Here, Karginov and Hannon explore the relationship between cellular stress and translational repression by the Ago2/miRNA machinery. Transcriptome-wide profiling reveals remodeling of Ago2 transcript occupancy in response to inhibition of translation initiation. Acute stress increases Ago2–mRNA interactions on most transcripts. In contrast, transcripts that are preferentially translated during stress exhibit significantly reduced Ago2–mRNA interactions. This study uncovers a critical role for Ago2 and mammalian miRNAs in mediating stress response and recovery.
Keywords: Argonaute, microRNA, stress, translation, CLIP
Abstract
When adapting to environmental stress, cells attenuate and reprogram their translational output. In part, these altered translation profiles are established through changes in the interactions between RNA-binding proteins and mRNAs. The Argonaute 2 (Ago2)/microRNA (miRNA) machinery has been shown to participate in stress-induced translational up-regulation of a particular mRNA, CAT-1; however, a detailed, transcriptome-wide understanding of the involvement of Ago2 in the process has been lacking. Here, we profiled the overall changes in Ago2–mRNA interactions upon arsenite stress by cross-linking immunoprecipitation (CLIP) followed by high-throughput sequencing (CLIP-seq). Ago2 displayed a significant remodeling of its transcript occupancy, with the majority of 3′ untranslated region (UTR) and coding sequence (CDS) sites exhibiting stronger interaction. Interestingly, target sites that were destined for release from Ago2 upon stress were depleted in miRNA complementarity signatures, suggesting an alternative mode of interaction. To compare the changes in Ago2-binding patterns across transcripts with changes in their translational states, we measured mRNA profiles on ribosome/polysome gradients by RNA sequencing (RNA-seq). Increased Ago2 occupancy correlated with stronger repression of translation for those mRNAs, as evidenced by a shift toward lighter gradient fractions upon stress, while release of Ago2 was associated with the limited number of transcripts that remained translated. Taken together, these data point to a role for Ago2 and the mammalian miRNAs in mediating the translational component of the stress response.
Eukaryotic organisms have evolved elaborate cellular response pathways to cope with environmental stresses, such as heat or cold shock, UV damage, nutrient deprivation, excess reactive oxygen species, or hypoxia. In a central and deeply conserved pathway, distinct stresses are sensed by specific kinases that converge on a common target: eukaryotic initiation factor 2α (eIF2α) (Wek et al. 2006; Sonenberg and Hinnebusch 2009). Phosphorylation of eIF2α leads to a shutdown of protein synthesis at the stage of translation initiation. To the same end, numerous stresses also inactivate the mammalian target of rapamycin (mTOR), a kinase that normally promotes initiation by phosphorylation of eIF4E-BP and other initiation factors (Sonenberg and Hinnebusch 2009; Sengupta et al. 2010). Thus, translational arrest provides a rapid stereotypic response to the changing environment of the cell. The resulting stalled initiation complexes of mRNAs and 40S ribosomal subunits, along with many RNA-binding proteins, assemble into microscopically visible foci termed stress granules in acutely stressed cells (Anderson and Kedersha 2008, 2009; Buchan and Parker 2009).
Against the general repression of protein synthesis, translation of certain mRNA subsets is up-regulated in stress. In a classic example, increased expression of the transcription factor ATF4 in stressed mammalian cells employs the differential use of upstream ORFs (uORFs) as a result of initiation inhibition (Harding et al. 2000; Vattem and Wek 2004). Without stress, translation proceeds through two short uORFs in the 5′ untranslated region (UTR) of ATF4, the latter uORF overlapping with and preventing translation of the main coding sequence. Under stress-induced conditions of lowered initiation rates, translation at the second uORF does not initiate and instead starts at the ATF4 coding region. In addition, circumventing inhibition of cap-dependent initiation through the use of internal ribosome entry sites has been suggested as a mechanism of translational up-regulation for some mRNAs in stress (Wouters et al. 2005). ATF4 and other transcription factors, such as XBP1 and ATF6, activated by noncanonical splicing and proteolysis, respectively (Ron and Walter 2007), mediate the downstream transcriptional response to specific stresses.
RNA-binding proteins impart another layer of regulation, particularly in higher eukaryotes. Cellular stress causes significant rearrangements in their interactions with specific mRNAs, leading to positive and negative effects on mRNA stability and translation (Abdelmohsen et al. 2008). Among other participants, HuR translocates from the nucleus to the cytoplasm upon stress and binds AU-rich mRNA elements to inhibit their decay or promote translation. During amino acid starvation, the TIA-1/TIAR proteins bind to and repress translation of 5′ terminal oligopyrimidine tract (TOP) mRNAs, which themselves encode many proteins involved in translation (Damgaard and Lykke-Andersen 2011).
