Abstract
A recent study published in Nature by Keestra and colleagues addresses how the immune system detects the pathogenic potential of microbes and provides evidence that one strategy involves NOD1, which monitors the activation state of the RhoGTPases that are targeted by virulence effectors produced by pathogenic microbes. Interestingly, their findings reveal striking similarities with previous observations made in flies and plants, establishing the evolutionary conservation of this detection system in the innate immune arsenal in many taxa.
The discovery that Drosophila Toll, and the homologous Toll-like receptors (TLRs) in animals, are pattern recognition receptors (PRRs) that act as cellular sensors of microbes has attracted considerable attention during the last two decades1. The PRR system is based on the detection of conserved molecular motifs, microbe-associated molecular patterns (MAMPS), that are shared by most microbes, virulent or not. Given the conserved nature of MAMPS, this model does not explain how the host discriminates between harmful pathogens and beneficial commensal microbes. An elegant hypothesis is that, in addition to the PRR system, the host is able to monitor the pathogen-induced disruption of cellular homeostasis. This type of immune surveillance system has been demonstrated in plants and termed “effector-triggered immunity” (ETI)2. Recently, the concept of ETI has been extended to metazoans and proof of its importance as an innate immune mechanism has now been provided in Drosophila melanogaster, Caenorabditis elegans and mammals3.
The ETI model is of particular relevance when considering that most major pathogenic bacteria have evolved many protein effectors commonly referred to as virulence factors. These effectors are either directly injected into host cells by cell-bound bacteria, or are secreted toxins endowed with the ability to bind to and translocate into the host cell cytosol. Once within the host cell, these bacterial effectors perturb cellular homeostasis by modifying the activity of critical regulators. Common amongst the arsenal of numerous pathogenic bacteria are virulence factors that target the small RhoGTPases of the host4. This predilection for targeting RhoGTPases is probably because it allows bacteria to hijack the many cellular functions that contribute to immunity including phagocytosis, apoptosis, as well as production of reactive oxygen species (ROS) and inflammatory mediators5. In this regard, the RhoGTPases represent a common target and vulnerability in the host cell, which explains why their aberrant activity can often indicate pathogen invasion.
Providing the foundation for the work of Keestra et al.6, previous studies have shown that effectors that activate RhoGTPase can induce unusual immune responses in the host. Both Salmonella typhimurium and Shigella spp. are enteric pathogens that invade host cells using a type III secretion system that is able to inject effectors in the cytosol of host cells. Earlier work by the group of Jorge Galan showed that Salmonella effectors could activate epithelial cells through a PRR-independent mechanism that was dependent on the GEF activity of certain effectors and involving target GTPases in the host7. Similarly, the Shigella effector, GEF-H1 was shown to augment NF-κB-dependent immune responses in a NOD1-dependent manner also after modifying RhoGTPases8. Finally, the activation of RacGTPase by the CNF1 toxin of uropathogenic Escherichia coli also triggers an immune response9. In this case the response is via an innate immune signaling pathway conserved in Drosophila and mammals involving IMD in flies and the related Rip proteins, RIP1 and RIP2, in mammalian cells9. Moreover, this response can be beneficial for the host and help clear the bacteria as shown in an in vivo fly model. Now, using a mouse model of Salmonella typhimurium infection, Keestra et al.6 further define the mechanism of detection of effectors that target RhoGTPase in vertebrates. They show that mammals detect the activity of the injected effector SopE once it is active in the host cytosol. Using cell-based assays, they demonstrate that this detection mechanism is through NOD1, a NOD-Like Receptor (NLR) protein, in a molecular complex containing HSP90. Together this complex detects the activation of the RhoGTPases Rac1 and Cdc42 and transduces a danger signal though RIP2 kinase. Furthermore, using NOD1-deficient mice, they show that SopE-triggered inflammation is markedly reduced.
