The DNA damage response kinase ATR coordinates cellular responses to replication stress. Using selective ATR inhibitors, Couch et al. show that acute ATR inhibition leads to DNA replication fork collapse and rapid cell death. They define a mechanism by which ATR ensures repair of stalled replication forks via phosphorylation of a conserved site on the SMARCAL1 DNA translocase. This study provides critical insight into the action of ATR inhibitors being developed as cancer therapeutics.
Keywords: ATR, SMARCAL1, DNA damage response, DNA replication, cell cycle checkpoint, HARP
Abstract
The DNA damage response kinase ataxia telangiectasia and Rad3-related (ATR) coordinates much of the cellular response to replication stress. The exact mechanisms by which ATR regulates DNA synthesis in conditions of replication stress are largely unknown, but this activity is critical for the viability and proliferation of cancer cells, making ATR a potential therapeutic target. Here we use selective ATR inhibitors to demonstrate that acute inhibition of ATR kinase activity yields rapid cell lethality, disrupts the timing of replication initiation, slows replication elongation, and induces fork collapse. We define the mechanism of this fork collapse, which includes SLX4-dependent cleavage yielding double-strand breaks and CtIP-dependent resection generating excess single-stranded template and nascent DNA strands. Our data suggest that the DNA substrates of these nucleases are generated at least in part by the SMARCAL1 DNA translocase. Properly regulated SMARCAL1 promotes stalled fork repair and restart; however, unregulated SMARCAL1 contributes to fork collapse when ATR is inactivated in both mammalian and Xenopus systems. ATR phosphorylates SMARCAL1 on S652, thereby limiting its fork regression activities and preventing aberrant fork processing. Thus, phosphorylation of SMARCAL1 is one mechanism by which ATR prevents fork collapse, promotes the completion of DNA replication, and maintains genome integrity.
Every cell division cycle, cells must accurately and completely replicate their genome. Errors in replication result in mutations and chromosomal rearrangements that contribute to tumorigenesis. Replication stress caused by DNA lesions, insufficient nucleotides, or even collisions of replication and transcriptional machineries increases the chance of errors. However, replication stress also activates a DNA damage response (DDR) that slows cell cycle progression and promotes DNA repair to ensure accurate duplication of the genome.
The DDR kinase ataxia telangiectasia and Rad3-related (ATR), a member of the phosphoinositol 3-kinase-like kinase (PIKK) family, coordinates much of the cellular response to replication stress (Cimprich and Cortez 2008; Nam and Cortez 2011). ATR is activated upon replication fork stalling and uncoupling of the replicative helicase and polymerase (Byun et al. 2005). Once activated, ATR phosphorylates hundreds of substrates to induce the replication checkpoint and promote fork repair.
ATR is essential for cell viability, and hypomorphic ATR mutations cause the rare disease Seckel syndrome, characterized by growth retardation, microcephaly, and other developmental problems (O'Driscoll et al. 2003). ATR is thought to be a good drug target for cancer therapy because its function is especially critical in replicating tumor cells, which have elevated levels of replication stress due to activated oncogenes and frequent loss of the G1 checkpoint (Reaper et al. 2011; Toledo et al. 2011b; Schoppy et al. 2012). The mechanism by which ATR-selective inhibitors kill cells is unknown but is likely linked to the replication fork stabilization and repair activities of ATR instead of its G2 checkpoint function (Nam et al. 2011; Toledo et al. 2011a). Defining these mechanisms is important for the development of ATR pathway inhibitors for cancer treatment.
Replication fork repair is a complex process that can proceed through multiple pathways depending on the cause, persistence, and genomic context of the replication stress. These mechanisms include fork stabilization to allow completion of replication by a converging replication fork, lesion bypass, template switching through recombination or fork reversal, and double-strand break (DSB)-mediated restart (Branzei and Foiani 2010). Many enzymes participate in these activities, including helicases, DNA translocases, nucleases, and specialized polymerases. ATR can phosphorylate many of these enzymes; however, the mechanisms by which it promotes fork stabilization and repair and cell viability remain largely unknown.
One ATR substrate that acts at stalled forks is SMARCAL1 (also known as HARP) (Bansbach et al. 2009; Postow et al. 2009). SMARCAL1 binds branched DNA structures and can catalyze DNA annealing, branch migration, fork regression, and fork restoration (Yusufzai and Kadonaga 2008; Betous et al. 2012, 2013; Ciccia et al. 2012). SMARCAL1 is recruited to stalled forks through an interaction with replication protein A (RPA) (Bansbach et al. 2009; Ciccia et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009), which directs it to regress stalled forks with a leading strand gap and restore a normal fork structure (Betous et al. 2013). Both overexpression and siRNA silencing of SMARCAL1 cause replication-associated DNA damage (Bansbach et al. 2009). Furthermore, loss-of-function mutations in SMARCAL1 cause the human disease Schimke immunoosseous dysplasia, which is characterized by growth defects, renal failure, immune deficiencies, and predisposition to cancer (Boerkoel et al. 2002; Baradaran-Heravi et al. 2012; Carroll et al. 2013). How ATR phosphorylation of SMARCAL1 regulates its genome maintenance functions at a damaged fork has not been investigated.
Using a selective ATR inhibitor (ATRi), we demonstrate that acute inhibition of ATR kinase activity perturbs the timing of replication initiation, impairs fork elongation rates, and causes rapid lethality in S-phase cells experiencing replication stress. Stalled forks collapse when ATR is inhibited due to SLX4-dependent endonuclease cleavage, which yields DSBs and the CtIP-dependent appearance of single-stranded template and nascent DNA strands. Excessive SMARCAL1 activity is partly responsible for this aberrant fork processing. ATR phosphorylation of a conserved SMARCAL1 serine regulates SMARCAL1 and is one mechanism by which ATR maintains genome integrity during DNA replication. Thus, our results provide a mechanistic description of fork collapse in mammalian cells and define a specific enzymatic pathway responsible for this collapse. They also explain why both too much and too little SMARCAL1 causes replication-associated DNA damage, emphasizing the need to properly regulate this replication fork repair enzyme. Finally, these data provide insights into the mechanism of action of ATRis that are being developed to treat cancers with high levels of dependency on this replication stress response pathway.
Results
Acute ATR inhibition causes rapid cell death in cells experiencing replication stress
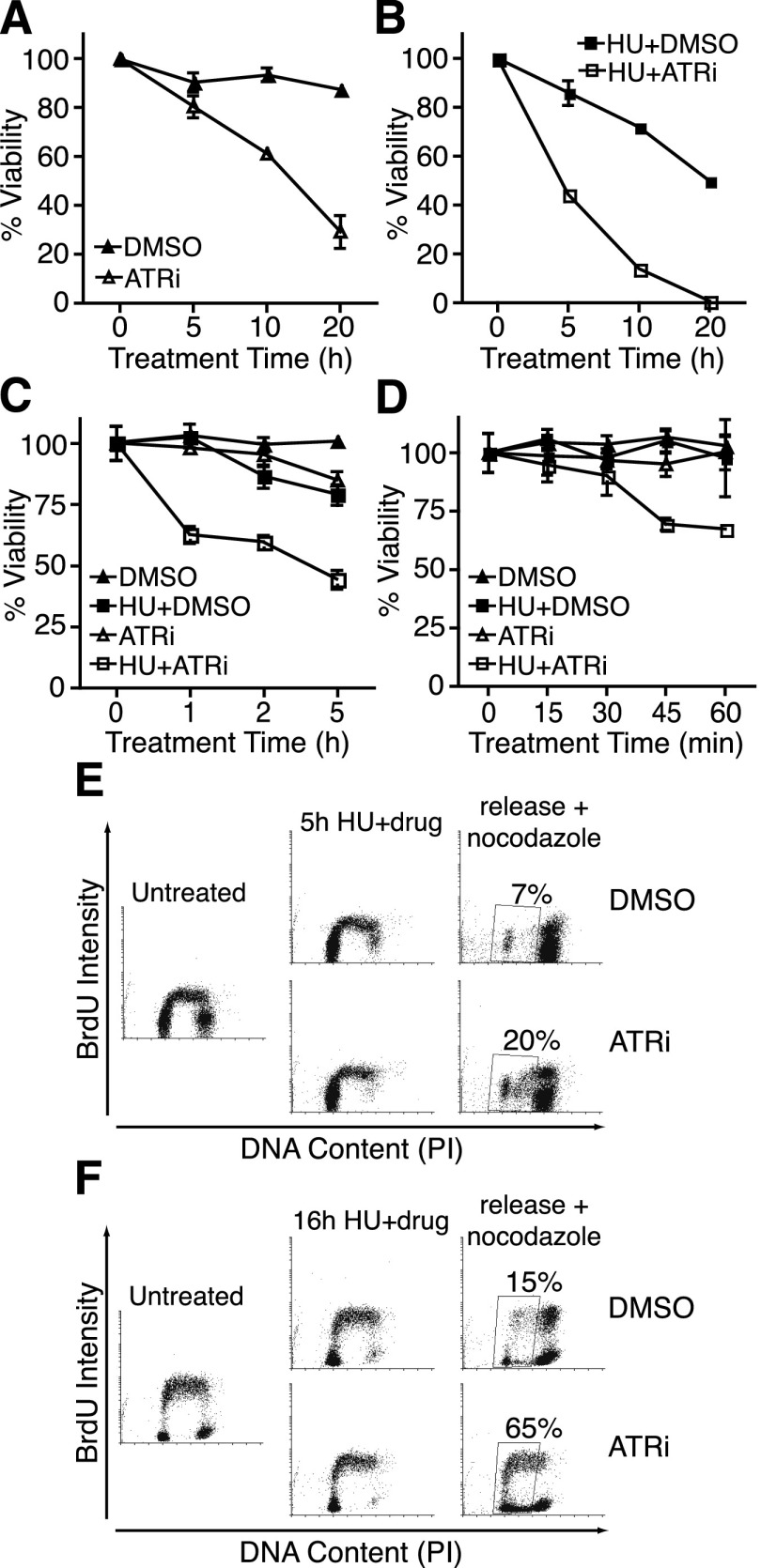
Conditional deletion of ATR in dividing mouse or human cells causes cell death as the mRNA and protein levels decay over time (Brown and Baltimore 2000; de Klein et al. 2000; Cortez et al. 2001). The gradual nature of these genetic loss-of-function experiments precludes an analysis of the immediate effects of ATR deficiency. To overcome this technical challenge, we used a selective ATR kinase inhibitor to examine how cells respond to acute and transient ATR inhibition. In this analysis, 39% of asynchronous cultured U2OS cells were no longer able to form colonies after a 10-h treatment with a concentration of ATRi sufficient to block phosphorylation of CHK1, and 71% of cells were inviable after 20 h of treatment (Fig. 1A).