Argonaute (Ago) proteins and their associated microRNAs (miRNAs), as core parts of the RNA-induced silencing complex (RISC), play important roles in post-transcriptional gene regulation by inducing mRNA decay and repressing translation (Bartel 2009). Although mRNA decay has been measured to contribute most of the repression at steady cellular states (Baek et al. 2008; Selbach et al. 2008; Guo et al. 2010), experiments with kinetic resolution have shown that the effect of Ago proteins on translational repression may be important during transitions between cellular states (Bazzini et al. 2012; Bethune et al. 2012; Djuranovic et al. 2012). In animals, the canonical model posits that the ∼22- to 23-nucleotide (nt) miRNAs guide their Ago partners to target sites in mRNA 3′ UTR and coding sequences (CDSs) through imperfect sequence complementarity. Complementarity in the “seed” region of the miRNA, nucleotides 2–8, is a key determinant of the interaction. However, pairing elsewhere in the miRNA (Brennecke et al. 2005; Grimson et al. 2007) as well as seed interactions involving bulges in the target (Chi et al. 2012) are also functionally relevant.
The function of Ago proteins has not been studied in great detail in the context of stress. Interestingly, miR-122-guided repression of CAT-1 mRNA by Ago2 has been shown to be relieved upon stress, up-regulating the mRNA's translation (Bhattacharyya et al. 2006). Here, we investigate the overall involvement of Ago2 in mediating the translational response to acute stress.
Results
Arsenite stress leads to rearranged and increased associations of Ago2 with the coding and 3′ UTRs of mRNAs
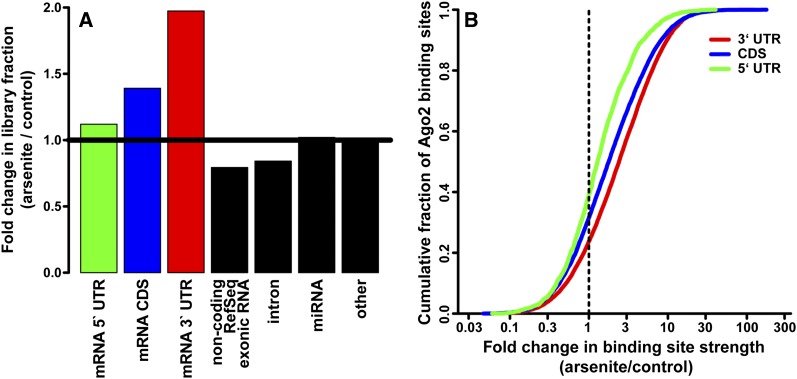
To evaluate the changes in Ago2–mRNA interactions upon stress, we performed Ago2 cross-linking immunoprecipitation (CLIP) followed by high-throughput sequencing (CLIP-seq or HITS-CLIP) from 293S cells treated with 1 mM arsenite for 30 min as well as untreated cells. The sequenced reads were mapped to unique positions in the human genome, and overlapping/adjacent reads were assembled into peaks representing Ago2-binding sites. A simple examination of the read annotations immediately indicated that the fraction of the libraries mapping to mRNA 3′ UTRs increased twofold upon arsenite stress (Fig. 1A). An example of the increased Ago2 association with the 3′ UTR of β-actin is shown in Supplemental Figure 1. The CDS category similarly increased 1.4-fold. In contrast to these known classes of Ago targets, mRNA 5′ UTRs and other RNA categories were not dramatically increased in representation.
Figure 1.
Ago2 association with mRNA 3′ UTR and CDS sites increases upon arsenite stress. (A) Fold changes in the fraction of CLIP-seq libraries annotated by the indicated categories. (B) Cumulative distribution plot of Ago2-binding sites in 5′ UTR, CDS, and 3′ UTR mRNA regions as a function of arsenite-induced fold change in normalized Ago2 site strength.
Detailed quantitative analysis of CLIP-seq data crucially depends on the normalizations applied to the data sets. To account for sequencing depth, we chose to normalize the read counts in each library to the sum of miRNA reads, a known, bona fide Ago2-bound component of the CLIP data whose fraction did not change in relation to the overall library upon stress (Fig. 1A). Furthermore, the observed CLIP signal at each Ago2-binding site is expected to depend on the associated miRNA abundance and mRNA abundance and the inherent strength of interaction. To enable us to factor out the mRNA abundance, we performed RNA sequencing (RNA-seq) on the same samples and divided the CLIP signal by the corresponding mRNA levels.
Using the above normalizations and filtering for peaks with at least 10 counts across replicates yielded ∼24,000 Ago2 interaction loci. At these distinct binding sites, arsenite treatment induced a broad range of changes in signal intensity (Fig. 1B), including complete loss or gain of observed peaks. This suggested a significant remodeling of Ago2–mRNA interactions during the early phase of the stress response. Substantiating the increases in library fraction (above), the majority of 3′ UTR- and CDS-binding sites became more occupied upon stress, with a median fold change of 2.4 and 1.8, compared with a 1.3-fold increase in 5′ UTR peak occupancy (Fig. 1B).
