP (indeed, we found that, like DTT, TCEP can reduce the ebselen-derivatized peptides such as glutathione in a separate experiment). Furthermore, [ALY+H]+, [GIVE+H]+, [VEAL+H]+, [FVNQ+ H]+, [YQLE+H]+, [YTPKA+H]+, and [QLENY+H]+ were detected at m/z 366.1, 417.2, 431.3, 507.4, 552.2, 579.2, and 666.4, respectively (shown in the inset of Figure 2a). These ions correspond to seven additional peptides, which do not have free cysteine residues and, as a result, no corresponding ebselen-derivatization ions were detected for these peptides. Thus, in this insulin digest, a total of 18 peptides were observed (listed in the Figure 2b inset), covering 100% of the sequence of insulin. Meanwhile, [2(ebselen)+H]+ was also detected at m/z 550.9. Due to the multi-component nature of the sample, the ESI-MS spectrum in Figure 2a has multiple peaks and appears complicated. Using the Q-trap instrument, PIS was employed to screen those peptides containing cysteine residues. By monitoring the product ion of m/z 276, all of the ebselen-derivatized peptide ions mentioned above were selectively detected corresponding to 11 thiol peptides, as shown in Figure 2b (inset also shows the list of detected thiol peptides). This experiment provides a simple and rapid method to selectively identify thiol peptides in complex protein digest mixtures. This method also complements our previous method for selective identification of disulfide bond-containing peptides in protein digests based on their response to electrochemical reduction [29]. Furthermore, this selective strategy is also applicable to thiol proteins. In another experiment, a protein mixture containing β-lactoglobulin A, α-lactalbumin, lysozyme, and insulin was derivatized via the selective reaction with ebselen and then analyzed by ESI-MS. As illustrated in Figure 3a, the ESI-MS spectrum shows the multiply charged ions of α-lactalbumin, lysozyme, and insulin, together with one ebselen-derivatized β-lactoglobulin A ions. PIS was then applied to screen the thiol protein ions. As shown in Figure 3b, the ebselen-derivatized β-lactoglobulin A ions were selectively detected by monitoring the product ion of m/z 276. Thus, it can be seen that the ebselen derivatization in conjunction with PIS is powerful in the analysis of both thiol peptides and proteins in mixtures.
Figure 2.
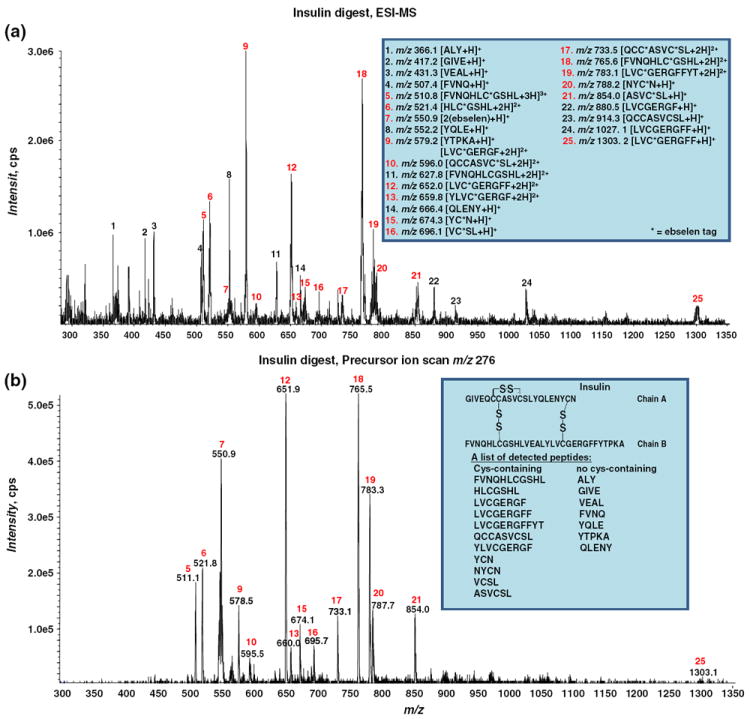
(a) ESI-MS mass spectrum showing the reaction of the reduced peptic digested insulin with ebselen. The inset shows the list of all the peptide peaks; (b) PIS based on the monitoring of the characteristic fragment ion of m/z 276 upon CID shows the selective detection of thiol peptides in the reduced peptic digested insulin. The insulin sequence and a list of detected cysteine-containing and non-cysteine containing peptides are shown in the inset
Figure 3.
