Abstract
The μ-opioid receptor (MOR) is known to undergo extensive alternative splicing as numerous splice variants of MOR have been identified. However, the functional significance of MOR variants, as well as how splice variants other than MOR-1 might differentially regulate HIV-1 pathogenesis in the CNS, or elsewhere, has been largely ignored. Our findings suggest that there are specific differences in the MOR variant expression profile among CNS cell types, and that the expression levels of these variants are differentially regulated by HIV-1. While MOR-1A mRNA was detected in astroglia, microglia and neurons, MOR-1 and MOR-1X were only found in astroglia. Expression of the various forms of MOR along with the chimeric G protein qi5 in HEK-293T cells resulted in differences in calcium/NFAT signaling with morphine treatment, suggesting that MOR variant expression might underlie functional differences in MOR-effector coupling and intracellular signaling across different cell types. Furthermore, the data suggest that the expression of MOR-1 and other MOR variants may also be differentially regulated in the brains of HIV infected subjects with varying levels of neurocognitive impairment. Overall, the results reveal an unexpected finding, that MOR-1 may not be the predominant form of MOR expressed by some CNS cell types and that other splice variants of MOR-1, with possible differing functions, may contribute to the diversity of MOR-related processes in the CNS.
Keywords: MOR/OPRM1, splice variant, HIV-1, opiate drug abuse, astroglia, microglia
Introduction
There is evidence to suggest that the μ-opioid receptor-1 (MOR-1) may have a direct role in human immunodeficiency virus (HIV) type-1 (HIV-1) replication and pathogenesis (Li et al, 2003; Peterson et al, 1993; Peterson et al, 1994; Peterson et al, 1998; Peterson et al, 1990). Prior studies suggest that opioids suppress immune function (Adler et al, 1993; Bidlack, 2000; Donahoe and Vlahov, 1998; Peterson et al, 2001; Roy et al, 2006; Sharp et al, 1998). In addition to immune suppression, we and others have proposed that opioids might affect the neuropathogenesis of HIV-1 in the central nervous system (CNS) through direct actions of opiate drugs on opioid receptor-expressing neurons and glia within the CNS (Gurwell et al, 2001; Hauser et al, 2005; Nath et al, 2000; Nath et al, 2002). The reported interactions between MOR-1 activation and HIV-1 are thought to influence the disease in the CNS, affecting the course of HIV-associated neurocognitive disorders (HAND), and enhancing HIV encephalitis (HIVE) in opioid drug abusers (Anthony et al, 2005; Arango et al, 2004; Bell et al, 1998). Additionally, the HIV-1 envelope glycoprotein 120 (gp120) has been shown to up-regulate MOR-1 in TPA-differentiated HL-60 cells and human vascular endothelium (Beltran et al, 2006; Cadet et al, 2001). Thus, increases in MOR-1 subsequent to HIV infection may intrinsically modify (and perhaps even augment) the sensitivity and consequences of opioid co-exposure. MOR-1 can also undergo heterologous cross-sensitization with chemokine receptors that are involved in HIV infectivity and inflammation (such as CCR5 and CCR2), which could affect processes such as HIV-cellular binding and entry (Chen et al, 2004; Rogers et al, 2000).
Despite the identification of numerous splice variants of MOR, most studies examining MOR in pain, addiction, and HIV interactions have focused on the canonical MOR-1 (for a recent review on MOR-HIV interactions see (Regan et al, 2011)). However, a recent report found that alternative splicing of MOR underlies the pruritic versus antinociceptive effects of morphine (Liu et al, 2011), suggesting MOR-1 splice variants may also differentially mediate HIV-related effects. More importantly, it is not known if or how HIV might regulate MOR variants expressed on particular CNS cell types, and this could be of critical importance to understanding HIV-related neurological complications. To address this gap in understanding, we began to profile MOR splice variants expressed among human CNS cell types, and made the novel observations that: (1) there are cell-type specific differences in the MOR variants that are expressed by human astrocytes, microglia, and neurons; (2) the expression levels of these variants may be differentially regulated by HIV-1; and, (3) there may be differences in morphine-induced cellular signaling among these variants. Furthermore, our profiling of MOR variants in individual CNS cell types suggests the surprising finding that MOR-1A, rather than MOR-1, is the predominant form of MOR on microglia and neurons. Astrocytes appear to co-express MOR-1, MOR-1A, and MOR-1X, and all three variants are also expressed in human brain samples. In addition, our results from human brain tissue of HIV infected subjects with varying levels of neurocognitive impairment suggests that the expression levels of MOR-1 and other MOR variants may be differentially regulated during the progression of disease. We hypothesize that these variants may mediate MOR-related HIV effects such as morphine-induced viral replication and MOR-dependent increases in inflammation by a mechanism separate from that of MOR-1. These findings should help us to more accurately define the mechanisms by which opioid abuse exacerbates the progression of HIV-related complications. Expression of specific MOR variants may make cells more vulnerable to HIV-opiate interactions in the CNS and elsewhere.
Results
MOR is expressed on astrocytes, microglia, and neurons
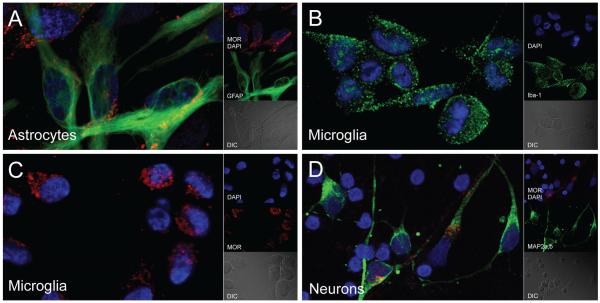
To confirm that MOR was expressed on primary human CNS cell types, we performed immunocytochemistry with an antibody whose epitope is within amino acids 1-15 of the N-terminus of human MOR, and that will recognize most MOR splice variants, which differ largely in the C-terminus. We found that MOR was indeed expressed on astrocytes, microglia, and neurons (Fig. 1). However, as expected, most astrocytes and microglia expressed MOR while only 5-10% of neurons were MOR-positive (data not shown). These results confirm that MOR containing exon 1 is expressed on CNS cell types, but they do not determine which specific C-terminal splice variants might be expressed.
Fig. 1.