We chose a short stress treatment window to focus on the direct rearrangements of Ago2–mRNA interactions and minimize the confounding effects of miRNA or mRNA level changes on the CLIP peak signal. Indeed, individual miRNA abundances did not show any statistically significant changes after the 30-min stress (Supplemental Fig. 2A), in general agreement with the robustness and stability of Ago–miRNA binding. Similarly, Western blots indicated that the levels of Ago2 remained constant after the treatment. Some mRNAs exhibited changes (Supplemental Fig. 2B), the majority of which were small in magnitude and were accounted for by the RNA-seq normalization. Since the levels of participating components were relatively fixed, we conclude that the observed changes primarily arose from dynamic adjustments in interaction strength between Ago2 and mRNAs at specific sites.
The increased association of Ago2 with mRNAs is triggered upstream of translational arrest and stress granule assembly
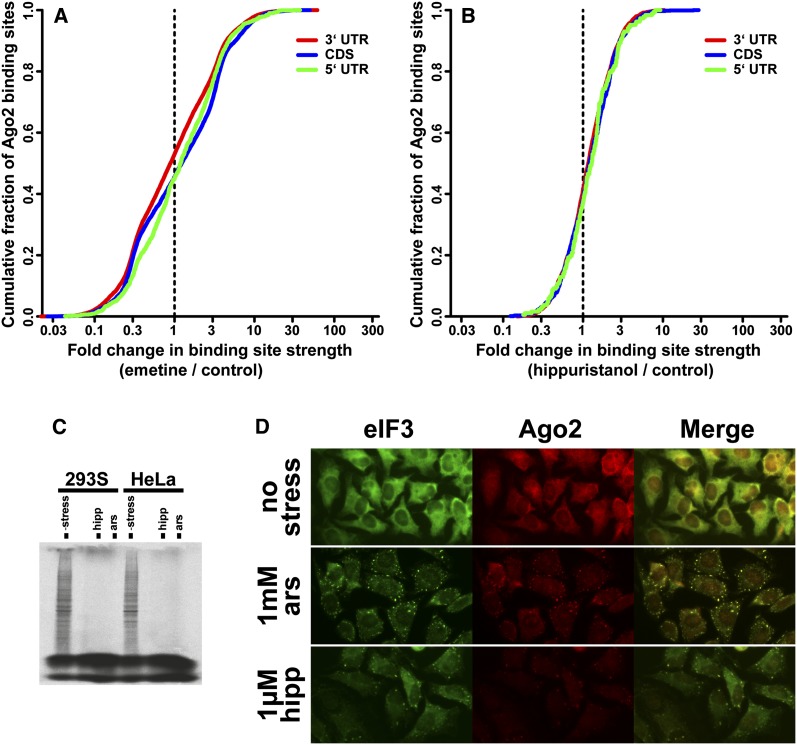
Cellular stress results in translational arrest and stress granule formation, either of which could indirectly influence the association of Ago2 with mRNAs through changes in ribosome occupancy or colocalization in the granules. To address these possibilities, we elicited the cellular responses by artificial means that bypass the upstream signaling pathways and assayed the Ago2–mRNA interactions by CLIP-seq. Emetine inhibits translation elongation by stabilizing ribosomes on mRNA and does not cause stress granule assembly (Kedersha et al. 2000). In contrast to arsenite stress, emetine induced a slight decrease in the 3′ UTR and slight increases in 5′ UTR and CDS CLIP library fractions (Supplemental Fig. 3A). Similarly, fold changes for individual Ago2 peaks did not amount to significant net strengthening of 3′ UTR and CDS sites (Fig. 2A). Therefore, translation arrest at the elongation step does not reproduce the tighter association of Ago2 with mRNAs caused by stress.
Figure 2.
Inhibition of translation and stress granule assembly are not sufficient to significantly increase Ago2–mRNA binding. (A) Cumulative distribution of Ago2-binding sites in 5′ UTR, CDS, and 3′ UTR mRNA regions as a function of fold change in normalized Ago2 site strength upon treatment with 20 μg/mL emetine for 60 min. (B) Same as in A, except with 1 μM hippuristanol treatment for 30 min. (C) Hippuristanol and arsenite result in near-complete inhibition of translation. 293S or HeLa cells were treated with 1 mM arsenite or 1 μM hippuristanol for 30 min (along untreated controls), followed by addition of 35S-labeled methionine and cysteine. After an additional 10 min, cells were harvested, and nascent protein synthesis was assessed by SDS-PAGE. (D) Immunofluorescence of HeLa cells with eIF3 (a stress granule marker) and Ago2 indicates stress granule formation upon 1 mM arsenite or 1 μM hippurstanol treatment for 30 min.
Hippuristanol mimics the downstream effects of physiological stress more closely by blocking translation initiation, which results in assembly of stress granules (Mazroui et al. 2006). However, this inhibition specifically targets eIF4A to disrupt formation of initiation complexes, while upstream signaling events such as phosphorylation of eIF2α are not engaged. We confirmed that in our conditions, addition of hippuristanol blocks nascent protein synthesis to near completion, similar to arsenite stress (Fig. 2C). In addition, immunofluorescence staining of hippuristanol- and arsenite-treated cells showed induction of stress granules containing the eIF3 marker and Ago2 (Fig. 2D, HeLa cells; Supplemental Fig. 4, 293S cells). Nevertheless, Ago2 CLIP-seq under hippuristanol presented only modest increases in 5′ UTR, 3′ UTR, and CDS library fractions (Supplemental Fig. 3B) as well as slight per-peak strengthening (Fig. 2B). Unlike those in arsenite treatment, these shifts did not differentiate between 5′ UTR and 3′ UTR/CDS categories.
