Abstract
Lipid solvation provides the primary driving force for the insertion and folding of integral membrane proteins. Although the structure of the lipid bilayer is often simplified as a central hydrophobic core sandwiched between two hydrophilic interfacial regions, the complexity of the liquid-crystalline bilayer structure and the gradient of water molecules across the bilayer finetune the energetic contributions of individual amino acid residues to the stability of membrane proteins at different depths of the bilayer. The tryptophan side-chain is particularly interesting because despite its widely recognized role in anchoring membrane proteins in lipid bilayers, there is little consensus on its hydrophobicity among various experimentally determined hydrophobicity scales. Here we investigated how lipid-facing tryptophan residues located at different depths in the bilayer contribute to the stability of integral membrane proteins using the outer membrane protein A (OmpA) as a model. We replaced all lipid-contacting residues of the first transmembrane β-strand of OmpA with alanines and individually incorporated tryptophans in these positions along the strand. By measuring the thermodynamic stability of these proteins, we found that OmpA is slightly more stable when tryptophans are placed in the center of the bilayer and that it is somewhat destabilized as tryptophans approach the interfacial region. However, this trend may be partially reversed when a moderate concentration of urea rather than water is taken as the reference state. The measured stability profiles are driven by similar profiles of the m-value, a parameter that reflects the shielding of hydrophobic surface area from water. Our results indicate that knowledge of the free energy level of the protein’s unfolded reference state is important for quantitatively assessing the stability of membrane proteins, which may explain differences in observed profiles between in vivo and in vitro scales.
The thermodynamic stability of polytopic membrane proteins is determined by a delicate balance between lipid solvation and packing of amino acid side-chains within cell membranes1,2. Favorable transfer of side-chains into the hydrophobic core of a lipid bilayer is crucial for the topological localization and stabilization of membrane proteins. Thus, hydrophobicity scales, which represent quantitative measures of individual side-chains to partition between water and hydrophobic environments, are powerful tools not only to identify the membrane spanning polypeptide segments, but also to predict the stability of membrane proteins3.
Among the many existing hydrophobicity scales, four experimentally determined scales are particularly useful to understand the contribution of lipid solvation to the thermodynamic stability of membrane proteins: the Wimley-White whole-residue scales based on water-octanol and water-membrane interface partitioning of model pentapeptides4,5, the Hessa-von Heijne scale based on translocon-endoplasmic reticulum (ER) membrane partitioning of model transmembrane segments6,7, and the Moon-Fleming scale based on the folding equilibrium of the β-barrel membrane protein OmpLA between water and the lipid bilayer8. In these scales, unique transfer free energy values were determined for each amino acid side-chain using organic solvent, the membrane interface or the center of the hydrophobic core of a bilayer as the reference hydrophobic environments.
However, the latter two studies also showed that the transfer free energies of amino acid side-chains strongly depend on the depth at which the side-chain is embedded in the bilayer7,8. Computational and statistical analyses on the structures of membrane proteins revealed a strong positional dependence of side-chain distributions along the bilayer normal7,9–11. These findings suggest that the complexity of the liquid-crystalline bilayer structure adjusts the energetic contribution of individual side-chains to membrane protein stability at different depths. However, it is still not clear for most side-chains how this depth-dependence is related to the thermodynamic stability of membrane proteins.
Here, we determined the energetic contribution of the tryptophan (Trp) side-chain to the thermodynamic stability of the β-barrel outer membrane protein A (OmpA) at different depths in a fluid lipid bilayer. Tryptophan is particularly interesting because it is known to play an important role in anchoring membrane proteins in lipid bilayers12–15. Although there is considerable consensus among the different hydrophobicity scales at least on the qualitative hydrophobic ranking of most side-chains, there is a surprisingly wide spread of values found for the hydrophobicity of Trp. On the Nozaki-Tanford16 and Wimely-White octanol and interface scales, Trp is the most hydrophobic residue, but it is only moderately hydrophobic on the Kyte-Doolittle17, Eisenberg-Weiss18, Engelman-Steiz-Goldman19, Hessa-von Heijne6, and Moon-Fleming8 scales. One possibility, which we are exploring in the present study, is that Trp’s contribution to the stability of membrane proteins is particularly dependent on membrane-depth and its context in the protein sequence. A better knowledge of factors that contribute to this stability is therefore crucial for understanding Trp’s role in membrane protein folding.
It has been recognized by several groups that the folding of β-barrel membrane proteins can be used as a powerful model system to study the energetics of lipid-protein interactions. In a few cases, including OmpA, β-barrel membrane proteins have been shown to reversibly unfold in urea or guanidinium solutions from their folded state in lipid bilayers 8,20,21. Thus, by mutating the lipid-contacting residues, the contribution of lipid solvation of specific residues to the thermodynamic stability of membrane proteins can be estimated8,22. In this work, we replaced all lipid-contacting residues of the first β-strand of OmpA with alanines (Ala) and individually incorporated Trps in these positions along the strand. By measuring the thermodynamic stability of these proteins, we found that OmpA is slightly more stable when Trp is placed in the center of the lipid bilayer and that it is somewhat destabilized as Trp approaches the interfacial regions of the bilayer. However, interfacial Trps may be more stabilizing than central Trps when 3 M urea rather than buffered water is taken as the reference state, i.e., a condition that may better reflect folding conditions in vivo.
Materials and Methods
DNA construct
All site-directed mutagenesis was carried out by using QuickChange kit (Agilent). First, the quadruple mutant W7A/T9A/L13A/W15A of full-length mature OmpA was generated and named W0. The plasmid pET1113 encoding wild-type proOmpA gene was used as a template23. Next, using W0 as a new template, Ala7, Ala9, Ala11, Ala13, and Ala15 were individually mutated to tryptophans to generate W7, W9, W11, W13, and W15, respectively. The E. coli BL21(DE3) [ΔlamB ompF::Tn5 ΔompA ΔompC]24 cells were transformed with the mutant plasmids for expression of OmpA. In this E. coli strain, chromosomally encoded OmpA and all other major outer membrane porin genes were deleted so that mutant OmpA could be purified without the interference from intrinsic wild-type OmpA.