Appearance of MOR across human CNS cell types. Representative images of primary human a astrocytes, b, c microglia, and d neurons immunolabeled for the N-terminus of MOR (red) and their respective cell-type specific markers (green) (GFAP, Iba-1, and MAP2a,b, respectively). DAPI (blue) staining indicates cell nuclei. DIC, differential interference contrast microscopy
MOR-1, MOR-1A, and MOR-1X are differentially expressed across CNS cell types
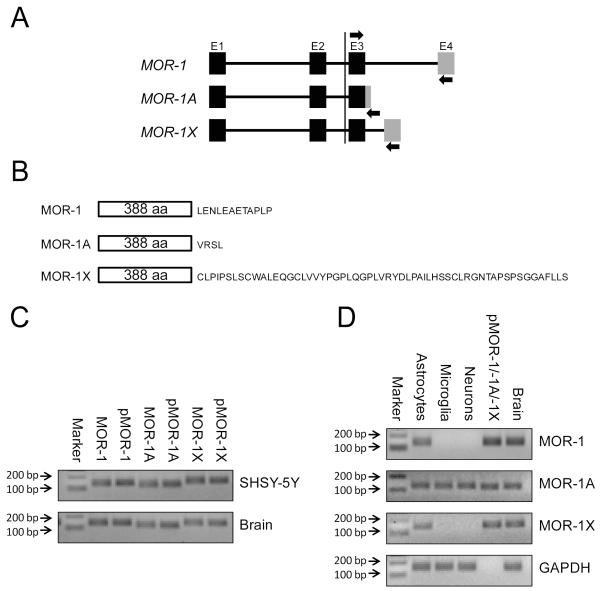
Having established that MOR containing exon 1 was expressed on CNS astrocytes, microglia, and neurons, we next selected three specific C-terminal MOR splice variants to examine in these cells. We assayed for the presence of one of the shortest (MOR-1A) and longest (MOR-1X) C-terminal splice variants, as well as MOR-1, using specific PCR primer sets (Bare et al, 1994; Pan et al, 2003) (Fig. 2a, b). Antibodies that detect these individual variants are not commercially available. We first confirmed that MOR-1, MOR-1A, and MOR-1X were present in the human neuroblastoma cell line SHSY-5Y (which is heavily enriched in MORs), in a sample of whole human brain tissue, and using plasmids containing the human cDNA of these variants as a positive control. The results showed that these variants are expressed in human brain and that the primer sets used in our study (Table 1) amplified PCR fragments of the expected relative sizes of the different variants examined (Fig. 2c).
Fig. 2.
Expression of MOR-1, MOR-1A, and MOR-1X in human brain and individual CNS cell types. a Schematic representation of the 4 exons comprising the MOR-1, MOR-1A, and MOR-1X genes. Arrows denote the locations of PCR primers used in this study in which a common forward primer targeted exon 3 and the reverse primers targeted each variant-specific exon 4 (sequences of the primers are listed in Table 1). b Schematic representation of the MOR-1, MOR-1A, and MOR-1X proteins showing the differences in the amino acid sequence encoded by the C-terminal exon 4. c PCR products amplified using primer sets for the indicated MOR variants in SHSY-5Y cells and a sample of whole human brain tissue (p, human plasmid cDNA of the specific variants served as a positive control). Note the expected relative sizes of the bands. d MOR variants detected in primary human astrocytes, microglia, and neurons by PCR with human brain tissue and plasmid cDNA serving as controls. GAPDH served as a loading control.
Table 1.
Primer sets used for PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| MOR-1 | 5′-CCCAACCTCTTCCAACATTGAGCAA-3′ | 5′-AACGGAGCAGTTTCTGCTTCCAGAT-3′ |
| MOR-1A | 5′-CCCAACCTCTTCCAACATTGAGCAA-3′ | 5′-CTAGAGACTGCGTACCTGATGATTAG-3′ |
| MOR-1X | 5′-CCCAACCTCTTCCAACATTGAGCAA-3′ | 5′-GCTCTAGAGCCCAGCAAGACAG-3′ |
| GAPDH | 5′-CATGGCACCGTCAAGGCTGAGAA-3′ | 5′-CAGTGGACTCCACGACGTACTCA-3′ |
| TBP | 5′-GCTGCGGTAATCATGAGGATAAGA-3′ | 5′-TGAGCACAAGGCCTTCTAACCTTA-3′ |
Having established the presence of the variants of interest in a human brain-derived cell line and in whole human brain tissue, we next determined the expression profile of these variants in individual primary human CNS cell types. Our results revealed that primary human astrocytes co-express MOR-1, MOR-1A, and MOR-1X, while we were only able to detect MOR-1A in microglia and neurons (Fig. 2d).
MOR-1, MOR-1A, and MOR-1X expression is differentially regulated by HIV-1 in individual CNS cell types
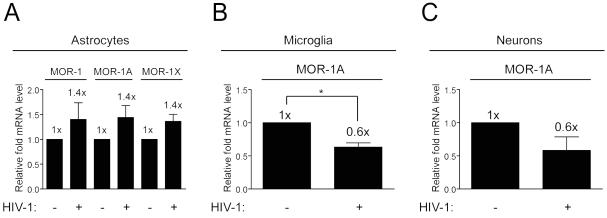
Having determined the expression profile of MOR-1, MOR-1A, and MOR-1X in astrocytes, microglia, and neurons, we next determined if the mRNA expression levels of the variants we detected in the individual cell types might be regulated by HIV-1 treatment. We found that HIV-1 treated astrocytes showed a trend towards increasing expression of MOR-1, MOR-1A, and MOR-1X compared to untreated controls, whereas in microglia and neurons we found a consistent downward trend in MOR-1A expression levels in response to HIV-1 (Fig. 3). However, these findings were only statistically significant in microglia. These results suggest that in addition to MOR-1, the expression levels of other MOR variants can also be regulated by HIV-1, which may be of critical importance to HIV-opioid interactions in the CNS, and especially those with microglia.
Fig. 3.
HIV-1 regulation of MOR variant expression in CNS cell types. qRT-PCR detection of expression for the indicated MOR variants in untreated and HIV-1 treated primary human a astrocytes, b microglia, and c neurons. Expression levels were determined by the normalization to GAPDH mRNA and compared to levels in untreated control samples which were set to a value of “1”. Error bars show SEM from at least 3 individual lots of cells. For astrocytes, p=0.3537 for MOR-1, p=0.2121 for MOR-1A, and p=0.1251 for MOR-1X expression levels when untreated and HIV-1 treated cells were compared. p=0.0295 for microglia and p=0.1336 for neurons when MOR-1A expression levels were compared between untreated and HIV-1 treated cells; *p< 0.05.
