Eukaryotes can have thousands of 45S ribosomal RNA (rRNA) genes, many of which are silenced during development. Using fluorescence-activated sorting, Pontvianne et al. show that active and silenced rRNA genes in Arabidopsis thaliana are segregated into different subnuclear compartments. Mutations that disrupt rRNA gene chromatin modifications or copy number abrogate nuclear partitioning. This study provides functional insight into the significance of rRNA gene segregation between alternative nuclear territories, showing that this physical dichotomy depends on epigenetic state.
Keywords: transcription, gene silencing, DNA methylation, histone deacetylation, chromatin assembly, RNA polymerase I, ribosomal RNA gene
Abstract
Eukaryotes can have thousands of 45S ribosomal RNA (rRNA) genes, many of which are silenced during development. Using fluorescence-activated sorting techniques, we show that active rRNA genes in Arabidopsis thaliana are present within sorted nucleoli, whereas silenced rRNA genes are excluded. DNA methyltransferase (met1), histone deacetylase (hda6), or chromatin assembly (caf1) mutants that disrupt silencing abrogate this nucleoplasmic–nucleolar partitioning. Bisulfite sequencing data indicate that active nucleolar rRNA genes are nearly completely demethylated at promoter CGs, whereas silenced genes are nearly fully methylated. Collectively, the data reveal that rRNA genes occupy distinct but changeable nuclear territories according to their epigenetic state.
In eukaryotes, 45S ribosomal RNA (rRNA) genes are tandemly arrayed at nucleolus organizer regions (NORs) (see Fig. 1A). Their transcripts, generated by RNA polymerase I (Pol I) in the nucleolus, are processed into the 18S, 5.8S, and 25-28S (depending on species) catalytic RNAs of ribosomes (Moss et al. 2007; Kressler et al. 2010; Hannan et al. 2013). The number of active 45S rRNA genes changes with the physiological needs of the cell (McStay and Grummt 2008; Tucker et al. 2010). For instance, Arabidopsis thaliana has ∼1500 rRNA genes per diploid genome (Copenhaver et al. 1995; Copenhaver and Pikaard 1996), with subtypes distinguishable by insertions/deletions at their 3′ ends (Fig. 1A). All subtypes are expressed immediately following germination, but by ∼10 d of seedling growth, the variant 1 subtype (Fig. 1A), accounting for ∼50% of all rRNA genes, is silenced via epigenetic mechanisms that include changes in DNA methylation and histone modification (Earley et al. 2006, 2010; Pontvianne et al. 2010, 2012). Chromatin modifications mediate rRNA gene dosage control in yeast, mouse, and human cells as well (Sandmeier et al. 2002; McStay and Grummt 2008; Grummt and Langst 2013).
Figure 1.
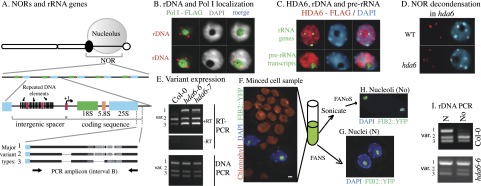
Partitioning of active and silent rRNA genes between the nucleolus and nucleoplasm. (A) Relationships between the nucleolus, NORs, and 45S rRNA gene repeats. The drawing at the top depicts a metaphase chromosome with the remnants of a nucleolus associated with the secondary constriction, the location of active rRNA genes in the preceding interphase. The black oval represents a chromomere in which rRNA genes are assembled into dense heterochromatin. In A. thaliana, insertions/deletions in the 3′ external transcribed region define rRNA gene variant types. (B) Localization of rRNA genes (rDNA) and Pol I. DNA-FISH using an rRNA gene probe (red signals) and immunolocalization of Flag-tagged Pol I using an anti-Flag antibody (green signals) were performed in A. thaliana interphase nuclei. DNA was counterstained with DAPI (gray signals). (C) Subnuclear localization of rRNA genes, pre-rRNA transcripts, and Flag-tagged HDA6. rRNA gene or transcript FISH signals are shown in green, immunolocalized HDA6 is in red, and DAPI-stained DNA is in blue. Merged signals are shown in the right column. (D) DNA-FISH detection of rRNA genes in wild-type (Col-0) and hda6 nuclei. rRNA gene FISH signals are shown in red and are merged with the DAPI (blue) image in the right column. (E) Detection of rRNA gene variant types and their transcripts by PCR using genomic DNA or reverse-transcribed (RT) cDNA of wild-type (Col-0) or hda6 plants. The amplified region is shown in A. (F) Leaf cell homogenate of a FIB2:YFP plant stained with DAPI and subjected to fluorescence microscopy. Chloroplasts fluoresce red, DAPI-stained DNA is blue, and nucleolar FIB2:YFP is green. (G) Purified nuclei obtained by FANS. (H) Purified nucleoli obtained by FANoS. (I) PCR detection of rRNA gene variant types in DNA of purified nuclei (N) or nucleoli (No) of wild-type (Col-0) or hda6 plants. The PCR amplicon is shown in A.