Expression and purification of OmpA
The detailed procedure for expression and purification of OmpA was described previously22. Briefly, 20 ml of transformed E. coli cells grown overnight were inoculated in 1 L LB media containing 0.1 g/L ampicilin and cultured at 37 °C. The expression of OmpA was induced by addition of 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) when OD600nm reached approximately 0.6. The cells were further grown for 4 to 5 hours and harvested. The wet cell paste was resuspended in 15 ml/L culture of solution A (0.75 M sucrose, 10 mM TrisHCl (Tris(hydroxymethyl)aminomethane hydrochloride), pH 5.0, and 0.05 % β-mercaptoethanol (v/v). Solution B (10 ml/L culture; 40 mM EDTA (ethylenedinitrilotetraacetic acid), 10 mM TrisHCl pH7.0, 50 µg/ml lysozyme, and 0.05 % β-mercaptoethanol) was then added while the mixture was stirred and ice cooled for 1 hour. After the concentration of β-mercaptoethanol was increased to 0.2 %, the resuspension was French-pressed two times. Solid urea (Sigma, SigmaUltra grade) was sequentially added to the stirred lysate at room temperature until the final concentration reached 4 M. Titanium dust and cell debris were removed by centrifugation at 2,000 g (4 °C, 30 minutes). The supernatant was ultra-centrifuged at 32,000 rpm (4 °C, for a minimum of 6 hours) in a 45 Ti rotor (Beckman-Coulter) to spin down the pre-extracted total membrane fraction.
The pre-extracted membranes were resuspended in 10 ml/L culture of 8 M urea solution (SigmaUltra grade) containing 20 mM TrisHCl pH 8.0 and 0.1 % β-mercaptoethanol by the use of a Potter homogenizer at room temperature. A nominal 10 ml of pure isopropanol was added to the resuspension in a glass vial and the whole mixture was incubated at 55 °C in a preheated water bath for exactly 30 minutes. This step is critical to extract OmpA from the outer membrane. After the extraction, the sample was rapidly cooled down in ice until a temperature below 10 °C was reached. The mixture was ultra-centrifuged at 28,000 rpm at 4 °C for at least 90 minutes in a 45 Ti rotor. The unfolded OmpA solubilized in the supernatant was further purified by anion-exchange column chromatography (25 ml Q-Sepharose, fast flow, GE Healthcare) using a FPLC (Fast protein liquid chromatography) system (Biorad). The column was equilibrated with isopropanol mixed with an equal volume of buffer (15 mM TrisHCl pH 8.5, 0.1 % β-mercaptoethanol, and 8 M urea (reagent grade, Sigma)). After the sterile-filtered sample was loaded, the protein-bound column was washed with a buffer solution containing 15 mM TrisHCl pH 8.5, 0.05 % β-mercaptoethanol, and 8 M urea. Then, OmpA was eluted by a linear salt gradient from 0 to 100 mM NaCl (15 mM TrisHCl pH 8.5, 0.05 % β-mercaptoethanol, and 8 M urea). Combined fractions of high-purity OmpA were concentrated in a stirred ultrafiltration cell (Amicon; 10 kDa cut-off filter) to a final concentration of 20~50 mg/ml as estimated by the Bradford protein assay (Biorad).
Small unilamellar vesicles
Stock solutions of 1,2-dipalmitoleoyl-sn-glycro-3-phosphcholine (DPoPC, Avanti Polar Lipids) and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phosphor-rac-(1-glycrol)] (POPG, Avanti Polar Lipids) dissolved in chloroform were mixed to a molar ratio of 9:1 (DPoPC:POPG). A total of 12 µmol of lipid was dried under a stream of nitrogen gas and further in a high-vacuum dessicator for 2 hours. The dried lipid was thoroughly mixed with 1.2 ml of 10 mM glycine buffer (pH 9.2 and 1.0 mM EDTA) to yield a final lipid concentration of 10 mM. The lipid dispersion was sonicated for 50 minutes in an ice-water bath using a Branson ultrasonifier microtip at 50 % duty cycle. Titanium dust was removed by centrifugation at 8,000 rpm for 15 minutes twice, and the resulting SUVs were equilibrated overnight at 4 °C.
Refolding and urea-induced equilibrium unfolding of OmpA
Concentrated OmpA unfolded in 8 M urea was diluted 50~100 fold in 10 mM SUVs to a final protein concentration of 12 µM. The refolding reaction was incubated for 3 hours at 37 °C. Minor residues of aggregated protein formed during refolding were removed by centrifugation at 6,000 rpm for 10 minutes twice. Refolded OmpA-lipid complexes were further diluted in aliquots of different urea concentrations in 10 mM glycine buffer (1 mM EDTA, pH 9.2). The dilution was 10 fold for fluorescence and 2.5 fold for SDS-PAGE experiments. The unfolding reactions were incubated with desired concentrations of urea at 37 °C overnight to reach equilibrium.
Fluorescence spectroscopy
Fluorescence spectra were collected in a Fluorolog-3 spectrofluorometer (Horiba). The excitation wavelength was 290 nm and Trp fluorescence of OmpA at different urea concentrations was measured over 300 nm to 400 nm range. 4.2 nm slits were used for both excitation and emission. All spectra were background-subtracted with a proper reference sample of identical composition but without protein.
SDS-PAGE shift assay
The equilibrated samples were mixed with the same volume of SDS sample buffer. The mixtures were loaded on 12.5 % SDS-PAGE without heating.