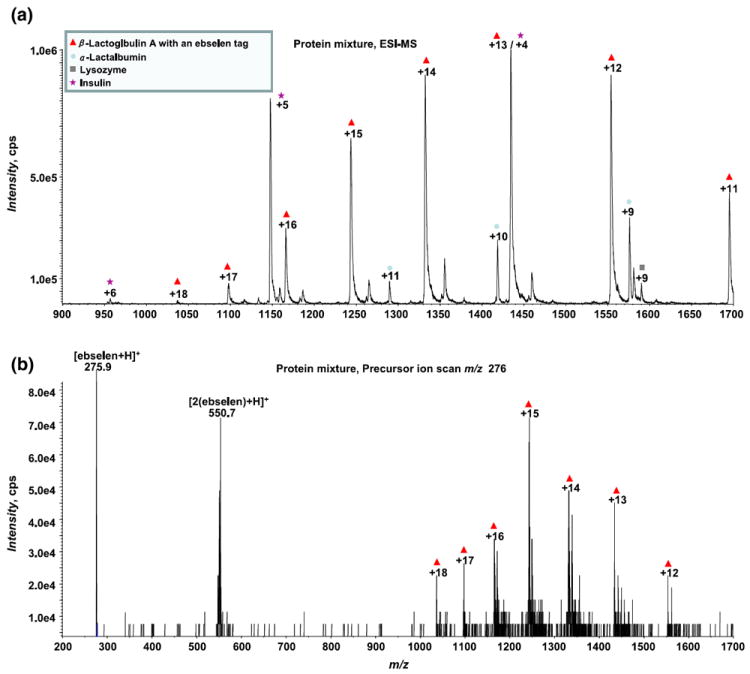
(a) ESI-MS mass spectrum showing a mixture of β-lactoglobulin A, α-lactalbumin, lysozyme and insulin after reaction with ebselen; (b) precursor ion scanning based on the monitoring of the characteristic fragment ion of m/z 276 upon CID shows the selective detection of thiol containing protein β-lactoglobulin A in a protein mixture
In stark contrast to ebselen tagging, peptide ions with phenylselenenyl tags arising from NPSP derivatization display drastically different CID dissociation behaviors, in which the tags are preserved and consecutive backbone cleavages occur (eq 1, Scheme 2). This is probably due to the lack of intramolecular nucleophilic attack of selenenyl sulfide bond by adjacent amide nitrogen of the selenium tag as in the case of ebselen. As shown in Figure 1d, extensive b and y ions (b2’, b4’-NH3, b5’, b5’-NH3, y1, y1-NH3, y2, y2-NH3, y3, y4, y5’) were seen (the single prime indicates one phenylselenenyl tag). Again, similar phenomena were observed for CID of the NRCSQGSCWN ion with two phenylselenenyl tags, [NRCSQ’GSC’WN+ 2 H]2+ (m/z 733.8, Figure 1e), and of the insulin chain B ion with two phenylselenenyl tags, [FVNQHLC’GSHLVEALYLVC’GERGFFYTPKA+3 H]3+ (m/z 1238.0, Figure 1f). For the former peptide ion, mainly b2, b3’, b4’, b5’, b6’, b7’, b8”, b9”, b9”2+, y2, y3’, y4’, y5’, y6’, y7’, and y8” were observed (Figure 1e; the double prime indicates two phenylselenenyl tags in the fragment ions). For the latter, b3, b4-NH3, b5, b6, b14’2+, b15’, b15’2+, b16’2+, b25”2+, b26”2+, b27”2+, b27”-H2O2+, b27”3+, y4, y8, y9, y11, y14’, y15’, y15’-NH3, y22’-NH32+, y23’2+, y24”2+, y25”2+, y26”2+, and y28”3+ were detected (Figure 1f). It turns out that the phenylselenenyl tag could facilitate peptide sequencing and be valuable in pinpointing the location of cysteine residues of peptides. For example, based on the distinct selenium isotope distribution of y5’ which is absent for y4 (Figure 1d inset), it is clear that y5’ contains the tag and thus a cysteine residue while y4 does not. This result is in agreement with the peptide structure (the cysteine is located in the second residue of the peptide HCKFWW) and also emphasizes the high selectivity of the NPSP derivatization toward cysteine. The derivatization selectivity was further confirmed by the appearance of fragment ion pairs of b2/b3’ and b7’/b8” in the case of peptide NRCSQGSCWN (having Cys-3 and Cys-8) and the generation of the fragment ion pairs of y11/y14’ and y23’2+/y24”2+ in the case of insulin chain B (having Cys-7 and Cys-19). In particular, the pair y11/y14’ reveals that one of the two cysteines is located between the 17th and 19th residues of the chain B.