Morphine stimulates differential calcium/NFAT signaling of MOR splice variants
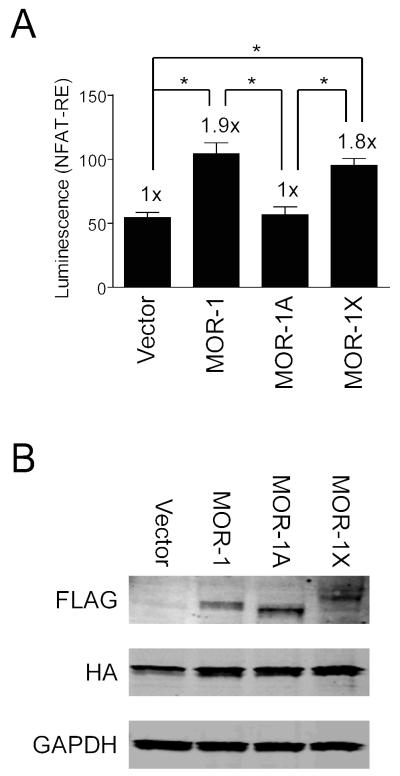
We next determined whether there might be a physiological significance to differential expression and HIV-1 regulation of the identified MOR splice variants. HEK-293T cells were co-transfected with plasmid constructs encoding MOR-1, MOR-1A, and MOR-1X or a control vector as well as the chimeric G protein qi5 and a luciferase reporter under control of a NFAT response element to serve as an indicator of calcium signaling (for a recent review on calcium/NFAT signaling see (Putney, 2012)). In response to morphine treatment, MOR-1 and MOR-1X each showed statistically significant (~2-fold) increases in NFAT signaling over vector control levels while MOR-1A signaling remained at control levels (Fig. 4a). This suggests a differential calcium response among the splice variants, since NFAT signaling is largely directed by changing calcium levels. To determine whether these differences might be attributed to varying MOR expression, western blotting was performed which showed relatively similar levels of expression for MOR-1, MOR-1A, and MOR-1X (Fig 4b). Thus, our results suggest that the MOR variant expression profile of an individual cell or a cell type can influence its response to morphine.
Fig. 4.
MOR-1A differs in NFAT-RE mediated calcium signaling compared to MOR-1 and MOR-1X. a HEK-293T cells co-transfected with plasmids encoding the indicated MOR variants or control vector, the qi5 chimeric G protein, and a luciferase reporter under control of a NFAT response element (RE) were examined for luciferase expression following morphine treatment. Error bars show SEM from 5 independent experiments in triplicate (n=15). F(3,56)=16.92, p<0.0001. *p< 0.05. b Western blotting of lysates from a for MOR (FLAG) expression levels. Note the expected relative sizes of the bands. qi5 (HA) served as a transfection control and GAPDH as a loading control.
Expression of MOR splice variants in the brains of HIV infected subjects with varying levels of neurocognitive impairment
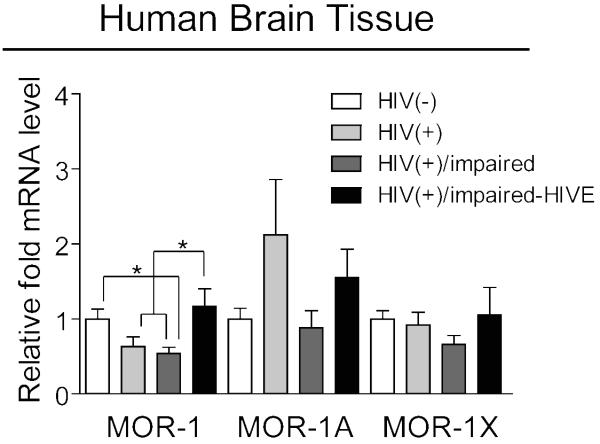
To determine whether there might be a disease-relevant in vivo correlation of the MOR variant heterogeneity that we observed in vitro, we obtained human brain tissue from subjects included in the NNTC Gene Array Project (details of the project are described in the Materials and methods). We found that there were statistically significant differences in MOR-1 mRNA expression levels across the four groups of subjects (Fig. 5). Expression was decreased in HIV infected subjects with neurocognitive impairment as compared to HIV-negative controls, with a similar decreasing trend occurring in infected subjects without impairment. Interestingly, HIV infected subjects with combined neurocognitive impairment and HIV encephalitis (HIVE) showed an increase in expression of MOR-1 compared to HIV infected subjects with and without neurocognitive impairment that was quite similar to HIV-negative controls. Although MOR-1X had a pattern of expression similar to that of MOR-1 across the four groups of subjects, while MOR-1A showed a trend of increased expression in infected subjects without neurocognitive impairment, we were unable to detect statistically significant differences in the expression levels of these variants.
Fig. 5.
MOR variant expression levels in HIV-1-infected human brain tissue. MOR-1, MOR-1A, and MOR-1X expression levels were examined by qRT-PCR across the indicated groups of subjects. Expression levels were determined by the normalization to TBP mRNA and presented as comparison with that in untreated samples which were set to a value of “1”. Error bars show SEM. F(3,24)=4.95, p=0.0081 for MOR-1; F(3,24)=1.87, p=0.1611 for MOR-1A; and F(3,24)=1.01, p= 0.4043 for MOR-1X. p=0.0805 when HIV-negative and -positive groups were compared for MOR-1 by Student’s unpaired, two-tailed t test. *p< 0.05. Details on the numbers and anatomical regions of samples analyzed for each group of subjects are listed in Supplementary Table 3
Discussion
In this study, we set out to determine whether the MOR-1A and MOR-1X C-terminal MOR-1 splice variants are differentially expressed in the CNS compared to MOR-1, how the expression of these variants might be regulated by HIV-1, and whether there might be differences in cellular signaling among MOR splice variants. We were prompted, in part, by the inherent diversity in μ-, δ-, and κ-opioid receptor expression among neurons, astroglia, and microglia (Stiene-Martin and Hauser, 1991; Stiene-Martin et al, 2001; Stiene-Martin et al, 1998; Turchan-Cholewo et al, 2008). Moreover, the range and diversity of responses among neurons, astroglia, and microglia to MOR agonists alone or following HIV-1 co-exposure suggest that more fundamental differences in MOR receptor-effector coupling might exist among CNS cell types (Hauser et al, 2007; Hauser et al, in press). Accordingly, we questioned whether opioid receptor splice variants might differ among cell types and whether this might contribute to the multiplicity of opiate drug actions in the CNS. Our analysis was limited to three MOR variants, and more extensive studies are needed to look at other variants expressed in the CNS. We chose to examine MOR-1A and MOR-1X because they possess some of the greatest differences in the length of their C-terminal tails compared to MOR-1, and because all three variants are completely different in terms of amino acid sequence at their C-termini. This approach was previously used to assess regional and functional differences of other MOR splice variants with similar discrepancies in their C-termini found in mice (Abbadie et al, 2000; Koch et al, 2001). Thus, the MOR-1A and MOR-1X variants are presumed to have some of the greatest potential differences in interactions with intracellular proteins, and in cellular signaling. Interestingly, MOR-1A was only detected in human microglia and neurons, while astrocytes co-expressed MOR-1, MOR-1A, and MOR-1X transcripts. However, it is possible that MOR-1 and MOR-1X are expressed at very low levels in microglia and neurons as well and were just below the level of the parameters used for detection in our study. Nonetheless, our results suggest that the variants we detected are the predominant forms expressed in the cell types examined and that MOR splice variants can be non-uniformly distributed across cell types.