Fitting of equilibrium unfolding curves
The fitting procedures to obtain the free energy of unfolding of OmpA were essentially the same as the procedure previously described in Hong and Tamm20 but described in more detail in the Supporting Information. Fluorescence spectra were parameterized by calculating an average emission wavelength, <λ>, defined as <λ>=Σ(Fi λ i)/ΣFi. λi and Fi are the wavelength and the corresponding fluorescence intensity, respectively, at the ith measuring step in the spectrum. The unfolding curves, <λ> versus [urea], were fitted to the following form of the two-state model using IgoPro software (Wavemetrics)20.
| (1) |
<λ>F and <λ>U are the average emission wavelengths of the folded and unfolded states, respectively, determined from linear extrapolations to 0 M urea of the plateau values of the two states. Cm is the urea concentration where the fractions of folded and unfolded states are equal. QR is the relative ratio of the total fluorescence intensity of the native state to that of the unfolded state and is needed for normalization when one uses <λ> values to represent species concentrations. The unfolding free energy was obtained from the fitted values of Cm and m25,
| (2) |
The contribution of the lipid-contacting Trp residues to the stability of OmpA was calculated at different membrane depths:
| (3) |
ΔGoUnfold,Wo([urea]) is the unfolding free energy of the W0 mutant with no Trp in the β1 strand at a specific urea concentration and ΔGoUnfold,Wx([urea]) corresponds to the unfolding free energy of Wx, in which X designates the position of the Trp in β1.
The unfolding free energy of a protein (ΔGoUnfold,Wx([urea])) at a certain urea concentration was obtained by:
| (4) |
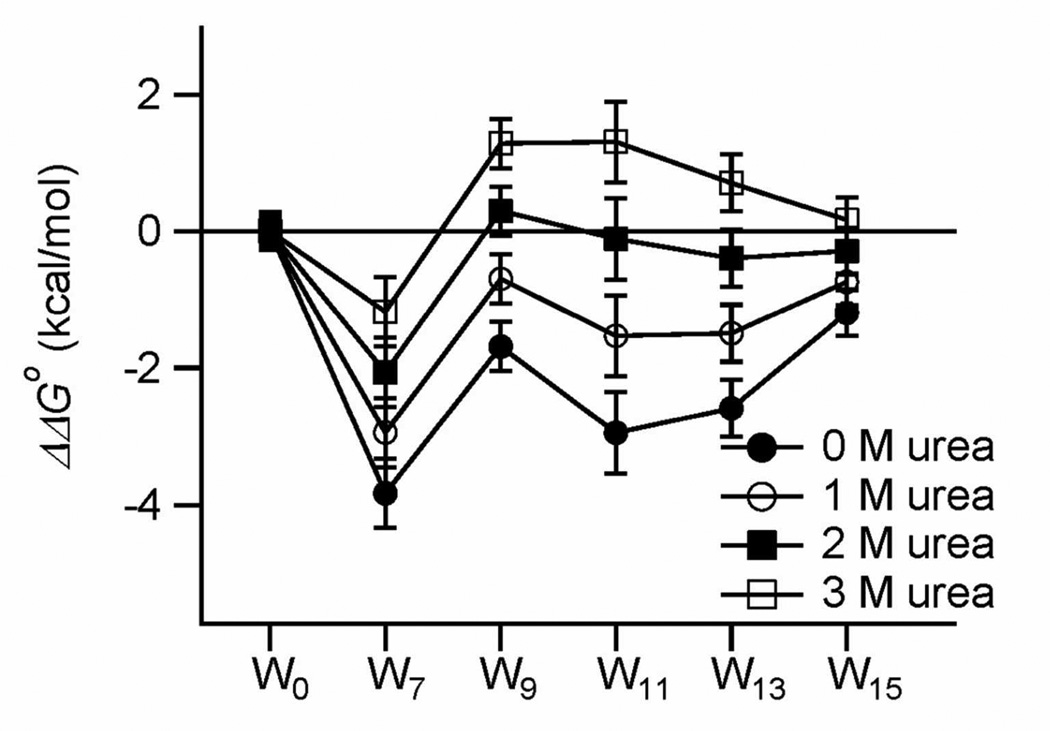
ΔGoUnfold,H2O is the unfolding free energy in 0 M urea concentration, which is obtained from Eq. 2 The statistical significance of the differences in ΔΔGoTrp-Ala (Figure 4) was evaluated by ANOVA test (Kirkman (1996) Statistics to use. http://www.physics.csbsju.edu/stats/ accessed on 5 March 2013). Each data set included the ΔΔGoTrp-Ala and the upper and lower standard error limits of ΔΔGoTrp-Ala for each mutant.
Figure 4.
Difference free energies (ΔΔGoTrp-Ala) versus Trp position for four concentrations of urea. The values at 0 M urea are the customary ΔΔGoUnfold,H2O values.
