Abstract
Quantitative measurements of diffusion can provide important information about how proteins and lipids interact with their environment within the cell and the effective size of the diffusing species. Confocal FRAP is one of the most widely accessible approaches to measure protein and lipid diffusion in living cells. However, straightforward approaches to quantify confocal FRAP measurements in terms of absolute diffusion coefficients are currently lacking. Here, we report a simplified equation that can be used to extract diffusion coefficients from confocal FRAP data using the half time of recovery and effective bleach radius for a circular bleach region, and validate this equation for a series of fluorescently labeled soluble and membrane-bound proteins and lipids. We show that using this approach, diffusion coefficients ranging over three orders of magnitude can be obtained from confocal FRAP measurements performed under standard imaging conditions, highlighting its broad applicability.
Keywords: fluorescence microscopy, FRAP, half time of recovery, diffusion coefficient, confocal laser scanning microscope
Introduction
Diffusion is a fundamental process that is relevant over all scales of biology. How rapidly diffusion occurs is characterized by the diffusion coefficient D, a parameter that provides a measure of the mean squared displacement per unit time of the diffusing species. At the cellular level, measurements of D can provide important insights into how proteins and lipids interact with their environment, such as the binding of transcription factors with DNA and of proteins and lipids with membrane domains (1,2). In addition, especially for the case of soluble proteins, an accurate measure of D can provide important information about the effective size of the diffusing species (3).
Currently, biologists have at their disposal a wide range of fluorescence-based techniques to monitor the diffusion of biomolecules in cells, including single particle tracking (SPT), fluorescence correlation microscopy (FCS), photoactivation, and fluorescence recovery after photobleaching (FRAP) (4). Of these approaches, many require specialized equipment, analytical tools, or probes. In contrast, confocal FRAP is one of the most accessible methods to measure apparent diffusion (or effective diffusion). In FRAP, the diffusion of a population of fluorescently labeled molecules can be studied by photobleaching the molecules contained within a user-defined region of interest (ROI), and then monitoring fluorescence recovery due to the exchange of bleached molecules within the bleach ROI with the surrounding reservoir of unbleached molecules. Most modern confocal laser scanning microscopes (CLSMs) are equipped to perform FRAP measurements, and FRAP measurements are straightforward to carry out. In addition, FRAP can be performed using many fluorescently tagged proteins or on directly fluorescently labeled molecules (4,5).
Despite the relative ease of performing confocal FRAP measurements, straightforward methods to quantitatively analyze the resulting data are currently lacking. Most current approaches to calculate D’s from confocal FRAP data require fitting of recovery curves with analytical diffusion models using customized programs, a process which often demands a significant level of knowledge of FRAP theory (Table 1). Alternatively, FRAP measurements can be quantified simply in terms of the half time of recovery (τ1/2), defined as the time required for a bleach spot to recover half way between initial and steady state fluorescence intensities (6-9). An advantage of this approach is that τ1/2 can be readily extracted from FRAP recovery curves (6-9). However, τ1/2 is strongly dependent on experimental parameters such as the nominal radius of the user-defined bleach spot (rn) and the exact bleaching protocol, as well as the characteristic dynamics of the molecule under investigation. Therefore, τ1/2 is an empirical parameter that cannot be readily compared across studies. In contrast, D provides a quantitative measure of diffusion (10,11). Knowledge of the absolute magnitude of D has many advantages over measurements of τ1/2. For example, D is uniquely determined by the size of the diffusing molecule as well as its local environment (12). Therefore, discrepancies between the theoretically expected D and measured apparent (or effective) diffusion coefficients may indicate possible molecular interactions or more complex situations. For example, in cell membranes the presence of microdomains prevents proteins from undergoing free diffusion (2). Accurate estimates of D are also a necessary starting point for reaction-diffusion analysis (1,13).
Table 1.
Analytic 2D FRAP models:
| References | Bleaching Geometry | Laser Profiles | Postbleach Profile | Diffusion during photobleach | Data types | Microscopy | Target proteins |
|---|---|---|---|---|---|---|---|
| Kang et al. (17) | circle in R2 | Gaussian (rn) | Gaussian (re) | corrected | f(t) | CLSM/ wide-field static | Slow/Fast |
| Smisdom et al. (24) | circle in R2 | Gaussian (ρ) | Uniform with Gaussian edge | Not corrected | f(t) | CLSM | Slow |
| Axelrod et al. (10) | circle in R2 | Gaussian (rn)/Uniform (rn) | Gaussian (rn)/Uniform (rn) | Not corrected | f(t) / τ1/2 | wide-field static | Slow |
| Braga et al. (15) | circle in R2 | Uniform (rn) | Gaussian (re) | corrected | f(t) | CLSM | Slow/Fast |
| Soumpasis (11) | circle in R2 | Uniform (rn) | Uniform (rn) | Not corrected | f(t) / τ1/2 | wide-field static | Slow |
| Sprague et al. (25) | circle in R2 | Uniform (rn) | Uniform (rn) | Not corrected | f(t) | wide-field static | Slow/Fast |
| Waharte et al. (26) | circle in R2 | NA | Gaussian(re) | corrected | f(x, t) | CLSM | Slow/Fast |
| Current Study | circle in R2 | Gaussian (rn) | Gaussian (re) | corrected | f(t)/τ1/2 | CLSM/ wide-field static | Slow/Fast |
| Angelides et al.(21) | circle in a box | Gaussian (rn) | Gaussian (rn) | Not corrected | f(t) | wide-field static | Slow |
| Deschount et al. (22) | box in R2 | Gaussian (ρ) | Uniform with Gaussian edge | Not corrected | f(t) | CLSM | Slow |
| Seiffert & Oppermann (23) | circle/line in R2 | NA | Gaussian (re) | corrected | f(x,t)/re(t) | CLSM | Slow |
R2: infinite 2 dimensional plane, rn: bleaching spot radius= static laser radius, ρ: scanning laser radius, re: effective radius measured from a fluorescent intensity profile, f(t): fluorescence intensity from an ROI, f(x,t): time course of spreading radius of postbleach profiles.