To examine the subnuclear distribution of active and silenced rRNA genes, we adapted fluorescence-activated sorting technology to isolate whole A. thaliana nuclei or nucleoli liberated from sonicated nuclei. Exploiting sequence variation among differentially expressed rRNA gene subtypes and mutations that disrupt rRNA gene chromatin modifications or copy number, we show that rRNA genes occupy alternative subnuclear compartments depending on their activity state.
Results and Discussion
Partitioning of active versus inactive rRNA genes between the nucleolus and nucleoplasm
A. thaliana has NORs on chromosomes 2 and 4, each consisting of ∼375 rRNA genes and spanning ∼4 Mb (Copenhaver et al. 1995; Copenhaver and Pikaard 1996). Pol I, which transcribes 45S rRNA genes, localizes to the nucleolus of interphase cells (Fig. 1B, green signal). The nucleolus is rich in RNA and proteins but has little DNA, thus appearing as a black hole in nuclei stained with the DNA-binding dye DAPI (Fig. 1B, gray signal). 45S pre-rRNA transcripts are detected throughout the nucleolus by RNA-FISH (fluorescent in situ hybridization) (Fig. 1C, bottom row). However, the most prominent 45S rRNA gene DNA-FISH signals are external to the nucleolus (Fig. 1B [red signals], C [top row, green signals]; note that NOR associations can result in fewer than four signals). rRNA genes inside the nucleolus are decondensed and more difficult to detect by DNA-FISH, depending on the extent of their dispersal (e.g., cf. the two nuclei in Fig. 1B).
HISTONE DEACETYLASE 6 (HDA6) is required for uniparental rRNA gene silencing in hybrid Arabidopsis suecica and for developmentally regulated silencing of variant 1 rRNA genes in nonhybrid A. thaliana (Earley et al. 2006, 2010). HDA6 localizes throughout nuclei, including the nucleolus (Fig. 1C). In hda6 mutants, NORs decondense (Probst et al. 2004; Earley et al. 2006), and rRNA gene FISH signals inside the nucleolus increase (Fig. 1D). In leaves of wild-type plants (ecotype Col-0), variant 2 and 3 rRNA gene subtypes are expressed, and variant 1 genes are silenced (Fig. 1E, RT–PCR primer locations are shown in A). However, in hda6-6 or hda6-7 mutants, all variant subtypes are expressed (Fig. 1E).
To determine whether both active and silenced rRNA genes are associated with nucleoli, we performed fluorescence-activated sorting of whole nuclei or isolated nucleoli from plants expressing the nucleolar protein FIBRILLARIN2 fused to YFP (yellow fluorescent protein) (Barneche et al. 2000). FIB2:YFP localizes specifically within the nucleolus, as shown in Figure 1F. Fluorescence-activated nuclear sorting (FANS) of cell homogenates yielded homogeneous nuclei (Fig. 1G; Supplemental Fig. S1A). Alternatively, cell extracts were sonicated to disrupt nuclei and then subjected to fluorescence-activated nucleolar sorting (FANoS), yielding nucleoli free of intact nuclei, other organelles, or cellular debris (Fig. 1H; Supplemental Fig. S1B,C).
rRNA gene subtypes in isolated nuclei or nucleoli were identified by PCR amplification using primers flanking the variable region (see Fig. 1A). All variant types are present in nuclei of wild-type Col-0 or hda6 mutants, as expected (Fig. 1I). However, in nucleoli of wild-type plants, variant 2- and 3-type rRNA genes are enriched (Fig. 1I, top row), correlating with their selective expression (see Fig. 1E). In hda6 mutants, in which variant 1 gene silencing does not occur, variant 1 genes are also present in nucleoli (Fig. 1I, bottom row). Collectively, these results indicate that rRNA genes are present in nucleoli when active and are excluded from nucleoli when silenced.