Furthermore, nonspecific changes of Ago2 occupancy, such as those driven by differences in mRNA accessibility due to polysome release or localization to stress granules, would be expected to have coordinated effects on all of the sites within a given mRNA. However, examination of the arsenite CLIP data showed substantial variability in binding site response within the same transcript. These observations, considered together with the mild effects of emetine and hippuristanol, indicated that the increased association of Ago2 with mRNAs was largely a result of the signaling events upstream of stress granule formation and translational arrest.
Ago2-binding sites that are released upon arsenite stress are depleted in miRNA signature motifs
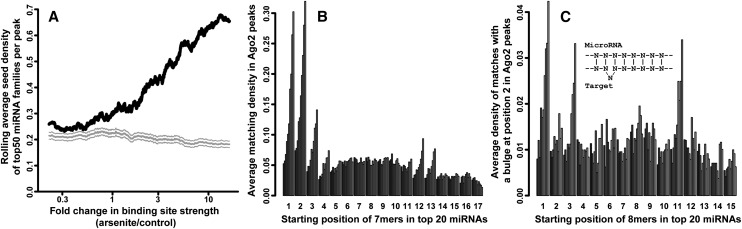
We next aimed to understand what features of the Ago2-binding sites determine changes in their occupancy. Major miRNA–target interaction modes include complementarity in the seed region, compensatory pairing elsewhere in the miRNA, and bulged interactions within the seed. Initially, we looked for the expected 7mer seed site complements for the top 50 miRNA families in the Ago2-binding sites. Strikingly, seed site complementarity had a very strong association with the behavior of the binding site upon stress: Sites that became more strongly bound upon stress were significantly more enriched in miRNA seed complements (Fig. 3A). Conversely, Ago2-binding sites that were present under normal conditions but were released (or unchanged) in stress were substantially less enriched, nearing the background level of shuffled controls. This severe ramping effect was not observed or was much less pronounced under hippuristanol or emetine treatment (Supplemental Fig. 5), suggesting that it is a specific part of the stress response.
Figure 3.
miRNA complementarity signatures are enriched in Ago2-binding sites that strengthened upon arsenite stress and, conversely, are depleted in Ago2 sites that weaken upon stress. (A) Occurrences of 7mer seed complements (nucleotides 2–8) for the 50 most abundant miRNA families in 293S cells were counted within Ago2-binding sites. The sites were ordered from weakening to strengthening, and a rolling average of the seed density (with a window of 1/20th of the number of sites) was plotted (black line). As a control, seed densities in dinucleotide-shuffled sites ± 1 SD are plotted in gray. (B) 7mer sequences starting from each position in the top 20 abundant miRNAs were used to look for complementary sites in Ago2 binding peaks. For each starting position (arranged along the X-axis), the peaks were binned into 10 equal-sized groups ordered from weakening to strengthening upon arsenite treatment, and the density of 7mer complements in Ago2 sites were plotted. See Supplemental Figure 7A for the corresponding control analysis with sequence-shuffled peaks. (C) Similar to B, except searching for complementarity involving a 1-nt bulge in the target mRNA after position 2 in the 7mer. See Supplemental Figure 7B for the corresponding control analysis with sequence-shuffled peaks.
We considered the hypothesis that changes in binding upon stress were driven by a particular subset of miRNAs. For example, we wondered whether there were individual miRNAs targeting sites destined for Ago2 release, against the general trend of increased binding. Scoring for seed interactions, none of the top 50 miRNA families exhibited preferential targeting of sites that were released from Ago2, compared with shuffled controls. On the other hand, many of them individually showed statistically significant preferential targeting to peaks that strengthened, reinforcing the aggregate tendency for the top 50 miRNA families (exemplified by the abundantly expressed miR-30 family in Supplemental Fig. 6).
We considered the possibility that our observations might indicate other modes of miRNA–target interactions that may explain the released sites. To address this question, we expanded the search for miRNA–target complementarity to all 7mers along the miRNA, not restricting ourselves to just the seed. All 7mers from the top 20 miRNAs starting at the indicated positions were extracted and used in separate searches for complementarity in Ago2 sites (Fig. 3B). For each search, the Ago2 sites were segregated into bins by their weakening/strengthening upon stress. Consistent with previous reports, Ago2-binding sites displayed evidence of pairing at the 3′ end of the miRNA (7mers at positions 12 and 13 in Figure 3B; shuffled controls in Supplemental Fig. 7A,B) in addition to the seed region (positions 1–4). However, this effect was also substantially more pronounced for peaks that were strengthened in arsenite (that is, substantially higher densities of 7mer matches were seen for bins of strengthened Ago2 sites). Furthermore, a search for interactions with miRNA seeds involving 1-nt bulges in the target demonstrated their presence (shown for bulges after nucleotide 2 in Fig. 3C). Again, this binding mode was markedly manifest in strengthened peaks. Finally, searches for less extensive and potentially weaker interactions involving 6mers (Supplemental Fig. 7C,D) and even 5mers similarly revealed their prevalence in the peaks that strengthen.