One potential solution to the problem of extracting quantitative D values from confocal FRAP data is to utilize the explicit forms of the classical 2 dimensional FRAP equations for small circular bleaching spots derived by Axelrod for a Gaussian laser (10) and by Soumpasis for a uniform laser (11). Both Axelrod and Soumpasis (10,11) reported equations that relate D, τ1/2, and rn for a pure isotropic diffusion model. For example, the Soumpasis equation is given by
| (1) |
where rn is the radius of the uniform bleach laser and the coefficient 0.224 was numerically determined (11). However, the derivation of both the Axelrod and Soumpasis FRAP equations assumed that diffusion during photobleaching is negligible. This is not necessarily the case for confocal FRAP. Due to the long scanning time of CLSMs (~s), significant diffusion during the photobleaching may occur in confocal FRAP. rn thus may not provide an accurate description of the initial conditions required for this equation to be valid. Failure to take this into account can lead to underestimation of D when the Soumpasis or Axelrod equations are used to analyze confocal FRAP data, especially for fast diffusing soluble proteins (14-17). This discrepancy can be resolved using a modified form of the conventional FRAP equations by using the postbleach fluorescence intensity profile to correct for any diffusion that occurs during the photobleaching by incorporating an empirically determined measure, the effective bleach radius re from the postbleach profile (17). However, this approach also requires full fitting of the FRAP recovery curves to calculate D. In the current study, we sought to further simplify this approach by deriving an equation to calculate D from confocal FRAP data obtained using circular bleach regions.
Theory
All the parameters and their meanings are summarized in Table 2. For simplicity, we assume that cell are flat enough (as in the case of COS 7 cells) so that both the plasma membrane as well as the cytosol can be treated as 2D objects. We also assume that bleaching spot size is small compared with the cell size, so that we can treat the cell as an infinite plane (R2). Having these, laser intensity profiles for photobleaching lasers in R2 are described as either a Gaussian laser:
or a uniform laser,
where H(·) is the Heaviside function. In both cases, rn is defined as a nominal radius, and the excitation lasers are represented as εIrn (x, y) for attenuation factor ε (≪ 1).
Table 2.
List of parameters and their definitions
| Parameter | Meaning | Parameter | Meaning | |
|---|---|---|---|---|
| D | Diffusion coefficient | Fi | Prebleach fluorescence intensity | |
| τ1/2 | Half time of recovery | F0 | Initial postbleach fluorescence intensity | |
| re | Nominal radius = bleaching spot radius =sampling region radius | F∞ | Postbleach steady state fluorescence intensity | |
| re | Effective radius = Spreading radius of post-bleach profile | F1/2 | Fluorescence intensity at the half of recovery = [F0 + F∞]/2 | |
| q | Quantum yield | Mf | Mobile fraction = [F∞ − F0]/[Fi − F0] | |
| ε | Attenuation factor for excitation laser | τD |
|
|
| K | Bleaching depth parameter | Drn | D from Soumpasis Eq. using | |
| γ | The ratio of the nominal and effective radius = rn/re | Dre | D from Soumpasis Eq. using | |
| Ci | Prebleach fluorescent protein concentration | DConfocal | D from rn, re and | |
| n | Number of independent experiments | N | Total number of cells for FRAP data collection |
If we assume the concentration of fluorescent proteins C(x, y, t) satisfies the diffusion equation
where D (μm2/s) is a diffusion coefficient and , then the solution can be computed from a convolution of the fundamental solution of the diffusion equation and an initial condition as
where the fundamental solution of the diffusion equation in R2 is defined as
Therefore, the fluorescence intensity from the bleach ROI can be computed (10, 17) from
where q is the quantum yield of the fluorophores and C(x, y, t) describes the concentration of fluorescent proteins within the confocal volume at time t.
It has been empirically demonstrated that a confocal postbleach profile can be described as a simple Gaussian function (constant minus Gaussian) (13);
| (2) |
where Ci is the prebleach fluorescent protein concentration, and re is the half width at the approximately 14% of bleaching depth from the top. We define re as the effective radius of a postbleach profile, in contrast to the nominal radius (rn) from a user-defined bleaching spot radius.
Following the computation in Axelrod et al. (10,17), for a solution of the diffusion equation with unknown diffusion coefficient D and an initial condition given by Eq. 2, a diffusion FRAP equation for confocal FRAP can be found as
| (3) |
Where and γ = rn/re. Mf is defined as
for prebleach fluorescence intensity (Fi), postbleach initial fluorescence intensity(F0) and postbleach steady state fluorescence intensity (F∞). K can be computed from Eq. 3 at t = 0 (i.e.F(0) = F0),
| (4) |
If we let , i.e. the fluorescence intensity at the half time of recovery then, by definition, F(τ1/2) = F1/2. Since F1/2 can be related to Mf by
by setting F(τ1/2) = F1/2:
where we applied the definition of K in Eq 4. By multiplying 2Fi to both the denominator and numerator in the right hand side, we obtain
Finally, by solving for D in , after applying γ = rn/re, we have
| (5) |
When re = rn in Eq. 4 (i.e., bleaching is instantaneous), we obtain which is essentially identical to the Soumpasis equation (Eq. 1). The approximately 3% difference in the proportionality constants in Eq. 1 and Eq. 5 is due to the different assumptions on either Gaussian or uniform laser profiles.
Results and discussion
To validate our new FRAP equations (Eqs. 3, 5), we used them to extract D values from confocal FRAP data obtained using an LSCM under standard imaging conditions for a series of proteins and lipid probes localized either to the plasma membrane, or within the cytosol (Fig. 1). The proteins and lipid probes studied included Alexa488 conjugated cholera toxin B-subunit (Alexa-CTxB), a glycolipid binding bacterial toxin commonly studied as a marker of lipid rafts; a model GPI-anchored protein (YFP-GL-GPI); a fluorescent lipid analog (DiIC16); and soluble EGFP in the cytosol (EGFP). Additionally, the protein Flotillin-1, which is diffusely distributed across the cell surface when overexpressed (19) was examined. Previously reported diffusion coefficients for these molecules span over three orders of magnitude (Table 3), covering a range of D values that are physiologically relevant for most studies of protein and lipid dynamics in cells.
Figure 1. Representative images of the subcellular distribution of the proteins and lipids studied.