Figure 1.
Acute ATR inhibition causes rapid cell lethality and an inability to complete DNA replication after a replication stress challenge. (A–D) U2OS cells were treated with DMSO, 5 μM ATRi, and/or 3 mM HU for the indicated times and released into fresh growth medium for 10–14 d. Colonies were visualized by methylene blue staining. Results shown are mean ± standard error of the mean (SEM) of at least two independent experiments. (E,F) RPE-hTERT cells were labeled with 20 μM BrdU for 20 min, treated with 3 mM HU for 5 h (E) or 16 h (F) in the presence or absence of 5 μM ATRi, and then released into fresh growth medium containing 1 μg/mL nocodazole for 24 h prior to harvesting. Cells were then fixed, acid-denatured, stained with BrdU antibodies and propidium iodide, and analyzed by flow cytometry. Plots were made using Cyflogic software.
Acute treatment with the combination of ATRi and replication stress induced by the ribonucleotide reductase inhibitor hydroxyurea (HU) caused a greater reduction in viability than either drug alone (Fig. 1A,B). Notably, 5 h of treatment with HU and ATRi caused a 55% reduction in viability. This percentage corresponds approximately to the proportion of cells in S phase at the onset of treatment plus the cells that enter S phase during the 5-h treatment. ATR inhibition is synergistic with HU: At the 20-h time point, the combination of HU+ATRi gave an experimental response (99% decrease in viability) greater than the additive effect predicted from HU alone and ATRi alone (89%).
To determine how rapidly the cell lethality occurs, we treated U2OS cells with or without HU and ATRi for up to 5 h. While HU alone or ATRi alone had only modest effects at earlier time points, the combination of HU and ATRi caused a 30% decrease in colony-forming ability within 45 min of treatment (Fig. 1C,D). Since HU treatment is expected to affect primarily S-phase cells, we tested whether these cells could recover from this treatment and complete DNA replication. By flow cytometry, at least 20% of cells treated with HU and ATRi for 5 h failed to complete DNA synthesis 24 h after release into fresh growth medium compared with only 7% of controls (Fig. 1E). After a 16-h HU treatment, this fraction of lagging cells increased to at least 65% of ATR-inhibited cells compared with only 15% of controls (Fig. 1F). Thus, ATR is required during an acute replication stress challenge to ensure that cells can recover, complete DNA synthesis, and retain long-term viability.
ATR inhibition deregulates replication timing control and causes replication fork collapse
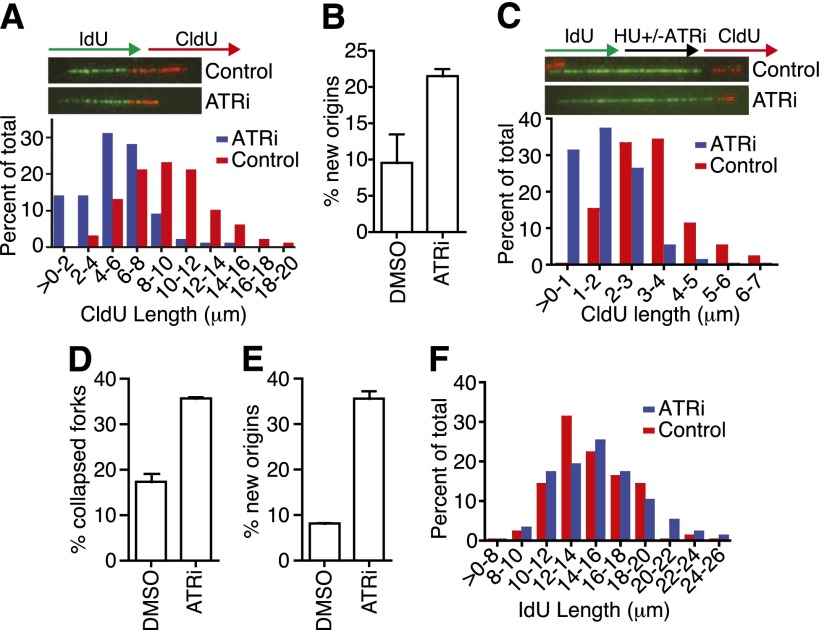
To understand the cause of the rapid lethality when ATR is inhibited, we used fiber labeling to examine DNA replication. Cells were labeled with IdU (green) for 20 min and then with CldU (red) for 20 min in the presence or absence of ATRi during both labeling periods. ATR inhibition caused the CldU fiber length to decrease to approximately half the length of controls, which indicates slower replication fork elongation (Fig. 2A). CHK1 inhibition causes a similar slowing of fork elongation, which was attributed to deregulation of origin firing (Seiler et al. 2007). This also appears to be the case after ATR inhibition, since we observed a large increase in the number of origins in the ATRi sample compared with controls (Fig. 2B).
Figure 2.
ATR regulates DNA replication initiation and elongation. (A,B) RPE-hTERT cells were labeled with IdU for 20 min and then with CldU for 20 min in the presence of DMSO (red bars) or 5 μM ATRi (blue bars) during both labeling periods before harvesting for fiber labeling. (A) Representative replication tracks and quantification of the length of CldU (red) tracks in dual-labeled tracks are shown. (B) Origin initiation was scored as the percentage of red-only tracks. (C–F) RPE-hTERT cells were labeled with IdU for 20 min, treated with 2 mM HU for 2 h in the presence of DMSO (red bars) or 5 μM ATRi (blue bars), and then labeled with CldU for 20 min before harvesting for fiber staining. Representative images and quantification of CldU (C) and IdU (F) track lengths in dual-labeled fibers are shown. Percentage of collapsed forks (green-only tracks) (D) and newly initiated origins (red-only tracks) (E) were quantitated. In all experiments, data was collected from several experimental samples with high-quality DNA fibers. Error bars are SEM.
Recovery of DNA synthesis after transient replication block is also dependent on ATR function. We labeled cells with IdU, blocked replication with high concentrations of HU in the presence or absence of ATRi, and then removed the drugs and labeled with CldU. The CldU-labeled replication tracks were significantly shorter in the ATR-inhibited condition compared with controls (Fig. 2C). Furthermore, ATR inhibition causes a large increase in the number of forks that cannot restart and collapse entirely during the HU treatment with no CldU incorporation after release (Fig. 2D). Origins were also derepressed under these conditions (Fig. 2E).
Similar experiments in cells lacking BRCA2 function found that the IdU-labeled tracks synthesized prior to HU addition undergo shortening, which indicates degradation of the newly synthesized DNA strands after fork stalling (Schlacher et al. 2011). We did not observe this phenotype in the ATRi samples, indicating that ATR is not required in this BRCA2–RAD51-dependent fork protection pathway (Fig. 2F).
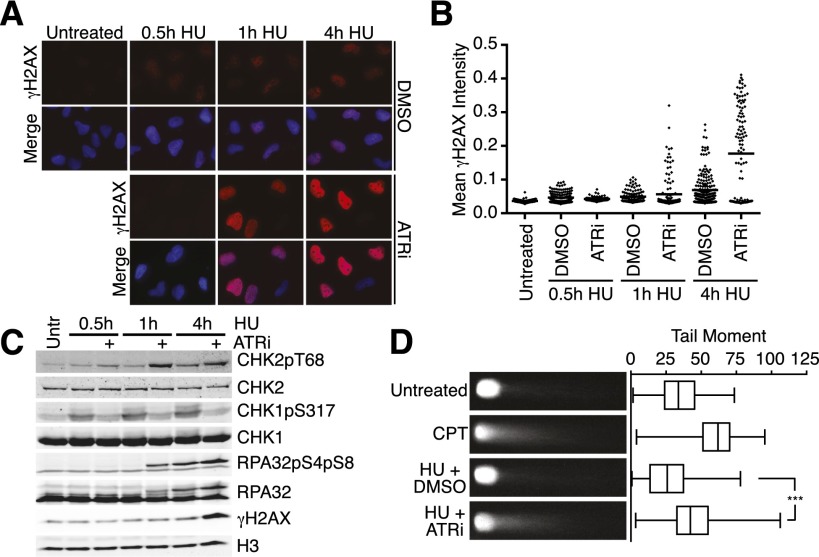
To examine what is happening to the stalled forks when ATR is inhibited, we first examined the levels of DNA damage in ATRi-treated cells. H2AX phosphorylation (γH2AX) provides an indirect marker for DNA damage, since it is phosphorylated at stalled forks and at DSBs. As expected, HU treatment alone causes low but detectable levels of γH2AX, which increase over time (Fig. 3A,B). In the presence of ATRi, γH2AX is absent at the earliest time points of HU treatment, indicating that ATR is required to phosphorylate H2AX near stalled replication forks, consistent with previous observations (Sirbu et al. 2011). However, by 1 h in HU, the γH2AX in ATR-inhibited cells exceeds that in controls. At this time point, a few cells display large, poorly defined γH2AX foci, but most contain a pan-nuclear staining pattern without discernible foci, suggesting rapid spreading through the chromatin. These data suggest the activation of the DSB-sensing kinases ATM and DNA-PKcs between 30 and 60 min of HU treatment when ATR is inhibited. Indeed, we detected elevated RPA32 S4/S8 phosphorylation (a DNA-PK-dependent phosphorylation site) and CHK2 T68 phosphorylation (an ATM-dependent phosphorylation site) in ATR-inhibited cells compared with controls with kinetics similar to γH2AX (Fig. 3C). Thus, a DSB may form at the stalled fork, generating the ATM- and DNA-PKcs-activating signal.
Figure 3.
Stalled replication forks collapse into DSBs when ATR is acutely inhibited. (A,B) U2OS cells were treated with 3 mM HU in the presence or absence of 5 μM ATRi for the indicated times before preparation for immunofluorescence using anti-γH2AX antibodies. Dot plot of mean γH2AX intensity per nucleus is shown in B. (C) U2OS were cells treated with 3 mM HU in the presence or absence of 5 μM ATRi for the indicated times. Following treatment, cell lysates were separated by SDS-PAGE and then immunoblotted to detect the indicated proteins and phosphorylation levels. (D) U2OS cells were treated for 1 h with 1 μM CPT or for 4 h with 3 mM HU in the presence or absence of 5 μM ATRi. Neutral COMET assay was performed, and at least 100 individual cells were scored for tail moment using CometScore software. Representative images and a box and whisker plot are shown. Samples were compared with one-way ANOVA (P < 0.0001). Bonferroni's multiple comparison test was used as a follow-up to compare untreated versus CPT (P < 0.0001) and HU+DMSO versus HU+ATRi (P < 0.0001).