Scheme 2.
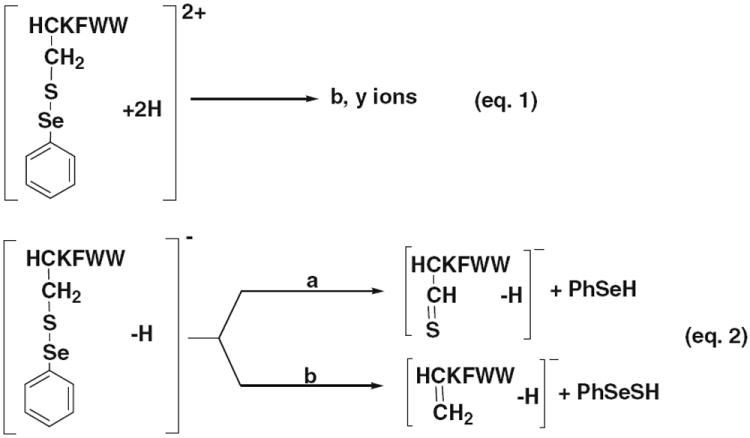
CID MS/MS processes for [HC’KFWW+2 H]2+ (eq 1) and [HC’KFWW – H]− (eq 2)
CID MS/MS of the Negative Ions of Selenamide-Derivatized Peptides
Selenium and sulfur, two elements in the Group VIB, exhibit many expected similarities in chemical properties [30, 31]. Indeed, they have similar electronegativities of 2.55 and 2.58 (Pauling scale), respectively. The Se–S bond is also close to the S–S bond, both of which can be electrolytically reduced [32]. It is known that S–S undergoes marginal cleavage under positive ion CID except with high energy or with metal ions [33]. But S–S undergoes facile cleavage under negative ion CID [3, 34]. Proton abstraction from the Cα and Cβ of the cysteine residue causes diverse fragmentation reactions including the cleavage of disulfide bonds. Persulfides, thioaldehydes, and dehydroalanine residues at the cysteine position are generated upon negative ion CID [35]. Therefore we also investigated the dissociation behavior of the selenamide-derivatized peptide ions containing Se–S bonds under negative ion CID conditions. The basic cleavages of Se–S bond in ebselen- and NPSP-derivatized peptide ions are outlined in eq 2 (Schemes 1 and 2). For the fragmentation of ebselen-derivatized peptide anions, it is similar to that of their cationic counterparts and the loss of neutral ebselen is the major dissociation pathway (eq 2, Scheme 1). As shown in the CID MS/MS mass spectrum of [HC*KFWW – H]− (m/z 1179.0, Figure 4a), m/z 904.5 corresponding to [HCKFWW – H]− was generated by loss of one ebselen. In the case of CID of the NPSP-derivatized peptide anion, the elimination of the proton from α and β carbons of the cysteine residue gives rise to the thioaldehyde and dehydroalanine moieties, respectively (eq 2, Scheme 2). These processes can be seen in the CID MS/MS mass spectrum of [HC’KFWW – H]− (m/z 1060.0, Figure 4b), which results in intense fragment ions of m/z 902.3 and 870.4. It does confirm that Se–S cleavage occurs in negative ion CID as expected. The negative ion CID might be useful in the analysis of acidic peptides as those peptides prefer to form anions.
Figure 4.