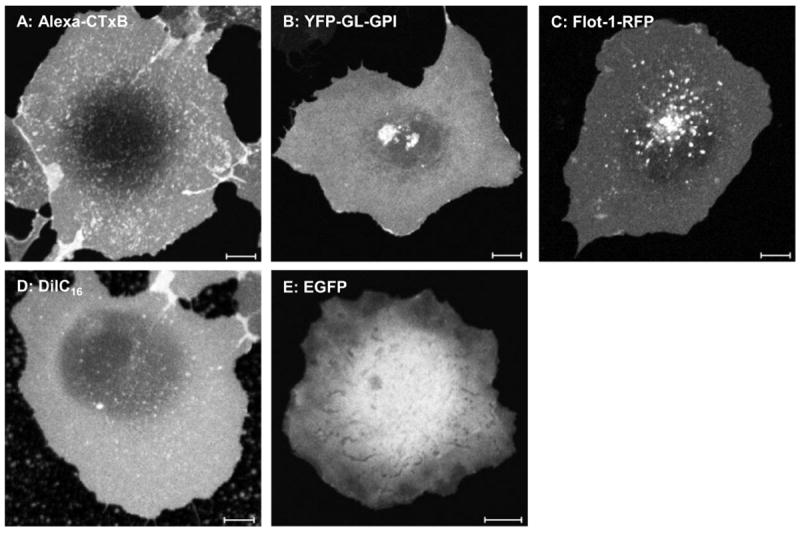
COS7 cells transfected or exogenously labeled with Alexa conjugated cholera toxin B-subunit (Alexa-CTxB), the model GPI-anchored protein (YFP-GL-GPI), Flotillin-1-RFP (Flot-1-RFP), a fluorescent lipid analog (DiIC16), or EGFP. Scale bar = 10 μm.
Table 3.
Diffusion coefficients in cells previously reported in the literature for the molecules examined in the current study by confocal FRAP
We performed confocal FRAP analysis of each of these molecules using a circular bleach region with a nominal bleach radius (rn) of 1.1 μm by repetitively scanning the bleach region. We verified that when diffusion was inhibited by fixing the sample prior to bleaching that the radius of the bleach region was similar to the user-defined nominal bleach radius (Fig. 2). Next, we calculated the effective bleach radius (re) and bleach depth (K) from the postbleach fluorescence profile (Fig. 3a,b). For slowly diffusing molecules such as CTxB, the effective bleach radius calculated from the postbleach profile was only slightly larger than the nominal bleach radius (Fig. 2). However, for the majority of molecules examined exchange of fluorescently tagged proteins outside of bleach region was evident. This effect was especially pronounced for the fastest diffusion molecules (Fig. 3a,b). Importantly, the postbleach profiles were well described by an exponential function for all of the molecules examined. This indicates that this approximation provides a reasonable description of the data even for molecules with widely ranging diffusional mobilities and thus that our FRAP equations are valid under these conditions.
Figure 2. Diffusion of Alexa-CTxB on live vs. fixed cells.
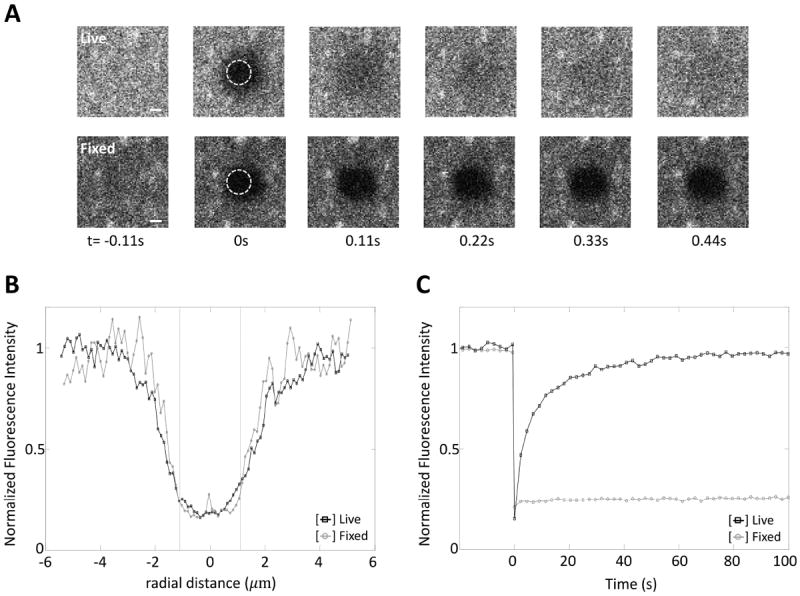
(A) Representative images of Alexa-CTxB on the plasma membrane during a FRAP experiment on either live or fixed cells for rn=1.1μm. Scale bar = 1 μm. (B) Postbleach profiles of Alexa-CTxB on the plasma membranes of live (□, n=12) and fixed (○, n=5) cells. (C) FRAP data of Alexa-CTxB on the plasma membranes of live (□, n=12) and fixed (○, n=5) cells. Since the bleach ROI is slightly off center in our system as seen in the images of (A) at t=0, a correction was made to align the center of the postbleach profile to determine re.
Figure 3. Confocal FRAP data of Alexa-CTxB, YFP-GL-GPI, Flot-1-RFP, DiIC16, and EGFP.
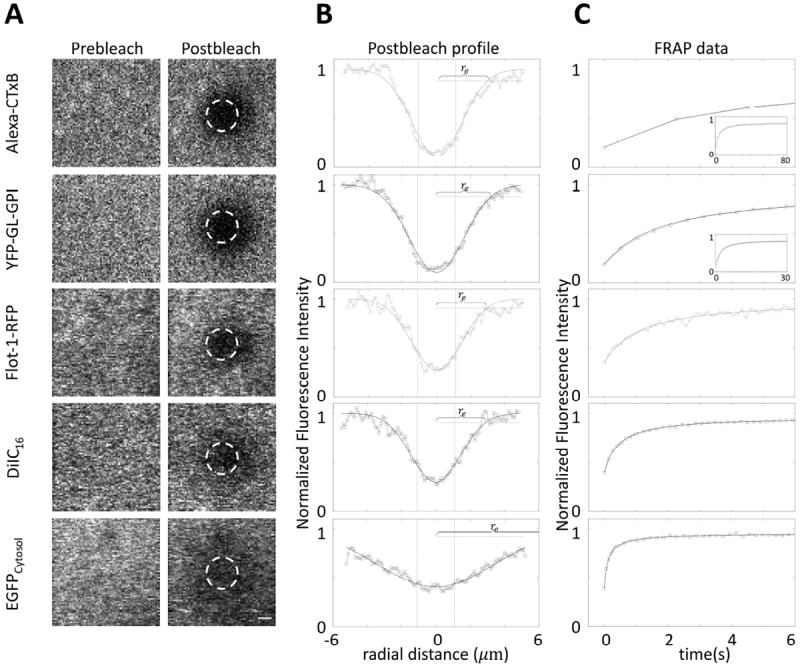
(A) Representative pre- and post-bleach images. Dashed circles represent user-defined bleaching spots with diameter 2.2 μm (i.e. rn= 1.1 μm). To speed data acquisition for the FRAP analysis, we cropped the imaging window to make it slightly larger than the bleaching spot size. Scale bar, 1 μm. (B) Averaged postbleach profiles for N=12~14 cells along the diagonals of postbleach images (□) and the best fitting Gaussian (Eq. 2, —). Dotted vertical lines show x=±rn (user defined bleach ROI). Dash-dot horizontal lines are 14% of bleaching depth from the top. (C) Representative FRAP recovery curves (□) and the best fit to Eq. 3 (—). Insets show full FRAP curves for Alexa-CTxB and YFP-GL-GPI. In (B) and (C), y- axes are normalized fluorescence intensities, i.e. Fi = 1. Since the bleach ROI is slightly off center in our system, a correction was made to determine re as described in the Materials and Methods.