In summary, all major miRNA interaction signatures were enriched in sites of increased Ago2 occupancy upon arsenite stress. However, remarkably, sites that weakened were not explained by any of the tested modes (that is, the bins corresponding to weakened/released Ago2 sites were not enriched in miRNA complementarity for any of the Nmer positions in Fig. 3B,C). Furthermore, we were unable to detect any sequence motifs—regardless of their complementarity to expressed miRNAs—that were enriched in weakened sites. While fewer in number, these sites were of comparable abundance and length with those that strengthened. Thus, it is possible that this class of Ago2–mRNA interactions is achieved in a miRNA sequence-independent manner.
mRNAs that are preferentially translated in stress are released from Ago2
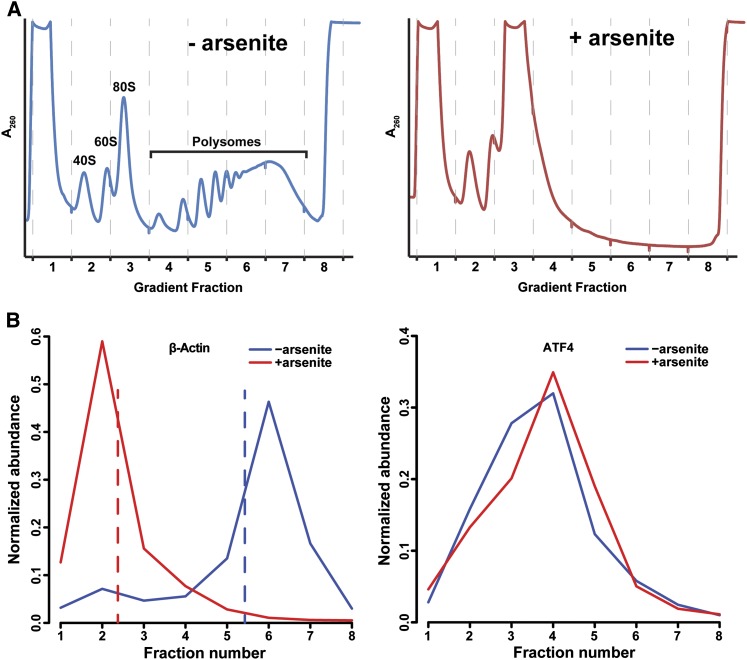
To assess the translational status of mRNAs under stress, we fractionated 293S cell lysates with and without arsenite treatment on sucrose gradients (Fig. 4A). Addition of arsenite lead to marked dissociation of polysomes, consistent with translational arrest at initiation. The resulting fractions were subjected to RNA-seq to uncover the distribution of mRNAs along the gradient. Addition of spike-ins allowed us to normalize the data across fractions and construct polysome profiles for each detected mRNA (Fig. 4B)—essentially a high-throughput version of Northern blotting across sucrose gradients. Most mRNAs, exemplified by β-actin, underwent translation on polysomes under normal conditions and were displaced to lighter, nontranslating fractions upon arsenite stress. However, mRNAs such as ATF4, known to maintain translation upon stress, remained in polysomal fractions or shifted slightly toward heavier polysomes (Fig. 4B). The RNA-seq-derived profiles were in excellent agreement with mRNA profiles measured by quantitative PCR (qPCR) for several transcripts (Supplemental Fig. 8). We quantified the stress-induced mRNA shifts by changes in each transcript's center of mass in the gradients (Fig. 4B, dashed lines); negative shifts signified movement toward lighter fractions.
Figure 4.
Analysis of mRNA translation by sucrose gradients and RNA-seq. (A) 293S cell lysates with (right) or without (left) arsenite treatment were fractionated on sucrose gradients. (B) Example profiles of β-actin (left) or ATF4 (right) mRNAs along the gradients, calculated from RNA-seq of gradient fractions with spiked internal controls. Shifts in mRNA profiles upon arsenite stress were quantified as the differences in the mRNA's center of mass (dashed lines) between conditions.