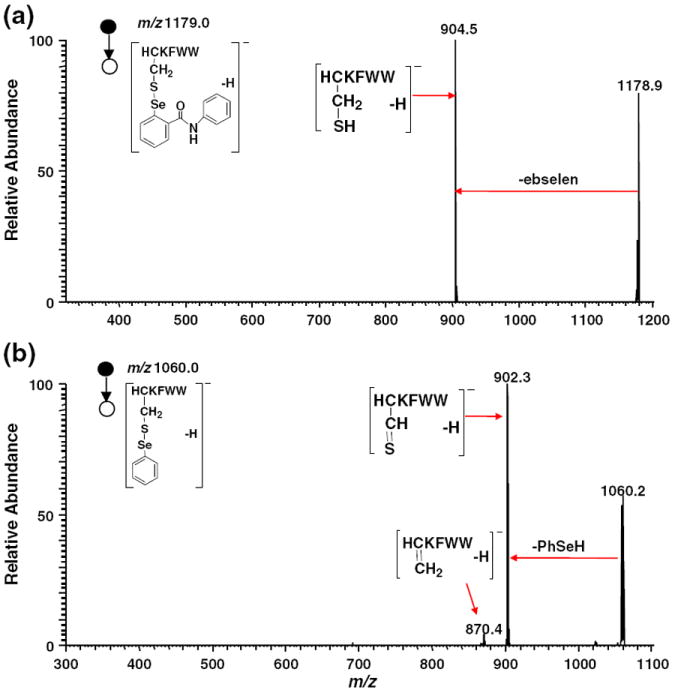
CID MS/MS mass spectra of the negative ion of (a) [HC*KFWW – H]− (m/z 1179.0) and (b) [HC’KFWW – H]− (m/z 1060.0)
ETD MS/MS of Selenamide-Derivatized Peptide Ions
Both ECD [21] and ETD [11] are powerful dissociation methods which often give rise to extensive backbone cleavages. In ECD experiments, low-energy electrons are reacted with peptide cations in the ICR cell located in the center of the magnetic field of a Fourier-Transfer Ion cyclotron resonance mass spectrometer (FT-ICR-MS) [21]. ETD utilizes ion/ion reactions [11, 36, 37] to transfer electron to peptide ions for dissociation but is regarded to have similar mechanism to ECD. An early proposed mechanism for ECD (also known as the Cornell mechanism [21]) starts with electron localization at positively charged ammonium or guanidinium groups of a peptide that generates a hypervalent center [28], which transfers a hydrogen atom H. to a carbonyl oxygen of an amide group to form an aminoketal radical intermediate. The aminoketal radical intermediate subsequently undergoes the cleavage of the N– Cα bond to form c/z ions [38]. It has been previously shown that disulfide bonds can be preferentially cleaved by ECD because the S–S bond has a higher affinity for the H. atom compared with the amide carbonyl [28]. However, the mechanism of ECD and ETD fragmentations are still not thoroughly understood [21-23, 39]. Even though the Cornell mechanism provided a reasonable picture for ECD, some backbone fragmentations were not easily explained [24, 40]. An alternative mechanism, now known as the Utah-Washington mechanism [22, 23], has been proposed to explain the origin of c and z fragment ions. In this theory, direct electron attachment to Coulomb-stabilized amide π* orbitals makes the amide group an exceptionally strong base with a proton affinity (PA) in the range of 1100–1400 kJ/mol [41]. The amide anion radical is able to abstract a proton to result in an intermediate identical to the aminoketyl cation radical proposed in the Cornell mechanism and can undergo the same N–Cα bond cleavage. Recently, one of the powerful approaches for the ECD/ETD mechanistic elucidation is to introduce various tags with different electron or proton affinities to model peptides and to test the effect of the tags on the dissociation upon electron capture/transfer [24-27]. In consideration of convenient preparation of selenamide-derivatized proteins/peptides and the structural similarity of the resulting Se–S bond to S–S bond, electron-based dissociation behaviors of the derivatized peptide ions were also examined, with the purpose of providing some insights into the mechanism of the electron-based dissociation.