While the pharmacological effects of μ-, δ-, and κ-opioid receptor agonists and antagonists on HIV-1 replication have been systematically characterized (Chao et al, 2001; Li et al, 2003; Peterson et al, 2001; Peterson et al, 1993; Peterson et al, 1994; Peterson et al, 1998; Sharp et al, 2001), surprisingly few studies have attempted to assess the molecular basis by which opioids affect the HIV-1 life-cycle. Cross-desensitization of MOR with HIV-1 co-receptors such as CXCR4 and CCR5 suggests direct or downstream interactions whereby MOR may affect viral entry (Chen et al, 2004; Grimm et al, 1998; Rogers and Peterson, 2003; Rogers et al, 2000). Moreover, no studies have attempted to examine the pharmacogenetic relationship between MOR splice variants and HIV-1 replicative fitness. Inherent differences in the contributions of MOR-expressing neurons, astroglia, and microglia in the neuropathologic effects of opiate drug and HIV-1 co-exposure indicates that the consequences of MOR activation are functionally distinct in each cell type (El-Hage et al, 2005; Turchan-Cholewo et al, 2009; Zou et al, 2011). Determination of the MOR variant expression profile in different cell types, and elucidation of the role of the identified variants in relation to HIV-1, may be of critical importance to further defining these processes.
Exposure to the CXCR4-tropic IIIB HIV-1 strain resulted in a trend of equally increased mRNA expression of MOR-1, MOR-1A, and MOR-1X ~30% in astrocytes, while MOR-1A expression decreased 40% in microglia, with a similar downward trend seen in neurons. However, the biological consequences of MOR up- or down-regulation in different cell types are only speculative at present, especially since the only statistically significant effect seen was MOR-1A down-regulation in microglia. Nevertheless, assuming all the above changes had been significant, we speculate that MOR-1 and MOR-1X driven increases in NFAT reporter activity caused by increases in calcium would promote pro-inflammatory signaling in opiate drug and HIV-1 co-exposed astrocytes (El-Hage et al, 2008). Alternatively, the down-regulation of MOR-1A (and possibly other MOR variants not examined in this study) may serve as a protective mechanism against the direct (e.g., increased viral replication) and indirect (non-infectious, bystander effects) affects of HIV-opiate drug interactions in microglia and neurons, respectively (Peterson et al, 1990; Turchan-Cholewo et al, 2009). Interestingly, a prior study found that exposure to HIV-1 Tat protein significantly increased MOR mRNA expression in primary rat microglia and a N9 microglial cell line, while MOR transcripts were unaffected by Tat alone in primary rat astroglia (Turchan-Cholewo et al, 2008). Collectively, the results suggest that there are more robust changes in MOR expression by microglia in response to HIV-1 or Tat protein than astroglia. Although we initially surmised that microglia might show a more pronounced response since they can be productively infected, the response to Tat suggests that MOR expression is likely to be influenced by both bystander effects and through HIV-1 infection.
Although MOR can interact with the HIV-1 co-receptors CXCR4 and CCR5 through downstream signaling, and may interact directly at the molecular level through the formation of G protein-coupled receptor heterodimers (Burbassi et al, 2010; Chen et al, 2004), the potential for heterologous CXCR4 interactions across different MOR splice variants has not been explored. Moreover, since the HIV-1 IIIB strain preferentially targets CXCR4, it is possible that CCR5-tropic strains of HIV-1 would affect MOR expression differently. Recent evidence from our laboratory indicates that neurotoxic interactions between morphine and gp120 differ significantly depending on HIV-1 strain (Podhaizer et al, 2011). As neurons are rarely infected by HIV, we attribute the effects seen on this cell type to predominantly reflect viral interactions with cell surface receptors such as the HIV-1 co-receptors CXCR4 and CX3CR1, and subsequent downstream signaling events (i.e., bystander effects subsequent to viral exposure and not infection) (Heinisch and Kirby, 2009; Suzuki et al, 2011; Tran et al, 2005).
Interestingly, we found that we were able to detect differences in calcium signaling with morphine treatment among the different variants examined. MOR-1 and MOR-1X showed similar increases in calcium/NFAT signaling while MOR-1A signaling did not increase above vector control levels. As MOR-1 and MOR-1X have longer C-terminal tails, they may be more readily able interact with G proteins to initiate downstream cellular signaling events compared to MOR-1A with its short C-terminal tail. Amino acid sequence differences may also play a role in G protein interactions. As MOR-1A was found to be the predominant form of MOR in microglia and neurons among the variants examined in this study, this suggests the interesting hypothesis that the effects of morphine may be of the greatest significance in astrocytes, or that the effects of morphine in microglia and neurons may be mediated by yet another MOR splice variant, with MOR-1A acting as a “decoy” receptor. Additional signaling pathways need to be examined to determine whether MOR-1A may have roles in other cascades and if there is differential signaling between MOR-1 and MOR-1X. Lastly, differences in the expression of MOR splice variants among glial cell types may contribute to the occasional report suggesting that glia do not express opioid receptors (Kao et al, 2012; Schwartz et al, 1994). As previously noted, astrocytes are heterogeneous in their expression of MOR and a majority of astroglia lack the receptor. When expressed by astroglia, for example, MOR is highly plastic and differs among brain regions, stage of maturation, and in a cell cycle-dependent manner (Stiene-Martin et al, 1998).
Our analysis of MOR variants in HIV infected human brain tissue suggests that the expression levels of MOR-1 vary at different stages of disease progression and/or with varying levels of neurocognitive impairment, and that the same may be true for other MOR variants as well. We found that MOR-1 mRNA expression was decreased in HIV infected subjects with neurocognitive impairment compared to uninfected control subjects, with a similar trend in unimpaired HIV infected subjects. Interestingly, HIV infected subjects with combined neurocognitive impairment and HIVE expressed MOR-1 at levels near those of uninfected subjects. These in vivo results suggest that differences in the expression of MOR-1, and perhaps other MOR variants, might correlate with different states of neurocognitive impairment and/or inflammatory indices associated with the HIV disease process. However, more samples need to be analyzed to verify these preliminary indications for MOR-1A and MOR-1X. Such differences in expression might be due to a number of factors, including altered populations of cells or activation states during the course of the disease process (for example, increased microglial and/or astroglial inflammation), as well as to up- or down-regulation of specific variants within individual cell types. We noted relatively high variation in our samples, which likely influenced the statistical significance of the analyses. These could be related to differences across the brain regions analyzed, as well as inter-subject variability due to gender, length of disease, HAART regimen, and other influences. We propose that more well-matched samples may be needed to reveal the true relationships between HIV and MOR splice variants in the HIV infected brain. Since MOR-1 and MOR-1X were only detected in astrocytes in vitro, our in vivo results with these variants are most likely to reflect expression within this cell type. As MOR-1A was found in all three CNS cell types, our results with this variant probably represent the combined effects of astrocytes, microglia, and neurons. Since astrocytes are generally thought to outnumber other cell types (and especially neurons) in the human cortex, the changes in MOR variant expression in this cell type could mask effects that are occurring in other cell types (Bass et al, 1971; Nedergaard et al, 2003). However, it is also possible that the in vivo response of individual cell types to HIV infection/exposure may differ from that of isolated cell types in culture. Other cells in the CNS, such as oligodendroglia, neuronal and glial progenitor cells, and endothelial cells, may also contribute modestly to levels of MOR variants in the brain and reactions to HIV. Yet another factor to consider is heterogeneity of expression among individual astroglia as well as regional heterogeneity, which has been shown to effect numerous measures including chemokine/cytokine responses to HIV-1 Tat exposure (Fitting et al, 2010). Overall, tracking changes in vivo during HIV disease progression may provide information that is not reflected in transient, in vitro HIV models with isolated cell types because of temporal changes due to dynamic interactions between cell types in vivo.