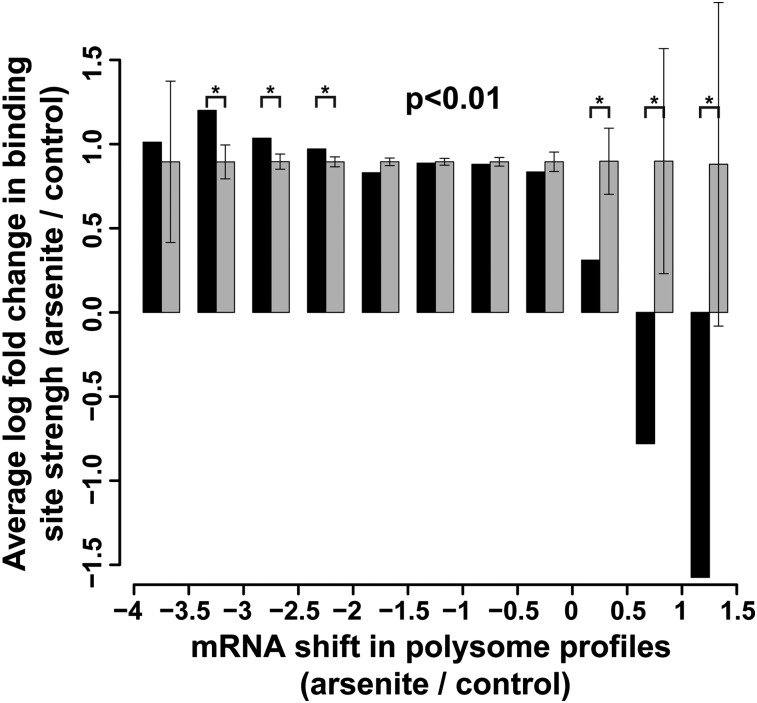
To examine the relationship between the translation status of mRNAs and their Ago2 occupancy as a function of stress, we binned mRNAs by the extent of their polysomal profile shifts and averaged the arsenite-induced log fold change of Ago2-binding sites on the mRNAs in each bin (Fig. 5, black bars). The general trend of increased Ago2 binding agreed with the translational repression of most mRNAs. Remarkably, mRNAs that were preferentially translated in stress (shifts of 0–1.5 fractions toward heavier polysomes) showed a substantially milder increase (0–0.5 natural log units) or decreases (0.5–1.5 natural log units) in Ago2 binding. These results were statistically significant (P < 0.01, permutation test, n = 10000) compared with log ratio-permuted controls (Fig. 5, gray bars). At the opposite end of the shift axis, mRNAs that were strongly repressed by stress (shifts of −3.5 to −2 U) displayed higher Ago2 binding compared with permuted controls. Interestingly, gene ontology enrichment analysis indicated that mRNAs coding for ribosomal proteins were more strongly bound by Ago2 upon stress as well as more translationally repressed compared with other mRNAs. This observation parallels the TIA-1/TIAR-dependent translational repression of 5′-TOP mRNAs upon starvation (Damgaard and Lykke-Andersen 2011) and suggests that Ago2 may present an additional mechanism of their regulation. Overall, per-mRNA statistics without binning reflected a weak but significant negative correlation (P = 2 × 10−5, Pearson test) between gradient shifts and average Ago2 binding log fold changes. As a control, we compared arsenite-induced polysome shifts and changes in Ago2 occupancy upon hippuristanol treatment. Unlike the above results, we observed neither the significant and coherent deviations from permuted expectation controls in binned tests (P > 0.01) nor any per-mRNA correlation (P = 0.5727), confirming that the arsenite-triggered Ago2 rearrangements on mRNAs are not a consequence of translation arrest. Thus, release of Ago2 inhibition appears to contribute to the preferential translation of a subset of mRNAs in stress, while continued or increased Ago2 targeting is correlated with translational repression. A list of mRNAs with 0–1.5 shifts in polysome profiles is presented in Supplemental Table 1.
Figure 5.
The translational fate of mRNAs upon arsenite stress correlates with changes in their Ago2 occupancy. mRNAs were binned by their shifts in polysome profiles, and the average log ratio of Ago2 site changes were computed for each bin (black bars). As a control, permuted log ratio averages ± 1 SD are shown in gray.
Discussion
We addressed the potential role of Ago2 in the stress response by assaying Ago2–mRNA interactions using CLIP-seq. This approach demonstrated a significant remodeling of the binding sites within the short time frame of acute stress. Recent analyses of mouse Ago2 target-binding kinetics in vitro (Wee et al. 2012) have shown that these interactions are sufficiently dynamic, and in vivo modifications to the RISC are likely to further modulate the rates upon cellular perturbations. The remodeling indicated a general tendency toward increased interactions, an effect that appeared to be a specific response to stress pathways and not a result of changes in mRNA binding availability due to translational arrest or stress granule assembly. The general strengthening of Ago2–target interactions correlated with and may have contributed to the translational shutdown of most mRNAs.
We confirmed and extended this observation by experimentally measuring the polysomal profiles of mRNAs and comparing them with changes in Ago2 binding. The translational fate of mRNAs tracked with stress-induced effects on Ago2 occupancy: mRNAs that were the most repressed were more strongly bound by Ago2. On the other end, for those few mRNAs that remained translated in stress, Ago2-binding sites were released. This indicated that release from Ago2 repression may play a role in the preferential translation of those mRNAs. Intriguingly, the ATF4 mRNA also contained binding sites that disappeared or diminished in stress. However, since the released sites mostly resided in a rarely included exon in the 5′ UTR (Supplemental Fig. 9), the contribution of these binding events to the translational regulation of ATF4 needs further investigation.