To provide direct evidence of DSB formation, we used a neutral COMET assay. The combination of HU and ATRi treatment caused an increase in COMET tail moment compared with controls, similar to what is caused by high-dose camptothecin (CPT) treatment (Fig. 3D), confirming DSB formation and replication fork collapse in ATR-inhibited cells.
ATR inhibition causes nascent-strand ssDNA formation
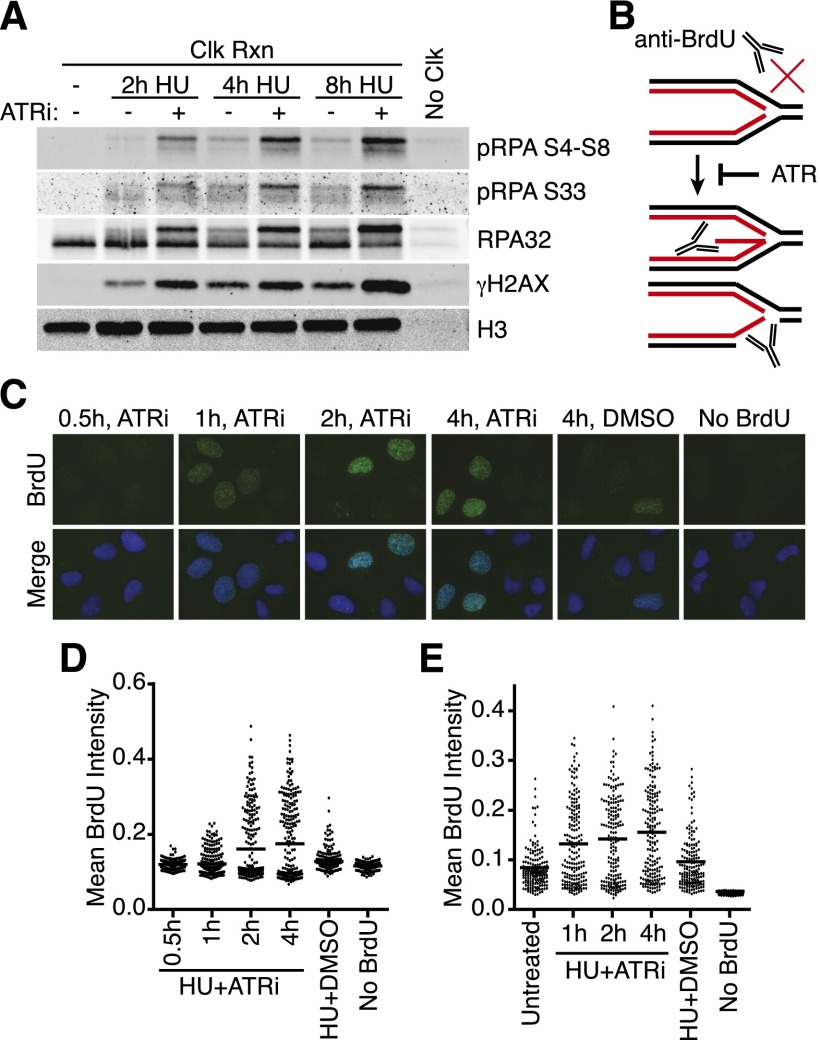
To confirm that the γH2AX and RPA32 hyperphosphorylation that happens in cells treated with HU and the ATRi occur at replication forks, we used iPOND (Sirbu et al. 2011, 2012). iPOND uses click chemistry to conjugate biotin to a nucleoside analog (EdU) incorporated in newly synthesized DNA, thereby permitting a single-step purification of proteins near the replication fork. As expected, the γH2AX is associated with the newly synthesized DNA strands (Fig. 4A). Additionally, we observed a striking accumulation of hyperphosphorylated RPA purified with the nascent DNA when ATR is inhibited (Fig. 4A). Quantitative mass spectrometry following the iPOND purification also confirmed an increase in total RPA in the HU+ATRi conditions (B Sirbu and D Cortez, unpubl.). iPOND only purifies RPA bound to or immediately adjacent to the EdU-labeled nascent strand and therefore does not detect an increase in RPA in cells treated with HU alone (Sirbu et al. 2011). Thus, the increased abundance of RPA in the iPOND samples may suggest that the EdU-labeled nascent strand becomes single-stranded and bound to RPA when ATR is inhibited.
Figure 4.
ATR inhibition causes both nascent and parental ssDNA accumulation at stalled replication forks. (A) 293T cells were labeled with EdU for 10 min prior to addition of 3 mM HU and 5 μM ATRi as indicated. Samples were processed for iPOND, and captured proteins were separated by SDS-PAGE and then immunoblotted. (B) Model for nascent-strand ssDNA assay. Black and red lines indicate template and nascent DNA strands, respectively. Without DNA denaturation, BrdU antibodies will not recognize intact replication forks but will recognize the labeled, nascent DNA when single-stranded. (C,D) The newly synthesized DNA in replicating U2OS cells was labeled for 10 min with 10 μM BrdU before addition of 3 mM HU and 5 μM ATRi as indicated. “No BrdU” sample is 4-h HU+ATRi treatment without BrdU prelabeling. DMSO samples were labeled with BrdU and treated with 3 mM HU for 4 h. After the indicated treatment times, cells were fixed and stained with antibodies against BrdU without DNA denaturation to selectively detect nascent-strand ssDNA. Representative images are shown in C, and a dot plot of mean BrdU intensity per nucleus is shown in D. (E) Parental DNA in replicating U2OS cells was labeled by the addition of 10 μM BrdU for 20 h followed by a chase into normal growth medium for 2 h before addition of 3 mM HU and 5 μM ATRi for the indicated times. DMSO samples were labeled with BrdU and treated with 3 mM HU for 4 h. Cells were fixed and stained with antibodies against BrdU without DNA denaturation to selectively detect parental-strand ssDNA. Dot plot of mean BrdU intensity per nucleus is shown.
To directly test whether the nascent strand becomes single-stranded at stalled forks in the absence of ATR activity, we developed an assay to selectively detect nascent-strand ssDNA using a short (10 min) BrdU incubation immediately before stalling forks with HU. Under nondenaturing conditions, the BrdU antibody selectively recognizes ssDNA (Fig. 4B). Treatment with HU and DMSO vehicle resulted in little BrdU staining (Fig. 4C, see “4h, DMSO” image). Conversely, treatment with HU and ATRi resulted in robust BrdU staining, indicating that the nascent strand becomes ssDNA (Fig. 4C,D). The nascent-strand ssDNA became detectable after 1 h of treatment and increased dramatically after 2 and 4 h (Fig. 4C,D). The timing of nascent-strand ssDNA formation correlates with the cellular lethality and γH2AX hyperphosphorylation caused by ATR inhibition.
We also tested whether ATR inhibition caused the appearance of parental-strand ssDNA at the replication fork. After labeling overnight with BrdU, we released cells into fresh growth medium for 2 h before treating with HU with or without ATRi. As expected, some parental-strand ssDNA formed during HU treatment alone compared with untreated; however, much more parental-strand ssDNA formed in the ATR-inhibited condition, indicating that ATR inhibition results in increased nascent- and parental-strand ssDNA (Fig. 4E). Thus, fork collapse when ATR is inactivated is characterized by DSBs and a large increase in ssDNA, consisting of both the template and newly synthesized DNA strands.
ATR prevents SLX4- and CtIP-dependent formation of DSBs and nascent-strand ssDNA at stalled forks
We reasoned that the nascent-strand ssDNA could be generated from template-strand resection after DSB formation, branch migration of the stalled fork to yield a reversed fork structure with unequal length nascent DNA strands, or both (Fig. 4B). The MUS81 structure-specific endonuclease generates DSBs during persistent exposure to replication stress agents or in response to silencing or inhibition of replication fork repair proteins such as WRN, SMARCAL1, and CHK1 (Hanada et al. 2007; Franchitto et al. 2008; Forment et al. 2011; Betous et al. 2012). Therefore, we tested whether siRNA depletion of MUS81 affects DSB formation in HU/ATRi-treated cells. Surprisingly, MUS81-depeleted cells showed no difference in γH2AX or DSB formation compared with controls (Fig. 5A; data not shown). We also failed to observe a change in HU/ATRi-induced ssDNA formation with MUS81 knockdown or in MUS81-deficient HCT116 cells (Supplemental Fig. S1).
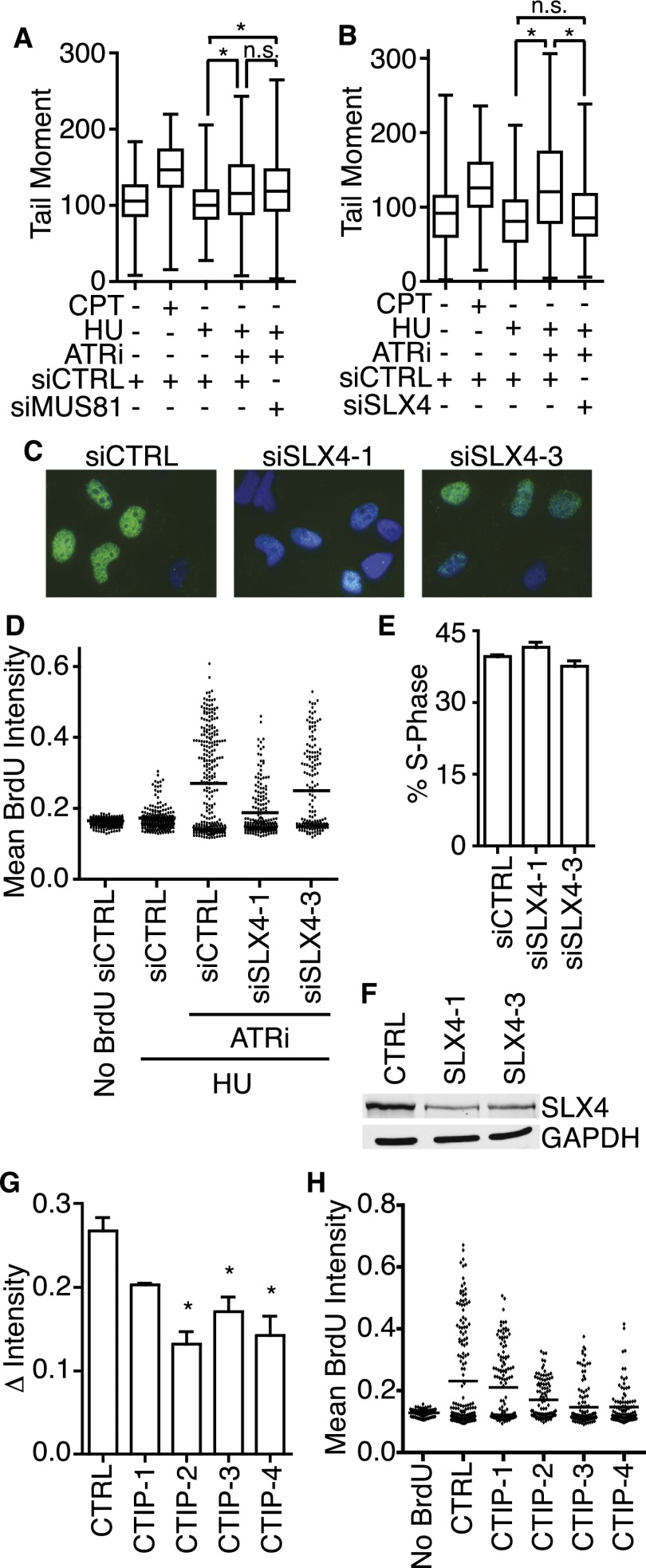
Figure 5.