MET1-dependent CG methylation is implicated in rRNA gene subtype silencing
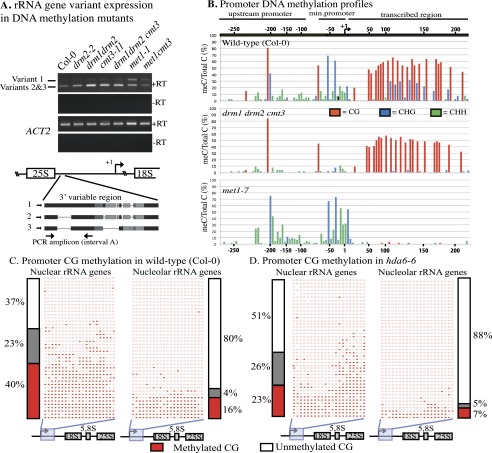
In A. thaliana, cytosine methylation at CG motifs is maintained by MET1 (the ortholog of mammalian DNMT1), CHG methylation (where H is A, T, or C) is maintained by CMT3, and RNA-directed CHH methylation is mediated by DRM2, whose paralog, DRM1, may function in some cells (Law and Jacobsen 2010). Variant 1 rRNA gene silencing fails to occur in met1 mutants (Fig. 2A), corresponding with the loss of promoter region CG methylation (Fig. 2B). In contrast, drm1-, drm2-, or cmt3-null mutations, alone or in combination, reduce promoter CHG and CHH methylation (Fig. 2B) but have negligible effects on variant 1 silencing (Fig. 2A).
Figure 2.
MET1-dependent CG methylation is required for variant 1-type rRNA gene silencing. (A) rRNA gene variant expression in wild type (Col-0) or drm2-2, drm1 drm2, cmt3-11, drm1 drm2 cmt3, met1-1, or met1 cmt3 mutants. RT–PCR using primers that discriminate variant 1- from variant 2- and 3-type rRNA genes was conducted (see the diagram for primer locations). RT–PCR of ACTIN2 (ACT2) mRNA serves as a positive control. Reactions lacking reverse transcriptase (−RT) serve as negative controls. (B) Frequencies at which individual cytosines are methylated between −316 and +243 relative to the transcription start site (+1), determined by bisulfite sequencing. Wild-type Col-0, drm1 drm2 cmt3 triple mutants, and met1-7 mutants are compared. Approximately 40 independent promoter clones were sequenced for each genotype. Cytosine-depleted regions are compressed on the X-axis. (C,D) Cytosine methylation in the downstream promoter region of rRNA genes in purified nuclei or nucleoli from wild-type or hda6 leaves, determined by bisulfite sequencing. Positions of methylated (filled circles) or unmethylated (open circles) cytosines in CG motifs of 43 independent promoter clone sequences are shown. Methylation haplotypes are grouped according to methylation density. Histograms show frequencies of hypomethylated haplotypes (white), haplotypes with intermediate methylation (gray), or heavily methylated haplotypes (red).
Active rRNA genes in the nucleolus are CG hypomethylated, as in met1 mutants
MET1's involvement in rRNA gene silencing prompted a comparison of CG methylation among nucleolar versus nuclear rRNA genes. In Figure 2, C and D, 21 CG positions in the downstream promoter region (same region as in Fig. 2B) are shown as filled (methylated) or open (unmethylated) circles, with each row representing an independent clone following bisulfite sequencing. In wild-type nuclei, 37% of promoter clones are unmethylated or lightly methylated (fewer than three meCGs), a similar number of clones (40%) is heavily methylated (11 or more meCGs), and 23% show intermediate levels of methylation (four to 10 meCGs) (Fig. 2C). In nucleoli, however, 80% of rRNA gene promoter sequences are unmethylated or lightly methylated, with only 16% heavily methylated.