Besides alterations to the MOR C-terminus, it was recently shown that the variant MOR-1K, lacking the N-terminal extracellular and first transmembrane domains, couples G proteins and functions differently from MOR-1 (Gris et al, 2010). This underscores the point that both the N- and C-termini of MOR may have important and varying functions, which could be related to altered G protein coupling and/or other molecular interactions. Having identified the variants MOR-1A and MOR-1X in different cell types, questions still remain regarding the exact biological functions of these variants and how they differ from MOR-1, and in particular MOR-1A as our data suggests that this is the predominant form of MOR expressed in microglia and neurons. As PCR was used to detect the variants examined in this study, future studies will be needed to confirm our findings using MOR variant-specific antibodies. Most commercially available MOR antibodies are either raised to the N-terminus of MOR or to an epitope specifically recognizing the C-terminal tail of MOR-1. An antibody to the variant MOR-1C was reported to be cross-reactive with an unknown protein (Schnell and Wessendorf, 2009). Development of antibodies with a high degree of specificity to diagnostic amino acid sequences will be critical for the future evaluation of specific MOR-1 splice variants.
In conclusion, while it has been long recognized that specific receptors underlie heterogeneous cell responses to opioid agonists and antagonists, our results suggest an additional level of complexity. Expression of splice variants of MOR appear to differ among cell types, and to result in differential activation of signaling pathways downstream of calcium, and perhaps other regulators. These findings underscore the novel concept that combined HIV and opiate co-exposure may cause cell-specific effects in particular cells or cell types through interactions with specific MOR-1 variants, and that the interactions of individual MOR variants with HIV may influence the progression of neuroAIDS. C-terminal MOR-1 splice variants (and particularly MOR-1A) may mediate MOR-related HIV effects in the CNS by a mechanism separate from that of MOR-1. Differences in the distribution and expression of MOR splice variants across cell types and among individuals may contribute to the differential susceptibility of some human sub-populations to opiate abuse or opiate-HIV co-morbid interactions.
Materials and methods
Cell culture and virus treatment
Primary human astrocytes (catalog number 1800), microglia (catalog number 1900-f1), and neurons (catalog number 1520) were obtained from ScienCell Research Laboratories (Carlsbad, CA) and cultured according to the manufacturer’s instructions. HEK-293T cells (catalog number Q401) were obtained from GenHunter Corporation (Nashville, TN). HIV-1 direct-pelleted virus, CXCR4-tropic strain IIIB (catalog number 10-124-000) was obtained from Advanced Biotechnologies, Inc (Columbia, MD). Cells were treated with HIV-1 at a p24 equivalent of ~100 ng/well of a 24-well cell culture plate for 48 h.
RT-PCR
Total RNA was isolated using the miRNeasy Mini Kit (Qiagen, Inc.; Valencia, CA) and used to generate cDNA templates by reverse transcription using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Carlsbad, CA) according to the manufacturer’s instructions. PCR reactions were performed in a total volume of 20 μL containing SensiMix SYBR qPCR reagents (Bioline USA, Inc.; Tauton, MA) using a Corbett Rotor-Gene 6000 real-time PCR system (Qiagen, Inc.). PCR conditions consisted of an initial hold step at 95 °C for 10 min followed by 40 amplification cycles of 95 °C for 5 sec, 55 °C for 10 sec, and 72 °C for 20 sec. Sequences of the primer sets used are listed in Table 1. The specificity of the amplified products was verified by melting curve analysis and agarose gel electrophoresis. Agarose gels were stained with ethidium bromide and images were taken using a Kodak Image Station 440 (Kodak; Rochester, NY). qRT-PCR data were calculated as relative expression levels by normalization against GAPDH or TATA-binding protein (TBP) mRNA using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Plasmids
Cloning of human MOR-1, MOR-1A, and MOR-1X cDNAs were reported previously (Pan et al, 2003; Ravindranathan et al, 2009). The coding regions of these genes were sub-cloned into the pPTuner IRES2 vector (Clontech Laboratories, Inc.; Mountain View, CA) with the N-terminal addition of a 3xFLAG epitope tag sequence using In-Fusion Cloning reagents (Clontech Laboratories, Inc.). pcDNA1-qi5-HA vector encoding Gq alpha with the C-terminal amino acids changed from Gq alpha to Gi alpha residues and an internal hemaglutinin epitope tag (catalog number 24501) was obtained from Addgene, Inc. (Cambridge, MA) and has been described previously (Conklin et al, 1993). pGL4.30[luc2P/NFAT-RE] vector encoding the luciferase reporter gene under control of a Nuclear Factor of Activated T-cell response element (catalog number E8481) was obtained from Promega Corporation (Madison, WI).
Immunocytochemistry
Cells were fixed with 3.7% paraformaldehyde, permeabilized with 0.5% Triton X-100, and immunolabeled. Cells were lastly stained with DAPI to reveal their nuclei. Primary antibodies to MOR (Novus Biologicals, LLC; Littleton, CO; catalog number NBP1-31180), GFAP (Millipore; Billerica, MA; catalog number MAB360), Iba-1 (Wako Chemicals USA, Inc.; Richmond, VA; catalog number 019-19741), and MAP2a,b (Millipore, catalog number MAB378) were all used at a 1:200 dilution. MOR was visualized with Alexa Fluor 594 (Molecular Probes; Carlsbad, CA; catalog number A-21207), GFAP and MAP2a,b with Oregon Green 488 (Novex; Carlsbad, CA; catalog number O-6380), and Iba-1 with Alexa Fluor 488 (Molecular Probes; catalog number A-11034) conjugated secondary antibodies, all used at a 1:500 dilution. Samples were imaged using a Zeiss LSM 700 laser scanning confocal microscope equipped with a 63x oil immersion objective. Images were collected using ZEN 2009 Light Edition software (Carl Zeiss, Inc.; Thornwood, NY) and edited using Adobe Photoshop CS3 Extended 10.0 software (Adobe Systems, Inc.; San Jose, CA).