SLX4 is required to generate DSBs and nascent-strand ssDNA at stalled replication forks when ATR is inhibited. (A,B) U2OS cells were transfected with MUS81, SLX4, or control siRNA prior to treatment for 1 h with 1 μM CPT or for 4 h with 3 mM HU in the presence or absence of 5 μM ATRi. A neutral COMET assay was performed. Samples were compared with one-way ANOVA (P < 0.0001). (A) Bonferroni's multiple comparison test was used as a follow-up to compare siCTRL HU+DMSO versus siCTRL HU+ATRi (P < 0.0001), siCTRL HU+DMSO versus siMUS81 HU+ATRi (P < 0.0001), and siCTRL HU+ATRi versus siMUS81 HU+ATRi (P > 0.05). (B) Bonferroni's multiple comparison test was used as a follow-up to compare siCTRL HU+DMSO versus siCTRL HU+ATRi (P < 0.0001), siCTRL HU+DMSO versus siSLX4 HU+ATRi (P > 0.05), and siCTRL HU+ATRi versus siSLX4 HU+ATRi (P < 0.0001). (C,D) U2OS cells were transfected with nontargeting or SLX4 siRNA and then labeled with 10 μM BrdU for 10 min before addition of 3 mM HU and 5 μM ATRi for 4 h. Samples were then processed to quantitate nascent-strand ssDNA. (C) Representative images of nascent-strand ssDNA assay in transfected cells. (D) Representative dot plot of the mean BrdU intensity per nucleus. (E) U2OS cells transfected with nontargeting or SLX4 siRNA were labeled for 40 min with 10 μM BrdU before harvesting for ethanol fixation, acid denaturation, and staining with BrdU antibodies and propidium iodide to measure the percentage of cells in S phase by flow cytometry. Values represent mean ± SEM of three replicates. (F) U2OS cells transfected with nontargeting or SLX4 siRNA were harvested and lysed. Lysates were separated by SDS-PAGE and immunoblotted with antibodies to detect SLX4 or GAPDH. (G,H) U2OS cells were transfected with nontargeting or CtIP siRNA. Transfected cells were labeled with 10 μM BrdU for 10 min before addition of 3 mM HU and 5 μM ATRi for 4 h. Samples were then processed to detect nascent-strand ssDNA. (G) Quantitation of the percentage of nuclei containing nascent-strand ssDNA; mean ± SEM of three experiments; (*) P < 0.05. (H) Dot plot of mean BrdU intensity per nucleus for a representative experiment is shown.
Multiple other structure-specific nucleases exist in mammalian cells. SLX4 is a molecular scaffold that functions as the structural subunit of the SLX4–SLX1 Holliday junction resolvase and coordinates the SLX1, XPF, and MUS81 nucleases (Fekairi et al. 2009; Svendsen et al. 2009). In contrast to MUS81, silencing SLX4 by siRNA largely abrogated DSB formation in HU+ATRi-treated cells (Fig. 5B). Furthermore, SLX4 depletion also resulted in a large decrease in nascent-strand ssDNA formation (Fig. 5C,D). This result is not due to a change in the S-phase population (Fig. 5E). Furthermore, the response in the nascent-strand ssDNA assay correlates with SLX4 knockdown efficiency, indicating that it is not an off-target effect of the siRNA (Fig. 5F). Silencing SLX1, XPF, MUS81, or the GEN1 nuclease individually had no consistent effect on nascent-strand ssDNA formation (Supplemental Figs. S1–S4). Thus, it is likely that multiple SLX4-coordinated nucleases function redundantly to process HU-stalled replication forks when ATR is inhibited.
Once the fork is cleaved, ssDNA could be generated by DNA end resection. CtIP promotes resection at DSBs through multiple exonucleases (Paull 2010). Silencing CtIP significantly reduced the amount of nascent-strand ssDNA with three of four siRNAs, and the fourth siRNA decreased nascent-strand ssDNA in all experiments but did not reach statistical significance (Fig. 5G,H). CtIP protein levels decreased to nearly undetectable levels with all four siRNAs, and the change in nascent-strand ssDNA intensity was not due to a change in the S-phase population of these cells (Supplemental Fig. S5). These results suggest that at least some of the ssDNA that accumulates at stalled forks when ATR function is inhibited is generated by end resection. Knockdown of individual exonucleases (including EXO1 and DNA2) alone did not result in any change in nascent-strand ssDNA, consistent with redundancy in end resection mechanisms (Supplemental Fig. S6).
Nascent-strand ssDNA formation involves replication fork remodeling
We next asked whether any fork processing steps occur upstream of SLX4-dependent cleavage. Specifically, we hypothesized that fork remodeling enzymes may be needed to generate a reversed replication fork structure that looks like a Holliday junction (or “chicken foot”), which is a preferred substrate for SLX4-dependent endonucleases. Therefore, we tested whether nascent-strand ssDNA formation depends on enzymes known to catalyze replication fork regression. SMARCAL1 is a DNA translocase that can anneal DNA strands and perform replication fork regression at stalled forks with leading strand gaps (Betous et al. 2012, 2013). Silencing SMARCAL1 with four siRNAs decreases nascent-strand ssDNA-positive cells compared with controls (Fig. 6A–C; Supplemental Fig. S7). Silencing other enzymes capable of catalyzing fork regression, including FANCM, HLTF, ZRANB3, and BLM, did not decrease nascent-strand ssDNA (Supplemental Figs. S8, S9).
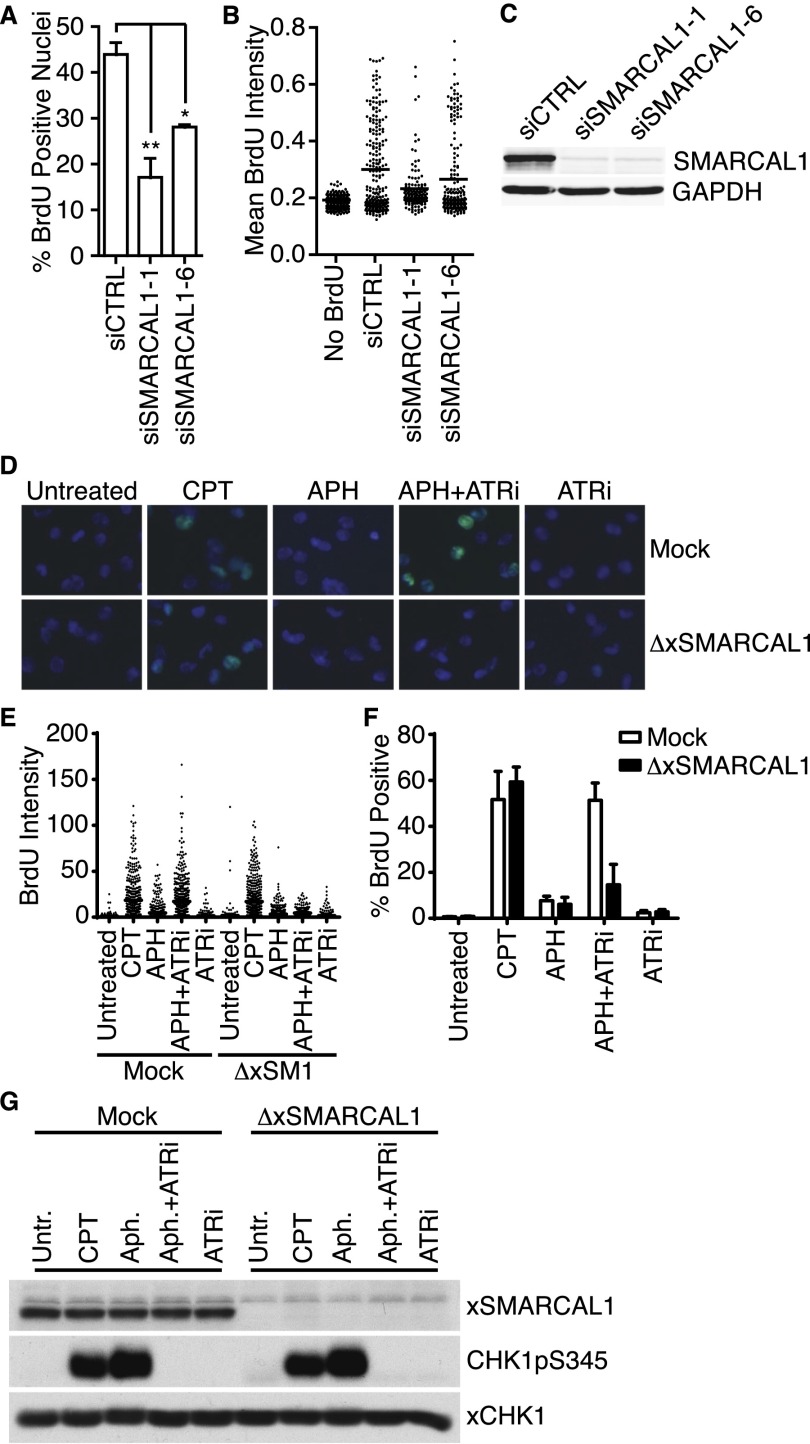
Figure 6.