Results
Strategy to measure the depth-dependent contribution of Trp to the stability of membrane proteins
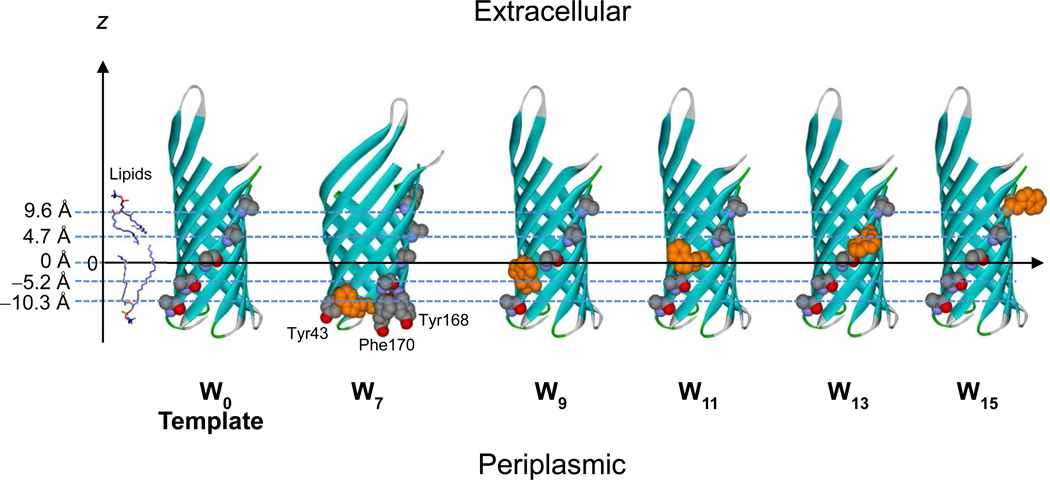
In order to study the energetics of the lipid solvation of Trp side-chains in the context of a native membrane protein structure, it is important to maximize the exposure of Trp to the surrounding lipids while minimizing its interactions with neighboring side-chains. Thus, we chose the first transmembrane β-strand (β1) and replaced all lipid-contacting residues to the small nonpolar side-chain Ala (Figure 1, template). Choosing β1 for this purpose had two advantages. First, β1 has fewer aromatic residues in the neighboring β2 and β8 than any other strand. This is important because we have previously shown that some Trps can favorably interact with other closely located aromatic residues and thereby enhance the protein’s stability, while simultaneously reducing the maximal lipid solvation of the side-chain22. β2 and β8 harbor only three aromatic residues, i.e., Tyr43 (β2), Tyr168 (β8) and Phe170 (β8), which are all located in the periplasmic interface region of the membrane26. The other lipid-contacting residues in β2 (Leu35, Ala37, Ala39, and Gly41) and β8 (Leu162, Leu164, and Val166) are mostly composed of small- and moderate-sized nonpolar side chains. Therefore, Trp7 is the only Trp in β1 that may interact with those nearby aromatic residues. The second reason for choosing β1 for mutagenesis was that the number of required point mutations was small compared to other strands and that two of the tested Trp positions were native Trps, which we reasoned would little perturb the correct folding of the mutant protein. The lipid-contacting residues in β1 are Trp7, Thr9, Ala11, Leu13, and Trp15. Thus, the quadruple mutant, W7A/T9A/L13A/W15A converted all lipid-facing residues of β1 to Ala. The resulting OmpA mutant (also referred to W0) served as a template for all Trp scanning mutations of this work. Based on this template a series of OmpA Trp mutants termed W7, W9, W11, W13 and W15 were generated as depicted in Figure 1.
Figure 1.
Scheme for Trp scanning to study the effect of Trps at different membrane depths on the thermodynamic stability of OmpA. The template mutant W0 with all alanines in the β1 strand of the OmpA structure (1QJP) is shown at the left. Single Trps (orange) are alternately substituted in positions 7 (W7), 9 (W9), 11 (W11), 13 (W13), and 15 (W15) of β1. Tyr43 in the β2-strand and Tyr168 and Phe170 in the β8-strand, which may participate in aromatic interactions with Trp, are highlighted in the W7 mutant. The distances of the Cα positions of the lipid-contacting residues in β1 from the virtual central plane of a bilayer as obtained from the PDB-TM database27 are shown on the z-axis.
To estimate the distances of individual Trp side-chains from the membrane center we used the PDB-TM database (http://pdbtm.enzim.hu) and applied it to the crystal structure of OmpA 27. According to this analysis the central plane of the lipid bilayer passes through OmpA very close to the Cα position of residue 11 (Figure 1). The distances of the Cα atoms of residues 7, 9, 11, 13 and 15 from the bilayer center are listed in Table 1.
Table 1.
Fitted parameters of urea-induced equilibrium unfolding of OmpA monitored by tryptophan fluorescence.
| Cα-bilayer center distance (Å) |
Cm (M) |
m (kcal/mol/M) |
ΔGoUnfold,H2O (kcal/mol) |
|
|---|---|---|---|---|
| W0 | none | 2.0 ± 0.1 | 1.2± 0.1 | 2.5 ± 0.2 |
| W7 | −10.3 | 3.0 ± 0.1 | 2.1 ± 0.2 | 6.3 ± 0.5 |
| W9 | −5.2 | 1.9 ± 0.1 | 2.2 ± 0.1 | 4.2 ± 0.4 |
| W11 | 0 | 2.1 ± 0.1 | 2.7 ± 0.3 | 5.4 ± 0.7 |
| W13 | 4.7 | 2.2 ± 0.1 | 2.3 ± 0.2 | 5.1 ± 0.5 |
| W15 | 9.6 | 2.2 ± 0.1 | 1.7 ± 0.1 | 3.7 ± 0.4 |
Folding efficiency of OmpA mutants with Trp at different membrane depths
First, we tested the refolding efficiency of the template and the five single β1-Trp mutants of OmpA in small unilamellar vesicles (SUVs) composed of DPoPC/POPG (9:1) using the SDS-PAGE shift assay. Without sample heating, correctly folded and membrane-inserted full-length OmpA migrates as a 30 kDa form while surface-adsorbed or unfolded proteins migrate as 35 kDa forms on SDS-PAGE gels28,29. As shown in the Supplementary Figure S1A, refolding was generally efficient for all mutants with yields varying from 87~98 %. Tryptophan fluorescence spectra of refolded mutants in SUVs were also similar to each other with the maximum emission wavelengths ranging between 334 and 336 nm (Supplementary Figure S1B), clearly different from the emission maxima of the respective unfolded proteins (Supplementary Figure S1C). These observations indicate that the rather invasive quadruple mutations were tolerated by the robust folding and membrane-insertion capability of OmpA. A calculation of the difference free energy changes of water-to-membrane transfer of the four mutations using the Wimley-White water-octanol scale for the core residues 9 and 13 (ΔΔGoThr9-Ala9 and ΔΔGoLeu13-Ala13) and the water-interface scale for the interfacial residues 7 and 15 (ΔΔGoTrp7-Ala7 and ΔΔGoTrp15-Ala15) showed that the sum or energy penalty for all four mutations is ΔΔGo = 6.5 kcal/mol4,5. Since the stability of wild-type OmpA in DPoPC:POPG bilayers is ΔGoFold,H2O = −9.2 kcal/mol22, one might expect that membrane insertion of the quadruple mutant W0 would still be favorable by about −3 kcal/mol. Interestingly, the experimental thermodynamic stability of W0 mutant was ΔGoFold,H2O = −2.5 kcal/mol (see Table 1), i.e. 6.7 kcal/mol less stable than wild-type, which is close to the Wimley-White scales-based prediction. Unfolding was also efficient (80 to 100%) for most mutants, but somewhat less efficient for W0 and W9 (~70%) as judged from SDS-PAGE assay (Supplementary Figure S1A). We do not completely understand the impaired unfolding efficiency of some of the mutants, but this does not affect the subsequent fluorescence-based analysis, which only considers the fraction that does unfold.