Promoter cytosine methylation was next examined in hda6 mutants (Fig. 2D), in which all variants are expressed and nucleolar (see Fig. 1E,I). In hda6 nuclei, more rRNA gene promoter sequences are unmethylated/lightly methylated compared with wild type (51% vs. 37%), and fewer are heavily methylated (23% vs. 40%). In hda6 nucleoli, 88% of rRNA gene promoter clones had either zero or only one meCG (Fig. 2D).
Collectively, the data of Figures 1 and 2 show that approximately half of the total rRNA gene pool is silenced (the variant 1 subtype), that sorted nucleoli are enriched for active rRNA gene variants, and that mutants that disrupt silencing cause all variant types to be nucleolar. Whereas the total pool of nuclear rRNA genes includes heavily methylated and unmethylated promoter sequences in similar proportions (∼40% each), the nucleolar rRNA genes are mostly (at least 80%) demethylated, suggesting that the demethylated state is the active state. It then follows that the heavily methylated state is the inactive state. We further deduce that the small fraction of fully methylated rRNA gene promoter sequences detected in purified nucleoli represent silenced rRNA genes located in cis to active genes, thereby facilitating their nucleolar association.
Variant-specific silencing is disrupted when rRNA gene copy number is reduced
Because selective rRNA gene silencing is thought to be a form of dosage control, reducing the rRNA gene pool size is expected to increase the proportion of active rRNA genes, as in yeast (French et al. 2003). Arabidopsis thaliana FASCIATA 1 (FAS1) and FAS2 are subunits of chromatin assembly factor 1 (CAF1), a histone chaperone implicated in replication-dependent deposition of histones H3 and H4 (Ramirez-Parra and Gutierrez 2007). In fas1 or fas2 mutants, 45S rRNA genes are lost (Mozgova et al. 2010), as shown by DNA-FISH (Fig. 3A) or quantitative PCR (qPCR) (Fig. 3B). The latter shows that rRNA gene numbers fall to ∼40% of wild type by the second generation (G2) and to ∼5%–10% of wild type by G5 before stabilizing in number. Beyond G9, fas mutant lines cannot be perpetuated due to sterility resulting from genome instability.
Figure 3.
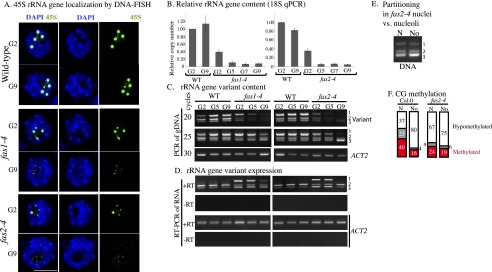
Reducing rRNA gene number in fas mutants disrupts variant 1 silencing, nucleolus–nucleoplasm partitioning, and CG methylation. (A) rRNA gene localization by DNA-FISH in nuclei of wild type or fas1 or fas2 mutants at G2 and G9. Nuclei were counterstained with DAPI. (B) qPCR analysis of relative rRNA gene numbers in wild type (WT) versus fas1-4 or fas2-4 at G2, G5, G7, or G9. (C) Semiquantitative PCR detection of rRNA gene variant types in genomic DNA of fas1 or fas2 mutants or lines derived from their wild-type siblings at G2, G5, or G9. Amplification products of rRNA gene variants after 20 or 25 cycles of PCR or of ACTIN2 after 30 cycles of PCR resolved by agarose gel electrophoresis. (D) RT–PCR amplification of rRNA gene variant transcripts or an ACTIN2 mRNA control. The lane order is the same as in C. (E) PCR detection of rRNA gene variant types in sorted nuclei or nucleoli of fas2-4 FIB2:YFP plants. (F) Frequencies of haplotypes showing CG hypomethylation, intermediate methylation, or heavy methylation in wild-type or fas2-4 nuclei (N) or nucleoli (No) within the rRNA gene promoter downstream region.
Semiquantitative analysis of rRNA gene variant abundance, assessed by agarose gel electrophoresis of genomic PCR products, shows that all variant types decrease from G2 to G9 in fas1 and fas2 mutants (Fig. 3C). The ∼40% reduction in rRNA gene number that occurs by G2 (refer to Fig. 3B) is sufficient to abrogate dosage control such that all variant types are expressed (Fig. 3D). In contrast, variant 1 genes are not expressed in wild-type siblings at G2, G5, or G9 (Fig. 3D).
