Abstract
The balance between the innate immunity of the host and the ability of a pathogen to evade it strongly influences pathogenesis and virulence. The two nonstructural (NS) proteins, NS1 and NS2, of respiratory syncytial virus (RSV) are critically required for RSV virulence. Together, they strongly suppress the type I interferon (IFN)-mediated innate immunity of the host cells by degrading or inhibiting multiple cellular factors required for either IFN induction or response pathways, including RIG-I, IRF3, IRF7, TBK1 and STAT2. Here, we provide evidence for the existence of a large and heterogeneous degradative complex assembled by the NS proteins, which we named “NS-degradasome” (NSD). The NSD is roughly ∼300-750 kD in size, and its degradative activity was enhanced by the addition of purified mitochondria in vitro. Inside the cell, the majority of the NS proteins and the substrates of the NSD translocated to the mitochondria upon RSV infection. Genetic and pharmacological evidence shows that optimal suppression of innate immunity requires mitochondrial MAVS and mitochondrial motility. Together, we propose a novel paradigm in which the mitochondria, known to be important for the innate immune activation of the host, are also important for viral suppression of the innate immunity.
Keywords: NS, IFN, mitochondria, RSV infection, MAVS, innate immunity
Introduction
Mitochondria are essential for triggering the cellular innate immune responses against invading pathogens, such as viruses. In the predominant pathway, virally generated RNA acts as a pathogen-associated molecular pattern (PAMP) and activates cytosolic RIG-I-like RNA helicases (RLRs), which then dock to the mitochondria via mitochondrial antiviral signaling adaptor (MAVS; also known as IPS-1, Cardif or VISA)1,2,3,4. This subsequently activates a signaling cascade that results in the phosphorylation and nuclear translocation of the interferon (IFN) regulatory factors, IRF3 and IRF7, leading to the induction of type I IFN (IFN-α and -β) expression, the major antiviral defense weapon of the cell5,6. The released IFN activates the IFN response pathway via Jak/STAT signaling, leading to the induction of the expression of a large family of cellular antiviral genes7,8,9.
Viruses, especially RNA viruses, whose RNA products can potentially function as PAMPs, have also evolved strategies to antagonize the type I IFN signaling pathways; this allows them to grow robustly and generate the immunopathology that is a characteristic of the viral infection10,11,12. Of particular importance are viruses of the Paramyxoviridae family that includes many clinically important viruses such as measles virus, mumps virus, parainfluenza viruses and respiratory syncytial virus (RSV). With the sole exception of pneumoviruses, members of this family produce IFN-suppressor proteins such as V, W and C through co-transcriptional RNA editing of the viral P gene10,13,14,15. Pneumoviruses, represented by RSV, do not use RNA editing; instead, they uniquely encode two nonstructural (NS) proteins, NS1 and NS2, which strongly suppress both IFN induction and IFN-response pathways by inhibiting or degrading a number of signaling proteins involved in these two pathways16,17,18,19,20,21,22,23,24,25,26,27,28,29, and thus act as essential virulence factors30,31,32. In pursuing the mechanism by which the NS proteins target such a diverse array of immune proteins that share little or no sequence identity, we wondered whether there is a common location of NS proteins and their targets. This led to the current discovery that NS proteins assemble a large degradative complex on the mitochondria, in which all the NS targets could be found. Furthermore, we studied the properties and dynamics of this complex and documented that the mitochondria, specifically mitochondrial MAVS, play a cardinal role in viral suppression of the innate immunity in addition to their traditional role in immune activation, which constitutes a novel paradigm in immune regulation upon viral infection.
Results
An expanding list of cellular innate immune proteins targeted by RSV NS proteins
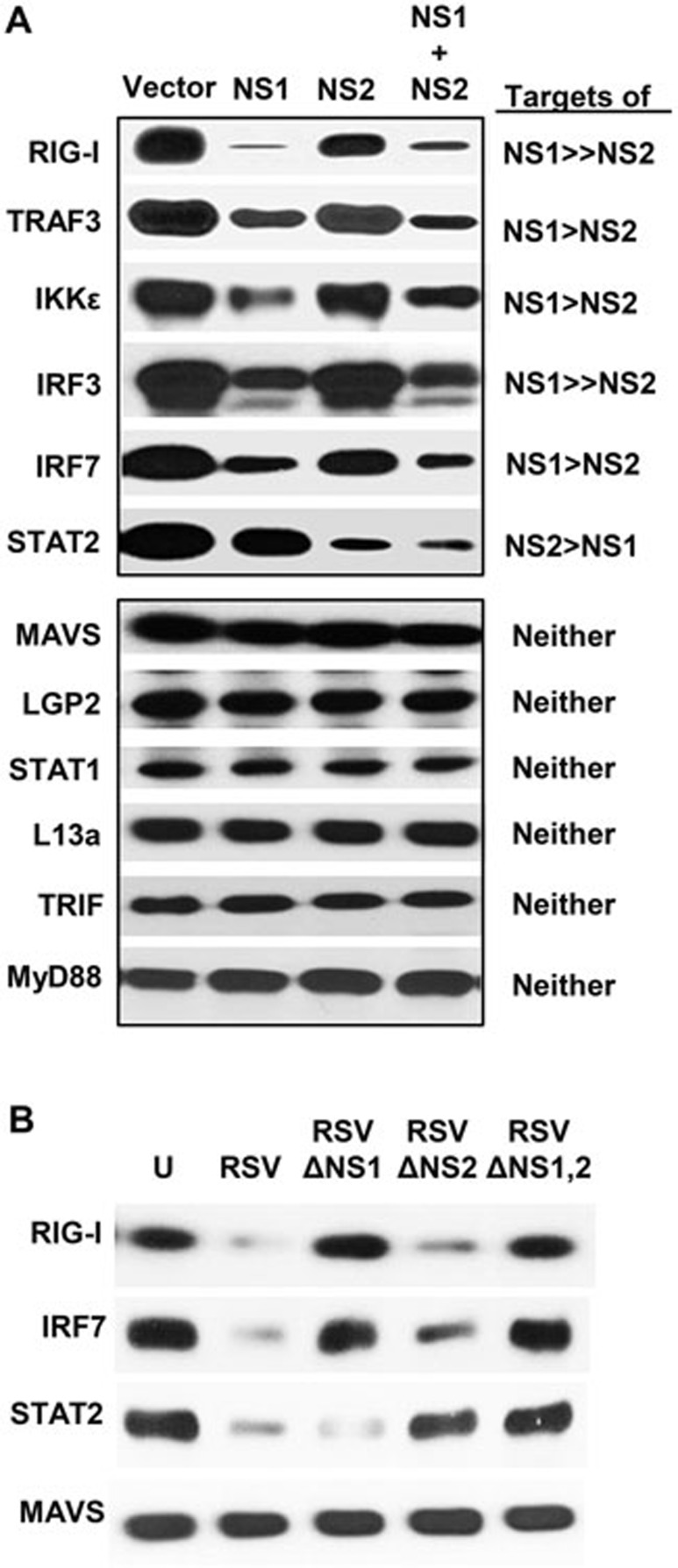
We and others16,17,18,19,20,21,22,23,24,25,26,27,28,29 have recently shown that NS1 and NS2 inhibit and/or degrade a number of proteins of the IFN induction and response pathways. We have since screened other members of these pathways, confirmed and extended the repertoire of NS targets to include: RIG-I, TRAF3, IKKε, IRF3, IRF7 and STAT2 (Figure 1A). These results also reveal that although NS1 and NS2 display some preferences, there is a significant degree of overlap in their targets. The inhibitory activity is nonetheless specific, as the expression levels of MAVS, LGP2, STAT1, TRIF, MyD88 and numerous IFN signaling-unrelated cellular proteins that we have tested, such as L13a (Figure 1A), actin28, GAPDH and SP6 (data not shown), were not affected by the ectopic expression of NS1 and/or NS2. We proposed that this is indicative of the ability of NS proteins to form specific degradative complex(s), which we set out to study.
Figure 1.
Degradation of various innate immune proteins by NS1 and NS2 overexpression. (A) The experiment was performed as described previously28. A549 cells in 24-well plates were transfected with 0.6 μg of the indicated recombinant plasmids of various targets (shown on left) and 0.6 μg of either the vector (pCAGGS), or pCAGGS-NS1 or pCAGGS-NS2, or both. At 24 h post transfection, total proteins were analyzed by immunoblotting with one of these primary antibodies: V5 (IRF3), HA (MAVS, L13a), LGP2, and FLAG for all the rest. The relative degradation degree of each target by NS1 or NS2 is indicated on the right. (B) Degradation of representative targets upon the infection of WT or NS-deleted RSV. The A549 cells were infected with WT RSV or various NS-deleted RSV strains at an m.o.i. of 3, and the total cell extracts were subjected to immunoblotting as in A. U = uninfected cells.
To verify that the degradation of NS targets is not an artifact resulting from the overexpression of the recombinant NS proteins, we performed similar experiments in cells infected with RSV and mutant RSV lacking NS1, NS2 or both. Indeed, results of immunoblotting (Figure 1B) were consistent with the roles of NS1 and NS2 deduced from experiments using recombinant proteins (Figure 1A). For example, wild-type (WT) RSV, which has both NS genes, degraded all three targets examined, namely RIG-I, IRF7 and STAT2. However, RSVΔNS2, which contains NS1, but not NS2, degraded RIG-I and IRF7 but not STAT2. Reciprocally, RSVΔNS1, which contains NS2, but not NS1, degraded STAT2, but left RIG-I and IRF7 intact. RSV infection exhibited no effect on MAVS, which was also shown to be a non-target in Figure 1A.