Figure 5a and d show the ETD MS/MS mass spectra of [HC*KFWW+2 H]2+ and [HC’KFWW+2 H]2+, respectively. In both cases, preferential cleavage of selenium tag takes place, giving rise to the main fragment ion of m/z 906.5, the protonated peptide ion without the tag. Also, m/z 1063.3 corresponding to the radical cation [HC’ KFWW+ 2 H]+▪ as a result of one electron capture is shown in Figure 5d. Few c ions with very low abundances from backbone cleavage were observed. The ETD process for the doubly charged ion of HCKFWW with one selenium tag is illustrated in eq 1, Scheme 3. The cleavage of the Se–S bond can be expedited by direct electron-capture as stabilized by the Coulomb effect (by the positively charged arginine or terminal amino of the peptide) followed with bond cleavage, leading to the loss of radical RSe▪ and thiolate anion. Subsequent proton transfer occurring to the thiolate anion forms singly charged HCKFWW. This proposed mechanism follows the Utah-Washington pattern and was further confirmed by ETD of the corresponding alkali metal-containing peptide ion. Figure 6 shows ETD MS/MS mass spectrum of [HC’KFWW+Na+K]2+ (m/z 562.2), and preferential cleavage of the phenylselenenyl tag was also observed to take place to form the fragment ion of m/z 967.3 ([HCKFWW+Na+K – H]+) (see the proposed mechanism in eq 2, Scheme 3). Also, m/z 1124.3 corresponding to [HC’KFWW+Na+K]+▪ is shown in Figure 6 as the charge reduced species. We also tested ETD of [C*QDSETRTFY+2Na]2+ (m/z 784.8) and [C’QDSETRTFY+2Na]2+ (m/z 725.3) and the losses of the selenium tags were observed as well (Figure S–3, Supporting Information). Again, as shown in Figure 5b and e, the NRCSQGSCWN peptide ions with two ebselen or phenylselenenyl tags also display similar ETD mass spectra. No c or z ions were observed, instead only few b/y ions with low abundances were detected. In both cases (Figure 5b and e), preferential cleavage of one selenium tag occurs predominantly, generating the main fragment ions of m/z 1429.4 and 1310.4, respectively. The ETD mass spectra of [FVNQHLC*GSHLVEALYLVC*GERGFFYTPKA+3 H]3+ and [FVNQHLC’GSHLVEALYLVC’GERGFFYTPKA+ 3 H]3+ were shown in Figure 5c and f, respectively. Likewise, there are no c or z ions observed, either. The cleavage of one ebselen tag occurs to either Cys-7 or Cys-19, giving rise to the fragment ion of m/z 1838.0 (shown in Figure 5c). Because of the presence of two selenenyl sulfide bonds in selenamide-derivatized NRCSQGSCWN and insulin chain B, the electron transfer to the tag completely suppresses the backbone cleavage resulting in no formation of c/z ions. These results indicate that the selenium tags can efficiently quench the formation of c/z ions in the ETD of peptide ions, and the mechanism can be accounted for by the Utah-Washington mechanism. On the other hand, although the prohibition of backbone cleavage occurs to derivatized peptide ions, extensive c/z ions can still be seen for ECD of selenamide-derivatized protein ions, as we will report in due course.
Figure 5.
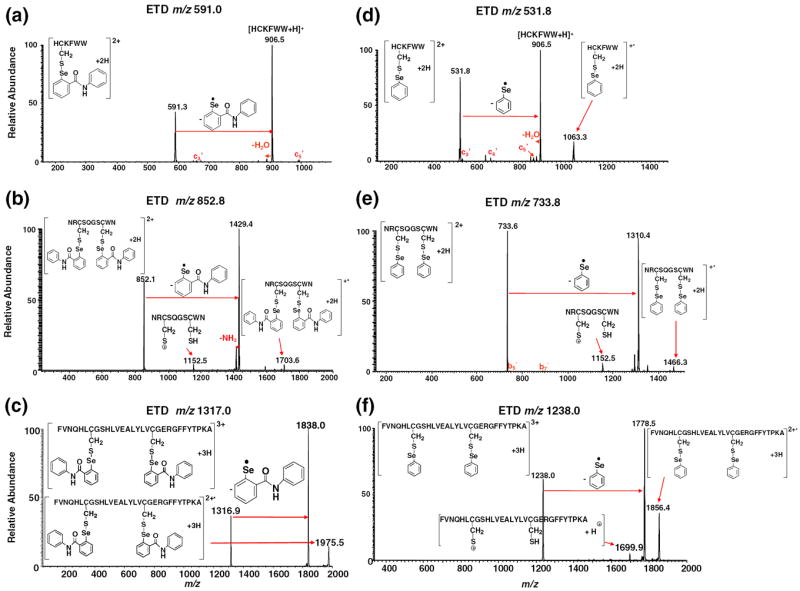
ETD MS/MS mass spectra of (a) [HC*KFWW+2 H]2+ (m/z 591.0); (b) [NRC*SQGSC*WN+2 H]2+ (m/z 852.8); (c) [FVNQHLC*GSHLVEALYLVC*GERGFFYTPKA+3 H]3+ (m/z 1317.0); (d) [HC’KFWW+2 H]2+ (m/z 531.8); (e) [NRC’SQGSC’WN+ 2 H]2+ (m/z 733.8); (f) [FVNQHLC’GSHLVEALYLVC’GERGFFYTPKA+3 H]3+ (m/z 1238.0)
Scheme 3.