Equilibrium unfolding of OmpA mutants
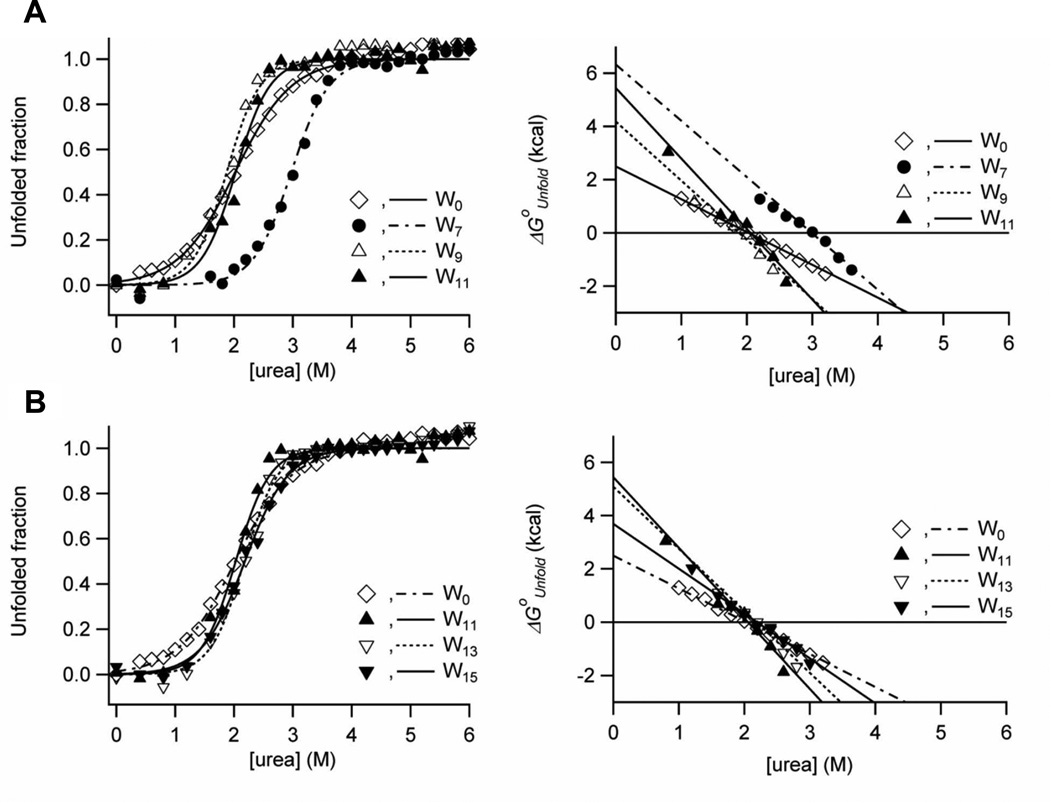
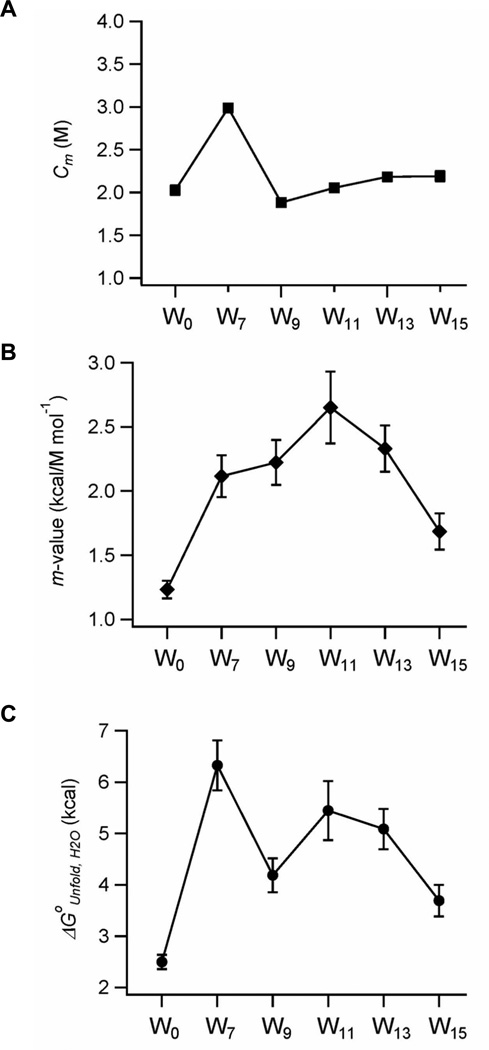
The thermodynamic stabilities of the OmpA mutants were measured by urea-induced equilibrium unfolding monitored by Trp fluorescence in bilayers composed of DPoPC/POPG (9:1) (Figure 2). Sigmoidal curves were observed for all mutants and the linearization of the unfolding free energy ΔGoUnfold versus urea concentration according to the two-state model (see Materials and Methods) yielded straight lines with a slope of m and an intercept of ΔGoUnfold,H2O. The fitted midpoints of the unfolding transition Cm and the linear dependence of ΔGoUnfold on urea concentration (m-value) are plotted against the Trp position along the β1-strand in Figures 3A and B. The product of Cm and m yields the unfolding free energy in water, ΔGoUnfold,H2O (Eq. 2)25, which is plotted against the Trp position in Figure 3C. These best fit thermodynamic parameters are listed in Table 1 for each of the mutants.