SMARCAL1 is required for the generation of nascent-strand ssDNA when ATR is inactivated. (A,B) U2OS cells were transfected with control or SMARCAL1 siRNA and then labeled with 10 μM BrdU for 10 min before addition of 3 mM HU and 5 μM ATRi for 4 h. Samples were then processed to quantitate nascent-strand ssDNA. (A) Bars represent mean ± SEM of the percentage of BrdU-positive cells across at least five experiments. (B) Representative dot plots of mean BrdU intensity per nucleus. (C) U2OS cells transfected with control or SMARCAL1 siRNA were harvested and lysed. Lysates were separated by SDS-PAGE and immunoblotted with antibodies to detect SMARCAL1 or GAPDH. (D–G) Sperm chromatin (4000 nuclei per microliter) was replicated in low-speed Xenopus extract containing DMSO or ATRi. After 10 min, extracts were labeled with 50 μM BrdU for 20 min prior to addition of DMSO, 50 μM CPT, or 100 μM aphidicolin (APH) as indicated. Sixty minutes after addition of chromatin, nuclei were fixed and spun down onto coverslips through a glycerol cushion. Where indicated, extracts were either mock- or xSMARCAL1-depleted. (D–F) Nuclei from extracts were processed to quantitate nascent-strand ssDNA. (D) Representative images of BrdU staining from each sample. (E) Representative dot plots of BrdU intensity per nucleus, measured using ImageJ. (F) Quantitation (mean ± SEM) of the percentage of BrdU-positive nuclei across three independent experiments. (G) Extracts were harvested, separated by SDS-PAGE, and immunoblotted with antibodies to detect xSMARCAL1, CHK1 pS345, or xCHK1.
To further confirm the requirement of SMARCAL1 in the fork collapse associated with ATR inactivation, we examined whether a similar phenotype could be observed in the Xenopus cell-free replication system. In this system, replication of sperm chromatin in the presence of the DNA polymerase inhibitor aphidicolin causes ATR activation due to polymerase and helicase uncoupling (Byun et al. 2005). Prelabeling the newly synthesized DNA strands with BrdU allowed us to observe the appearance of nascent-strand ssDNA when either CPT was added to promote fork collapse or ATR was inhibited in the presence of aphidicolin (Fig. 6D–F). Depleting xSMARCAL1 from the replicating extracts prevented the nascent-strand ssDNA generation only in the context of ATR inhibition with aphidicolin (Fig. 6D–F). xSMARCAL1 depletion had no effect on CPT-induced ssDNA or the activation of the ATR checkpoint (Fig. 6G). Thus, just as in mammalian cells, xSMARCAL1 is at least partially responsible for the aberrant replication fork processing in the Xenopus cell-free system when ATR is inactivated.
These data suggest that when ATR is not active, SMARCAL1 remodels stalled replication forks and thereby provides a substrate for endonuclease cleavage and DNA end resection. Silencing SMARCAL1 in the mammalian system only yielded a partial suppression of the ssDNA phenotype, indicating that other mechanisms may also be important.
Replication stress induces SMARCAL1 phosphorylation after it accumulates at stalled forks and binds DNA
One mechanism to explain these results is that ATR prevents aberrant fork processing by directly regulating SMARCAL1. The lack of effect of xSMARCAL1 depletion on CPT-induced ssDNA formation would be consistent with this mechanism since ATR is active in these circumstances. ATR can phosphorylate SMARCAL1 (Bansbach et al. 2009), but the functional consequences have not been described. Therefore, we investigated whether SMARCAL1 phosphorylation is a mechanism by which ATR stabilizes stalled replication forks.
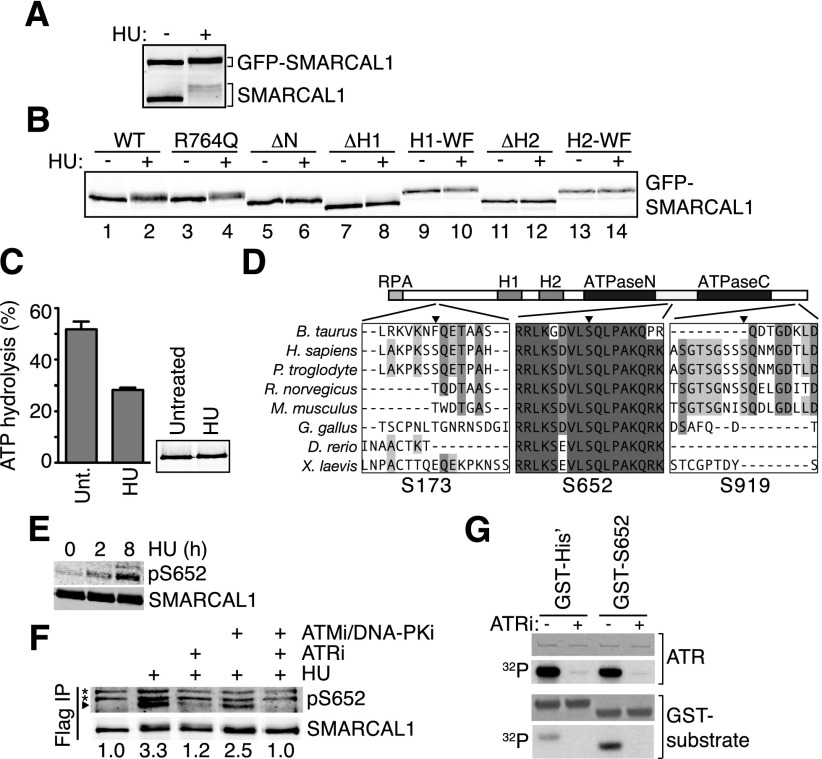
SMARCAL1 phosphorylation in response to replication stress can be monitored by a pronounced gel mobility shift of the endogenous protein (Fig. 7A; Bansbach et al. 2009). GFP-SMARCAL1 protein expressed at near endogenous levels in HEK293T cells also is phosphorylated in response to HU (Fig. 7A). SMARCAL1 localizes to stalled forks via an interaction between its N terminus and RPA2 (Bansbach et al. 2009; Ciccia et al. 2009; Yuan et al. 2009; Yusufzai et al. 2009). However, abrogation of stalled fork localization with a mutant that does not bind RPA (ΔN) caused a loss of damage-dependent phosphorylation, indicating that SMARCAL1 must be in proximity to the stalled fork to be phosphorylated (Fig. 7B). Deletion or point mutations in the SMARCAL1 HARP1 domain decrease but do not eliminate DNA binding (Betous et al. 2012) and decrease damage-dependent SMARCAL1 phosphorylation (Fig. 7B). Deletion or mutation of the HARP2 domain eliminates DNA binding (Betous et al. 2012) and eliminates phosphorylation (Fig. 7B). These HARP mutants retain the ability to localize to stalled forks (Supplemental Fig. S10), indicating that both localization and DNA binding are required for damage-dependent phosphorylation. Thus, phosphorylation occurs after SMARCAL1 is positioned to catalyze fork remodeling. However, the ability of SMARCAL1 to translocate on DNA is not a prerequisite for phosphorylation, since the ATPase-deficient R764Q mutant exhibits the same mobility shift as the wild-type protein in response to HU (Fig. 7B).
Figure 7.
ATR phosphorylates SMARCAL1 on S652 after SMARCAL1 binds to DNA at stalled forks. (A,B) HEK293T cells were transfected with small amounts of GFP-SMARCAL1 wild-type (WT) and mutant expression vectors to minimize overexpression. ΔH1 and ΔH2 are deletions of the HARP1 and HARP2 domains, respectively, while H1-WF and H2-WF are point mutants in each HARP domain (Betous et al. 2012). Cells were treated with 2 mM HU for 16 h where indicated prior to lysis, separation by SDS-PAGE, and immunoblotting with SMARCAL1 antibody. (C) SMARCAL1 was purified from untreated (unt.) or HU-treated (2 mM, 16 h) HEK293T cells and was used to measure ATPase activity in the presence of 5 nM forked DNA substrate. Error bars are standard deviation from three experiments (P = 0.0007, two-tailed unpaired t-test). The inset is an immunoblot showing equal amounts of protein used in each condition. The purified protein was treated with λ phosphatase before immunoblotting to eliminate the gel mobility shift and allow more accurate quantitation of protein concentration. (D) ClustalW was used to align SMARCAL1 from the indicated organisms. The position of the phosphorylated SQ residues S173, S652, and S919 relative to protein domains is depicted. (E,F) HEK293T cells were transfected with Flag-SMARCAL1 and treated with HU for the indicated times. Kinase inhibitors were added as indicated. Flag-SMARCAL1 was immunoprecipitated from cell lysates, separated by SDS-PAGE, and immunoblotted with either total SMARCAL1 antibody or pS652-specific antibody. (*) Nonspecific bands. Images were captured and quantitated relative to total SMARCAL1 using an Odyssey imaging system. (G) Purified ATR–ATRIP (ATR-interacting protein) complex phosphorylates a GST-S652 peptide in vitro. ATRi was added where indicated to ensure specificity of the kinase in the reaction. Shown are images of a Coomassie-stained gel to visualize the amount of ATR and GST protein in the reactions or an autoradiogram (32P) of the gel to visualize phosphorylation.
S652 phosphorylation reduces SMARCAL1 activity
To test the functional significance of damage-dependent phosphorylation, we isolated SMARCAL1 before and after exposure to HU. Phosphorylated SMARCAL1 isolated from HU-treated cells retains only 50% activity compared with SMARCAL1 isolated from untreated cells (Fig. 7C). This suggests that HU-dependent post-translational modifications inactivate SMARCAL1. This difference in activity is not due to a difference in cell cycle stage induced by HU, since the cells were synchronized and released into S phase prior to the experiment.
Next, we used mass spectrometry and phosphopeptide mapping to identify damage-induced phosphorylation sites. Mass spectrometry of SMARCAL1 purified from HU-treated cells identified phosphorylation of S652, which lies within a consensus sequence (SQ) for the ATR kinase (Supplemental Fig. S11) on a linker between the two lobes of the SMARCAL1 ATPase domain (Fig. 7D). We identified two additional SQ phosphorylation sites (S173 and S919) through phosphopeptide mapping (Supplemental Fig. S11). These phosphorylation sites are within poorly conserved, unstructured regions near the N and C termini, respectively (Fig. 7D).
Since S652 is highly conserved, we raised phosphopeptide-specific antibodies to this phosphorylation site. These antibodies show that S652 phosphorylation is increased in response to HU (Fig. 7E). Phosphorylation is largely ATR-dependent in cells (Fig. 7F), and purified ATR can phosphorylate S652 in an in vitro kinase assay (Fig. 7G).