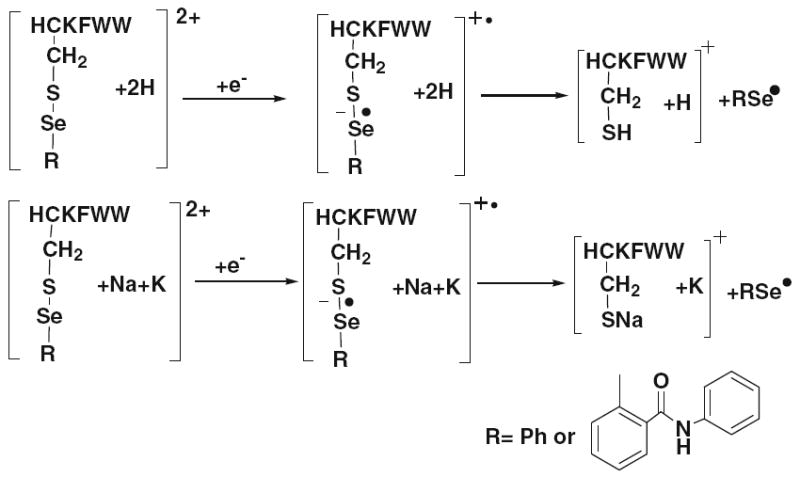
Dissociation processes for the doubly charged HCKFWW carrying two protons and one selenium tag (eq 1) and for the doubly charged HCKFWW carrying one sodium, one potassium, and one selenium tag (eq 2) upon ETD
Figure 6.
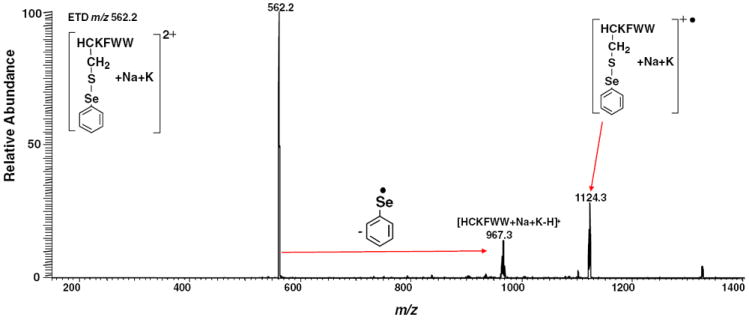
ETD MS/MS mass spectrum of [HC’KFWW+Na+K]2+(m/z 562.2)
Conclusions
In conclusion, unimolecular ion dissociation behaviors of selenamide-labeled thiol peptide ions upon CID and ETD were investigated. It is evident that derivatized peptide cations undergo tag-dependent CID dissociation pathways. Ebselen-derivatized peptide cations display the unique fragment ion of m/z 276 upon dissociation, which is useful for selective identification of thiol peptides and proteins in mixture. The robust phenylselenenyl tag is useful in peptide sequencing and locating of cysteine residues in peptides. In the negative ion mode CID, both types of tags are preferentially lost via the Se–S cleavage, similar to S–S bond. In addition, the preferential cleavage of Se–S bond occurs over the formation of c/z ions during ETD activation to both protonated and alkaliated peptides, following the Utah-Washington mechanism. Given the significance of thiol residues in proteins/peptides, the selective derivatization by selenamides and the rich ion dissociation chemistry revealed in this study would be useful in the proteomics research.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support of this work by NSF (CHE-0911160). Y.Z. and H.C. are thankful to Professor Michael L. Gross for the access to the NIH/NCRR Mass Spectrometry Resources at Washington University in St. Louis (grant number 2P41RR000954).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-011-0170-4) contains supplementary material, which is available to authorized users.
References
- 1.Basford RE, Huennekens FM. Studies on thiols. I. Oxidation of Thiol groups by 2,6-Dichlorophenol Indophenol. J Am Chem Soc. 1955;77:3873–3877. [Google Scholar]
- 2.Gorman JJ, Wallis TP, Pitt JJ. Protein Disulfide Bond Determination by Mass Spectrometry. Mass Spectrom Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 3.Bilusich D, Bowie JH. Fragmentations of (M – H)− Anions of Underivatized Peptides. Part 2: Characteristic Cleavages of Ser and Cys and of Disulfides and Other Post-Translational Modifications, Together with Unusual Internal Processes. Mass Spectrom Rev. 2009;28:20–34. doi: 10.1002/mas.20206. [DOI] [PubMed] [Google Scholar]
- 4.Diedrich JK, Julian RR. Site-Specific Radical Directed Dissociation of Peptides at Phosphorylated Residues. J Am Chem Soc. 2008;130:12212–12213. doi: 10.1021/ja8023719. [DOI] [PubMed] [Google Scholar]
- 5.Gunawardena HP, O’Hair RAJ, McLuckey SA. Selective Disulfide Bond Cleavage in Gold(I) Cationized Polypeptide Ions Formed Via Gas-Phase Ion/Ion Cation Switching. J Proteome Res. 2006;5:2087–2092. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- 6.Kim HI, Beauchamp JL. Identifying the Presence of a Disulfide Linkage in Peptides by the Selective Elimination of Hydrogen Disulfide from Collisionally Activated Alkali and Alkaline Earth Metal Complexes. J Am Chem Soc. 2008;130:1245–1257. doi: 10.1021/ja075698w. [DOI] [PubMed] [Google Scholar]
- 7.Chrisman PA, McLuckey SA. Dissociations of Disulfide-Linked Gaseous Polypeptide/Protein Anions: Ion Chemistry with Implications for Protein Identification and Characterization. J Proteome Res. 2002;1:549–557. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Shefcheck K, Callahan J, Fenselau C. Extension of Microwave-Accelerated Residue-Specific Acid Cleavage to Proteins with Carbohydrate Side Chains and Disulfide Linkages. Int J Mass Spectrom. 2008;278:109–133. doi: 10.1016/j.ijms.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiao L, Bi H, Busnel JM, Liu B, Girault HH. In-Source Photocatalytic Reduction of Disulfide Bond During Laser Desorption Ionization. Chem Commun. 2008:6357–6359. doi: 10.1039/b813283f. [DOI] [PubMed] [Google Scholar]