NFAT-RE based calcium assay
HEK-293T cells were co-transfected with MOR, qi5, and luc2P/NFAT-RE plasmids using polyethylenimine (Polysciences, Inc.; Warrington, PA; catalog number 23966) with induction of MOR protein stabilization using 1000 nM Shield1 (Clontech Laboratories, Inc.; catalog number 632189). Twenty-four hours post-transfection cells were treated with 500 nM morphine and harvested 18 h later in Glo Lysis Buffer (Promega Corporation; catalog number E2661), mixed in a 1:1 ratio with Bright-Glo Assay Reagent (Promega Corporation; catalog number E2610), and luminescence was measured on a PHERAstar FS microplate reader (BMG LABTECH, Inc.; Durham, NC).
Western blotting
Lysates from calcium assays were separated by SDS-PAGE for western blotting. Primary antibodies to FLAG (Sigma-Aldrich, Inc.; Saint Louis, MO; catalog number F3165), HA (Abcam, Inc.; Cambridge, MA; catalog number ab9110), and GAPDH (Santa Cruz Biotchnology, Inc.; Santa Cruz, CA; catalog number sc-47724) were all used at a 1:1000 dilution. FLAG and GAPDH were visualized with Alexa Fluor 680 (Molecular Probes; catalog number A-21057) and HA with IRDye 800CW (LI-COR Biosciences, Inc.; Lincoln, NE; catalog number 926-32211) conjugated secondary antibodies, all at a 1:3000 dilution, using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Human brain tissue
Human brain tissue was obtained from the National NeuroAIDS Tissue Consortium (NNTC) Gene Array Project (Morgello et al, 2001). Briefly, the array project consists of four groups of subjects (HIV-negative, n=6; HIV-positive, n=6; HIV-positive with neurocognitive impairment (HIV-positive/impaired), n=7; and HIV-positive with combined neurocognitive impairment and HIV encephalitis (HIVE) (HIV-positive/impaired-HIVE), n=5) with samples taken from three brain regions (frontal cortex, frontal lobe white matter, and basal ganglia). Further details on the subjects and project can be found in Supplementary Tables 1 and 2 and at http://www.nntc.org/. Details on the samples used in this study can be found in Supplementary Table 3.
Statistics
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc.; La Jolla, CA). Data derived from human brain tissue and luciferase assays were analyzed by one-way ANOVA with Student Neuman-Keuls post-hoc tests. Data derived from individual cell types was analyzed by Student’s paired, two-tailed t tests. A value of p<0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Michael F. Miles for providing us with the SHSY-5Y cells. MOR-1 and MOR-1X plasmid cDNAs were kind gifts of Gavril W. Pasternak and MOR-1A plasmid cDNA was a kind gift from Raymond L. White. We also thank Yuntao Wu for helpful discussions and are most grateful to the subjects who provided samples that were analyzed in this study. We gratefully acknowledge the support of the National Institutes of Health (NIH)—National Institutes on Drug Abuse grants T32 DA007027, R01 DA024461, R01 DA018633 and K02 DA027374. The human tissue provided by the National NeuroAIDS Tissue Consortium (NNTC) for this publication was made possible from NIH funding through the National Institute of Mental Health and National Institute of Neurological Disorders and Stroke by the following grants: Manhattan HIV Brain Bank: U01MH083501, R24MH59724; Texas NeuroAIDS Research Center: U01MH083507, R24NS45491; National Neurological AIDS Bank: 5U01MH083500, NS38841; California NeuroAIDS Tissue Network: U01MH083506, R24MH59745; Statistics and Data Coordinating Center: U01MH083545, N01MH32002. This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official view of the NNTC or the NIH.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Seth M. Dever and Ruqiang Xu contributed equally to this work.
Contributor Information
Seth M. Dever, Department of Pharmacology & Toxicology, Virginia Commonwealth University School of Medicine, 1217 East Marshall Street, Richmond, VA 23298-0613, USA
Ruqiang Xu, Department of Anatomy & Neurobiology, Virginia Commonwealth University School of Medicine, Richmond, VA 23298, USA.
Sylvia Fitting, Department of Pharmacology & Toxicology, Virginia Commonwealth University School of Medicine, 1217 East Marshall Street, Richmond, VA 23298-0613, USA.
Pamela E. Knapp, Department of Pharmacology & Toxicology, Virginia Commonwealth University School of Medicine, 1217 East Marshall Street, Richmond, VA 23298-0613, USA; Department of Anatomy & Neurobiology, Virginia Commonwealth University School of Medicine, Richmond, VA 23298, USA; Institute for Drug & Alcohol Studies, Virginia Commonwealth University School of Medicine, Richmond, VA 23298, USA
Kurt F. Hauser, Department of Pharmacology & Toxicology, Virginia Commonwealth University School of Medicine, 1217 East Marshall Street, Richmond, VA 23298-0613, USA; Institute for Drug & Alcohol Studies, Virginia Commonwealth University School of Medicine, Richmond, VA 23298, USA.
References
- Abbadie C, Pan Y, Drake CT, Pasternak GW. Comparative immunohistochemical distributions of carboxy terminus epitopes from the mu-opioid receptor splice variants MOR-1D, MOR-1 and MOR-1C in the mouse and rat CNS. Neuroscience. 2000;100:141–53. doi: 10.1016/s0306-4522(00)00248-7. [DOI] [PubMed] [Google Scholar]
- Adler MW, Geller EB, Rogers TJ, Henderson EE, Eisenstein TK. Opioids, receptors, and immunity. Adv Exp Med Biol. 1993;335:13–20. doi: 10.1007/978-1-4615-2980-4_3. [DOI] [PubMed] [Google Scholar]
- Anthony IC, Ramage SN, Carnie FW, Simmonds P, Bell JE. Does drug abuse alter microglial phenotype and cell turnover in the context of advancing HIV infection? Neuropathol Appl Neurobiol. 2005;31:325–38. doi: 10.1111/j.1365-2990.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- Arango JC, Simmonds P, Brettle RP, Bell JE. Does drug abuse influence the microglial response in AIDS and HIV encephalitis? AIDS. 2004;18(Suppl 1):S69–74. [PubMed] [Google Scholar]
- Bare LA, Mansson E, Yang D. Expression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994;354:213–6. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- Bass NH, Hess HH, Pope A, Thalheimer C. Quantitative cytoarchitectonic distribution of neurons, glia, and DNa in rat cerebral cortex. J Comp Neurol. 1971;143:481–90. doi: 10.1002/cne.901430405. [DOI] [PubMed] [Google Scholar]