To test whether fas mutations affect rRNA gene nucleoplasmic–nucleolar partitioning, we crossed a fas2-4 mutant (G2) with a FIB2:YFP transgenic plant, collected seeds from their F1 offspring, and identified fas2-4 homozygotes in the F2 generation. We then used FANS or FANoS to isolate purified nuclei or nucleoli, respectively. All rRNA gene variant types are present in nucleoli of fas2 mutants (Fig. 3E), consistent with the failure to silence variant 1 genes (Fig. 3D).
Bisulfite sequencing of fas2-4 nuclear rRNA genes showed that CG hypermethylated sequences are reduced by half compared with wild type (Fig. 3F). However, in isolated nucleoli, the rRNA genes of wild-type and fas2 plants are similarly demethylated. Collectively, the data indicate that reducing rRNA gene numbers in fas mutants abrogates the dosage control system such that variant 1 genes are no longer silenced and hypermethylated, and virtually all rRNA genes become nucleolus-associated.
Methylation density versus site-specific methylation
Our results suggest that active rRNA genes, which localize within the nucleolus, are demethylated, or nearly so, at >20 CG positions adjacent to the transcription start site. In contrast, silenced rRNA genes are deduced to be heavily methylated based on the fact that heavily and lightly methylated rRNA promoter sequences are detected at similar frequency in whole nuclei and that silenced variant 1 type genes account for ∼50% of the total rRNA gene pool. These data suggest that quantitative, essentially all-or-nothing, methylation correlates with rRNA gene on or off states in A. thaliana, as opposed to specific methylation of single cytosines, as indicated by studies in mice (Santoro and Grummt 2001).
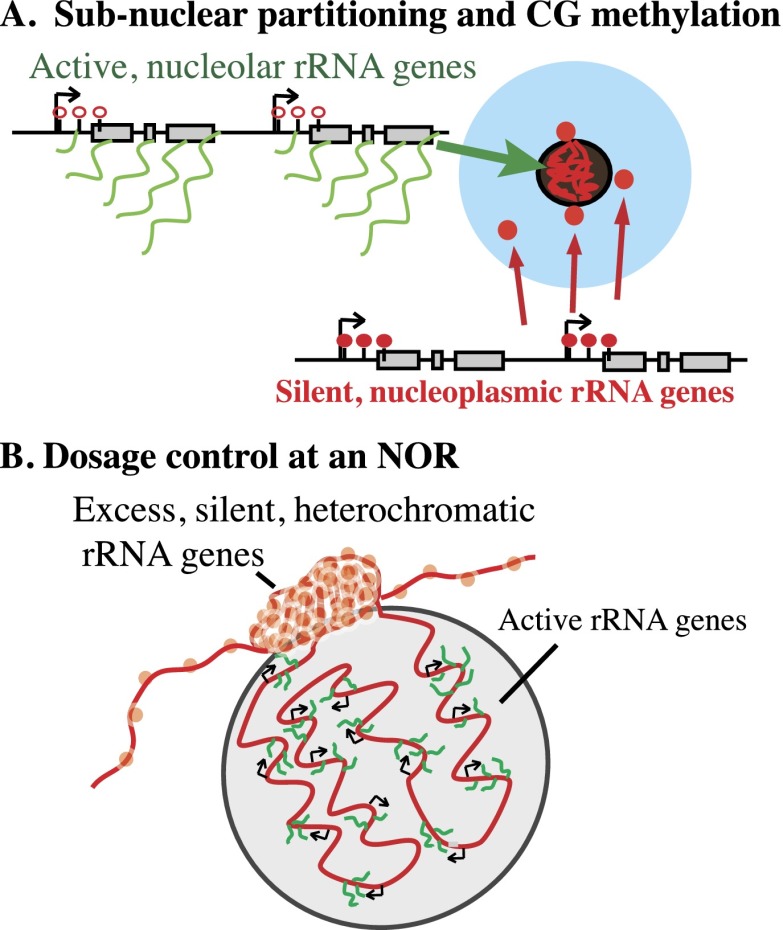
A model for dynamic subnuclear rRNA gene localization
rRNA gene DNA-FISH signals external to the nucleolus are observed in both plant and animal interphase cells. In Arabidopsis, these external signals colocalize with histone modifications typical of silent heterochromatin (Lawrence et al. 2004; Earley et al. 2006). In yeast, which have a single NOR, electron microscopy has shown that active and inactive rRNA genes are interspersed with one another (French et al. 2003). However, in plants and mammals, there is evidence that active and silenced rRNA genes can occupy different portions of an NOR. On metaphase chromosomes, secondary constrictions at NORs stain intensively with silver (Goodpasture and Bloom 1975; Roussel and Hernandez-Verdun 1994) due to persistent binding of Pol I transcription factors on rRNA genes that were active in the preceding interphase (Prieto and McStay 2008). Importantly, secondary constrictions and silver-stained regions do not necessarily encompass an entire NOR. Instead, condensed portions of an NOR can be adjacent to a decondensed, silver-stained portion of the NOR (Caperta et al. 2002). Collectively, these cytological observations, combined with our results with sorted nucleoli, suggest that NORs can span the nucleoplasm–nucleolus boundary, with