- 10.Ueberheide BM, Fenyo D, Alewood PF, Chait BT. Rapid Sensitive Analysis of Cysteine Rich Peptide Venom Components. Proc Natl Acad Sci U S A. 2009;106:6910–6915. doi: 10.1073/pnas.0900745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasicek L, Brodbelt JS. Enhanced Electron Transfer Dissociation Through Fixed Charge Derivatization of Cysteines. Anal Chem. 2009;81:7876–7884. doi: 10.1021/ac901482s. [DOI] [PubMed] [Google Scholar]
- 13.Diedrich JK, Julian RR. Site Selective Fragmentation of Peptides and Proteins at Quinone Modified Cysteine Residues Investigated by ESI MS. Anal Chem. 2010;82:4006–4014. doi: 10.1021/ac902786q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simek P, Husek P, Zahradnickova H. Gas Chromatographic-Mass Spectrometric Analysis of Biomarkers Related to Folate and Cobalamin Status in Human Serum after Dimercaptopropanesulfonate Reduction and Heptafluorobutyl Chloroformate Derivatization. Anal Chem. 2008;80:5776–5782. doi: 10.1021/ac8003506. [DOI] [PubMed] [Google Scholar]
- 15.Williams DK, Jr, Meadows CW, Bori ID, Hawkridge AM, Comins DL, Muddiman DC. Synthesis, Characterization, and Application of Iodoacetamide Derivatives Utilized for the ALiPHAT Strategy. J Am Chem Soc. 2008;130:2122–2123. doi: 10.1021/ja076849y. [DOI] [PubMed] [Google Scholar]
- 16.Lane A, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Desorption Electrospray Ionization Mass Spectrometry Reveals Surface-Mediated Antifungal Chemical Defense of a Tropical Seaweed. Proc Natl Acad Sci U S A. 2009;106:7314–7319. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiwert B, Hayen H, Karsta U. Differential Labeling of Free and Disulfide-Bound Thiol Functions in Proteins. J Am Soc Mass Spectrom. 2008;19:1–7. doi: 10.1016/j.jasms.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Jha SK, Udgaonkar JB. Exploring the Cooperativity of the Fast Folding Reaction of a Small Protein Using Pulsed Thiol Labeling and Mass Spectrometry. J Biol Chem. 2007;282:37479–37491. doi: 10.1074/jbc.M706714200. [DOI] [PubMed] [Google Scholar]
- 19.Sevcikova P, Glatz Z, Tomandl J. Determination of Homocysteine in Human Plasma by Micellar Electrokinetic Chromatography and In-Capillary Detection Reaction with 2,2’-Dipyridyl Disulfide. J Chromatogr A. 2003;990:197–204. doi: 10.1016/s0021-9673(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 20.Xu K, Zhang Y, Tang B, Laskin J, Roach PJ, Chen H. Study of Highly Selective and Efficient Thiol Derivatization Using Selenium Reagents by Mass Spectrometry. Anal Chem. 2010;82:6926–6932. doi: 10.1021/ac1011602. [DOI] [PubMed] [Google Scholar]
- 21.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 22.Chen XH, Turecek F. The Arginine Anomaly: Arginine Radicals are Poor Hydrogen Atom Donors in Electron Transfer Induced Dissociations. J Am Chem Soc. 2006;128:12520–12530. doi: 10.1021/ja063676o. [DOI] [PubMed] [Google Scholar]
- 23.Sawicka A, Skurski P, Hudgins RR, Simons J. Model Calculations Relevant to Disulfide Bond Cleavage Via Electron Capture Influenced by Positively Charged Group. J Phys Chem B. 2003;107:13505–13511. [Google Scholar]