Figure 2.
Urea-induced equilibrium unfolding of OmpA mutants in DPoPC/POPG (9:1) lipid bilayers monitored by Trp fluorescence emission. (A) Unfolding transition curves for W0, W7, W9 and W11 mutants with Trps in the periplasm-facing monolayer (left) and corresponding plots of unfolding free energies versus urea concentration (right). (B) Unfolding transition curves for W0, W11, W13 and W15 mutants with Trps in the extracellular-space-facing monolayer (left) and corresponding plots of unfolding free energies versus urea concentration (right).
Figure 3.
Plots of fitted parameters describing the equilibrium unfolding of OmpA depending on the position of Trp residue along the β1-strand. (A) Midpoints of transition (Cm) and (B) m–values versus Trp position. (C) The free energy of unfolding (ΔGoUnfold,H2O) depending on the position of Trp residue
Among the mutants tested, W0 with no lipid-contacting Trp in β1 showed the lowest Cm and m-values (2.0 M and 1.2 kcal/mol/M, respectively) and thus the lowest thermodynamic stability (ΔGoUnfold,H2O = 2.5±0.2 kcal/mol). When a Trp residue was substituted at position 7, Cm increased significantly to 3.0 M. This mutant will be further discussed below. All other mutants had rather moderate shifts of Cm’s, which were still comparable to the level of W0 (Figure 3A). On the other hand, the m-values exhibited continuous changes generating a bell-shaped curve as a function of Trp position (Figure 3B). A maximal value of 2.7 kcal/mol/M was reached when Trp was placed near the center of bilayer (W11 mutant) and the m-values gradually decreased (to 2.1 kcal/mol/M for W7 and 1.7 kcal/mol/M for W15) as Trp was moved toward both interfacial regions.
Membrane depth-dependent contribution of Trp to the stability of OmpA
The contribution of lipid solvation of Trps to the stability of OmpA was evaluated by calculating the difference unfolding free energy (ΔΔGoTrp-Ala [Wx] = ΔGoUnfold[W0] – ΔGoUnfold[Wx]: x=7, 9, 11, 13 and 15) between two mutants in the presence and absence of individual Trps in β1 (Eqs. 3 and 4; Figure 4). Large negative values of ΔΔGoTrp-Ala[Wx] indicate favorable contributions of Trps to OmpA’s thermodynamic stability and excellent solvation of these Trps by membrane lipids. In general, stabilization of OmpA by lipid-solvated Trps depended quite strongly on membrane depth, structural context, and the concentration of the denaturant urea. At 0 M urea, Trps in all lipid-contacting positions along β1 stabilized OmpA (ΔΔGoTrp-Ala[Wx] < 0). OmpA was more stable when Trp was located near the center of the lipid bilayer (Trp11) and gradually destabilized as Trp was moved toward both interfaces. For example, the central Trp11 was 1.8 ± 0.7 kcal/mol more stable than the interfacial Trp15 (Figure 4 and Table 1). An Anova test with the four ΔΔGoTrp-Ala values for W9, W11, W13 and W15 mutants at 0 M urea yielded p=0.0042, which is below the significance threshold of p=0.01 and indicates that this trend is statistically significant. Apart from Trp7, the symmetric trend of ΔΔGoTrp-Ala around Trp11 further indicates that the curvature difference between the inner and outer leaflets of SUVs appears to not significantly affect the stability contribution of Trps at different depths. However, due to the limited number of sites investigated, we do not exclude the possibility that membrane curvature may have more subtle effects on the contribution of aromatic residues to the folding of membrane proteins.
Contrary to all other lipid-contacting Trps in β1, Trp7 did not follow this general rule and had an unusually strong stabilization effect (statistical significance of p=0.0002 with ΔΔGoTrp-Ala values for W7, W9, W11, W13 and W15 mutants). This Trp is located in the periplasmic interface, where it potentially interacts with other close-by aromatic residues. W7 was the most stable mutant among the mutants tested (ΔGoUnfold,H2O = 6.3±0.5 kcal/mol, Figure 3C and Table 1). When compared to the comparable Trp15, which is located at the other membrane interface, Trp7 had an excess degree of stabilization of −2.6±0.6 kcal/mol at 0 M urea (Figure 4). This particularly favorable contribution by Trp7 is most likely due to its aromatic interaction with Tyr43 on β2. Based on the crystal structure of OmpA, the inter-centroid distance between the aromatic cores of Trp7 and Tyr43 is 4.8 Å (Figure 1). We previously showed by double-mutant cycle analysis that aromatic-aromatic interactions stabilize OmpA on the order of ΔΔGo = −1 to −2 kcal/mol when the inter-centroid distance between two neighboring aromatic rings is smaller than 7 Å22. The interaction energy between Trp7 and Tyr43 is ΔΔGo = −1.0 kcal/mol22. Although the inter-centroid distances between Trp7 and Tyr168 and between Trp7 and Phe170 are 8.3 Å and 13.4 Å, respectively, in the crystal structure (Figure 1), it is possible that Trp7 also favorably interacts with Tyr168 and/or Phe170 on β8 since these residues likely sample multiple conformations30. Although it is conceivable that Trp9 may also interact with the aromatic residues in β2 and β8, our results (Figure 3C and Figure 4) do not indicate any interference by this potential interaction.