active rRNA genes inside the nucleolus and inactive genes primarily outside (Fig. 4). Silent rRNA genes at the boundary between these sub-NOR domains could account for the minority (<20%) of heavily methylated rRNA genes present in flow-sorted nucleoli. Importantly, our results show that localization inside or outside of the nucleolus is not an absolute property of a given rRNA gene variant type but reflects transcriptional and epigenetic states that are changeable according to the needs of the cell. We reason that excess rRNA genes coalesce into dense heterochromatic structures at the external edge of the nucleolus but spool out into the nucleolus as more rRNA genes are needed. Conversely, excess rRNA genes would be reeled into the external reservoir such that rRNA gene partitioning between the nucleolus and nucleoplasm is the cytological manifestation of dynamic rRNA gene dosage control.
Figure 4.
Model for the organization of rRNA genes in interphase nuclei. (A) The blue circle represents a nucleus visualized by DAPI staining, with the black hole representing the nucleolus. Results of FANS or FANoS experiments indicate that condensed rRNA gene DNA-FISH signals in the nucleoplasm correspond to silent rRNA genes that are heavily methylated at promoter CG motifs. In contrast, active rRNA genes are decondensed, localized within the nucleolus, and CG-demethylated. (B) A single NOR can be composed of condensed, silent rRNA genes external to the nucleolus as well as decondensed, active rRNA genes dispersed within the nucleolus. Changing the number of rRNA genes that spool out from, or are reeled into, the reservoir of rRNA genes at the external periphery of the nucleolus can account for changes in the number of active versus silenced genes during development.
Materials and methods
RT–PCR
RNA was isolated from 2- to 4-wk-old leaves of A. thaliana using Plant RNAeasy kits (Qiagen) and treated with Turbo DNase (Ambion) for 60 min. Semiquantitative RT–PCR was performed using random-primed cDNA generated from 1.5 μg of total RNA and SuperScript III reverse transcriptase (Invitrogen). PCR primers for the rRNA gene variable region were CTCGAGGTTAAATGTTATTACTTGGTAAGATTCCGG (interval A forward), TGGGTTTGTCATATTGAACGTTTGTGTTCATATCACC (interval A reverse), GACAGACTTGTCCAAAACGCCCACC (interval B forward), and CTGGTCGAGGAATCCTGGACGATT (interval B reverse). ACTIN2 PCR primers were AAGTCATAACCATCGGAGCTG (forward) and ACCAGATAAGACAAGACACAC (reverse).
Cytosine methylation analyses
Genomic DNA was extracted using Illustra DNA phytopure extraction kits (GE Healthcare). After digestion with BamHI, 2 μg of DNA was bisulfite-treated using an EpiTect Bisulfite kit (Qiagen). The rRNA gene promoter region was PCR-amplified as described previously (Pontvianne et al. 2010) using primers GGATATGATGYAATGTTTTGTGATYG (forward) and CCCATTCTCCTCRACRATTCARC (reverse). PCR products were cloned into pGEM-T-Easy (Promega) and sequenced. Methylation was analyzed using CyMATE (Hetzl et al. 2007) and graphed using a custom Perl script and Microsoft Excel.