- 24.Sohn CH, Chung CK, Yin S, Ramachandran P, Loo JA, Beauchamp JL. Probing the Mechanism of Electron Capture and Electron Transfer Dissociation Using Tags with Variable Electron Affinity. J Am Chem Soc. 2009;131:5444–5459. doi: 10.1021/ja806534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iavarone AT, Paech K, Williams ER. Effects of Charge State and Cationizing Agent on the Electron Capture Dissociation of a Peptide. Anal Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamot-Rooke J, Malosse C, Frison G, Turecek F. Electron Capture in Charge-Tagged Peptides. Evidence for the Role of Excited Electronic States. J Am Soc Mass Spectrom. 2007;18:2146–2161. doi: 10.1016/j.jasms.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Xia Y, Gunawardena HP, Erickson DE, McLuckey SA. Effects of Cation Charge-Site Identity and Position on Electron-Transfer Dissociation of Polypeptide Cations. J Am Chem Soc. 2007;129:12232–12243. doi: 10.1021/ja0736764. [DOI] [PubMed] [Google Scholar]
- 28.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron Capture Dissociation of Gaseous Multiply-Charged Proteins is Favored at Disulfide Bonds and Other Sites of High Hydrogen Atom Affinity. J Am Chem Soc. 1999;121:2857–2862. [Google Scholar]
- 29.Zhang Y, Dewald HD, Chen H. On-Line Mass Spectrometric Analysis of Proteins/Peptides Following Electrolytic Reduction of Disulfide Bonds. J Proteome Res. 2011;10:1293–1304. doi: 10.1021/pr101053q. [DOI] [PubMed] [Google Scholar]
- 30.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. Wiley; New York: 1980. [Google Scholar]
- 31.Bagnall KW. The Chemistry of Selenium, Tellurium, and Polonium. Elsevier; Amsterdam: 1966. [Google Scholar]
- 32.Killa HMA, Rabenstein DL. Determination of selenols, Diselenides, and Selenenyl Sulfides by Reversed-Phase Liquid Chromatography with Electrochemical Detection. Anal Chem. 1988;60:2283–2287. doi: 10.1021/ac00171a025. [DOI] [PubMed] [Google Scholar]
- 33.Kim HI, Beauchamp JL. Mapping Disulfide Bonds in Insulin with the Route 66 Method: Selective Cleavage of S–C Bonds Using Alkali and Alkaline Earth Metal Enolate Complexes. J Am Soc Mass Spectrom. 2009;20:157–166. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Andreazza HJ, Bowie JH. The Application of Negative Ion Electrospray Mass Spectrometry for the Sequencing of Underivatized Disulfide-Containing Proteins: Insulin and Lysozyme. Phys Chem Chem Phys. 2010;12:13400–13407. doi: 10.1039/c0cp00717j. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Kaltashov IA. Mapping of Protein Disulfide Bonds Using Negative Ion Fragmentation with a Broadband Precursor Selection. Anal Chem. 2006;78:4820–4829. doi: 10.1021/ac060132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron Transfer Ion/Ion Reactions in a Three-Dimensional Quadrupole Ion Trap: Reactions of Doubly and Triply Protonated Peptides with SO2−▪. Anal Chem. 2005;77:1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion Dependence in the Partitioning Between Proton and Electron Transfer in Ion/Ion Reactions. Int J Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 38.Zubarev RA. Reactions of Polypeptide Ions with Electrons in the Gas Phase. Mass Spectrom Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 39.Simons J. Mechanisms for S–S and N–Ca Bond Cleavage in Peptide ECD and ETD Mass Spectrometry. Chem Phys Lett. 2010;484:81–95. [Google Scholar]
- 40.Hudgins RR, Håkansson K, Quinn JP, Hendrickson CL, Marshall AG. Electron Capture Dissociation of Peptides and Proteins Does Not Require a Hydrogen Atom Mechanism. Proceedings of the 50th ASMS Conference on Mass Spectrometry and Allied Topics; Orlando, FL. June 2002. [Google Scholar]
- 41.Yao CX, Turecek F. Hypervalent Ammonium Radicals: Competitive N–C and N–H Bond Dissociations in Methyl Ammonium and Ethyl Ammonium. Phys Chem Chem Phys. 2005;7:912–920. doi: 10.1039/b414764b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.