We next fit the recovery curves to obtain τ1/2 (Fig. 3c, Table 4), and computed D either by fitting the FRAP data with Eq. 3 (Fig. 3c) or by plugging these numbers directly into the simplified DConfocal equation (Eq. 5). As illustrated in Figure 4, essentially identical D values were obtained using Eq. 3 (Dfitting) and Eq. 5 (DConfocal) (Student’s t-test, p>0.05) demonstrating that the two approaches yield similar results as expected. In addition, the D values obtained in this way were in good agreement with the range of values previous reported from studies using more elaborate approaches to quantify D (Table 3). Moreover, the estimated D’s obtained using DConfocal and Dfitting are independent of experimental conditions as assessed by comparing measurements of EGFP diffusion in the cytoplasm using different bleaching spot size and photobleaching times (Fig. 5 and Table 5, Student’s t-test, p>0.05).
Table 4.
FRAP parameters and measured D’s of various markers.
| Markers | Alexa-CTxB (n=5, N=56) |
YFP-GL-GPI (n=3, N=36) |
RFP-Flot (n=3,N=33) |
DiIC16 (n=4, N=58) |
EGFP (n=3, N=37) |
|---|---|---|---|---|---|
| Parameter | |||||
| re (μm) | 2.7±0.2 | 3.2±0.1 | 2.3±0.1 | 3.0±0.1 | 7.1±0.2 |
| τ1/2 (s) | 4.1±0.7 | 1.8±0.1 | 0.97±0.05 | 0.43±0.28 | 0.16±0.03 |
| K | 0.95±0.04 | 0.97±0.02 | 0.79±0.06 | 0.64±0.06 | 0.61±0.09 |
| Mf | 0.98±0.04 | 0.99±0.02 | 0.95±0.04 | 0.93±0.02 | 0.95±0.04 |
| Dfitting (μm2/s) | 0.26±0.03 | 0.73±0.04 | 0.72±0.08 | 2.7±0.7 | 41.3±8.2 |
re (μm): effective radius measured from the averaged postbleach profiles. τ1/2 (s): Halftime-of-recovery of a FRAP curve. K: Photobleaching depth parameter. Mf: Mobile fraction estimated by data fitting. Dfitting: Diffusion coefficient determined by data fitting, n: number of independent experiments, N: total number of cells. Mean ± SD over n.
Figure 4. Comparison of diffusion coefficients determined by various schemes.
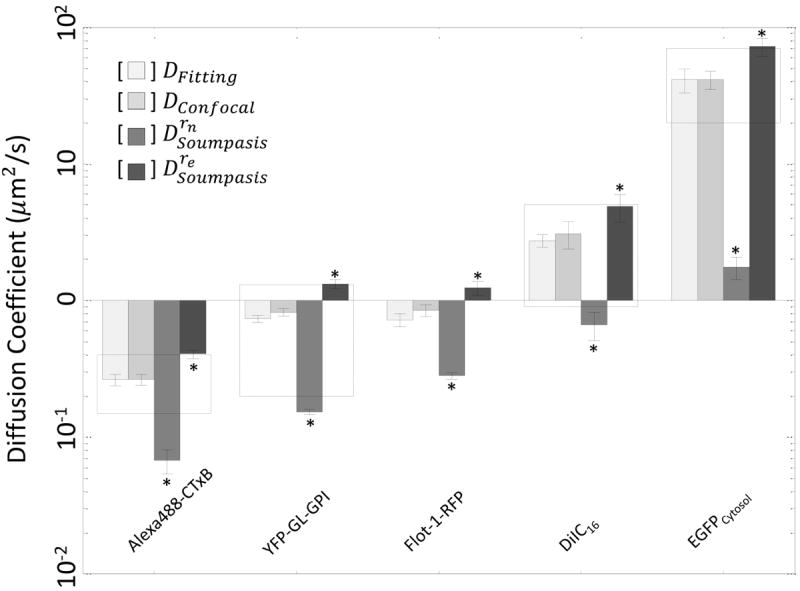
Comparison of diffusion coefficients determined by FRAP data fitting (DFitting, Eq. 3), versus the DConfocal equation (Eq. 5), or the Soumpasis equation using either rn (Drn) or re (Dre) in log scale. D’s were found from averaged FRAP curves (N≥12 cells per experiment) for three or more separate experiments (n≥3). Error bars represent standard errors. Dashed boxes show D’s reported in the literature (Table 3). *, p<0.05 compared to DFitting, Student’s t-test.
Figure 5. Comparison of diffusion coefficients for EGFP in the cytosol obtained by confocal FRAP under different experimental conditions as calculated using the Soumpasis equation or DConfocal equation.