Several studies have detected miRNA targeting signatures, such as seed complements, 3′ compensatory interactions, and bulged seed targeting, in bulk Ago2 CLIP-seq data. Here, examining the Ago2–mRNA interactions in two states (with and without arsenite stress) allowed us to identify a distinct class of binding. Stratifying sites by their differential response to stress indicated that sites that are present under normal conditions but show little to negative changes upon stress are strongly depleted of miRNA signatures. This suggested a miRNA-independent mode of Ago2 binding to mRNAs. In a related phenomenon, robust Ago2 interactions have been observed in mouse embryonic stem cells devoid of canonical miRNAs (Leung et al. 2011b). In addition, previous biochemical studies have demonstrated that human RISC is capable of binding ssRNA in a sequence-independent manner (Ameres et al. 2007). Presumably, the specificity and/or affinity of this binding mode are aided by additional factors. Although the functional consequences of such binding events are currently not understood, the fact that loss of these binding sites accompanies preferential translation of certain mRNAs argues for their repressive function in that context.
What is the molecular mechanism behind arsenite-induced changes in Ago2 binding to mRNAs? Since the observed strengthening is not correlated with translational arrest or stress granule assembly, it is likely caused by upstream signals in the stress response, such as post-translational modification of Ago2 or association with additional factors. In this regard, several stress-related Ago2 modifications have been observed. Phosphorylation of Ser 387 occurs at a basal level in human Ago2 and is up-regulated in arsenite stress through the p38 MAPK pathway (Zeng et al. 2008). This modification is required for localization to P bodies under normal conditions but not under stress. A separate study identified a number of additional phosphorylation sites (Rudel et al. 2011), with mutations at Y529 affecting P-body localization. The effect of stress on these phosphorylation events remains to be further investigated.
Ago2 also undergoes hydroxylation of Pro 700, a modification that regulates its stability and may be required for P-body localization but not for arsenite-induced Ago2 localization to stress granules (Qi et al. 2008). Interestingly, in hypoxic stress, Ago2 hydroxylation levels are increased, and localization to stress granules is reduced in a P700 mutant (Wu et al. 2011). Finally, poly-ADP-ribosylation of Ago2, present in basal conditions, is up-regulated in stress (Leung et al. 2011a). To account for the strengthening of most Ago2–mRNA interactions, it will be crucial to assess the fraction of the Ago2 pool that is affected by the above modifications.
Considered together, our data suggest a model in which stress-induced signaling affects global Ago2–mRNA interactions, possibly through post-translational modifications or additional binding factors. The range of changes includes strengthening of the majority of sites, which contribute to the additional down-regulation of most repressed mRNAs, as well as release from other sites, which allows for preferential translation of some mRNAs. The differential behavior of Ago2-binding sites upon stress may be dictated by their binding mode, with miRNA-independent sites preferentially being released in stress. The determinants of specificity for this class of Ago2 sites as well as the mechanism of their dissociation upon stress await further studies.
Materials and methods
Cell treatment and sequencing protocols
293S and HeLa cells were grown in DMEM + 10% FBS + penicillin/streptomycin in adherent monolayer to ∼70% confluency and treated with 1 mM sodium arsenite for 30 min, 1 μM hippuristanol for 30 min, or 20 μg/mL emetine for 60 min. For immunofluorescence imaging, cells were grown on glass coverslips in six-well plates and stained according to the Kedersha and Anderson protocol (Kedersha and Anderson 2007), except blocking and incubation were done in 0.5% BSA in PBS. Anti-eIF3η (N-20, Santa Cruz Antibodies) and anti-Ago2 (clone 11A9, Sigma) antibodies were used. For nascent protein synthesis determination by 35S labeling, 5 μL of EasyTag Express35S Protein Labeling Mix (Perkin Elmer) was added to treated cells in six-well plates for an additional 10 min; the cells were washed once in PBS, harvested, and analyzed by 10% SDS-PAGE. For CLIP-seq, treated samples along with untreated controls were processed using a modification of the Darnell procedure (see detailed protocol at the end of the Supplemental Material; Chi et al. 2009). For each replicate/condition, two to five 15-cm plates were used. For arsenite treatment, four biological replicates were used with the Abnova Ago2 antibody (clone 2E12-1C9). For hippuristanol treatment, two biological replicates were split and used with the Abnova antibody, a Sigma Ago2 antibody (clone 11A9), and a Santa Cruz Biotechnology Ago2 antibody (clone 4F9). For emetine treatment, one biological replicate was split and used with the above three antibodies. During the isolation of Ago2–RNA cross-links from the protein membrane in the emetine experiment, two adjacent regions of the membrane containing the Ago2–mRNA “HI” smear from 130 kD and up were excised and sequenced separately but showed similar sequence content and were analyzed together. For RNA-seq, total RNA from aliquots of cells harvested for CLIP was extracted with Trizol. The aqueous phase was further extracted twice with acid phenol and once with chloroform, ethanol-precipitated, and