- Bell JE, Brettle RP, Chiswick A, Simmonds P. HIV encephalitis, proviral load and dementia in drug users and homosexuals with AIDS. Effect of neocortical involvement. Brain. 1998;121(Pt 11):2043–52. doi: 10.1093/brain/121.11.2043. [DOI] [PubMed] [Google Scholar]
- Beltran JA, Pallur A, Chang SL. HIV-1 gp120 up-regulation of the mu opioid receptor in TPA-differentiated HL-60 cells. Int Immunopharmacol. 2006;6:1459–67. doi: 10.1016/j.intimp.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Bidlack JM. Detection and function of opioid receptors on cells from the immune system. Clin Diagn Lab Immunol. 2000;7:719–23. doi: 10.1128/cdli.7.5.719-723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbassi S, Sengupta R, Meucci O. Alterations of CXCR4 function in mu-opioid receptor-deficient glia. Eur J Neurosci. 2010;32:1278–88. doi: 10.1111/j.1460-9568.2010.07402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet P, Weeks BS, Bilfinger TV, Mantione KJ, Casares F, Stefano GB. HIV gp120 and morphine alter mu opiate receptor expression in human vascular endothelium. Int J Mol Med. 2001;8:165–9. doi: 10.3892/ijmm.8.2.165. [DOI] [PubMed] [Google Scholar]
- Chao CC, Gekker G, Sheng WS, Hu S, Peterson PK. U50488 inhibits HIV-1 expression in acutely infected monocyte-derived macrophages. Drug Alcohol Depend. 2001;62:149–54. doi: 10.1016/s0376-8716(00)00185-x. [DOI] [PubMed] [Google Scholar]
- Chen C, Li J, Bot G, Szabo I, Rogers TJ, Liu-Chen LY. Heterodimerization and cross-desensitization between the mu-opioid receptor and the chemokine CCR5 receptor. Eur J Pharmacol. 2004;483:175–86. doi: 10.1016/j.ejphar.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature. 1993;363:274–6. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- Donahoe RM, Vlahov D. Opiates as potential cofactors in progression of HIV-1 infections to AIDS. J Neuroimmunol. 1998;83:77–87. doi: 10.1016/s0165-5728(97)00224-5. [DOI] [PubMed] [Google Scholar]
- El-Hage N, Bruce-Keller AJ, Yakovleva T, Bazov I, Bakalkin G, Knapp PE, Hauser KF. Morphine exacerbates HIV-1 Tat-induced cytokine production in astrocytes through convergent effects on [Ca(2+)](i), NF-kappaB trafficking and transcription. PLoS One. 2008;3:e4093. doi: 10.1371/journal.pone.0004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Gurwell JA, Singh IN, Knapp PE, Nath A, Hauser KF. Synergistic increases in intracellular Ca2+, and the release of MCP-1, RANTES, and IL-6 by astrocytes treated with opiates and HIV-1 Tat. Glia. 2005;50:91–106. doi: 10.1002/glia.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting S, Zou S, Chen W, Vo P, Hauser KF, Knapp PE. Regional heterogeneity and diversity in cytokine and chemokine production by astroglia: differential responses to HIV-1 Tat, gp120, and morphine revealed by multiplex analysis. J Proteome Res. 2010;9:1795–804. doi: 10.1021/pr900926n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm MC, Ben-Baruch A, Taub DD, Howard OM, Resau JH, Wang JM, Ali H, Richardson R, Snyderman R, Oppenheim JJ. Opiates transdeactivate chemokine receptors: delta and mu opiate receptor-mediated heterologous desensitization. J Exp Med. 1998;188:317–25. doi: 10.1084/jem.188.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gris P, Gauthier J, Cheng P, Gibson DG, Gris D, Laur O, Pierson J, Wentworth S, Nackley AG, Maixner W, Diatchenko L. A novel alternatively spliced isoform of the mu-opioid receptor: functional antagonism. Mol Pain. 2010;6:33. doi: 10.1186/1744-8069-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwell JA, Nath A, Sun Q, Zhang J, Martin KM, Chen Y, Hauser KF. Synergistic neurotoxicity of opioids and human immunodeficiency virus-1 Tat protein in striatal neurons in vitro. Neuroscience. 2001;102:555–63. doi: 10.1016/s0306-4522(00)00461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, El-Hage N, Buch S, Berger JR, Tyor WR, Nath A, Bruce-Keller AJ, Knapp PE. Molecular targets of opiate drug abuse in neuroAIDS. Neurotox Res. 2005;8:63–80. doi: 10.1007/BF03033820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, El-Hage N, Stiene-Martin A, Maragos WF, Nath A, Persidsky Y, Volsky DJ, Knapp PE. HIV-1 neuropathogenesis: glial mechanisms revealed through substance abuse. J Neurochem. 2007;100:567–86. doi: 10.1111/j.1471-4159.2006.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch S, Kirby LG. Fractalkine/CX3CL1 enhances GABA synaptic activity at serotonin neurons in the rat dorsal raphe nucleus. Neuroscience. 2009;164:1210–23. doi: 10.1016/j.neuroscience.2009.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SC, Zhao X, Lee CY, Atianjoh FE, Gauda EB, Yaster M, Tao YX. Absence of mu opioid receptor mRNA expression in astrocytes and microglia of rat spinal cord. Neuroreport. 2012 doi: 10.1097/WNR.0b013e3283522e1b. DOI: 10.1097/WNR.0b013e3283522e1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch T, Schulz S, Pfeiffer M, Klutzny M, Schroder H, Kahl E, Hollt V. C-terminal splice variants of the mouse mu-opioid receptor differ in morphine-induced internalization and receptor resensitization. J Biol Chem. 2001;276:31408–14. doi: 10.1074/jbc.M100305200. [DOI] [PubMed] [Google Scholar]
- Li Y, Merrill JD, Mooney K, Song L, Wang X, Guo CJ, Savani RC, Metzger DS, Douglas SD, Ho WZ. Morphine enhances HIV infection of neonatal macrophages. Pediatr Res. 2003;54:282–8. doi: 10.1203/01.PDR.0000074973.83826.4C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Liu ZC, Sun YG, Ross M, Kim S, Tsai FF, Li QF, Jeffry J, Kim JY, Loh HH, Chen ZF. Unidirectional cross-activation of GRPR by MOR1D uncouples itch and analgesia induced by opioids. Cell. 2011;147:447–58. doi: 10.1016/j.cell.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Morgello S, Gelman BB, Kozlowski PB, Vinters HV, Masliah E, Cornford M, Cavert W, Marra C, Grant I, Singer EJ. The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol. 2001;27:326–35. doi: 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- Nath A, Anderson C, Jones M, Maragos W, Booze R, Mactutus C, Bell J, Hauser KF, Mattson M. Neurotoxicity and dysfunction of dopaminergic systems associated with AIDS dementia. J Psychopharmacol. 2000;14:222–7. doi: 10.1177/026988110001400305. [DOI] [PubMed] [Google Scholar]
- Nath A, Hauser KF, Wojna V, Booze RM, Maragos W, Prendergast M, Cass W, Turchan JT. Molecular basis for interactions of HIV and drugs of abuse. J Acquir Immune Defic Syndr. 2002;31(Suppl 2):S62–9. doi: 10.1097/00126334-200210012-00006. [DOI] [PubMed] [Google Scholar]