Nuclear and nucleolar DNA purification
Leaves (1 g) from 4-wk-old FIB2:YFP plants were fixed for 20 min in 4% formaldehyde in Tris buffer (10 mM Tris-HCl at pH 7.5, 10 mM EDTA, 100 mM NaCl). Leaves were washed twice for 10 min each in ice-cold Tris buffer and minced in 1 mL of 45 mM MgCl2, 20 mM MOPS (pH 7.0), 30 mM sodium citrate, and 0.1% Triton X-100 using a razor blade. The homogenate was filtered through 40-μm mesh (BD Falcon) and subjected to FANS or sonicated using a Bioruptor (three 5-min pulses, medium power; Diagenode) to liberate nucleoli that were then sorted by FANoS. Sorting of nuclei or nucleoli was triggered by the FIB2:YFP signal using a BD FACS Aria II. Sorted nuclei or nucleoli were treated with RNase A and proteinase K prior to DNA purification and PCR or bisulfite sequencing analyses.
DNA-FISH and qPCR
DNA-FISH and qPCR analyses of fas mutants were performed as previously described (Mozgova et al. 2010) using 18S rRNA gene primers CTAGAGCTAATACGTGCAACAAAC (forward) and GAATCGAACCCTAATTCTCCG (reverse) and UBIQUITIN 10 (UBQ10) control primers AACGGGAAAGACGATTAC (forward) and ACAAGATGAAGGGTGGAC (reverse). DNA-FISH, RNA-FISH, and protein immunolocalization of Flag-tagged proteins were performed as described previously (Pontes et al. 2003, 2006).
Acknowledgments
We thank Jim Powers and the Light Microscopy Imaging Center at Indiana University for microscopy support. This work was supported by the National Institutes of Health grant GM60380 to C.S.P. C.S.P. is an Investigator of the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation. T.B. was supported by an NIH Ruth L. Kirschstein National Research Service Award and funds from Howard Hughes Medical Institute. I.M., P.M., V.M., and J.F. were supported by the Czech Science Foundation (P501/11/0289) and project CEITEC-CZ.1.05/1.1.00/02.0068 from the European Regional Development Fund. C.C. did the bisulfite sequencing of Figure 2, T.B. did the DNA methylation analyses of Figure 2B, C.H. did the flow sorting, and O.P. did the FISH and immunolocalizations of Figure 1. I.M. generated consecutive fas generations and, with P.M., V.M., and J.F., did the analyses of Figure 3, A and B. F.P. designed and performed all other experiments. F.P. and C.S.P. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.221648.113.
References
- Barneche F, Steinmetz F, Echeverria M 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem 275: 27212–27220 [DOI] [PubMed] [Google Scholar]
- Caperta AD, Neves N, Morais-Cecilio L, Malho R, Viegas W 2002. Genome restructuring in rye affects the expression, organization and disposition of homologous rDNA loci. J Cell Sci 115: 2839–2846 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS 1996. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J 9: 259–272 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Doelling JH, Gens S, Pikaard CS 1995. Use of RFLPs larger than 100 kbp to map the position and internal organization of the nucleolus organizer region on chromosome 2 in Arabidopsis thaliana. Plant J 7: 273–286 [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS 2010. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24: 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpasture C, Bloom SE 1975. Visualization of nucleolar organizer regions in mammalian chromosomes using silver staining. Chromosoma 53: 37–50 [DOI] [PubMed] [Google Scholar]
- Grummt I, Langst G 2013. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829: 393–404 [DOI] [PubMed] [Google Scholar]
- Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB 2013. Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 1829: 342–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O 2007. CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J 2010. Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V 2007. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Mokros P, Fajkus J 2010. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Neves N, Silva M, Lee JH, Chen ZJ, Viegas W, Pikaard CS 2003. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc Natl Acad Sci 100: 11418–11423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS 2006. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. 2010. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Feng W, Stroud H, Jacobsen SE, Michaels SD, Pikaard CS 2012. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev 26: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B 2008. Pseudo-NORs: A novel model for studying nucleoli. Biochim Biophys Acta 1783: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. 2004. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C 2007. The many faces of chromatin assembly factor 1. Trends Plant Sci 12: 570–576 [DOI] [PubMed] [Google Scholar]
- Roussel P, Hernandez-Verdun D 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp Cell Res 214: 465–472 [DOI] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 21: 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I 2001. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Tucker S, Vitins A, Pikaard CS 2010. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol 22: 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]