The minimal NS degradasome is a heterogeneous complex of 300-750 kD
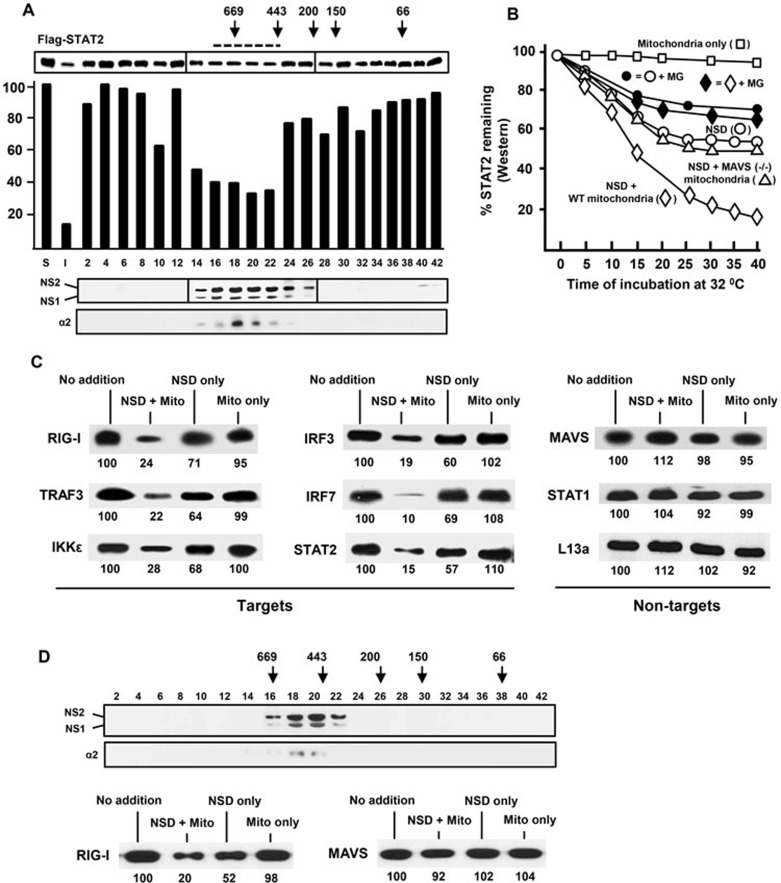
We co-transfected A549 cells with NS1- and NS2-expressing plasmids27,28, harvested cell-free extracts 20 h later using a non-denaturing buffer and subjected them to size fractionation using a Superdex 200 10/300 GL resin pre-calibrated with size markers, as described in the Materials and Methods. Substantial amounts of NS1/2 proteins (roughly 60% of the total) were found in the void volume, but we did not characterize the fraction of the void volume further due to its crude nature and the likely presence of multiple large cellular structures such as cell membranes and intracellular organelles. The column fractions were assayed in vitro for their degradative activity towards immunoprecipitated FLAG-tagged human STAT2 that was also overexpressed in the A549 cells. A broad range of fractions with obvious STAT2-degrading activities were indeed detected (#16-22, Figure 2A), indicating that the complex is relatively large with a size range of approximately 300-750 kD. No such activity could be detected in similarly processed extracts harvested from vector-transfected (no NS) cells (data not shown). Both NS1 and NS2 could be detected in the active fractions (#16-22, Figure 2A), with a small amount detected in the low-molecular-weight fractions (#40-42, Figure 2A), indicating that most of the NS proteins in the column fractions are part of the large complex. We named the pool of the active fractions (#16-22, Figure 2A) “NS degradasome” or NSD and proceeded to define its properties.
Figure 2.
Isolation and analysis of NSD. (A) Size-fractionation of NSD by Superdex chromatography. The column was pre-calibrated with the following marker proteins: thyroglobulin 669 kD; apoferritin 443 kD, β-amylase 200 kD, alcohol dehydrogenase 150 kD, BSA 66 kD; their elution positions are marked with arrows. Various fractions were analyzed by immunoblotting for FLAG-tagged NS1 and NS2. The bar graph represents the degradative activity of the fractions towards STAT2, plotted for the remaining amounts of STAT2 after the in vitro NSD degradation reaction that was performed for 20 min at 32 °C. The un-numbered bars at the extreme left (S, I) represent the following: “S”, no-NSD control reaction, taken as 100, and other bars were plotted as its percentage; “I”, reaction used 2 μl of the input extracts (pre-Superdex) instead of the fractions. The fractions pooled as “NSD” (#16-22) are indicated by the horizontal broken line. The 20S core proteasomal subunit α2 was detected by immunoblotting using B-4 antibody. (B) Enhanced degradative activity of NSD by mitochondrial supplementation. Where mentioned, mitochondria were prepared from MAVS KO (−/−) mouse embryonic fibroblasts (MEFs). Each reaction was sampled at 5 min intervals during incubation, starting at 5 min after the addition of the pooled NSD fractions obtained from A. The amounts of residual FLAG-STAT2 were quantified by immunoblotting. The NSD-only reaction (○) was performed as in A; addition of WT mitochondria (◊) enhanced NSD activity, whereas MAVS-deficient mitochondria (Δ) did not. The control, “no-NSD” reaction supplemented with just mitochondria (□) showed little STAT2 degradation. Parallel reactions (indicated by the corresponding filled symbols, ♦, •) contained MG132 (MG, 20 μM). (C) NSD substrate profile. A number of recombinant IFN signaling-related proteins overexpressed in the A549 cells were immunoprecipitated and used as substrates in “NSD + mitochondria” reaction. All reactions were incubated at 32 °C for 20 min. No addition = immunoprecipitated substrate only; NSD only = no mitochondria; Mito only = no NSD. Non-targets (right panel) are proteins whose expression was not affected by the ectopic expression of NS proteins. (D) NSD in RSV-infected cells. The A549 cells were infected with RSV at an m.o.i. of 3, and were lysed and analyzed 18 h post infection as in A. The NS proteins were detected by immunoblotting with an antibody against the C-terminal 11-mer peptide of NS226, which in fact reacts with both NS1 and NS2, likely due to their C-terminal homology (F/YDLNP). Proteasomal α2 subunit expression was analyzed as in A. The numbers under the blots in C and D are densitometric intensities of the bands, relative to the “no addition” lane set as 100.
NSD is metastable and its activity is enhanced by the presence of MAVS+ mitochondria
We routinely observe 60% - 90% degradation of endogenous STAT2 upon transient transfection of optimal amounts of NS plasmids (ex vivo) (Figure 1); however, in our initial screening of the degradative activities of the NSD fractions in vitro, NSD degraded only about 50% STAT2 (#16-22, Figure 2A). When we tested the kinetics of the degradation, the pooled NSD fractions (#16-22, Figure 2A) were found to be active for only about 15 min (Figure 2B). As we and others have recently found that a large portion of NS proteins are localized to the mitochondria17,27, we speculated that mitochondrial association may be required for optimal NS functionality. However, as the purified NSD complex is ∼300-750 kD in size, it cannot accommodate mitochondria; and thus, any mitochondria associated in vivo may have been dissociated from the complex during chromatography. This was confirmed by the absence of mitochondrial proteins Cox-1 and LetM1 in the NSD fractions as demonstrated by immunoblotting (data not shown). To test whether addition of mitochondria could enhance NSD activity, we supplemented the reaction with purified mitochondria and found that the supplementation indeed allowed the NSD degradative activity to last longer, leading to nearly 80% degradation of STAT2 over 30 min (Figure 2B). Mitochondria alone had no effect on STAT2 stability. Interestingly, mitochondria purified from MAVS knockout (KO) cells did not enhance NSD activity (Figure 2B) and neither did the affinity-purified MAVS proteins (data not shown). Thus, it appears that mitochondria enhance the degradative activity of a metastable NSD complex by employing the mitochondrially located MAVS as an adaptor or a scaffold.
To further establish the authenticity of the reconstituted mitochondria-NSD complex, we tested whether its target profile matches that of the NS proteins overexpressed in the A549 cells. Indeed, the in vitro reconstituted mitochondria-NSD complex degraded immunoprecipitated RIG-I, TRAF3, IKKε, IRF3, IRF7 and STAT2 but not MAVS, STAT1 and L13a (Figure 2C). A control fraction of the same size range obtained by similar chromatography of lysates of unmanipulated cells did not display degradative activity. Thus, the reconstituted mitochondria-NSD complex faithfully recapitulated the in vivo target and non-target repertoires observed in Figure 1.
To verify that the assembly of NSD is not an artifact resulting from the overexpression of recombinant NS proteins, we performed similar fractionation in RSV-infected cells. As shown in Figure 2D, an NSD complex of similar size was indeed detected; furthermore, its substrate specificity was also comparable to that of recombinant NS-assembled NSD, as shown by using RIG-I and MAVS as a representative substrate and non-substrate, respectively (Figure 2D). Finally, we detected the presence of alpha 2 (α2) subunit of the 20S core proteasome in both RSV- and recombinant NS-assembled NSDs by immunoblotting (Figures 2A and 2D). However, it is interesting to note that the NSD degradative activity was only partially sensitive to MG132, a specific proteasome inhibitor. As shown in Figure 2B, MG132 reduced the activities of NSD by about 50% (compare the filled circles and diamonds with the unfilled ones). Thus, it is possible that the complex contains non-proteasomal proteases, which may contribute to the optimal degradative activity of NSD.