- Nedergaard M, Ransom B, Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci. 2003;26:523–30. doi: 10.1016/j.tins.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Xu M, Gilbert AK, Pasternak GW. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301:1057–61. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- Peterson PK, Gekker G, Lokensgard JR, Bidlack JM, Chang AC, Fang X, Portoghese PS. Kappa-opioid receptor agonist suppression of HIV-1 expression in CD4+ lymphocytes. Biochem Pharmacol. 2001;61:1145–51. doi: 10.1016/s0006-2952(01)00574-3. [DOI] [PubMed] [Google Scholar]
- Peterson PK, Gekker G, Schut R, Hu S, Balfour HH, Jr., Chao CC. Enhancement of HIV-1 replication by opiates and cocaine: the cytokine connection. Adv Exp Med Biol. 1993;335:181–8. doi: 10.1007/978-1-4615-2980-4_26. [DOI] [PubMed] [Google Scholar]
- Peterson PK, Hu S, Anderson WR, Chao CC. Nitric oxide production and neurotoxicity mediated by activated microglia from human versus mouse brain. J Infect Dis. 1994;170:457–60. doi: 10.1093/infdis/170.2.457. [DOI] [PubMed] [Google Scholar]
- Peterson PK, Molitor TW, Chao CC. The opioid-cytokine connection. J Neuroimmunol. 1998;83:63–9. doi: 10.1016/s0165-5728(97)00222-1. [DOI] [PubMed] [Google Scholar]
- Peterson PK, Sharp BM, Gekker G, Portoghese PS, Sannerud K, Balfour HH., Jr. Morphine promotes the growth of HIV-1 in human peripheral blood mononuclear cell cocultures. AIDS. 1990;4:869–73. doi: 10.1097/00002030-199009000-00006. [DOI] [PubMed] [Google Scholar]
- Podhaizer EM, Zou S, Fitting S, Samano KL, El-Hage N, Knapp PE, Hauser KF. Morphine and gp120 Toxic Interactions in Striatal Neurons are Dependent on HIV-1 Strain. J Neuroimmune Pharmacol. 2011 doi: 10.1007/s11481-011-9326-z. DOI: 10.1007/s11481-011-9326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney JW. Calcium Signaling: Deciphering the Calcium-NFAT Pathway. Curr Biol. 2012;22:R87–9. doi: 10.1016/j.cub.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Ravindranathan A, Joslyn G, Robertson M, Schuckit MA, Whistler JL, White RL. Functional characterization of human variants of the mu-opioid receptor gene. Proc Natl Acad Sci U S A. 2009;106:10811–6. doi: 10.1073/pnas.0904509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan PM, Dave RS, Datta PK, Khalili K. Epigenetics of micro-opioid receptors: Intersection with HIV-1 infection of the central nervous system. J Cell Physiol. 2011 doi: 10.1002/jcp.24004. DOI: 10.1002/jcp.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TJ, Peterson PK. Opioid G protein-coupled receptors: signals at the crossroads of inflammation. Trends Immunol. 2003;24:116–21. doi: 10.1016/s1471-4906(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Rogers TJ, Steele AD, Howard OM, Oppenheim JJ. Bidirectional heterologous desensitization of opioid and chemokine receptors. Ann N Y Acad Sci. 2000;917:19–28. doi: 10.1111/j.1749-6632.2000.tb05369.x. [DOI] [PubMed] [Google Scholar]
- Roy S, Wang J, Kelschenbach J, Koodie L, Martin J. Modulation of immune function by morphine: implications for susceptibility to infection. J Neuroimmune Pharmacol. 2006;1:77–89. doi: 10.1007/s11481-005-9009-8. [DOI] [PubMed] [Google Scholar]
- Schnell SA, Wessendorf MW. Lack of evidence for the mu-opioid receptor splice variant MOR1C in rats. J Comp Neurol. 2009;517:452–8. doi: 10.1002/cne.22175. [DOI] [PubMed] [Google Scholar]
- Schwartz JP, Nishiyama N, Wilson D, Taniwaki T. Receptor-mediated regulation of neuropeptide gene expression in astrocytes. Glia. 1994;11:185–90. doi: 10.1002/glia.440110212. [DOI] [PubMed] [Google Scholar]
- Sharp BM, McAllen K, Gekker G, Shahabi NA, Peterson PK. Immunofluorescence detection of delta opioid receptors (DOR) on human peripheral blood CD4+ T cells and DOR-dependent suppression of HIV-1 expression. J Immunol. 2001;167:1097–102. doi: 10.4049/jimmunol.167.2.1097. [DOI] [PubMed] [Google Scholar]
- Sharp BM, Roy S, Bidlack JM. Evidence for opioid receptors on cells involved in host defense and the immune system. J Neuroimmunol. 1998;83:45–56. [PubMed] [Google Scholar]
- Stiene-Martin A, Hauser KF. Glial growth is regulated by agonists selective for multiple opioid receptor types in vitro. J Neurosci Res. 1991;29:538–48. doi: 10.1002/jnr.490290415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiene-Martin A, Knapp PE, Martin K, Gurwell JA, Ryan S, Thornton SR, Smith FL, Hauser KF. Opioid system diversity in developing neurons, astroglia, and oligodendroglia in the subventricular zone and striatum: impact on gliogenesis in vivo. Glia. 2001;36:78–88. [PMC free article] [PubMed] [Google Scholar]
- Stiene-Martin A, Zhou R, Hauser KF. Regional, developmental, and cell cycle-dependent differences in mu, delta, and kappa-opioid receptor expression among cultured mouse astrocytes. Glia. 1998;22:249–59. [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, El-Hage N, Zou S, Hahn YK, Sorrell ME, Sturgill JL, Conrad DH, Knapp PE, Hauser KF. Fractalkine/CX3CL1 protects striatal neurons from synergistic morphine and HIV-1 Tat-induced dendritic losses and death. Mol Neurodegener. 2011;6:78. doi: 10.1186/1750-1326-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PB, Ren D, Miller RJ. The HIV-1 coat protein gp120 regulates CXCR4-mediated signaling in neural progenitor cells. J Neuroimmunol. 2005;160:68–76. doi: 10.1016/j.jneuroim.2004.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchan-Cholewo J, Dimayuga FO, Ding Q, Keller JN, Hauser KF, Knapp PE, Bruce-Keller AJ. Cell-specific actions of HIV-Tat and morphine on opioid receptor expression in glia. J Neurosci Res. 2008;86:2100–10. doi: 10.1002/jnr.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchan-Cholewo J, Dimayuga FO, Gupta S, Keller JN, Knapp PE, Hauser KF, Bruce-Keller AJ. Morphine and HIV-Tat increase microglial-free radical production and oxidative stress: possible role in cytokine regulation. J Neurochem. 2009;108:202–15. doi: 10.1111/j.1471-4159.2008.05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Fitting S, Hahn YK, Welch SP, El-Hage N, Hauser KF, Knapp PE. Morphine potentiates neurodegenerative effects of HIV-1 Tat through actions at mu-opioid receptor-expressing glia. Brain. 2011;134:3616–31. doi: 10.1093/brain/awr281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.