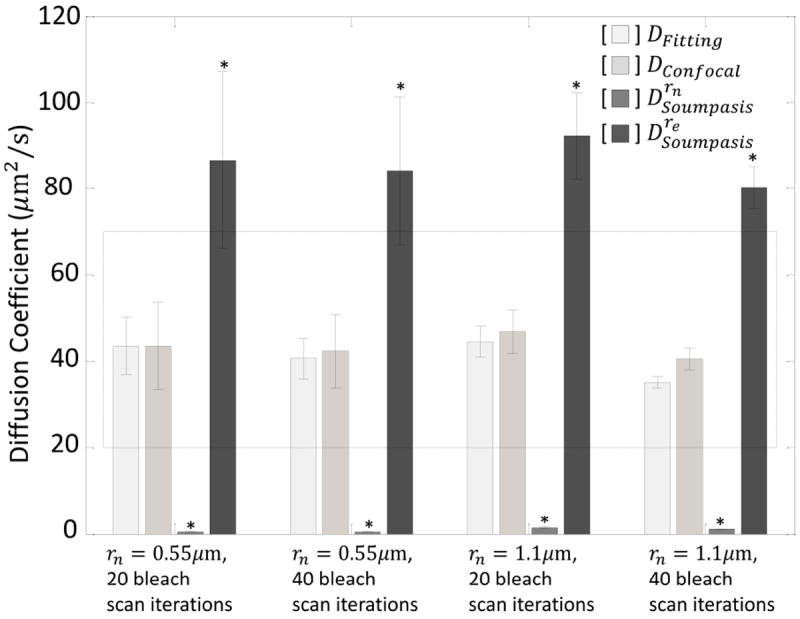
Diffusion coefficients were determined by FRAP data fitting (DFitting, Eq. 3), by the DConfocal equation (Eq. 5), by the Soumpasis equation using rn (Drn), and by the Soumpasis equation using re (Dre) for individual confocal FRAP curves (N≥6). Dashed box indicates the range of EGFP’s diffusion coefficients in the cytosol reported in the literature. re was measured from an averaged postbleach profile (n=1,N=10 cells) and D’s were obtained from individual FRAP data (n=1, N=8,10,10, and 8 cells). *, p<0.05 compared to DFitting, Student’s t-test.
Table 5.
Comparison of FRAP parameters for EGFP in the cytosol obtained by confocal FRAP under different experimental conditions
| Experimental Setup | 1.1 μm diameter ROI | 2.2 μm diameter ROI | ||
|---|---|---|---|---|
| Parameters | 20 iterations (N=8) |
40 iterations (N=10) |
20 iterations (N=10) |
40 iterations (N=8) |
| rn (μm) | 0.55 | 0.55 | 1.1 | 1.1 |
| re (μm) | 7.2 | 7.3 | 8.6 | 9.3 |
| τ1/2 (s) | 0.28±0.10 | 0.30±0.12 | 0.22±0.02 | 0.28±0.02 |
| K | 0.29±0.01 | 0.32±0.02 | 0.38±0.01 | 0.41±0.01 |
| Mf | 0.89±0.04 | 0.89±0.03 | 0.84±0.01 | 0.87±0.02 |
rn (μm): user defined nominal radius. re (μm): effective radius measured from the averaged postbleach profiles. τ1/2 (s): Halftime-of-recovery of an individual FRAP curve. K: Photobleaching depth parameter. Mf: Mobile fraction calculated by Eq. 3. Iterations: the number of cycles in photobleaching laser scans. FRAP curves (n=1) taken from (17) were re-analyzed without correction for background and photofading. Mf’s found in Table 4 were estimated from data fitting and are therefore larger than Mf’s in Table 5. N: total number of cells. Mean ± SE over N.
To illustrate the importance of using the corrected values of bleach radius to calculate D values, the same FRAP data were analyzed by Soumpasis equation (Eq. 1) using either rn (Drn) or re (Dre). This approach yielded D’s that were significantly different from those measured using Eq. 3 or Eq. 5 (Fig. 4, Student’s t-test, p<0.05). The relation between DConfocal and Drn can be easily seen from 0 < γ ≤ 1 and
| (6) |
which indicates that
| (7) |
Therefore, Drn and Dre can be understood as the upper and lower bounds for D. Importantly, Eqs. 6-7 provide the analytic relations between the equations that relate D and τ1/2 for conventional and confocal FRAP in terms of γ, the ratio rn/re. This relationship also indicates that τ1/2 will not scale proportionally with D under conditions where re > rn, providing further evidence that τ1/2 is not a good approximator of diffusional mobility. For example, depending on the exact imaging conditions used, molecules with similar values of D can demonstrate different values of τ1/2 (compare results for YFP-GL-GPI and RFP-Flot in Table 4).
The non-fitting method using Eq. 5 heavily hinges on obtaining accurate measurements of re and τ1/2, which can be achieved by analysis of large amount of FRAP data (n>3, and N>10). Here, the statistics of individual re, τ1/2, D, and Mf were carefully compared with the values from the averaged FRAP curve for a set of experiments. We found that there was no significant difference between the values found from averaged FRAP data and mean values of the parameters obtained over multiple individual FRAP data as long as the FRAP dataset size is large (Figure 6, Student’s t-test, p>0.05). Working with averaged FRAP data is more advantageous than using the mean value arising from individual measurements from a computational cost perspective, especially for noisy data from a small ROI size (rn) or fast diffusion. We also carefully compared D and Mf obtained from individual FRAP data with the values from the averaged FRAP curve for a set of experiments to check if the normalization of the FRAP curves incidently masks biological variability, such as expression level dependent diffusion coefficients (20). However, for the proteins and lipids examined here, D and Mf obtained from individual FRAP data did not show any significant differences from those obtained from the averaged FRAP curves (Figure 6, Student’s t-test, p>0.05).
Figure 6. Comparison of values obtained from averaged FRAP data vs. mean of values from individual FRAP data.