NSD-substrate complex associates with mitochondria in an MAVS-dependent manner
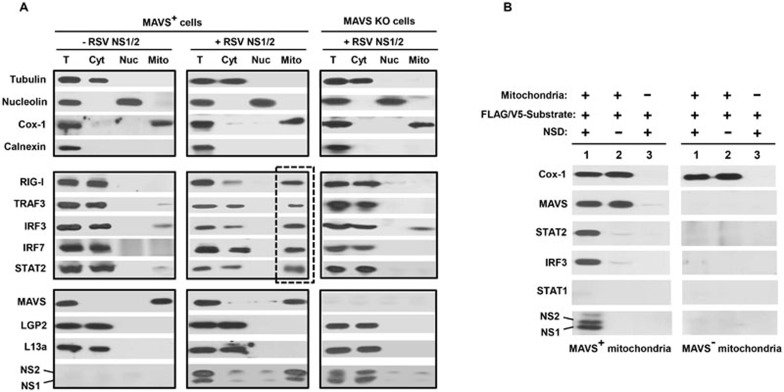
If mitochondria act as the site of NSD assembly and function, one would anticipate that NSD substrates need to associate with mitochondria prior to their destruction, even if transiently. To test this, we overexpressed various NSD substrates in the A549 cells, grew the cells in the presence of MG132 to reduce substrate degradation, fractionated the cell-free extracts and tested the presence of the NSD substrate proteins in the mitochondrial fraction versus in the cytosolic or nuclear fractions. Of note, the mitochondria-associated membrane (MAM) is recognized as a specialized membrane subcompartment, linking the mitochondria to the endoplasmic reticulum (ER)-peroxisome network33,34. Recent evidence suggested that the MAM-associated MAVS serves as an early docking platform for mitochondrial recruitment of RIG-I and TRAF3, and that this is important for IFN induction35. Calnexin is a specific marker for the MAM-ER-peroxisome complex and is absent in highly purified, MAM-free mitochondria35,36. We first confirmed the purity of the subcellular fractions through the use of these well-established markers35,36, i.e., tubulin for cytoplasm, nucleolin for nucleus, Cox-1 for mitochondria and calnexin for MAM-ER-peroxisome complex (Figure 3A, all top panels). In particular, the lack of calnexin in our mitochondrial fraction suggested the absence of MAM. We then found that a substantial portion of the examined NSD substrates, which are normally cytosolic proteins (Figure 3A, middle left panel), localized to the mitochondria when NS proteins were coexpressed (Figure 3A, middle center panel, the lane boxed with broken lines) and their cytosolic portions were concomitantly reduced. This cytosol-to-mitochondria relocation was not observed in MAVS KO cells (Figure 3A, the right panels). Interestingly, the majority of overexpressed NS proteins was also detected in the mitochondrial fraction in WT cells, whereas most of them were present in the cytosol in MAVS KO cells. Taken together, these results suggest that the NSD substrates, together with the NS proteins, may use MAVS to translocate to the mitochondria for the eventual degradation reaction.
Figure 3.
MAVS plays a key role in NSD-mitochondria association. (A) Intracellular (ex vivo) complex of NSD, its substrates and mitochondria. WT or MAVS KO cells were transfected with various NS targets (middle row panels) or non-targets (bottom row panels) together with NS1 and NS2 plasmids when indicated. Cells were then grown in the presence of 50 μM MG132. The top panel shows the relative purity of each subcellular fraction (T = total lysates; Cyt = cytosol; Nuc = nucleus; Mito = mitochondria) using fraction/organelle-specific markers: tubulin for cytosol, nucleolin for nucleus and Cox-1 for mitochondria. The lack of calnexin in the mitochondrial fraction excludes the presence of MAM, peroxisomes and ER and indicates its relative purity. Transfection of NS promotes the appearance of all examined NS targets in the mitochondrial fraction (box with broken line) in WT (MAVS+) cells, not in MAVS KO cells. MAVS expression was detected in the mitochondrial fraction; in contrast, LGP2 and L13a are never found in the mitochondria, regardless of NS overexpression (bottom row). NS proteins (lowest row) were largely detected in the mitochondrial fraction. A small, basal level of IRF3 is found associated with mitochondria in the absence of NS, likely reflecting its natural affinity for mitochondrial Bax37. (B) Intracellular (in vitro) complex of NSD, its substrates and mitochondria. Mitochondria purified from WT and MAVS KO MEFs were employed in reconstituted NSD reactions as described in Figure 2B in the presence of 20 μM MG132. Following incubation for 10 min at 32 °C, the mitochondria were pelleted by centrifugation at 10 000× g for 10 min at 4 °C, and the pellets were analyzed by SDS-PAGE immunoblotting. Only the complete reactions (lane 1) containing all three components (NSD, mitochondria and substrate) allowed the association of substrates with mitochondria. STAT1, a non-substrate, did not associate. Cox-1 served as the mitochondrial marker.
These findings encouraged us to test the MAVS-dependent recruitment of NSD to the mitochondria in vitro. We incubated the three components, namely mitochondria, NSD and affinity-purified epitope-tagged substrates, in the presence of the proteasome inhibitor, pelleted the mitochondria and the bound proteins by centrifugation, and analyzed the presence of specific proteins by immunoblotting. We tested two NSD substrates that are involved in the IFN induction and response pathways, respectively. The results show that both NSD substrates (IRF3 and STAT2) indeed associated with mitochondria in an NSD- and MAVS-dependent manner, whereas STAT1 did not associate with mitochondria regardless of the presence of NSD and/or MAVS (Figure 3B), thus recapitulating the intracellular (ex vivo) NSD substrate profile in the cell-free assay (in vitro).
IFN-signaling suppression by NS proteins requires mitochondrial MAVS
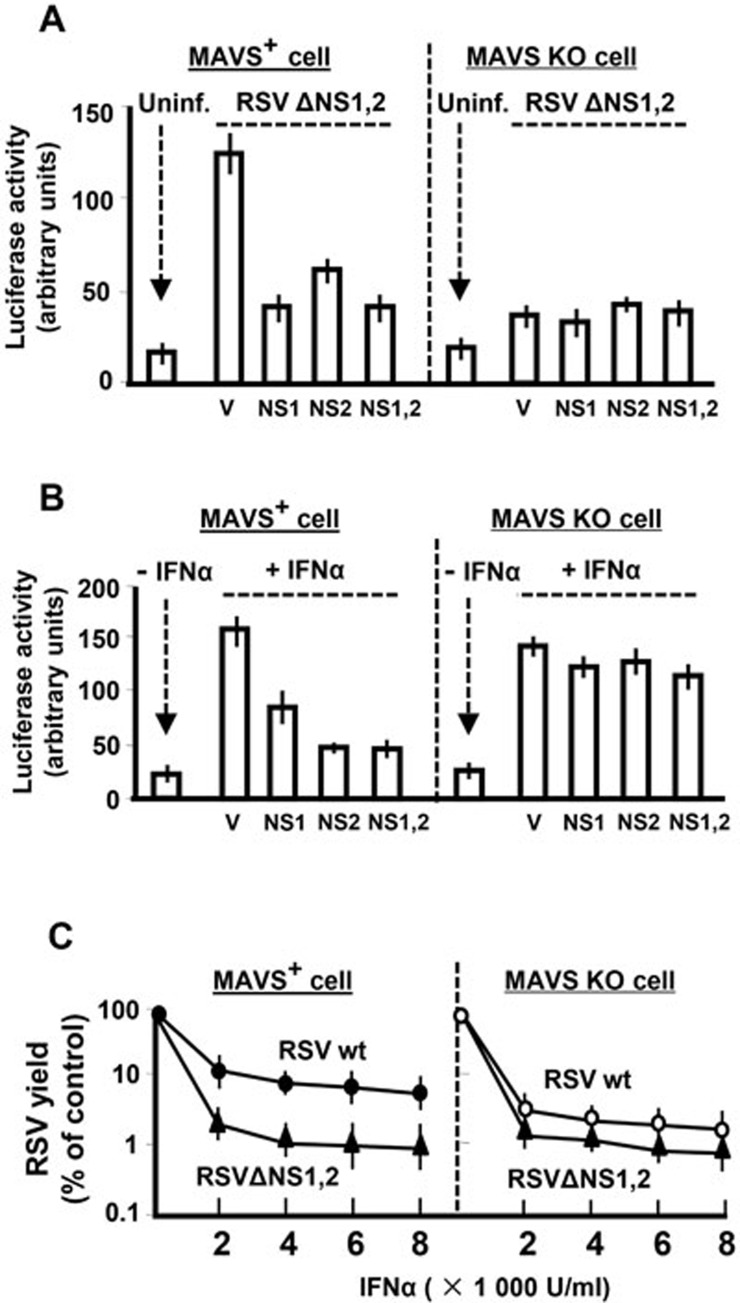
To ascertain the physiological relevance of the MAVS-mitochondria axis in immune suppression, we tested whether this axis has a bearing on type I IFN signaling. We performed two types of luciferase (Luc) assays using reporter plasmids that contain the IFNβ promoter and the IFN-responsive ISGF54 gene promoter, respectively. Whereas the former is a measure of IFN induction pathway, the latter measures IFN response, and we have previously shown that both reporter activities are suppressed by the NS proteins27,28. As before, we triggered the IFN induction pathway by infecting the cells with NS-deleted RSV (RSVΔNS1ΔNS2)27 and found that this induction is severely defective in the MAVS KO cells (Figure 4A), suggesting a cardinal role of MAVS in RSV-triggered IFN induction. To activate the IFN response pathway (Figure 4B), we treated the cells with recombinant IFNα27, which effectively increased IFN-responsive Luc activities in both WT and MAVS KO cells, suggesting that MAVS plays very little role, if any, in mediating cellular responses to type I IFN.
Figure 4.