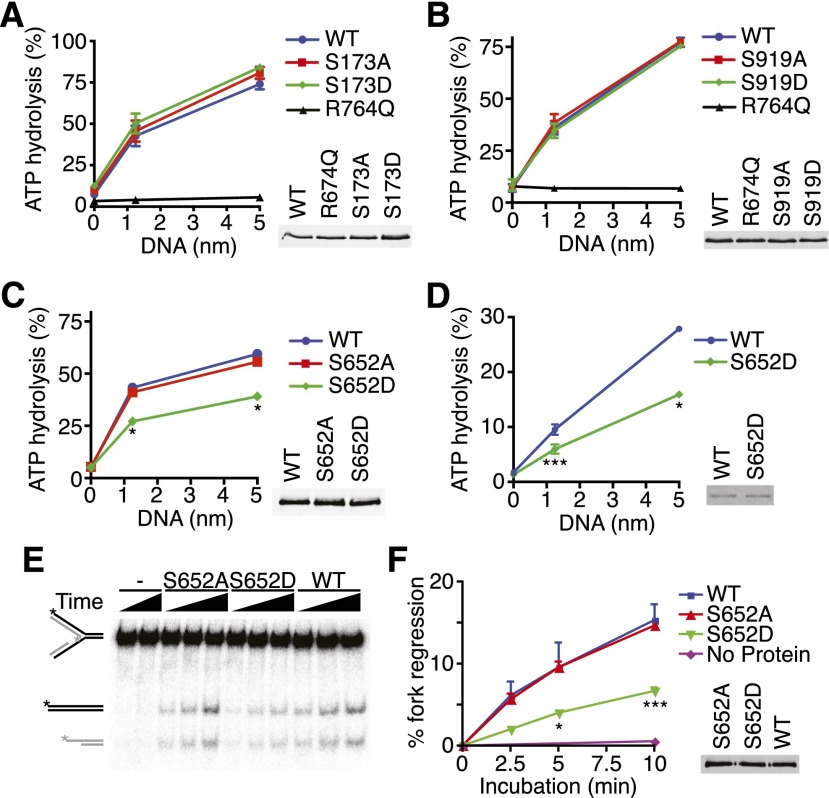
To determine whether these phosphorylation sites are functionally important, we tested whether phosphorylation alters SMARCAL1 localization or biochemical functions. Phosphorylation of S173, S652, or S919 does not regulate SMARCAL1 localization to stalled replication forks, since neither nonphosphorylatable nor phosphomimetic mutations in these residues alter their ability to colocalize with RPA foci in response to DNA damage (Supplemental Fig. S12). Mutations of S173 or S919 also did not affect the DNA-stimulated ATPase activity of SMARCAL1 (Fig. 8A,B). In contrast, the phosphomimetic S652D mutant is a much less active DNA-stimulated ATPase compared with the wild type (Fig. 8C,D), even though it has indistinguishable DNA-binding activity (Supplemental Fig. S13). The extent of ATPase inhibition caused by the phosphomimetic mutation is similar to the inhibition of SMARCAL1 observed after HU-treatment (Fig. 7C), suggesting that S652 phosphorylation is a major mechanism by which SMARCAL1 is inhibited in cells exposed to persistent replication stress. Combining the S173D, S652D, S919D mutations did not further alter SMARCAL1 activity (data not shown), and mutation of S652 to alanine had no effect on SMARCAL1 activity, indicating that the serine itself is not required for function.
Figure 8.
SMARCAL1 phosphorylation on S652 inhibits its ATP-dependent fork remodeling activity. (A–D) The indicated Flag-SMARCAL1 proteins were purified from HEK293T cells (A–C) or baculovirus-infected insect cells (D), and their ATPase activity was measured in the presence of increasing concentrations of splayed arm DNA substrate. The insets in A–C are representative immunoblots, and the inset in D is a Coomassie-stained gel showing equal amounts of wild-type and mutant SMARCAL1 proteins used. Error bars in all panels represent SEM (n = 3) and in many cases were smaller than the symbol. (*) P < 0.0002; (**) P < 0.002; (***) P < 0.05. (E,F) The fork regression activity of purified SMARCAL1 proteins was assayed on a model replication fork substrate schematized on the far left. (See Supplemental Table S1 for details.) A representative DNA gel (E) and quantitation of three independent experiments (F) (mean and SEM) are shown. The inset is a representative immunoblot showing equal amounts of wild-type and mutant SMARCAL1 proteins.
Consistent with its reduced ATPase activity, we also observed a significant reduction in the ability of the phosphomimetic S652D SMARCAL1 to catalyze fork regression compared with the S652A or wild-type proteins, indicating that SMARCAL1 phosphorylation at S652 limits its ability to perform branch migration (Fig. 8E,F). Combined with the observation that SMARCAL1 must first bind DNA before it can be phosphorylated, these data are consistent with a model in which SMARCAL1 initially is recruited to a damaged fork, binds the DNA to perform a fork remodeling activity, and then is phosphorylated on S652 to regulate this activity.
Too much SMARCAL1 activity in cells phenocopies the effect of ATR inhibition on fork collapse
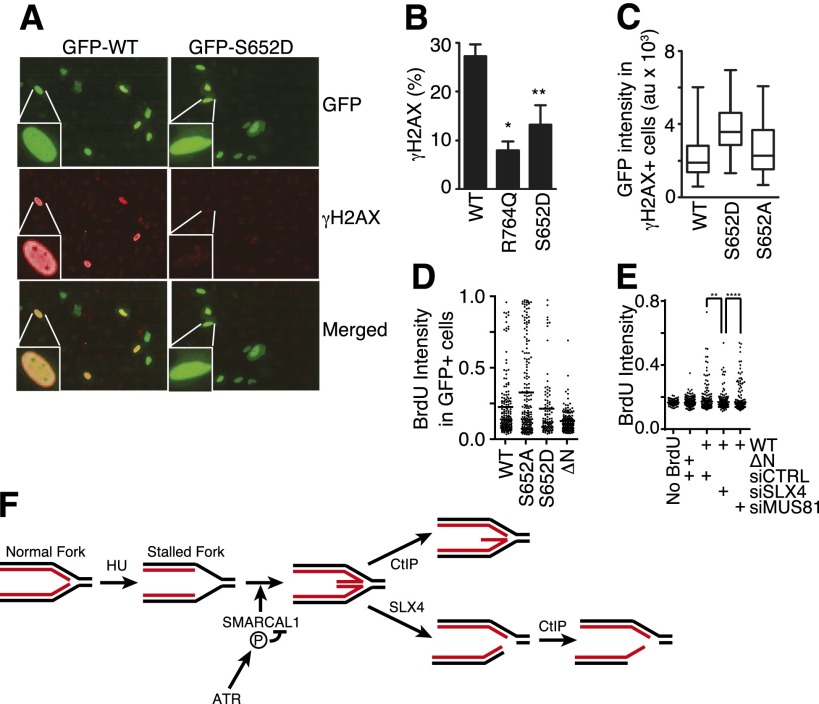
To confirm that ATR-dependent SMARCAL1 phosphorylation also inhibits SMARCAL1 activity in cells at stalled replication forks, we used an assay that measures the consequences of uncontrolled SMARCAL1 activity. Expression of wild-type SMARCAL1 from a strong promoter causes the induction of γH2AX throughout the nucleus in a pan-nuclear pattern (Fig. 9A–C; Bansbach et al. 2009) similar to what is observed when ATR is inhibited in HU-treated cells (Fig. 3A), supporting a functional link between ATR and SMARCAL1. The induction of pan-nuclear γH2AX by overexpressed SMARCAL1 is dependent on DNA replication, the ability of SMARCAL1 to localize to stalled forks, and its enzymatic activity (Bansbach et al. 2009). Like ATR inhibition, overexpression of SMARCAL1 causes increased levels of ssDNA at replication forks (Fig. 9D,E). Furthermore, just as is the case with ATR inhibition, knockdown of SLX4, but not MUS81, resulted in a decrease in ssDNA formation in cells expressing SMARCAL1 (Fig. 9E). Taken together, these data indicate that the phenotype of cells overexpressing SMARCAL1 is similar to that induced when ATR is inhibited, as predicted if ATR phosphorylates SMARCAL1 as a mechanism of limiting its activity and preventing fork collapse.
Figure 9.
Phosphorylation of SMARCAL1 at S652 decreases its activity at DNA replication forks in cells. (A–C) GFP-SMARCAL1 wild-type and mutant proteins were overexpressed in U2OS cells. Cells were stained with DAPI to mark the nucleus and antibodies to γH2AX. Images were acquired using an Opera automated confocal microscope, and the levels of GFP-SMARCAL1 and γH2AX levels were quantitated in each nucleus using Columbus software. (A) Representative images. (B) Data represent the percentage of cells expressing between 500 and 2500 arbitrary units of GFP-SMARCAL1 that contain a mean γH2AX intensity of >1000 arbitrary units. Error bars represent SEM from three independent experiments. (*) P = 0.0007; (**) P = 0.023. (C) The expression level of GFP-SMARCAL1 (as measured by GFP intensity) in each cell that had a γH2AX intensity of >1000 arbitrary units is plotted in box and whisker format; significantly higher GFP-SMARCAL1 S652D protein levels were needed to induce γH2AX than either wild-type or S652A protein (P < 0.0001). (D) GFP-SMARCAL1 proteins with the indicated mutations were expressed in U2OS cells. BrdU was added to the culture medium 16 h prior to fixation and staining in nondenaturing conditions to measure the total level of ssDNA. The mean intensity (arbitrary units) of BrdU staining per GFP-SMARCAL1-expressing cell is graphed. The line indicates the mean value in each population (Mann-Whitney test, S652A vs. S652D; P = 0.013). (E) GFP-SMARCAL1 proteins were expressed in U2OS. These cells were then transfected with control, MUS81, or SLX4 siRNA. BrdU was added to the culture medium 16 h prior to fixation and staining in nondenaturing conditions to measure the total level of ssDNA. The mean intensity (arbitrary units) of BrdU staining per cell is graphed. The line indicates the mean value in each population (Mann-Whitney test, siCTRL vs. siSLX4; P = 0.0012; siSLX4 vs. siMUS81; P < 0.0001). (F) Model for nascent-strand ssDNA generation at stalled forks. Black and red lines represent template and nascent strands, respectively. HU causes uncoupling of the replicative helicase and polymerases, resulting in template-strand ssDNA at the replication fork. ATR prevents aberrant fork remodeling by the SMARCAL1 enzyme. In the absence of ATR-dependent SMARCAL1 S652 phosphorylation, a Holliday junction-like structure may persist at the fork and is cleaved by SLX4-dependent nucleases, generating a DSB. CtIP-dependent nucleases then resect the break, yielding nascent-strand ssDNA. CtIP may also process a reversed fork structure prior to SLX4 cleavage, which could contribute to the nascent-strand ssDNA formation.
If ATR-catalyzed S652 phosphorylation decreases SMARCAL1 activity to prevent aberrant fork processing, then the S652D phosphomimetic mutation should yield a protein that is less capable of inducing the pan-nuclear γH2AX phenotype. Indeed, overexpression of GFP-SMARCAL1 S652D induces significantly less pan-nuclear H2AX phosphorylation compared with the wild-type protein (Fig. 9A,B). We did observe some cells with pan-nuclear γH2AX when S652D is overexpressed; however, those cells contained, on average, twice the amount of S652D protein overexpression compared with the level of the wild-type or S652A proteins needed to induce pan-nuclear γH2AX (Fig. 9C).