washed twice with 70% ethanol. Sequencing libraries were prepared from 1 μg of total RNA using a modified NSR primer protocol (Armour et al. 2009). Briefly, total RNA was annealed with 2 μL of 100 μM first strand NSR primer pool in a 10-μL volume for 5 min at 65°C. The first strand reverse transcription reaction mix (4 μL of 5× reaction buffer, 1 μL of 0.1 M DTT, 4 μL of 10 mM dNTPs, 1 μL of SuperScript III [Invitrogen]) was added. The reaction was incubated for 90 min at 40°C (in some replicates, the reaction was incubated for 30 min without apparent differences in the result), followed by 15 min at 70°C. RNase H (1 μL; Invitrogen) was added for a 20-min incubation at 37°C, followed by 15 min at 75°C. The DNA was purified with the QiaQuick PCR purification kit, eluted in 30 μL of EB buffer, and subjected to second strand synthesis: 25 μL of cDNA, 10 μL of 10× NEBuffer 2, 5 μL of 10 mM dNTPs, 4 μL of 5 U/μL exo- Klenow (New England Biolabs), 10 μL of 100 μM second strand NSR primer pool, and 46 μL of H2O. The reaction was incubated for 30 min at 37°C and purified over QiaQuick columns as above. Next, the PCR reaction was set up as follows: 25 μL of purified DNA, 20 μL of 5× buffer 2, 10 μL of 25 mM MgCl2, 5 μL of 10 mM dNTPs, 10 μL of 10 μM P5-SBS3T-NSR primer (see the oligonucleotide sequence table below), 10 μL of 10 μM P7-SBS8-NSR primer, 1 μL of Expand High-Fidelity Plus Polymerase (Roche), and 19 μL of H2O. The PCR reaction was cycled with the following program: 2 min at 94°C; two cycles of 10 sec at 94°C, 2 min at 40°C, and 1 min at 72°C; 25 cycles of 14 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C; and 5 min at 72°C. The PCR products were run on a 2% agarose gel, excised in the 200- to 500-bp range, and gel-purified. The above libraries were sequenced on one Illumina GAIIx lane each.
Polysome profiling and RNA-seq
One 15-cm plate of 293S cells at ∼70% confluency was used for each condition. Cells were washed twice with cold PBS + 100 μg/mL cycloheximide, harvested by scraping, centrifuged, and resuspended in 850 μL of hypotonic lysis buffer (5 mM Tris at pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl). Next, 12 μL of RNasin Plus (Promega) and cycloheximide and DTT to a final concentration of 100 μg/mL and 2 mM, respectively, were added and mixed by vortexing. Twenty-five microliters of 10% Triton X-100 and 25 μL of 10% sodium deoxycholate were added, and the cells were vortexed and centrifuged at 14,000 rpm for 2 min in a tabletop centrifuge. The supernatants were loaded onto 11 mL of 10%–50% sucrose gradients (10%–50% sucrose, 20 mM Hepes at pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT) and centrifuged at 38,000 rpm for 2 h at 4°C in an SW41 rotor. The gradients were fractionated into eight 1.5-mL fractions using a Teledyne Isco density gradient fractionator. From each fraction, 375 μL was spiked with 1 μL of a 1:10 dilution of ERCC RNA spike-ins (mix 1, Ambion), and total RNA was extracted by Trizol LS, with an additional acid phenol-chloroform and a chloroform extraction to exclude DNA contamination. RNA-seq libraries were constructed from 230 ng of starting total RNA material from each fraction using the above NSR protocol but with 3′ (reverse) PCR primers suitable for multiplexing and sequencing on Illumina HiSeq (see the oligonucleotide table in the Supplemental Material). Three to four individual libraries were multiplexed on a single HiSeq lane.
Detailed protocols are presented in the Supplemental Material.
Acknowledgments
We thank Marek Kudla, Aldo Mele, and Dr. Robert Darnell for assistance with the CLIP procedure; Dr. Jerry Pelletier for the generous gift of hippuristanol; and Assaf Gordon, Simon Knott, and Simon Anders for helpful discussions and bioinformatic support. We thank Jason Estep for assistance with experiments. F.V.K. was supported by a post-doctoral fellowship from the American Cancer Society (PF-07-058-01-GMC). All sequencing data are available through Gene Expression Omnibus, accession number GSE44404.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.215939.113.
References
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: Implications for cellular senescence. Biol Chem 389: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W 2012. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep 13: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J 2011. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 25: 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431: 61–81 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P 2011a. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA 2011b. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol Biol Cell 17: 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J 2008. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455: 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G 2011. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res 39: 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD 2012. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11 [DOI] [PubMed] [Google Scholar]
- Wouters BG, van den Beucken T, Magagnin MG, Koritzinsky M, Fels D, Koumenis C 2005. Control of the hypoxic response through regulation of mRNA translation. Semin Cell Dev Biol 16: 487–501 [DOI] [PubMed] [Google Scholar]
- Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, Lagna G, Hata A 2011. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol 31: 4760–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR 2008. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 413: 429–436 [DOI] [PubMed] [Google Scholar]