MAVS is required for optimal IFN suppression by NS protein. IFN induction and IFN response were quantified by the appropriate Luc reporter assays, and RSV growth was measured by plaque assay in Hep-2 cells as previously described28,38. NS1 and NS2 plasmids have also been described before28, and V is the empty vector control. WT and MAVS KO MEFs were infected with NS1/2-deleted RSV. Where indicated, NS1 and/or NS2 proteins were overexpressed. (A) IFN induction. IFN gene activation was evaluated by the luciferase assays using constructs containing IFNβ gene promoter. (B) IFN response. The ability of NS1/2 to suppress the host cell response to recombinant IFNα was tested by using the ISGF54-Luc reporter assays, and suppression was considerably reduced in MAVS KO cells. (C) RSV growth. Monolayers of WT and MAVS KO MEFs were infected at 1 m.o.i. in the presence of increasing amounts of recombinant IFNα as shown39, and infectious progeny virus, liberated after 24 h, was plaque assayed by serial dilution.
In testing the function of NS proteins in this system, we found that NS1 and NS2 suppressed both pathways in WT cells (Figures 4A and 4B, left panels), confirming our previous findings27. In the MAVS KO background, the role of NS proteins could not be determined for the IFN induction pathway, as this pathway could not be activated in the first place. Nonetheless, the IFN response pathway was operational in both WT and MAVS KO cells (Figure 4B, V lanes). We found that the NS proteins were almost completely ineffective in suppressing the IFN response pathway in the MAVS KO background. Overall, the evidence presented so far shows that mitochondrial MAVS is co-opted by NSD to degrade IFN signaling-related NSD substrates, which is likely essential for RSV to antagonize IFN production and action. The virological relevance of this finding was investigated by testing RSV replication, which revealed that NS-deleted RSV exhibited impaired virus replication compared with normal RSV in WT cells, although this discrepancy disappeared in the MAVS KO cells (Figure 4C). Overall, these results indicate that NS function is highly defective in MAVS-deficient cells.
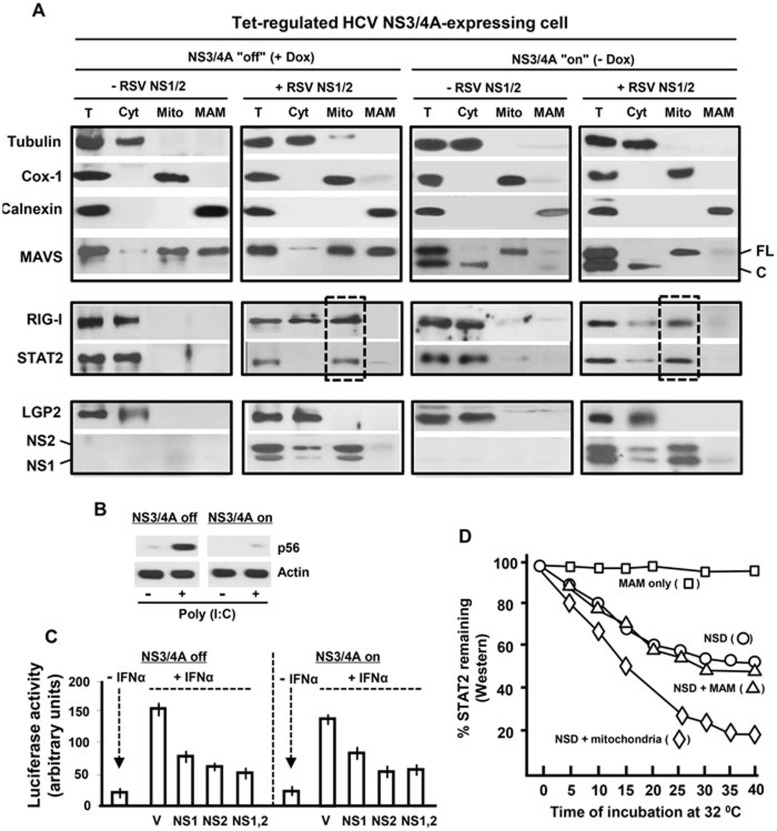
NS function is independent of MAM-associated MAVS
As mentioned earlier, besides mitochondrial MAVS, a subset of cellular MAVS binds to MAM, which serves as the major signaling molecule (platform) for RIG-I-mediated IFN induction35. Recent studies have also revealed that hepatitis C virus (HCV) is a robust suppressor of the IFN response pathway40 and that the viral NS3/4A protein, a serine protease, achieves this by specifically and efficiently degrading MAM-associated MAVS2,35,41. However, NS3/4A protease leaves the mitochondrial MAVS intact35. This provides us with an in vivo strategy of interrogating the role of MAM-MAVS in the RSV NS1/2-mediated immune suppression. We used the previously authenticated cell line in which NS3/4A expression could be induced by doxycycline withdrawal (−Dox)35. We confirmed that upon doxycycline withdrawal, nearly half of the total MAVS was cleaved and the MAM-associated MAVS disappeared, whereas the mitochondrial MAVS indeed survived (Figure 5A, top panel). Next, we inhibited the proteasomal activity by MG132 to better study the expression of RIG-I and STAT2 in these cells. For both proteins, induction of NS3/4A expression did not inhibit their NS-mediated recruitment to the mitochondria (Figure 5A; middle panel, the lane boxed with broken lines). Consistently, as a negative control, the non-substrate of NSD LGP2 did not translocate to the mitochondria under any condition; the mitochondrial localization of the majority, if not all, of NS1 and NS2 was also confirmed (Figure 5A; bottom panel). The loss of MAM-MAVS-dependant function was confirmed by the lack of poly(I:C)-triggered induction of p56, a member of the IFN-stimulated gene (ISG) family, in NS3/4A-on cells (−Dox), whereas this induction was normal in the control, NS3/4A-off cells (+Dox) (Figure 5B). We then tested the ability of NS1/2 to suppress the IFN response pathway using the ISGF54-Luc reporter assay in these cells. NS1 and NS2 were found to be equally active in +Dox and −Dox cells (Figure 5C). Lastly, results of the in vitro reconstituted NSD reactions show that, unlike purified mitochondria, purified MAM failed to enhance NSD degradative activity (Figure 5D). Together, these results and those shown in Figure 3 strongly suggest that in contrast to the innate immune signalosome, which assembles on MAM-MAVS, the NS-enlisted innate immune degradasome assembles and functions on the mitochondrial MAVS.
Figure 5.
NS function and mitochondrial NSD assembly in cells with degraded MAM-MAVS. HCV NS3/4A-expressing cells35 were grown as described in Materials and Methods, and the expression of NS3/4A, which leads to MAM-MAVS cleavage, was induced by replacing Dox-containing media with Dox-free media. Induction of NS3/4A expression was confirmed by immunoblotting (data not shown). (A) Immunoblotting analysis of cell fractions was carried out as in Figure 3. The full-length (70 kD) and cleaved bands of MAVS are indicated by FL and C, respectively. (B) Cells in 12-well plates were transfected with 20 μg of poly(I:C) per well, and 10 h later, the total cell extracts were immunoblotted for p56 using specific antibody42. Actin served as a loading control. (C) IFN-activated reporter assay was performed in cells with or without NS3/4 expression. (D) In vitro degradative activity of NSD towards STAT2, with or without supplementation of mitochondria or MAM (purified as in A), was tested as in Figure 2B.
Once we found that mitochondria played a critical role in immune suppression, it was pertinent to ask how this novel mitochondrial function intersects with the known properties of this organelle. First of all, mitochondria are well known for their energy-generating role through oxidative phosphorylation and ATP synthesis. Second, in live cells, the mitochondria undergo dynamic changes in length and shape through fusion and fission43,44. Lastly, the mitochondria are also highly motile and swim around the cytosol through the use of the contractile microtubule network45,46. In the following sections, we inquired whether these mitochondrial properties have any bearing on the NS-mediated IFN signaling antagonism.
Mitochondrial ATP synthesis and respiration are not required for NSD function
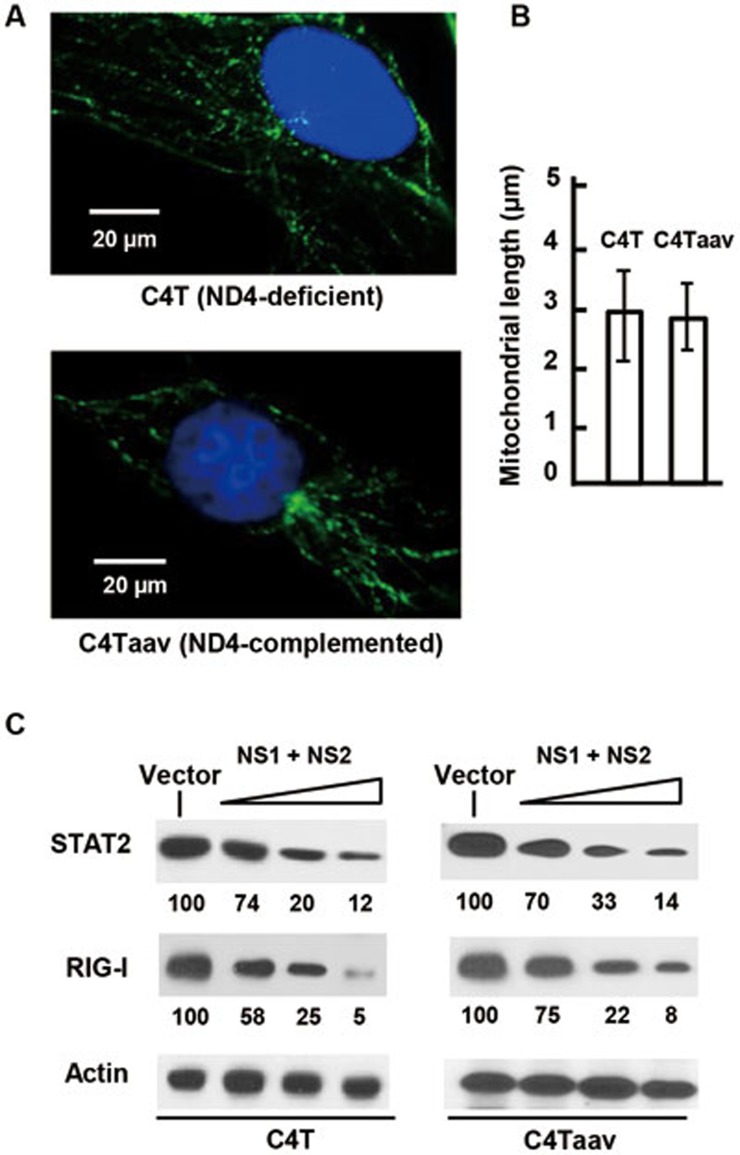
Mitochondrial oxidative phosphorylation is a significant source of cellular ATP under aerobic conditions. We now know that NSD is associated with mitochondria and that the microtubules also need ATP to drive mitochondrial movement. Moreover, it is possible that the in vivo assembly of NSD on mitochondria utilizes mitochondrial ATP. On the other hand, recent proteomic analysis of the A549 cells revealed that the expression of a subset of mitochondrial proteins suffered reductions upon RSV infection, which included the respiratory complex I protein and NADH dehydrogenase subunit 1047. As such reductions likely affect mitochondrial ATP synthesis, we inquired whether or not the mitochondrial ATP synthesis plays a role in NSD function. We took advantage of the human C4T cell line48, in which a frameshift mutation in the mitochondrial DNA (mtDNA)-encoded ND4 gene resulted in no assembly of NADH dehydrogenase, and a complete loss of enzyme activity, respiratory function and mitochondrial ATP synthesis. The primary source of ATP in these cells is glycolysis. In the control “WT” cell line, C4T-aav, the NADH dehydrogenase activity was fully restored by the introduction of the homologous Saccharomyces cerevisiae NDI1 gene48. In these two cell lines, mitochondrial length was essentially identical (Figures 6A and 6B) and so was NS degradative activity, as measured by using STAT2 and RIG-I as substrates (Figure 6C). Thus, NSD functionality does not depend on mitochondrial generation of ATP. In other words, if any step of NSD assembly or function is ATP-dependent, cytoplasmic ATP likely meets the requirement.