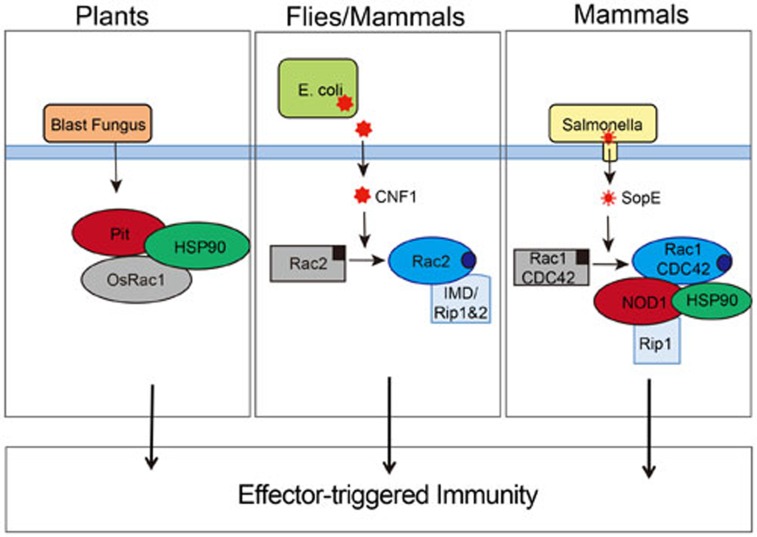
Together this emerging body of data support the idea that the activity of GTPases is monitored by NLR and related pathways, and used as a cue to augment ongoing immune responses during pathogen invasion. Interestingly, Kawano and colleagues have shown that resistance to the rice blast fungus also involves activation of Rac (OsRac1) downstream of Pit, a plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) receptor10. OsRac1 contributes to NBS-LRR-mediated production of ROS and induction of a hypersensitive response for the purpose of destroying infected tissue and preventing dissemination. Although the link between RhoGTPases and NLRs in mammals and NBS-LRR in plants is likely a consequence of convergent evolution, these data highlight some striking similarities between the two systems (Figure 1). Together these papers suggest that we should now consider the NOD proteins not only as intracellular PRR but also akin to plant NBS-LRRs that are able to sense the direct and indirect perturbations of host cell homeostasis. Moreover, these data place RhoGTPases as central players in the molecular cascades of ETI in many species, explaining why monitoring the activation state of RhoGTPases as a surrogate for the presence of virulent pathogens is an evolutionarily conserved strategy. Our current challenge will now be to determine how PRR- and effector-triggered immunity collaborate to confer optimal protection during infections with virulent pathogens.
Figure 1.
RhoGTPases are components of effector-triggered immune responses in different species. The role of the RhoGTPases OsRac1 in plants (left), Rac2 in flies and mammals (middle), and Rac1 and CDC42 in mammals (right) in ETI responses. Activation of Rac2 by CNF1 (middle), and Rac1 and CDC42 by SopE engages RIP kinase-dependent signaling pathways. In plants OsRac1 is activated downstream of the NBS-LRR, Pit, and the response to SopE (right panel) requires the NLR, NOD1. In contrast, flies lack NLRs and the ETI response occurs independently of NLRs but does require the RhoGTPase, Rac2. Middle and right panels — inactive GTPase bound to GDP is shown in grey (black square) and the active GTPase bound to GTP in blue (blue circle).
References
- Medzhitov R. Immunity. 2009. pp. 766–775. [DOI] [PubMed]
- Jones JD, Dangl JL. Nature. 2006. pp. 323–329. [DOI] [PubMed]
- Stuart LM, Paquette N, Boyer L. Nat Rev Immunol. 2013. pp. 199–206. [DOI] [PMC free article] [PubMed]
- Boquet P, Lemichez E. Trends Cell Biol. 2003. pp. 238–246. [DOI] [PubMed]
- Bokoch GM. Trends Cell Biol. 2005. pp. 163–171. [DOI] [PubMed]
- Keestra AM, Winter MG, Auburger JJ, et al. Nature. 2013. pp. 233–237. [DOI] [PMC free article] [PubMed]
- Bruno VM, Hannemann S, Lara-Tejero M, et al. PLoS Pathog. 2009. p. e1000538. [DOI] [PMC free article] [PubMed]
- Fukazawa A, Alonso C, Kurachi K, et al. PLoS Pathog. 2008. p. e1000228. [DOI] [PMC free article] [PubMed]
- Boyer L, Magoc L, Dejardin S, et al. Immunity. 2011. pp. 536–549. [DOI] [PMC free article] [PubMed]
- Kawano Y, Akamatsu A, Hayashi K, et al. Cell Host Microbe. 2010. pp. 362–375. [DOI] [PubMed]