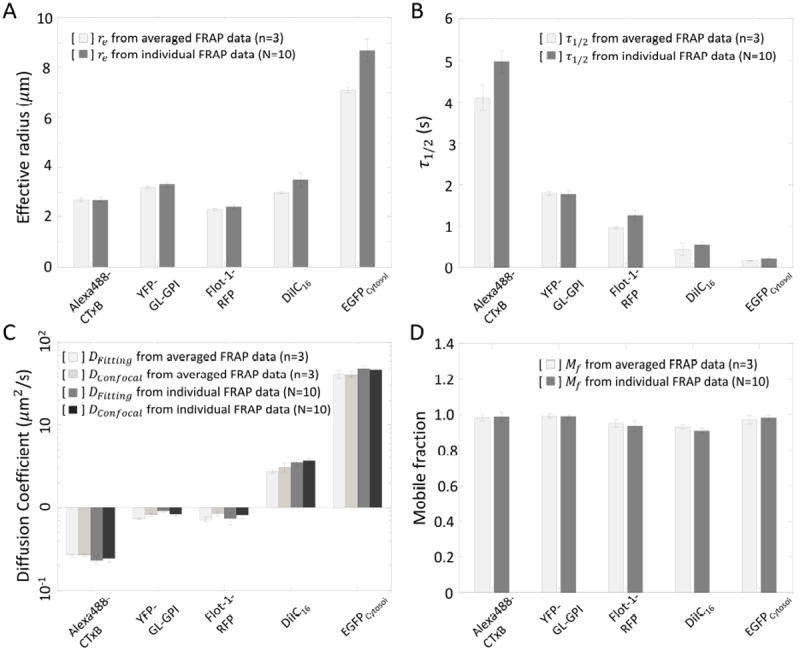
Comparison of Mean±SE of (A) re, (B) τ1/2, (C) D, and (D) Mf determined using averaged FRAP data from more than three independent experiements with 10 cells (n≥3, N=10) or means from 10 individual FRAP data in a single experiment (N=10). Error bars represent standard errors. p>0.05 Cross comparison, Student’s t-test.
In a previous study we found that re varies with both rn and bleaching time, and is more sensitive to rn than the bleaching time. For larger rn and longer bleaching time, a larger re is obtained (13, 17). In addition, the sensitivity of re to experimental definition of rn and bleaching time is higher for rapidly diffusing proteins (13,17). Nevertheless, variations of re and τ1/2 are constrained by the relation given by Eq. 5 for large range of K as long as the post bleach profiles can be described by a Gaussian (Eq. 2).
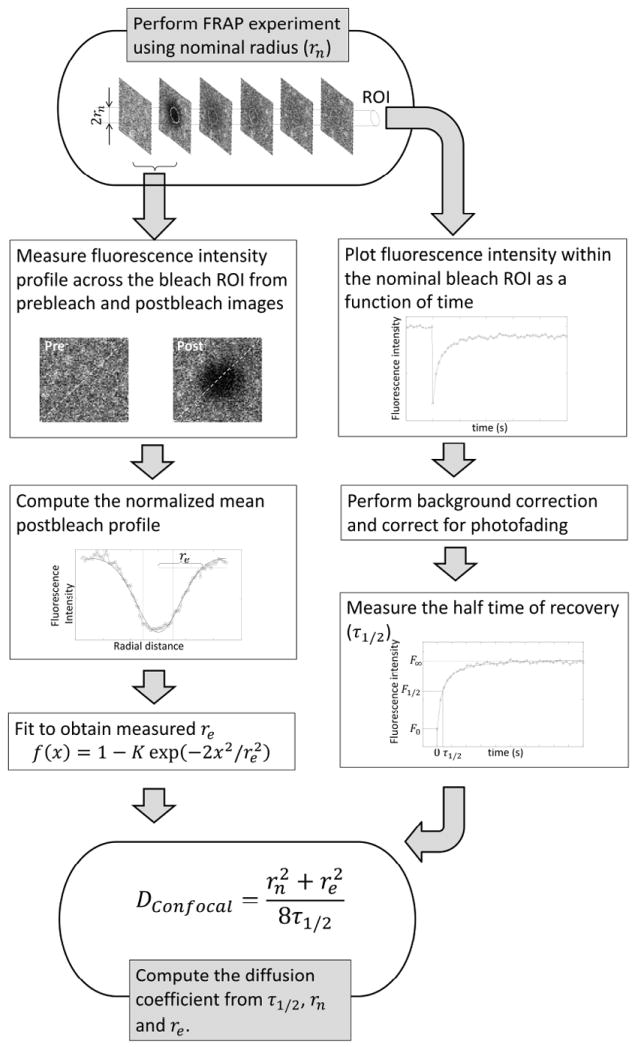
In summary, we derived and tested a new, simplified equation for the quantitative analysis of confocal FRAP data based on the pure isotropic diffusion model. An overall workflow to determine the apparent diffusion coefficient, D from τ1/2, rn and re is summarized in Figure 7. Taking advantage of widely available technology, this new approach should be accessible to many biologists, since most researchers already have access to CLSMs. Importantly, this method does not require generation of new constructs or the use of specialized fluorescent dyes or labels, and can be applied to both soluble and plasma membrane-associated molecules as illustrated here. Finally, it should be possible to readily incorporate this method into automated approaches to perform and analyze FRAP (18), and thus provide quantitative values for use in systems biology.
Figure 7. Workflow of applying the DConfocal equation.
Materials and Methods
Cell labeling and reagents
COS-7 cells were acquired from ATCC (Manassas, VA). Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum at 37°C and 5% CO2. Cells were plated on coverslips two days prior to experiments. Cells were transfected one day prior to imaging with the model GPI-anchored protein (YFP-GL-GPI) (5), Flotillin-1-RFP (Flot-1-RFP, the gift of Dr. Ai Yamamoto, Columbia University) (21), or EGFP (Clontech, Mountain View, CA) using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). DiIC16 (1,1’-dihexadecyl -3,3,3’,3’-tetramethylindocarbocyanine perchlorate) and Alexa488 fluorophore conjugated cholera toxin B subunit from Vibrio cholerae were obtained from Invitrogen (Carlsbad, CA). For exogenous labeling the cells were rinsed twice with media and then incubated for 5 minutes at room temperature with 100 nM Alexa488-CTxB or 5 μg/ml DiIC16. Cells were then rinsed twice with media and imaged. Cells were maintained in DMEM supplemented with 1 mg/ml Bovine Serum Albumin and 25 mM HEPES buffer during imaging. For FRAP on fixed samples, cell were labeled live with Alexa488-CTxB (as above). Immediately after labeling, cells were fixed with 3.4% paraformaldehyde for 15 minutes at room temperature. Cells were then rinsed with 1x PBS and mounted in Fluoromount G supplemented with 25 mg/ml DABCO (1,4 diazabicyclo[2.2.2]octane) (Sigma-Aldrich) and allowed to solidify overnight prior to imaging.