Figure 6.
Lack of a requirement of mitochondrial ATP synthesis in NSD function. (A) Images of both C4T and its NADH oxidase-restored isogenic cell lines stained with mitotracker are shown, and (B) average mitochondrial length was measured. (C) These cell lines were transfected with FLAG-STAT2 or FLAG-RIG-I plasmids together with increasing amounts of NS1 and NS2 plasmids (0.2, 0.4, 0.6 μg of each per well in a 24-well plate; or with 0.8 μg of no-NS vector) as indicated. Cell lysates were subjected to immunoblotting. Actin served as loading control. The STAT2 and RIG-I bands were densitometrically scanned, normalized against the actin band intensity and the resultant numbers are shown relative to the vector-only (no NS) band.
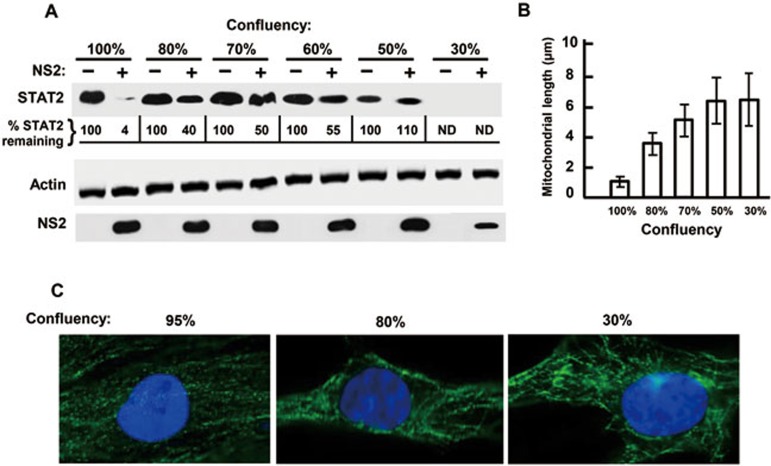
Shorter mitochondria correlate with better NSD function
In our laboratory, we routinely perform plasmid transfections at near 100% cell confluency of the monolayer. However, we made the serendipitous discovery that whenever confluency was lower, NSD function, as measured by degradation of transfected targets, seemed to be less efficient. To quantify this effect, we plated A549 cells at varying densities, transfected the monolayer with FLAG-STAT2 and NS2, and quantified the STAT2 protein levels. As shown in Figure 7A, NS2 degraded nearly 90% STAT2 in fully confluent monolayers, but the degradation started to drop at 80% confluency. At even lower confluency, we observed progressively lower expression of STAT2 (being essentially undetectable at 30%), perhaps due to poor transfection efficiency, but nonetheless, the inefficiency of NS2 in degrading STAT2 was clearly discernible as higher percentages of STAT2 remained undegraded. In our search for a mechanism of this defect, we came across a report49 revealing that cells in the G0/G1 phase have the shortest mitochondria, and we entertained the possibility that a near 100% cell confluency may have a similar effect on mitochondrial length. Indeed, when we measured the mitochondrial length at various cell densities, the average length at 100% confluency was only about 1 μm (Figure 7B and 7C), comparable to that reported for cells at G0/G1 phase. The length dramatically increased to about 4 μm at 80% confluency and continued to increase upon further reduction in cell density, eventually reaching a plateau of 6 μm at 30% confluency. Thus, shorter mitochondria appear to be of greater assistance to NS function. Because of the noted confluency dependence, all our standard NSD-mediated degradation assays (unless specified otherwise) were performed using cultures of 100% confluency.
Figure 7.
Higher cell confluency correlates with better NSD function. An estimated volume of A549 cell suspension was seeded to achieve roughly 60% confluency upon seeding, and decreased percentages of this amount (e.g., 80%, 70% etc as shown) were seeded in other wells. Next morning, when the first well reached full confluency, the confluency of the other wells was confirmed by microscopy. (A) NSD function assay. Once a series of cell confluency was achieved, all wells were transfected with FLAG-STAT2 and FLAG-NS2 plasmids; 24 h later, cell lysates were harvested and analyzed by immunoblotting as in Figure 1. Intensities of the band of remaining STAT2 were quantified and shown as percentage of the no-NS lane for each confluency. ND = band intensity was too faint to be determined. (B, C) Mitochondrial length. Mitochondria in live cells at different confluency were stained with Mitotracker and their average length was measured.
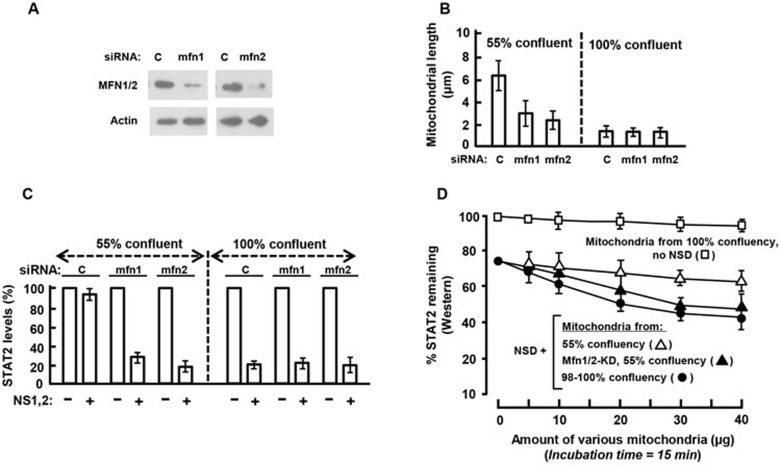
To further validate this finding, we used a genetic approach. Mitochondrial fusion in higher eukaryotes requires at least two essential GTPases, mitofusin1 (Mfn1) and mitofusin2 (Mfn2), which are ∼60% identical in amino acid sequences50,51,52,53. Loss-of-function of either protein shifts the dynamic equilibrium from fusion to fission, leading to shortening of the mitochondrial filaments54. We first silenced the expression of either Mfn gene using specific siRNA and then tested NSD function by immunoblotting in cells transfected with FLAG-STAT2 and NS plasmids. Microscopic length measurement showed that in a 100% confluent monolayer, where the mitochondria are already at their shortest states (Figures 7B and 8B), Mfn siRNAs did not cause any further shortening (Figure 8B). However, at 55% confluency, where the mitochondria are much longer in average, mitochondrial shortening induced by Mfn1 or Mfn2 depletion was evident (Figure 8B). When NSD function was tested, mitochondrial shortening was found to correlate with increased NSD activity, i.e., a higher percentage of STAT2 was degraded either at higher cell confluency or upon Mfn depletion (Figure 8C). In conclusion, in both growth-related (confluency) and genetic (Mfn-based) studies, shorter mitochondria seemed to facilitate NSD function.
Figure 8.
Shortening of mitochondria by Mfn knockdown facilitates NSD activity. (A) Mfn knockdown efficiency. Mfn1 and Mfn2 expression was silenced by using ON-TARGETplus siRNAs as shown by immunoblotting using an antibody recognizing both Mfn1 and Mfn2. (B) Mitochondrial length measurement. Lengths of stained mitochondria in live cells were determined in cells at the indicated confluency. (C) NSD function assay. Degradative activity of transfected NS1/NS2 towards STAT2 was assayed in normal and Mfn-deficient cells, and the remaining STAT2 levels were determined as described in Figure 7A. (D) In vitro demonstration of higher efficiency of shorter mitochondria in facilitating NSD activity. Mitochondria were purified from the following A549 cells: 98% - 100% confluency (●); 55% confluency (Δ); Mfn1/2 knockdown in 55% confluent monolayer (▴). Varying amounts of these mitochondria were employed in NSD reactions with fixed amounts of immune-purified FLAG-STAT2 and NSD, as described in Figure 2B, but the reaction was incubated for 15 min only. In reactions with lower amounts of mitochondria, the total volume was made up with the mitochondria storage buffer. As before, mitochondria alone (□) had little or no degradative activity.