We were interested to know whether the energetic contributions of the different Trps depended on the folded reference state, i.e., whether different results would be obtained when ΔGoUnfold was compared at a specific denaturant concentration (Eq. 4) rather than back-extrapolated to buffered water. Interestingly, the membrane depth-dependent contribution of Trp was highly sensitive to the concentration of urea (Figure 4). The most favorable contribution of the membrane-central Trp11 to the stability of OmpA gradually decreased as the urea concentration was increased and eventually was reversed at 3 M urea. However, the bell-shaped energy profile across the bilayer observed at 3 M urea may not be statistically significant (p=0.037 with ΔΔGoTrp-Ala values for W9, W11, W13 and W15 mutants) due to the relatively large standard errors compared to the small differences in ΔΔGoTrp-Ala values. However, the systematic trends observed as urea increases let us believe that the shape of the energy profile indeed reverses when the high urea reference states are compared with the low ones. At 3 M urea, which denatures OmpA significantly but not completely, the interfacial Trp15 appears to be more stabilizing than the central Trp11 by ~1.1±0.7 kcal/mol. All lipid-contacting Trps of β1 were moderately destabilizing at 3 M urea (ΔΔGoTrp-Ala[Wx] > 0) (Figure 4). While the data at 0 M urea are clearly statistically significant, we are more cautious about the data observed at the higher urea concentrations. Regardless, we still find the observed systematic trends for folding in more destabilizing conditions interesting and perhaps biologically meaningful as will be further discussed below.
Discussion
Aromatic residues play a unique role in protein-lipid interactions. Based on statistical analysis of membrane proteins of known structure, Trp and Tyr are significantly enriched in the water-bilayer interface implying a crucial role of these residues in anchoring membrane proteins in lipid bilayers9–11,31. Their strong membrane-partitioning character is also important for the translocation of signaling and fusion proteins to lipid bilayers32–35. Physico-chemical properties such as aromaticity, electric quadrupole moment, and large hydrophobic surface area are thought to be responsible for these phenomena36. Among aromatic residues, Trp has a high propensity to interact with phospholipids both in the interfacial and hydrophobic core regions31.
Here, we systematically studied the effect of Trp placed at different depths in a lipid bilayer on the stability of the β-barrel membrane protein OmpA. Lipid solvation of Trp stabilized OmpA at all depths, but was most stabilizing when placed near the center of the bilayer. Stabilization by lipid solvation of Trp was gradually weakened as this residue was moved toward either membrane interface.
Our result that the central Trp is most stabilizing agrees with predictions from the Wimley-White (WW) whole-residue hydrophobicity scales4,5. While Trp is the most hydrophobic amino acid on both the water-octanol and water-interface scales, the partition of Trp into octanol that mimics the hydrocarbon region of the bilayer is more favorable than its partition into lipid bilayer interfaces. Indeed, the free energy differences between the two WW scales can be used to distinguish between helical segments of membrane proteins that span the lipid bilayer and those that partition to the interface37. We can further use these scales to estimate the net preference of Trp for partitioning into the core of the bilayer relative to the bilayer interface according to
| (5) |
and we find ΔΔGoOctanol-interface = −0.57 ± 0.18 kcal/mol, which implies a mild preference of Trp for the bilayer interior. For comparison, our result of the contribution of the interior Trps (W9, W11, and W13) to the stability of OmpA in lipid bilayers relative to interfacial Trps (W15)
| (6) |
where x=9, 11, and 13, ranges from ΔΔGoInterior-interface = −0.5 ± 0.5 to −1.4 ± 0.5 and −1.8 ± 0.7 kcal/mol for W9, W13, and W11, respectively. These values are on the same order or a little more than estimated from the WW scales. In this comparison, we did not include W7 because its true energetic contribution is masked by cooperative interaction with other aromatic residues in the neighboring strands22,38 (see Results).
Contrary to the free energies derived from these membrane protein folding experiments and Wimley and White’s peptide partitioning studies, recent work from the von Heijne group showed that the transfer of Trp from the translocon to the lipid bilayer of the ER is less favorable in the center than in both membrane interfaces by ΔΔGoapp = 0.8~1.0 kcal/mol 7, 39. Öjemalm et al. further tested various non-natural amino acid analogues of Trp, Tyr and Phe and concluded that the hydrogen-bonding capability of aromatic side-chains is an important factor that determines the preference of polar aromatic residues for the interfaces over the interior of the ER membrane39. As already mentioned, many statistical analyses on the structures of a-helical and β-barrel membrane proteins also indicate a substantial enrichment of Trps at interfaces relative to the bilayer center 9–11,31,40, although Adamian et al. find Trp similarly prevalent in the lipid hydrocarbon (ΔG = −0.3 kcal/mol) and headgroup (ΔG = −0.5 kcal/mol) regions31.
What could be the reasons for the apparent discrepancy between these different scales? The thermodynamic stability of proteins is defined as the free energy difference between the folded and unfolded states25. Therefore, the free energy of the unfolded state is also critically important to define the thermodynamic stability of proteins, including membrane proteins. Could it be that we are seeing here a manifestation of different unfolded states in the in vitro biochemical and more biological cell-free systems? We think that this is actually quite likely. We know that OmpA has only negligible residual structure at 8 M urea20. Another β -barrel membrane protein of similar size, OmpX, also has only little residual structure at high concentration of denaturant41. The state of nascent proteins almost certainly is different from random coil in the translocon. Nascent proteins in the translocon channel are likely somehow ordered although this order is of course very far from the native state of the protein and certainly different from the disorder found in an unfolded membrane protein in solution.
Even if no structure can be detected in intrinsically disordered proteins in vitro, they are not completely random either42. In addition to depth-dependent bilayer effects in the folded state, non-random unfolded states of our proteins could contribute to the depth-dependent folding profiles that we observed in this work. For example, it is conceivable that the central Trps (W9, W11, and W13 mutants) of OmpA destabilize the unfolded states more than the interfacial Trps (W7 and W15 mutants). The notion that this may indeed be the case is supported by our observation that the Trp contribution to OmpA’s stability was remarkably more sensitive to solvent perturbation by urea for Trps near the bilayer center than for interfacial Trps (Figure 4). Another interesting observation is the large and systematic variation of the m-values of the mutants (Figure 3B). They are the principle players that drive the strong depth- and urea-dependence of the stability contribution of the different Trps in membranes (Figures 3B, 3C and 4). In soluble protein folding, m-values are generally believed to reflect how much hydrophobic surface area gets shielded from water43,44. It is therefore not surprising that residues which are more deeply buried in the bilayer feature higher m-values. However, non-randomness of unfolded states also needs to be taken into account in discussions of the sequence dependence of m-values45. In this context it is interesting to note that a Trp residue contributes to forming a single tiny cluster of local residual structure in the unfolded membrane protein OmpX in 8 M urea 41.