Finally, consistent with our model, we also found that overexpression of the S652D SMARCAL1 protein yielded less ssDNA than the S652A mutant, although it does remain slightly more active than a SMARCAL1-ΔN protein that cannot localize to stalled forks because it lacks an RPA-binding domain (Fig. 9D). These data suggest that S652 of SMARCAL1 is one of the ATR phosphorylation targets necessary to prevent fork collapse and emphasize the need for properly regulating SMARCAL1 to achieve successful stalled fork stabilization and repair.
Discussion
Our data are consistent with a specific model of how ATR prevents fork collapse in the context of replication stress (Fig. 9F). ATR maintains the integrity of the replication fork through regulation of several proteins, including direct phosphorylation of S652 of SMARCAL1. If properly regulated, SMARCAL1 participates in maintaining fork stability and promoting fork restart. However, in the absence of ATR regulation, SMARCAL1 catalyzes excessive replication fork reversal or other DNA remodeling that generates a substrate for the SLX4-dependent Holliday junction endonucleases. These enzymes cleave the reversed replication fork into a one-ended DSB, which CtIP-dependent endonucleases further process into ssDNA of both the parental and nascent strands. Consistent with this model, fork reversal is observed in replication checkpoint-deficient Saccharomyces cerevisiae cells in response to replication stress (Lopes et al. 2001), although no responsible yeast enzymes have been identified. In addition, nucleases process stalled forks in checkpoint-defective yeast cells (Cotta-Ramusino et al. 2005). This model also explains why both too little and too much SMARCAL1 in a cell causes replication fork collapse (Bansbach et al. 2009). Finally, the activity of unregulated SMARCAL1 in causing fork collapse is not limited to mammalian cells, since we also observed it in a Xenopus cell-free replication system when ATR is inhibited.
ATR as a drug target
The essential function of ATR to maintain cell viability is linked to its requirement to complete DNA replication and separable from its G2 checkpoint activity (Nam et al. 2011). Defining how ATR promotes the completion of DNA replication is essential to understanding the major pathway that controls genome integrity in S phase and identifying the mechanism of action of ATR-directed drugs. Our data indicate that acute ATR inhibition results in a rapid decrease in the rate of fork elongation. This effect likely results from loss of checkpoint control of origin firing, leading to a decrease in elongation rates due to depletion of essential replication factors such as nucleotides or replication proteins. The deregulation of origin timing is unlikely to be cell-lethal by itself, since it happens within 20 min of adding the ATRi, but loss of cell viability requires several hours without additional genotoxic agents. Furthermore, suppression of origin firing by the addition of CDK2 or CDC7 inhibitors did not improve the viability of ATRi-treated cells (Supplemental Fig. S14). More likely the essential function of ATR is to stabilize or repair stalled replication forks, which collapse when ATR is inhibited. Deregulated origin firing would exacerbate this problem by creating additional stalled forks. Consistent with this interpretation, viability is lost within 45 min in cells treated with both ATRi and HU. The increase in irreversible replication fork collapse, concomitant formation of DSBs, and excess ssDNA when ATR is inactivated suggest improper enzymatic processing of the stalled fork DNA. These fork processing events may be largely unsuccessful attempts to repair the damaged fork.
Aberrant stalled fork processing when ATR is inactivated
Previous studies implicated the MUS81 endonuclease as an enzyme that cleaves persistently stalled replication forks after several hours of HU treatment, leading to the appearance of high levels of γH2AX (Hanada et al. 2007). MUS81 was also reported to generate DNA damage in cells treated with a CHK1 inhibitor (Forment et al. 2011). However, we found no effect of MUS81 on the generation of DSBs in cells treated with the ATRi. Instead, we observed a strong dependency on SLX4. Furthermore, we found that SLX4 is required for nascent-strand ssDNA formation. SLX4 is a scaffolding protein for several endonucleases, including MUS81, SLX1, and XPF (Fekairi et al. 2009; Svendsen et al. 2009). Since we did not observe a significant effect of silencing any of these nucleases individually, our data suggest that more than one of them may function redundantly to cleave stalled forks when ATR is inhibited. CtIP-dependent exonucleases then process the cleaved fork to generate nascent-strand ssDNA. SLX4 depletion yields a complete loss of DSBs as measured by the COMET assay but only a partial suppression of the nascent-strand ssDNA. Therefore, it is possible that CtIP also directs resection of a stalled, regressed fork when ATR is inhibited prior to DSB formation, thereby generating a nascent-strand ssDNA overhang with or without SLX4-dependent cleavage.
A preferred substrate of SLX4-dependent endonucleases resembles a Holliday junction, which can form at a stalled fork due to either torsional stress or enzymatic processing. Our data are consistent with enzymatic processing of the fork by the SMARCAL1 fork-regressing enzyme to generate a preferred substrate for SLX4-dependent cleavage when ATR is inhibited. ATR phosphorylates SMARCAL1, and previous studies indicate that excess SMARCAL1 activity causes replication fork problems (Bansbach et al. 2009). Thus, the fork remodeling activities of SMARCAL1 promotes fork repair in some cases but can threaten genome stability if not properly regulated. SMARCAL1 is not needed for fork collapse in all circumstances. For example, our data indicate that CPT treatment induces fork collapse and ssDNA formation independently of xSMARCAL1. Since ATR would be active in CPT-treated cells, SMARCAL1 would be appropriately regulated.
Consistent with our model, we found that phosphorylated SMARCAL1 purified from cells treated with HU is significantly less active than SMARCAL1 purified from untreated cells. We identified three damage-induced phosphorylation sites on SMARCAL1, including S652, which is phosphorylated by ATR. Phosphomimetic mutations in S652 yield a protein that is significantly less active in catalyzing ATP hydrolysis and fork reversal in vitro and also less active in cells. S652 lies within the linker between the two RecA lobes of the ATPase domain. Flexibility of this linker is needed for conformational changes required for catalysis (Durr et al. 2005; Sprouse et al. 2006; Lewis et al. 2008). Since S652 phosphorylation does not impair SMARCAL1 localization or DNA-binding activity, we suspect that phosphorylation of the linker reduces the ability of SMARCAL1 to undergo this conformational change and thereby inhibits its translocase activity. SMARCAL1 phosphorylation requires it to first localize to the stalled fork and bind DNA. Thus, SMARCAL1 likely acts at damaged forks prior to ATR-dependent phosphorylation, which serves as a mechanism of limiting its activity.
Overexpression of active SMARCAL1 causes pan-nuclear γH2AX staining. This effect requires localization of SMARCAL1 to replication forks and its enzymatic activity. The exact source of this staining pattern is not clear, although it forms well before there is any evidence of apoptosis, is chromatin-associated (data not shown), and is also observed in cells treated with ATRi and HU. Thus, our data suggest that it is linked to the aberrant fork processing and collapse that occurs when ATR cannot regulate SMARCAL1. Consistent with this interpretation, the phosphomimetic S652D SMARCAL1 protein is less capable of inducing this phenotype and is less active as a fork regression enzyme. Furthermore, like ATR inhibition, SMARCAL1 overexpression also induces SLX4-dependent excess ssDNA formation at stalled forks, and this effect is alleviated by a phosphomimetic mutation in S652.
Another prediction of our model is that the S652A mutant should retain too much activity in HU-treated cells, perhaps generating some fork collapse events even when ATR is not inhibited (assuming S652 phosphorylation is not completely redundant with other mechanisms). Unfortunately, since SMARCAL1 overexpression causes significant perturbations to DNA replication on its own and the S652 site does not appear to be regulated properly on the exogenously expressed SMARCAL1 wild-type protein (Supplemental Fig. 11), we were unable to do effective genetic complementation experiments to examine the effect of the S652A mutant on fork collapse and ssDNA generation. Future experiments—either knocking in the mutation into the genome or using expression systems with native promoter and gene structures—will be needed.
Neither SMARCAL1 nor SLX4 silencing completely prevents nascent-strand ssDNA formation caused by ATR inhibition, suggesting that additional aberrant fork processing events remain to be identified. The lack of complete suppression by SMARCAL1 silencing may also be due to the competing increase in ssDNA generated due to the need for some properly regulated SMARCAL1 protein to protect stalled forks. In addition, it seems likely that additional mechanisms regulate the activity of SMARCAL1. Our phosphopeptide maps indicate that S652, S173, and S919 are the major HU-induced phosphorylation sites on SMARCAL1; however, mutation of all three serines to alanines does not eliminate the HU-induced mobility shift of the protein on SDS-PAGE gels (data not shown). Thus, there must be additional regulatory post-translational modifications that could also contribute to SMARCAL1 regulation. Finally, the ATRi may not completely block all ATR activity, so the results observed may not be equivalent to a complete loss of ATR function.
Conclusions
ATR signaling has long been known to regulate DNA replication and prevent fork collapse, based largely on studies in yeast. However, the molecular mechanisms of what ATR does to prevent fork collapse and even what fork collapse is in mammalian cells are not well understood. Our data define an ATR-dependent replication fork protection mechanism in human cells that is critical for genome maintenance and cell viability. These studies also help define the mechanism of action of ATR targeted therapeutic agents. Specifically, combinations of selective ATR pathway inhibitors with either intrinsic or added replication stress provide a rapid mechanism of cancer cell killing due at least in part to aberrant processing of stalled replication forks.
Materials and methods
Cell culture, clonogenic survival, and ATRi
U2OS cells were maintained in DMEM with 7.5% FBS. RPE-hTERT cells were maintained in DMEM/F12 medium with 10% FBS, 0.348% sodium bicarbonate, and 2 mM L-glutamine. MUS81−/− HCT116 cells (Shimura et al. 2008) were maintained in McCoy's 5A medium with 10% FBS. For clonogenic assays, U2OS cells were seeded onto 60-mm cell culture plates at 200–5000 cells per plate in triplicate. The following day, cells were treated with drug, washed, and released into fresh growth medium for 10–14 d prior to staining with methylene blue. The VE-821 ATR-selective inhibitor (Reaper et al. 2011) was synthesized by the Vanderbilt Institute for Chemical Biology Chemical Synthesis facility and used in most experiments. ATR inhibition in Xenopus extracts was achieved with 64 μM 4-{4-[(3R)-3-methylmorpholin-4-yl]-6-[4-(methylsulfonyl)piperidin-4-yl]pyrimidin-2-yl}-1H-indole (Foote et al. 2010), synthesized by CheminPharma. The minimum concentration needed to achieve a near-complete block of ATR-dependent CHK1 phosphorylation was determined and used in all experiments. This concentration did not inhibit ATM, DNA-PKcs, or mTOR kinases (Reaper et al. 2011; data not shown). siRNA transfections were performed using either HiPerfect (Qiagen) or Dharmafect 1 (Dharmacon) at a final siRNA concentration of 10 nM. siRNAs were purchased from Dharmacon or Ambion.