As the role of mitochondria in NSD-mediated degradation could be recapitulated in vitro (Figures 2B and 3B), we wondered whether mitochondria of various sizes could also differentially regulate NSD activity in vitro. To this end, we purified mitochondria from the A549 cells that were either fully confluent, or only 55% confluent, and from cells in which Mfn1/2 have been knocked down by RNAi (Figures 8A-8C). Various amounts of these mitochondria were then used to reconstitute NSD-mediated FLAG-STAT2 degradation assay in vitro. Results clearly show that when same amounts of mitochondria were added to the reaction, the shorter mitochondrial populations (those isolated from fully confluent or Mfn KO cells) are much more proficient in assisting NSD than the longer populations (those isolated from less confluent, Mfn-positive cells) (Figure 8D). These results suggest that the surface-to-weight ratio of mitochondria is an important factor in regulating NSD activity.
Mitochondrial motility is essential for NSD function
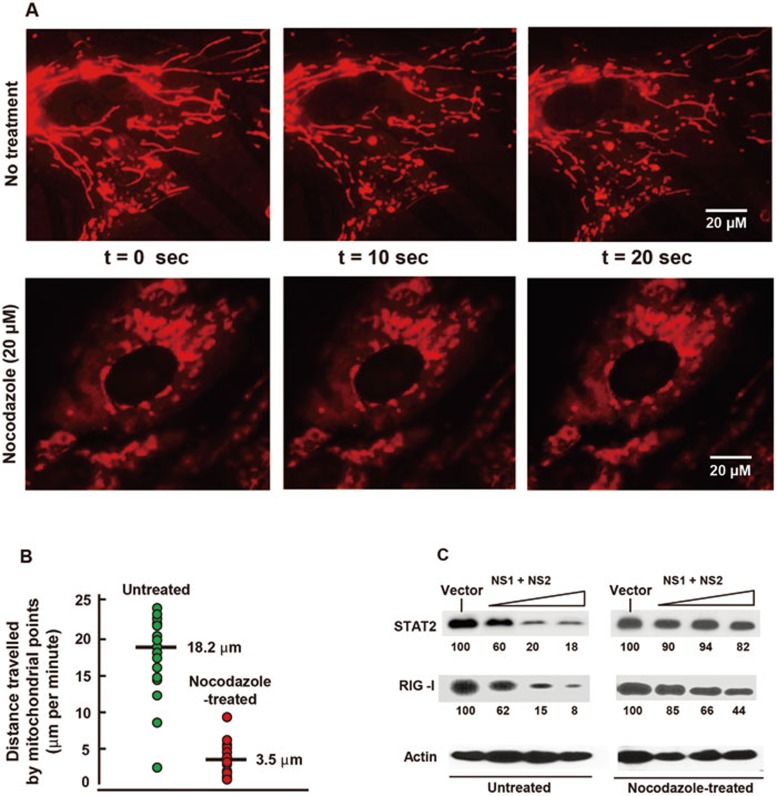
Mitochondria utilize the contractile force of the microtubules for their intracellular motility, but its significance and regulation remain elusive45. It is also currently unknown whether such movements have any impact in innate immunity. To investigate this possibility, we tested NSD function in cells treated with nocodazole, a microtubule-depolymerizing agent known to stop mitochondrial movement46. The inhibition was ascertained by live cell imaging of mitotracker-stained cells and by measuring multiple point-to-point distances over a time period46. Indeed, nocodazole treatment slowed down the average speed of mitochondrial movement to nearly one-sixth of that of the control cells (Figures 9A and 9B). Interestingly, in nocodazole-treated cells, degradation of STAT2 and RIG-I by NS1/2 was highly inhibited (Figure 9C). These results suggest a correlation between optimal mitochondrial motility and immune suppression by viral NS proteins, although alternative explanations such as a direct role of the microtubule network in NSD assembly cannot be ruled out.
Figure 9.
Motile mitochondria facilitate NSD function. (A) Inhibition of mitochondrial movement by nocodazole treatment. Live imaging of mitotracker-stained mitochondria in the A549 cells was performed starting from 4 h after the addition of nocodazole (20 μM) or control solvent45. Three frames at 10 s intervals are shown. (B) Quantification of mitochondrial movement. The movement was measured and the representative data from 15 mitochondrial spots are plotted. (C) NSD functional assay. This was performed in A549 cells transfected with the indicated plasmids. Nocodazole (20 μM) was added 5 h after plasmid transfection and the same medium was maintained until the cells were harvested for immunoblotting 20 h later. There was no obvious cytopathy in the treated cells. The intensities of STAT2 and RIG-I protein bands were normalized against those of actin and expressed relative to the vector-only (no NS) band.
Discussion
The mitochondria act as hubs for a multitude of chemical reactions that are essential for the life and death of all eukaryotic cells. The behavior of these highly dynamic organelles has remained enigmatic; implications of mitochondrial fission, fusion and motility, and events that control them have remained largely unknown. The most recent addition to the list of mitochondrial functions is their role in pathogen-activated innate immunity5,55. A critical discovery is that the ∼70 kD mitochondrial membrane protein MAVS serves as an adaptor for activated RLRs, which then sets off a cascade of signaling reactions, ultimately leading to the induction of antiviral and inflammatory responses mediated by type I IFN and NF-κB pathways. Our results here show that mitochondria and MAVS in fact play dual roles: they are essential not only for induction but also for suppression of the innate immune responses. A conjectural structure of the mitochondria-assembled NS degradative complex is presented as a working model in Figure 10.
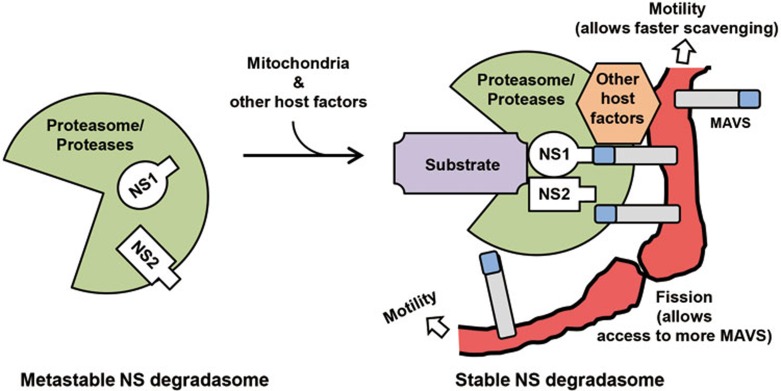
Figure 10.
A working model for mitochondria-assembled NSD. The core NSD, as purified by gel filtration, is shown on left (green), in which the subunits may be only loosely attached. NSD is stabilized by recruitment to MAVS on motile mitochondria, perhaps allowing recruitment of other host factors, such as chaperones and ubiquitination pathway proteins18. Both NS proteins may have the capability to interact with MAVS, if in proper spatial juxtaposition. We propose that mitochondrial motility allows rapid scavenging in the cytosol to collect and assemble all the subunits of the complex, and that mitochondrial fission provides a larger mitochondrial surface area, allowing more efficient use of a greater number of MAVS and other membrane structures if needed, free of space constraints.
The precise composition of the NSD needs to be determined, but it appears that the 300-750 kD fractions (Figure 2) harbor the core enzymes containing proteases or proteasomal activity. The mammalian proteasome consists of a 20S core enzyme that is roughly 600-700 kD in size56. Thus, the higher-molecular-weight fractions of NSD may be large enough to accommodate the 20S core, whereas the lower-molecular-weight fractions (300-500 kD) may represent various subsets of the larger complex, which are formed possibly due to the dissociation of various subunits, reflecting weak associations among these subunits. The core complexes may be stimulated or stabilized through their association with mitochondria to form the holoenzyme. The large amounts of NS1/NS2 that were excluded from the column fractions could be associated with mitochondria. We have not further characterized this pool of NS proteins due to the overall complexity of this pool that also includes other cellular organelles and structures. Nonetheless, the separation of some NSD from the mitochondria during chromatography seems to indicate that this mitochondrial association is labile. The overall metastable nature of the complex and the variable associations of various subunits may constitute important aspects of NSD regulation in vivo. Attempts are currently being made to immunoprecipitate the NSD complex using the FLAG tag attached to NS1/NS2 to determine its protein composition and to find out whether ubiquitin pathway enzymes and/or proteasomal subunits are integral components of NSD or are adventitiously associated. Moreover, finding of non-proteasomal proteases in the NSD will help explain the incomplete inhibition of NSD degradative activity by MG132; in fact, complex roles of both proteasomal and non-proteasomal proteases have been recently inferred in the negative regulation of IRF3 during viral infection57.