FRAP methods
FRAP experiments were carried out on a Zeiss LSM510 confocal microscope (Carl Zeiss MicroImaging, Jena, Germany) using filter sets provided by the manufacturer. Imaging was performed using a 40 × 1.3 NA Zeiss Plan-Neofluar objective at 4 × digital zoom. The confocal pinhole was set between 1.78 and 1.81 Airy units. The imaging window was further narrowed to a square observation ROI of 70 × 70 pixel (7.7 μm × 7.7 μm). The 70 × 70 window contained a circular bleach ROI 20 pixels (2.2 μm) in diameter. FRAP conditions were optimized for each molecule under study. Samples were imaged and bleached using a 30 mW Argon laser (488 and 514 laser lines) or 1.0 mW HeNe laser (543 nm laser line). Laser powers for prebleach and postbleach imaging were maintained between 6.8 and 17.1 nW (488 or 514 nm) or between 13.8 and 21.0 nW (543 nm). Bleaching of Alexa 488-CTxB was performed using the 488 nm line at 0.99 μW. EGFP and Flot-1-RFP were bleached using the 488 nm line at 2.28 μW, and YFP-GL-GPI was bleached using the 514 nm line at 1.38 μW. To bleach DiIC16 we combined the 488 nm line (40 nW), 514 line (360 nW), and 543. Bleaching regions were scanned 10 (Alexa-CTxB, YFP-GL-GPI) or 20 times (EGFP, Flot-1-RFP or DiIC16). Prebleach and postbleach images were collected with either no line averaging or with line averaging of 2. Time series images were collect using 2 s delay (Alexa-CTxB), 0.25 s delay (YFP-GL-GPI), or no delay (Flot-1-RFP, DiIC16, EGFP). This resulted in bleach times of 1.12 s (Alexa-CTxB), 0.69 s (YFP-GL-GPI), 0.56 s (Flot-1-RFP), 0.49 s (DiIC16), and 0.27 s (EGFP). During recovery, images were collected every 2.27 s (Alexa-CTxB), 0.52 s (YFP-GL-GPI), 0.14 s (Flot-1-RFP), 0.068 s (DiIC16), or 0.035 s (EGFP). All FRAP was performed at 37 °C using a stage heater.
Correction for background and photofading
To correct for background (Fbk), fluorescence from a cell-free area of the coverslip was measured under the same conditions as FRAP was performed. To correct for observational photofading (Ffading (t)), a time series was collected as for the FRAP studies, except no photobleaching step was performed. The raw FRAP data, Fraw (t) were corrected for background and photofading by
Finally, the corrected data were normalized by the prebleach intensity, as
τ1/2 measurements
To measure τ1/2 from the FRAP data a linear interpolation method was used. For the FRAP data {F(0), F(t1), F(t2),…, F(tn)} such that F(0) = F0 and F(tn) = F∞, the fluorescence intensity at half of recovery is defined as F1/2 = (F0 + F∞)/2. If F(tk) = F1/2 for some tk then we chose τ1/2 = tk. If F(tk) < F1/2 < F(tk+1) then we chose
re measurements
In order to obtain normalized initial postbleach profiles, fluorescence intensities were measured along a diagonal of the square observation ROIs of 70 × 70 pixel (7.7 μm × 7.7 μm) from the prebleach image obtained immediately prior to and after photobleaching. Between the profiles obtained from two diagonals, the dataset with better symmetry was chosen for analysis. The profiles were then normalized by dividing the postbleach profile by the prebleach profile. The normalized postbleach profiles were averaged over data sets (N=12~14). Since the bleach ROI was slightly off center in some cases, if necessary, the symmetry axis was found and set to be x = 0 from the normalized mean postbleach profile. The mean profiles were then fitted to
for K and re using a nonlinear least-squares fitting routine (nlinfit.m) available in MATLAB® (version 7.10, R2010a, The Mathworks, Inc.).
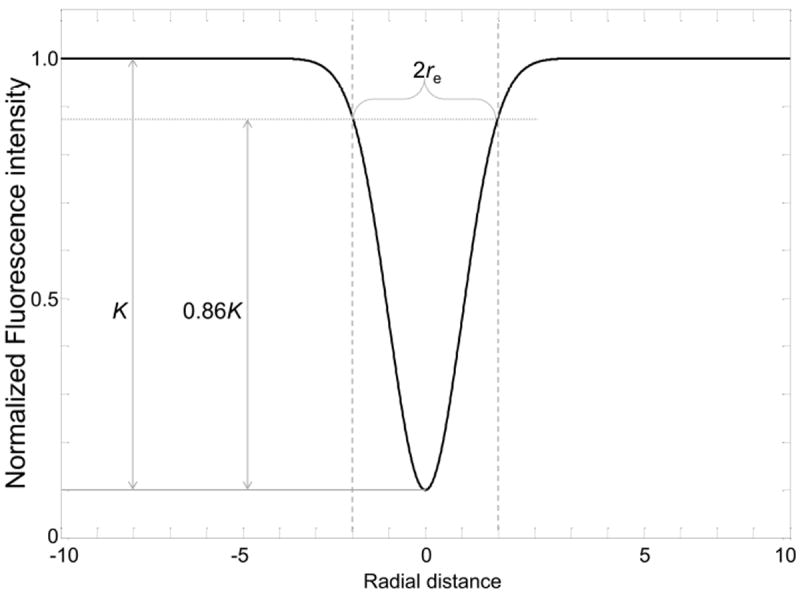
Alternatively, re can be determined by direct measurement following three steps. First, K can be determined from the bleaching depth in the nomalized postbleach profile as referred as in Figure 8. Then, the half width of cross-sections between the horizontal line at the height of 0.86K from the bottom of the postbleach profile (Fig. 8) and the postbleach profiles yields re without involving any fitting (Fig. 8).
Figure 8. Determination of re from a postbleach profile.
FRAP data fitting for D and Mf and weighted residual
The FRAP curves were normalized by the mean prebleach fluorescence intensity (i.e. Fi = 1) and the mean of multiple normalized FRAP data sets was used for data fitting. Data fitting was carried out for D and Mf by a nonlinear least-squares fitting routine (nlinfit.m) available in MATLAB® (version 7.10, R2010a, The Mathworks, Inc.) minimizing a weighted residual between averaged FRAP data from 10 experiments (FData (t)) and a theoretical FRAP curve (F(t), Eq. 3), where the weighted residual is defined by
If F(0) computed using K from postbleach profile fitting showed any discrepancy from FData (0), K was recalibrated using K = (1 + γ2)(1 − FData (0)) before data fitting.
Acknowledgments
Supported by R01 GM073846, NSF/DMS 0970008, and a Vanderbilt Discovery grant. The funding sources had no role in the study design, collection, analysis or interpretation of data, writing the report, or the decision to submit the paper for publication.