Finally, the changes in the unfolding free energy by mutation of lipid-exposed residues are designed in our study to capture differential lipid-protein interactions of the replaced residues8,22. However, although we designed our experiments to minimize energetic interferences from neighboring residues in the folded state, we cannot completely exclude the possibility that the apparent changes in the unfolding free energies are subject to subtle contributions from perturbed folded states of our OmpA mutants. However, such perturbations, if present, are likely only very minor as judged from the efficient refolding and spectroscopic characterization of all mutants. We also need to be cautious about the generality of our conclusions based on trends observed after mutation of just one strand of OmpA. Further mutational analysis including deletion of the aromatic residues in the neighboring strands β2 and β8 as well as targeting Trp residues in other β-strands would be interesting to pursue in future experiments.
In summary, our results suggest that lipid solvation of individual amino acid residues lining the membrane-embedded perimeter of integral membrane proteins is adjusted by the complex physico-chemical properties of the lipid bilayer that vary dramatically with membrane depth. This fine-tuning affects quite strongly the thermodynamic stability of not only OmpA, but most likely also all membrane proteins. For multipass α-helical or β-barrel membrane proteins, the depth-dependent lipid solvation is further modified by interactions with neighboring side-chains in the native structure and likely also by energetic contributions from unfolded states.
Supplementary Material
Acknowledgements
We thank Dr. Volker Kiessling at University of Virginia for providing the gel analysis program.
Abbreviations
- DPoPC
1, 2-dipalmitoleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-(1-rac-glycerol)]
- SUV
small unilamellar vesicle
- SDS-PAGE
SDS-PAGE sodium dodecylsulfate-polyacrylamide gel eletrophoresis
- LB
Luria-Bertani medium
Footnotes
This work was supported by grant R01 GM51329 from the National Institutes of Health (to LKT) and start-up funds from Michigan State University (to HH).
Supporting Information Available
Detailed data-fitting procedures to obtain the unfolding free energy of OmpA and supplementary figures are included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 2.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 3.Jayasinghe S, Hristova K, White SH. Energetics, stability, and prediction of transmembrane helices. J Mol Biol. 2001;312:927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- 4.Wimley WC, Creamer TP, White SH. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 5.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 6.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 7.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 8.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci U S A. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senes A, Chadi DC, Law PB, Walters RF, Nanda V, Degrado WF. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J Mol Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Schramm CA, Hannigan BT, Donald JE, Keasar C, Saven JG, Degrado WF, and Samish I. Knowledge-based potential for positioning membrane-associated structures and assessing residue-specific energetic contributions. Structure. 2012;20:924–935. doi: 10.1016/j.str.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 12.de Planque MR, Killian JA. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol Membr Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 13.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 14.Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 15.Gleason NJ, Vostrikov VV, Greathouse DV, Grant CV, Opella SJ, Koeppe RE. Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry. 2012;51:2044–2053. doi: 10.1021/bi201732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nozaki Y, Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971;246:2211–2217. [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 19.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 20.Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc Natl Acad Sci U S A. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 23.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 24.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 25.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 26.Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J Mol Biol. 2000;298:273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 27.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodionova NA, Tatulian SA, Surrey T, Jahnig F, Tamm LK. Characterization of two membrane-bound forms of OmpA. Biochemistry. 1995;34:1921–1929. doi: 10.1021/bi00006a013. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt JH, Tamm LK. Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- 30.Liang B, Arora A, Tamm LK. Fast-time scale dynamics of outer membrane protein A by extended model-free analysis of NMR relaxation data. Biochim Biophys Acta. 2010;1798:68–76. doi: 10.1016/j.bbamem.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamian L, Nanda V, DeGrado WF, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- 32.Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. Journal of Biological Chemistry. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 33.Ellena JF, Liang BY, Wiktor M, Stein A, Cafiso DS, Jahn R, Tamm LK. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci U S A. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popa A, Pager CT, Dutch RE. C-Terminal Tyrosine Residues Modulate the Fusion Activity of the Hendra Virus Fusion Protein. Biochemistry. 2011;50:945–952. doi: 10.1021/bi101597k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 36.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 37.Snider C, Jayasinghe S, Hristova K, White SH. MPEx: a tool for exploring membrane proteins. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez KM, Gable JE, Schlamadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the transmembrane protein OmpA. Biochemistry. 2008;47:12844–12852. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ojemalm K, Higuchi T, Jiang Y, Langel U, Nilsson I, White SH, Suga H, von Heijne G. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2011;108:E359–E364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wimley WC. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci. 2002;11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tafer H, Hiller S, Hilty C, Fernandez C, Wuthrich K. Nonrandom structure in the urea-unfolded Escherichia coli outer membrane protein X (OmpX) Biochemistry. 2004;43:860–869. doi: 10.1021/bi0356606. [DOI] [PubMed] [Google Scholar]
- 42.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shortle D. Staphylococcal Nuclease - a Showcase of M-Value Effects. Advances in Protein Chemistry, Vol 46. 1995;46:217–247. doi: 10.1016/s0065-3233(08)60336-8. [DOI] [PubMed] [Google Scholar]
- 44.Shortle D, Chan HS, Dill KA. Modeling the effects of mutations on the denatured states of proteins. Protein Sci. 1992;1:201–215. doi: 10.1002/pro.5560010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dill KA, Shortle D. Denatured states of proteins. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.