In the current study, we began with an apparent enigma of how the NSD recruits and targets proteins with little or no sequence similarity. Our finding of MAVS dependence of this event provides a potential integration point for this diversity. We hypothesize that NS1 and/or NS2 have a yet unknown MAVS-binding domain that serves to anchor the NSD to mitochondria and that additional interactions between NSD subunits and mitochondria may also form to stabilize an otherwise metastable core complex (Figure 10). A recent study has indeed shown a direct interaction of NS1 with MAVS17, but the domains of interaction have not been mapped. To date, nearly three dozen proteins with little or no discernible sequence similarity have been shown to bind MAVS, and the list seems to be growing5. These proteins regulate a diverse array of cellular events including, but not limited to, inflammation, apoptosis, autophagy and antiviral response (such as the NSD substrates presented here). Some of the NSD substrates are also known to bind MAVS. Specifically, the CARD domains of RIG-I and MDA5, RLR family members, interact with the single N-terminal CARD domain of MAVS by heterotypic CARD-CARD interaction, whereas LGP2, the third member of the RLR family, lacks the CARD domain and thus fails to bind MAVS1,3,4,10,12,58,59. TRAF3 interacts with at least one (aa 455-460) of the three TRAF-interacting motifs of MAVS; TBK1, IRF3 and IRF7 bind to the central region of MAVS; and IKKε binds to a region close to the C-terminal mitochondrial transmembrane (TM) domain of MAVS5. It is conceivable that tethering of these substrates to MAVS facilitates the recruitment of the NSD-substrate complex to mitochondria. This is consistent with the finding that LGP2 does not bind MAVS, is not recruited to the mitochondria (Figure 3A) and is not degraded by the NS proteins (Figure 1). Lastly, as MAVS appears to be the linchpin of both IFN signaling activation and suppression, it may also serve as a competing ground for the two events, such that occupancy of MAVS by the NSD-substrate complex in the RSV-infected cells may prevent the recruitment of the RIG-I-downstream IFN-activating factors such as TRAF2/6, RIP1 and TRAF3. Thus, NS may act to suppress innate immune activation even before the NSD substrates are degraded, with the following degradation leading to a more durable and irreversible suppression of innate immune responses.
The exact reason for the need of short and motile mitochondria for optimal NSD function will require further study. Our evidence suggests that mitochondrial length and motility are independent parameters, as neither Mfn knockdown nor cell confluency had a significant effect on mitochondrial motility (data not shown). We speculate that the mitochondrial fission likely increases the available mitochondrial surface area, allowing more MAVS to be available and more spaces for NSD to assemble. Mfn1 and Mfn2 are both involved in the IFN activation pathway, but in different ways. Mfn1 knockdown attenuates virus-activated RIG-I signaling35,55,60, whereas Mfn2 knockdown, which specifically disrupts MAM-mitochondria contacts61,62, enhances RIG-I signaling and increases the host-cell resistance to HCV and vesicular stomatitis virus35. Interestingly, the suppression of innate immunity by NS1/2 was enhanced by the loss of either Mfn (Figure 8), supporting that mitochondrial association is sufficient for NSD activity. This is further confirmed by our finding that methyl-β-cyclodextrin, a drug that disrupts MAM-mitochondria contacts via an Mfn-independent mechanism62, also did not inhibit NSD activity (data not shown).
Interestingly, there may be an intriguing relationship among mitochondrial length, cell confluency and RSV growth. First, mitochondrial length oscillates during the cell cycle, being the shortest at the G0/early G1 phase and the longest at the mitotic phase49. Second, cell cycle arrest and RSV growth seem to reinforce each other. In cell culture, cell cycle arrest facilitates RSV growth. Reciprocally, RSV infection of pulmonary epithelial A549 cells leads to significant cell cycle arrest at the G0/G1 phase63,64,65. This may be mediated in part by the viral matrix protein M that promotes p53 and p21 accumulation and decreases phosphorylation of the retinoblastoma protein (Rb)66. The ability of the M protein to traffic to the nucleus67,68 may underlie this proviral effect. In addition to the M protein, recent studies have revealed that NS1 itself may also promote cell cycle arrest69, which likely facilitates its IFN-suppressive function. Based on these, it is tempting to speculate that the ability of RSV M and NS1 proteins to hold the infected host cells at the G0/G1 phase may help virus growth and that the enhanced suppression of innate immunity by NS1/2, when the mitochondria are shorter at this phase, may in part contribute to the virus growth. Studies of mitochondrial size and NS function in synchronized cells should shed lights on this intriguing possibility.
Our results constitute the first evidence suggesting a role of mitochondrial movement in the regulation of innate immunity. The underlying mechanism is currently unknown. Nonetheless, it is reasonable to propose that mitochondrial movement throughout the cytosol allows them to access the various subunits of the NSD at a much faster rate than that of passive diffusion to a stationary organelle. In conclusion, our discovery of a central role of mitochondria in innate immune suppression has opened new directions for further studies in the future.
Materials and Methods
Cell culture, transfection and western blot analysis
These procedures were carried out as described previously27,28. MAVS (−/−) and its corresponding WT MEFs were kind gifts from Robert Silverman (Cleveland Clinic). The stable cell line, expressing HCV NS3/4A protease from a Tet-OFF promoter70, was a kind gift from Michael Gale (University of Washington); it was maintained in the presence of G418 (400 μg/ml), puromycin (1 μg/ml) and doxycycline (Dox) (1 μg/ml). To induce NS3/4A expression in these cells, the media were replaced by Dox-free media. C4T and C4Taav cells, generated in the Attardi laboratory47, were kindly provided by Anne Chomyn (California Institute of Technology, Pasadena, USA) and Yidong Bai (University of Texas Health Sciences Center, San Antonio, USA).
All recombinant plasmids used here contain FLAG tag, except that IRF3 contains a V5 tag and MAVS and L13a are HA-tagged. The FLAG-RIG-I and V5-IRF3 clones have been described37,71; all other clones were described previously27,28. Unless otherwise stated, all transfections were done in 100% confluent monolayers of cells (as stated earlier) using indicated amounts of plasmids. The A549 cells were transfected using Lipofectamine 2000, and all mouse cells were transfected using Lipofectamine LTX with PLUS reagent (Invitrogen/Life Technologies). All immunoblots were probed with the appropriate antibody, followed by the corresponding secondary antibody conjugated to horseradish peroxidase and developed using the ECL Prime kit (Pierce). All tag (FLAG, V5) antibodies were purchased from Millipore and non-tag antibodies from Santa Cruz. Bands were detected in the LI-COR Odyssey Fc imaging system, and the intensities were quantified with the integrated LI-COR software and normalized against actin.
In experiments requiring confluency measurement, the cells were grown in dishes with grids (Fisher Scientific) and low-magnification grayscale images of five different regions were captured in JPEG format using an AMG-EVOS digital inverted microscope. Each image was analyzed by NIH ImageJ software72, whereby the contour of the covered regions were manually marked by the mouse cursor and the fraction of the area covered by cells was calculated. Averaged confluency from the five regions was used as an estimate of the overall confluency.
Fractionation of cell lysates
For the isolation of NSD, approximately 106 cells in fully confluent A549 monolayer, grown in DMEM with 10% fetal bovine serum, were transfected with 30 μg each of codon-optimized pCAGGS-NS1 and pCAGGS-NS228. At 24 h post transfection, the monolayer was rinsed with PBS and lysed in a total volume of 200 μl Buffer A (50 mM Tris-acetate, pH 7.6; 150 mM NaCl; 0.5% NP-40; 15% glycerol; 0.5 mM DTT) by incubating in ice for 10 min, followed by scrapping and pipetting. Cell debris was removed by centrifugation at 2 000× g for 5 min in cold. The supernatant was loaded on a 10 × 300 mm Superdex 200 10/300 GL column (GE Healthcare) equilibrated with Buffer A minus NP-40 and developed in 80 ml of the same buffer. A total of 80 fractions (1 ml each) was collected, and aliquots of each fraction were dialyzed against the same buffer with 20% glycerol (instead of 15%) using a Rapid Equilibrium Dialysis (RED) Device (Thermo Scientific) in cold. The fractions were analyzed by immunoblotting and in vitro degradasome assay as described in the respective experiments.
Purification of subcellular fractions
Each subcellular fraction was purified from the equivalent of 2 × 107 cells grown in fully confluent monolayers. To isolate the nuclei and cytosol from various cells (A549, mouse WT MEF or MAVS KO MEF), we used the Nuclei PURE prep nuclei isolation kit NUC-201 (Sigma-Aldrich). After cell lysis, half of the lysate was centrifuged at 120 000× g for 30 min, and the supernatant was considered as the cytosolic fraction. The nuclei were purified from the other half by following the manufacturer's protocol.
Mitochondria were first purified using the “Mitochondria isolation kit for cultured cells” (Thermo Scientific; Product# 89874) using the “Reagent-based” procedure and the recommendation for higher purity (centrifuging at 3 000× g instead of 12 000× g). This preparation, containing mitochondria and MAM, was further subjected to Percoll gradient as follows35,36. The 3 000× g pellet from the kit procedure above was resuspended in 0.1 ml of mannitol buffer (0.25 M D-mannitol, 5 mM HEPES, 0.5 mM EGTA, pH 7.4). The suspension was layered on a 30% Percoll gradient and centrifuged at 95 000× g for 65 min. The visible series of mitochondrial bands in the middle section of the tube and the single MAM band just above them were collected by puncturing. Both fractions were washed twice with the same mannitol buffer as above (except that the HEPES concentration was increased to 25 mM) by centrifugation at 6 500× g, 10 min, resuspended in the same buffer plus 10% glycerol to a final concentration of 20 mg/ml protein (by Bradford assay) and stored frozen at −80 °C. The final yields of mitochondria and MAM from 2 × 107 cells were approximately 2 and 0.5 mg (protein), respectively.
Reconstitution of NSD-mediated degradation assay
The substrates for the assay were prepared by immuno-precipitation from transiently transfected A549 cells as follows. Roughly 106 cells in fully confluent monolayer were transfected with 30 μg of the substrate plasmid clone (e.g., FLAG-RIG-I, FLAG-STAT2, V5-IRF3, HA-MAVS, HA-L13a, etc), and 24 h later, washed with PBS and lysed with 0.5 ml of standard RIPA buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% each of Na-deoxycholate and NP-40, 0.05% SDS and manufacturer's recommended amount of protease inhibitor cocktail (Roche)), and 75 μl of a 60% slurry of protein A-Sepharose (PAS) was added to it, followed by rotatory mixing for 30 min in cold. The mixture was centrifuged at 10 000 rpm. for 5 min. To the cleared supernatant, appropriate amount of tag antibodies (against FLAG, V5 or HA) and the same amount of PAS as above were added, and mixing was performed for 2 h in cold. The mixture was centrifuged as before and the pellets were washed three times (200 μl each) with wash buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 20% glycerol) and were finally incubated in 200 μl of the same wash buffer containing the appropriate tag peptide (Sigma-Aldrich) at 100 μg/ml for 30 min at room temperature with gentle rotation. The mixture was centrifuged at 10 000× g for 10 min at room temperature and the supernatant containing the eluted tagged protein was used in the degradation assay described below.