Fiber labeling
hTERT-RPE cells were labeled with 20 μM IdU for 20 min, rinsed twice with equilibrated HEPES-buffered saline, treated with 2 mM HU or 3 μM ATRi as indicated, and then labeled with 100 μM CldU (Sigma) for 20 min. Cells were harvested and resuspended in ice-cold PBS. Two microliters of cell suspension was deposited on a microscope slide, and 10 μL of spreading buffer (200 mM Tris-HCl at pH 7.4, 0.5% SDS, 50 mM EDTA) was added to the cells on the slide for 6 min. Next, the slides were tilted to 15° to stretch the DNA fibers. Following fixation in a 3:1 solution of methanol:acetic acid, the DNA was denatured with 2.5N HCl, blocked with PBS containing 10% goat serum and 0.1% Triton X-100, and stained with rat anti-CldU (Abcam) and mouse anti-IdU (Becton Dickinson) diluted in PBS containing 10% goat serum and 0.1% Triton X-100 followed by secondary antibodies (Alexa-594 goat anti-rat IgG and Alexa-488 goat anti-mouse; Invitrogen).
Neutral COMET assays and iPOND
The neutral COMET assays were performed in accordance with the manufacturer's (Trevigen) instructions. The iPOND procedure was carried out as described previously (Sirbu et al. 2011, 2012).
ssDNA immunofluorescent assays
To detect parental-strand ssDNA, the cells were labeled for 20 h with 10 μM BrdU and then released into fresh growth medium for 2 h prior to addition of drugs. To detect nascent-strand ssDNA, the cells were labeled for 10 min with 10 μM BrdU immediately prior to addition of drugs. Three millimolar HU was added with 5 μM ATRi. Next, the cells were washed once with PBS, permeabilized with 0.5% Triton X-100 for 10 min at 4°C, fixed with 3% paraformaldehyde/2% sucrose solution for 10 min, and blocked for 15 min in 3% BSA-PBS. Fixed cells were then incubated with mouse anti-BrdU antibody (BD Pharmingen) for 60 min at 37°C followed by Alexa-488 goat anti-mouse (Invitrogen) secondary antibody. Images were collected using a Zeiss camera at a constant exposure time and quantitated with Cell Profiler (http://www.cellprofiler.org).
Xenopus cell-free replication experiments
Xenopus low-speed extract was prepared as described (Lebofsky et al. 2009). Immunodepletion of xSMARCAL1 was carried out by incubation of serum with nProtein A-Sepharose Fast Flow beads (GE Healthcare) at a 1:1 ratio for 1.5 h at 4°C, and the resulting beads were mixed with extract at a 3:1 extract:bead ratio for 1 h at 4°C. Rabbit IgG (10 mg/mL ; Sigma-Aldrich) was incubated with nProtein A-beads for mock depletions at a 1:10 ratio. For nuclei spin-downs, extracts (20 μL) were transferred to 1 mL of dilution buffer (30% glycerol, 150 mM KCl, 0.5% Triton X-100, 80 mM PIPES at pH 6.8, 1 mM MgCl2, 1 mM EGTA), and 1 mL of fixation buffer (30% glycerol, 0.5% Triton X-100, 80 mM PIPES at pH 6.8, 1 mM MgCl2, 1 mM EGTA, 4% formaldehyde) was added. The resulting 2 mL was then layered on top of a glycerol cushion (40% glycerol, 80 mM PIPES at pH 6.8, 1 mM MgCl2, 1 mM EGTA) and spun at 3500 rpm for 15 min in a JS-4.2 rotor. Nuclei were washed with TBS and blocked in TBS+1% BSA overnight. BrdU antibody (IIB5, Santa Cruz Biotechnology) was added for 5 h, followed by Alexa-594 goat anti-mouse (Invitrogen). Mean BrdU signal intensity was quantitated using ImageJ.
Protein purification
Flag-SMARCAL1 was purified from HEK293T cells following transient transfection or from insect cells after baculovirus infection as described previously (Betous et al. 2012). Endogenous SMARCAL1 was purified from HEK293T cells using SMARCAL1 909 antibodies bound to protein A-conjugated magnetic beads using the same procedure.
In vitro kinase assay
GST-S652 protein contained amino acids 645–661 of SMARCAL1 inserted into pBG101. GST-His protein was produced from the pBG101 vector without insertion of SMARCAL1 amino acids. It was larger than GST-S652, since there was no stop codon to prevent translation through the polylinker. Both proteins were purified from Escherichia coli using glutathione sepharose. Kinase reactions were performed with hyperactive ATR–ATRIP (ATR-interacting protein) purified from human cells and incubated with the TOPBP1 AAD protein as previously described (Mordes et al. 2008).
ATPase and fork reversal assays
ATPase assays were performed as described previously with a splayed-arm DNA substrate (Betous et al. 2012). Fork reversal assays were completed with 3 nM gel-purified, labeled fork reversal DNA substrate containing a leading strand gap and 3 nM SMARCAL1 protein in reaction buffer (40 mM Tris at pH 7.5, 100 mM KCl, 5 mM MgCl2, 100 μg/mL BSA, 2 mM ATP, 2mM DTT). Reactions were terminated by the addition of 3× stop buffer (0.9% SDS, 50 mM EDTA, 40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol). Samples were separated on 8% polyacrylamide (19:1) 1× TBE gels. The gels were dried and quantified using a Molecular Imager FX (Bio-Rad). Sequences of olignoucleotides used to make substrates are listed in Supplemental Table S1.
Immunoblotting and antibodies
Rabbit polyclonal SMARCAL1 909 antibody was described previously (Bansbach et al. 2009). The pS652 phosphopeptide-specific antibody was ordered from Bethyl Laboratories and made using the following peptide antigen: KSDVL(pS)QLPAK. Specificity was confirmed using phosphatase-treated lysates and a S652A mutant (data not shown). Rabbit polyclonal antibody against the C-terminal 890 amino acids of xSMARCAL1 was raised at Josman, LLC. Additional antibodies used included RPA2 (clone 9H8, Abcam); γH2AX (clone JBW301, Upstate Biotechnology); Flag M2 (Sigma); CHK1 pS317, CHK1 pS345, and CHK2 pT68 (Cell Signaling); CHK1and GAPDH (G4, Santa Cruz Biotechnology); RPA32 pS4-S8, RPA32, and SLX4 (Bethyl Laboratories); H3 and CtIP (Abcam); and HA (Covance). Quantitative immunoblotting was performed using an Odyssey instrument.
Pan-nuclear γH2AX immunofluorescent assay
U2OS cells were transfected with GFP-SMARCAL1 encoding vectors (pLEGFP-C1, Clontech) using Fugene HD (Promega) according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were seeded into 96-well CellCarrier plates (Perkin Elmer). Forty-eight hours after transfection, the cells were washed once with PBS, fixed with 3% paraformaldehyde solution, and permeabilized with 0.5% Triton X-100 for 10 min at 4°C. Fixed cells were then incubated with mouse anti-γH2AX antibody followed by Cy5-conjugated secondary antibody. After washing, the cells were incubated with DAPI and then imaged on an Opera automated confocal microscope (Perkin Elmer) using a 20× water immersion objective. Columbus software (Perkin Elmer) was used to define nuclei and calculate the mean intensity per nucleus for GFP and γH2AX.
Acknowledgments
We thank Miaw-Sheue Tsai for the production of baculoviruses, which was supported by the Structural Biology of DNA Repair grant P01 CA092584. We thank Noriko Hosoya, Kiyoshi Miyagawa, and Junko Oshima for supplying cell lines. This study was supported by NIH grants CA136933 and CA102792, with additional support from the Breast Cancer Research Foundation to D.C., and grants T32 CA09582 and F31 CA171586-01 to F.B.C. Funding for the Vanderbilt Proteomics Core was provided by the Center in Molecular Toxicology, P30 ES000267 and the Vanderbilt-Ingram Cancer Center. K.A.C. was supported by NIH grant ES016486 and a Glenn Foundation award, and R.D. was supported by a fellowship from the Jane Coffin Childs Memorial Fund.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214080.113.
References
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi A, Raams A, Lubieniecka J, Cho KS, DeHaai KA, Basiratnia M, Mari PO, Xue Y, Rauth M, Olney AH, et al. 2012. SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents in vivo. Am J Med Genet A 158A: 2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Couch FB, Mason A, Eichman BF, Manosas M, Cortez D 2013. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep doi: 10.1016/j.celrep.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. 2002. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 30: 215–220 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Badu-Nkansah A, Hunley T, Baradaran-Heravi A, Cortez D, Frangoul H 2013. Schimke immunoosseous dysplasia associated with undifferentiated carcinoma and a novel SMARCAL1 mutation in a child. Pediatr Blood Cancer doi: 10.1002/pbc.24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. 2012. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 47: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 10: 479–482 [DOI] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363–373 [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR III, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote K, Nissink J, Wilhelmus M 2010. Pyrimidine indole derivatives for treating cancer. Patent no. WO2010073034
- Forment JV, Blasius M, Guerini I, Jackson SP 2011. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE 6: e23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC 2009. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol 521: 229–252 [DOI] [PubMed] [Google Scholar]
- Lewis R, Durr H, Hopfner KP, Michaelis J 2008. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res 36: 1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D 2008. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Cortez D 2011. ATR signalling: More than meeting at the fork. Biochem J 436: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Zhao R, Cortez D 2011. Analysis of mutations that dissociate G(2) and essential S phase functions of human ataxia telangiectasia-mutated and Rad3-related (ATR) protein kinase. J Biol Chem 286: 37320–37327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- Paull TT 2010. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 9: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H 2009. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 284: 35951–35961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR 2011. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 7: 428–430 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ 2012. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest 122: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: Single-cell and -DNA fiber analyses. Mol Cell Biol 27: 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura T, Torres MJ, Martin MM, Rao VA, Pommier Y, Katsura M, Miyagawa K, Aladjem MI 2008. Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J Mol Biol 375: 1152–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Brenowitz M, Auble DT 2006. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J 25: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Fernandez-Capetillo O 2011a. Targeting ATR and Chk1 kinases for cancer treatment: A new model for new (and old) drugs. Mol Oncol 5: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O 2011b. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2008. HARP is an ATP-driven annealing helicase. Science 322: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT 2009. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 23: 2400–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]