References
- 1.Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol. 2010;22:403–411. doi: 10.1016/j.ceb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day CA, Kenworthy AK. Tracking microdomain dynamics in cell membranes. Biochim Biophys Acta. 2009;1788:245–253. doi: 10.1016/j.bbamem.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drake KR, Kang M, Kenworthy AK. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS One. 2010;5:e9806. doi: 10.1371/journal.pone.0009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyawaki A. Proteins on the move: insights gained from fluorescent protein technologies. Nat Rev Mol Cell Biol. 2011;12:656–668. doi: 10.1038/nrm3199. [DOI] [PubMed] [Google Scholar]
- 5.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J Cell Biol. 1984;99:2165–74. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A. 2002;99:3171–5. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lajoie P, Partridge EA, Guay G, Goetz JG, Pawling J, Lagana A, Joshi B, Dennis JW, Nabi IR. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J Cell Biol. 2007;179(2):341–56. doi: 10.1083/jcb.200611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–69. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soumpasis DM. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys J. 1983;41:95–97. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxton MJ. Modeling 2D and 3D diffusion. Methods Mol Biol. 2007;400:295–321. doi: 10.1007/978-1-59745-519-0_20. [DOI] [PubMed] [Google Scholar]
- 13.Kang M, Day CA, DiBenedetto E, Kenworthy AK. A quantitative approach to analyze binding diffusion kinetics by confocal FRAP. Biophys J. 2010;99:2737–47. doi: 10.1016/j.bpj.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss M. Challenges and artifacts in quantitative photobleaching experiments. Traffic. 2004;5:662–71. doi: 10.1111/j.1600-0854.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 15.Braga J, Desterro JM, Carmo-Fonseca M. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol Biol Cell. 2004;15:4749–60. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pucadyil TJ, Chattopadhyay A. Confocal fluorescence recovery after photobleaching of green fluorescent protein in solution. J Fluoresc. 2006;16:87–94. doi: 10.1007/s10895-005-0019-y. [DOI] [PubMed] [Google Scholar]
- 17.Kang M, Day CA, Drake K, Kenworthy AK, DiBenedetto E. A generalization of theory for two-dimensional fluorescence recovery after photobleaching applicable to confocal laser scanning microscopes. Biophys J. 2009;97:1501–11. doi: 10.1016/j.bpj.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad C, Wunsche A, Tan TH, Bulkescher J, Sieckmann F, Verissimo F, Edelstein A, Walter T, Liebel U, Pepperkok R, Ellenberg J. Micropilot: automation of fluorescence microscopy-based imaging for systems biology. Nat Methods. 2011;8:246–249. doi: 10.1038/nmeth.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto GP, Nichols BJ. The roles of flotillin microdomains--endocytosis and beyond. J Cell Sci. 2011;124:3933–40. doi: 10.1242/jcs.092015. [DOI] [PubMed] [Google Scholar]
- 20.Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. Cell Biol. 2002;157(5):865–72. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelides KJ, Elmer LW, Loftus D, Elson E. Distribution and lateral mobility of voltage-dependent sodium channels in neurons. J Cell Biol. 1988;106:1911–1925. doi: 10.1083/jcb.106.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deschout H, Hagman J, Fransson S, Jonasson J, Rudemo M, Lorén N, Braeckmans K. Straightforward FRAP for quantitative diffusion measurements with a laser scanning microscope. Opt Express. 2010;18:22886–905. doi: 10.1364/OE.18.022886. [DOI] [PubMed] [Google Scholar]
- 23.Seiffert S, Oppermann W. Systematic evaluation of FRAP experiments performed in a confocal laser scanning microscope. J Microsc. 2005;220:20–30. doi: 10.1111/j.1365-2818.2005.01512.x. [DOI] [PubMed] [Google Scholar]
- 24.Smisdom N, Braeckmans K, Deschout H, vandeVen M, Rigo JM, De Smedt SC, Ameloot M. Fluorescence recovery after photobleaching on the confocal laser-scanning microscope: generalized model without restriction on the size of the photobleached disk. J Biomed Opt. 2011;16:046021. doi: 10.1117/1.3569620. [DOI] [PubMed] [Google Scholar]
- 25.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–95. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waharte F, Steenkeste K, Briandet R, Fontaine-Aupart MP. Diffusion measurements inside biofilms by image-based fluorescence recovery after photobleaching (FRAP) analysis with a commercial confocal laser scanning microscope. Appl Environ Microbiol. 2010;76:5860–9. doi: 10.1128/AEM.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacia K, Scherfeld D, Kahya N, Schwille P. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys J. 2004;87:1034–1043. doi: 10.1529/biophysj.104.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day CA, Kenworthy AK. Mechanisms underlying the confined diffusion of cholera toxin B-subunit in intact cell membranes. PLoS One. 2012;7:e34923. doi: 10.1371/journal.pone.0034923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pautot S, Lee H, Isacoff EY, Groves JT. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nat Chem Biol. 2005;1:283–289. doi: 10.1038/nchembio737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson K, Hou Y, Derzko Z, Wojcieszyn J, Organisciak D. Lipid lateral diffusion in the surface membrane of cells and in multibilayers formed from plasma membrane lipids. Biochemistry. 1981;20:5268–5275. doi: 10.1021/bi00521a027. [DOI] [PubMed] [Google Scholar]
- 31.James PS, Hennessy C, Berge T, Jones R. Compartmentalisation of the sperm plasma membrane: a FRAP, FLIP and SPFI analysis of putative diffusion barriers on the sperm head. J Cell Sci. 2004;117:6485–95. doi: 10.1242/jcs.01578. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, SWang Z, Shah JV, Sun CH, Berns MW. Fluorescence correlation spectroscopy investigation of a GFP mutant-enhanced cyan fluorescent protein and its tubulin fusion in living cells with two-photon excitation. J Biomed Opt. 2004;9:395–403. doi: 10.1117/1.1646416. [DOI] [PubMed] [Google Scholar]
- 33.Dragestein KA, van Cappellen WA, van Haren J, Tsibidis GD, Akhmanova A, Knoch TA, Grosveld F, Galjart N. Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J Cell Biol. 2008;180:729–737. doi: 10.1083/jcb.200707203. [DOI] [PMC free article] [PubMed] [Google Scholar]