Unless otherwise stated, the NSD assay mixture consisted of the following in a reaction volume of 20 μl: 2-4 μl (depending on amount eluted) of the substrate as above, 2-8 μl (depending on activity) of each dialyzed fraction (or the pooled fraction, where mentioned) from the Superdex column and 2 μl of the purified mitochondria (40 μg protein) in buffer A (described earlier) plus 0.2 mM ATP. The reaction was scaled up based on the number of time samples had to be withdrawn for immunoblotting analysis. The mixture was incubated for various periods at 32 °C, and at the indicated times aliquots were taken and mixed with equal volume of 2× Laemmli sample buffer, boiled, cleared by brief centrifugation and analyzed by SDS-PAGE and immunoblot using appropriate tag antibody.
Mitochondrial measurements
Mitochondrial length and motility were measured by digital fluorescence microscopy of live cells49, stained with the MitoTracker Deep Red FM dye (Molecular Probes, Life Technologies). Cells were grown on coverslips, stained with MitoTracker and observed in a Nikon A1RSI Spectral Confocal Microscope System equipped with Ti-E inverted microscope with Perfect Focus system and NIS-Elements C image analysis software. To determine mitochondrial length, a stained cell was displayed on the monitor and the display was calibrated to correlate pixels to micron length. Length of each recognizable mitochondrion was determined by scrolling the whole length with the cursor. About 20 mitochondria per cell and in 10 cells were thus measured, and the average length was calculated.
The movement was also quantified in the same manner by capturing video frames every 5 s for 1-2 min. The frames were displayed on the screen and a recognizable spot on the mitochondria was followed over time. All motions (anterograde, retrograde, turning) in all the three axes (x, y, z) were measured and added together. Roughly 20-25 spots were tracked per cell, 10-15 cells were examined and the average motility was calculated.
Statistical analysis
Changes between treatment groups of cells or between sets of in vitro experiments were analyzed by one-way ANOVA and then by Student's t-test with Bonferroni correction. All numerical data were collected from at least three separate experiments. Results were expressed as mean ± SEM (error bars in graphs). Differences were considered to be significant at P < 0.05.
Acknowledgments
This research was supported in part by an Ohio Third Frontier Grant, cost-share funds from the Provost and the Research office of the Cleveland State University (CSU), NIH 1 S10 OD010381 (all to SB) and NIH grants (AI073303 and CA068782 to GCS). We are indebted to Chris Basler, Adolfo García-Sastre (Mount Sinai School of Medicine, NY, USA), Carl Ware (Sanford Burnham Medical Research Institute, La Jolla, USA) and Michael Gale (University of Washington, Seattle, USA) for various recombinant plasmids of the IFN pathway genes, and to Michael Gale for the NS3/4A-expressing cell line.
References
- Kawai T, Takahashi K, Sato S, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. Interferons and viral infections. Biofactors. 2009;35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- Bossert B, Marozin S, Conzelmann KK. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J Virol. 2003;77:8661–8668. doi: 10.1128/JVI.77.16.8661-8668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyapalle S, Wong T, Garay J, et al. Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS One. 2012;7:e29386. doi: 10.1371/journal.pone.0029386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Lynch OT, Suessmuth Y, et al. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol. 2007;81:3428–3436. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Zhou H, Cheng X, et al. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2–2 genes are attenuated in vitro and in vivo. Virology. 2000;273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy M, Shi L, Varga SM, Barik S, Behlke MA, Look DC. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology. 2006;344:328–339. doi: 10.1016/j.virol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Ren J, Liu T, Pang L, et al. A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J Gen Virol. 2011;92:2153–2159. doi: 10.1099/vir.0.032987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlender J, Bossert B, Buchholz U, Conzelmann KK. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J Virol. 74:8234–8242. doi: 10.1128/jvi.74.18.8234-8242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] J Virol. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Collins PL. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol. 2005;79:5353–5362. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedan S, Andrews J, Majumdar T, Musiyenko A, Barik S. Multiple functional domains and complexes of the two nonstructural proteins of human respiratory syncytial virus contribute to interferon suppression and cellular location. J Virol. 2011;85:10090–10100. doi: 10.1128/JVI.00413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedan S, Musiyenko A, Barik S. Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J Virol. 2009;83:9682–9693. doi: 10.1128/JVI.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MN, Collins PL. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol. 1999;73:466–473. doi: 10.1128/jvi.73.1.466-473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine. 2003;21:3647–3652. doi: 10.1016/s0264-410x(03)00426-2. [DOI] [PubMed] [Google Scholar]
- Teng MN, Whitehead SS, Bermingham A, et al. Recombinant respiratory syncytial virus that does not express the NS1 or M2–2 protein is highly attenuated and immunogenic in chimpanzees. J Virol. 2000;74:9317–9321. doi: 10.1128/jvi.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarcher JF, Furze J, Wyld S, Cook R, Conzelmann KK, et al. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J Virol. 2003;77:8426–8439. doi: 10.1128/JVI.77.15.8426-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol. 2009;41:1805–1816. doi: 10.1016/j.biocel.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozidis P, Williamson CD, Colberg-Poley AM.Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts Curr Protoc Cell Biol 2007. Chap 3:unit 3.27. [DOI] [PubMed]
- Chattopadhyay S, Marques JT, Yamashita M, et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010;29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke E, Mahoney NM, Almo SC, Barik S. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol. 2000;74:669–675. doi: 10.1128/jvi.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya PL, Kulkarni S. Respiratory syncytial virus strain A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology. 1999;261:227–241. doi: 10.1006/viro.1999.9835. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- Ball EH, Singer SJ. Mitochondria are associated with microtubules and not with intermediate filaments in cultured fibroblasts. Proc Natl Acad Sci USA. 1982;79:123–126. doi: 10.1073/pnas.79.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Mironov SL, Ivannikov MV, Schmidt J, Richter DW. Mitochondrial organization and motility probed by two-photon microscopy in cultured mouse brainstem neurons. Exp Cell Res. 2005;303:114–127. doi: 10.1016/j.yexcr.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Munday DC, Emmott E, Surtees R, et al. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol Cell Proteomics. 2010;9:2438–2459. doi: 10.1074/mcp.M110.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Hájek P, Chomyn A, et al. Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J Biol Chem. 2001;276:38808–38813. doi: 10.1074/jbc.M106363200. [DOI] [PubMed] [Google Scholar]
- Kennady PK, Ormerod MG, Singh S, Pande G. Variation of mitochondrial size during the cell cycle: A multiparameter flow cytometric and microscopic study. Cytometry A. 2004;62:97–108. doi: 10.1002/cyto.a.20091. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombès A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4254. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S, Gaume B, Herrler M, Youle RJ, Fuller MT. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- Ye J, Maniatis T. Negative regulation of interferon-β gene expression during acute and persistent virus infections. PLoS One. 2011;6:e20681. doi: 10.1371/journal.pone.0020681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol. 2008;8:11. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Onomoto K, Takamatsu S, et al. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Sano R, Annunziata I, Patterson A, et al. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca2+-dependent mitochondrial apoptosis. Mol Cell. 2009;36:500–511. doi: 10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Buckley S, Bui KC, et al. Cell cycle arrest in G0/G1 phase by contact inhibition and TGF-beta 1 in mink Mv1Lu lung epithelial cells. Am J Physiol. 1996;270:L879–L888. doi: 10.1152/ajplung.1996.270.5.L879. [DOI] [PubMed] [Google Scholar]
- Gibbs JD, Ornoff DM, Igo HA, Zeng JY, Imani F. Cell cycle arrest by transforming growth factor beta1 enhances replication of respiratory syncytial virus in lung epithelial cells. J Virol. 2009;83:12424–12431. doi: 10.1128/JVI.00806-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Munday DC, Howell G, Platt G, Barr JN, Hiscox JA. Characterization of the interaction between human respiratory syncytial virus and the cell cycle in continuous cell culture and primary human airway epithelial cells. J Virol. 2011;85:10300–10309. doi: 10.1128/JVI.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian T, Gibbs JD, Örvell C, Imani F. Respiratory syncytial virus matrix protein induces lung epithelial cell cycle arrest through a p53 dependent pathway. PLoS One. 2012;7:e38052. doi: 10.1371/journal.pone.0038052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildyal R, Baulch-Brown C, Mills J, Meanger J. The matrix protein of Human respiratory syncytial virus localises to the nucleus of infected cells and inhibits transcription. Arch Virol. 2003;148:1419–1429. doi: 10.1007/s00705-003-0112-y. [DOI] [PubMed] [Google Scholar]
- Ghildyal R, Ho A, Jans DA. Central role of the respiratory syncytial virus matrix protein in infection. FEMS Microbiol Rev. 2006;30:692–705. doi: 10.1111/j.1574-6976.2006.00025.x. [DOI] [PubMed] [Google Scholar]
- Wu W, Tran KC, Teng MN, et al. The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J Virol. 2012;86:7777–7789. doi: 10.1128/JVI.00460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Owen DM, Gale M., Jr Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense. J Biol Chem. 2007;282:10792–10803. doi: 10.1074/jbc.M610361200. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]