Abstract
Background
A T cell vaccine that lowers levels of HIV replication could significantly diminish the burden of the AIDS epidemic by attenuating disease progression and reducing the risk of HIV transmission. In order to learn which immune responses an effective HIV vaccine should elicit, we must first identify correlates of immune protection in vivo.
Objective
“Elite controllers” are rare HIV-infected individuals who are able to spontaneously control HIV replication without medication, maintaining viral loads that are consistently below the limits of detection by currently available commercial assays. The objective of this review is to examine studies of elite controllers that may help to elucidate mechanisms of HIV immune control that will be useful in designing a vaccine.
Methods
This review examines recent literature on HIV controllers as well as studies that have evaluated aspects of viral and host immunology that correlate with viral control.
Results/Conclusions
Although many elements of both innate and adaptive immunity are associated with control of HIV infection, the specific mechanism(s) by which HIV elite controllers achieve control remain undefined. Ongoing studies of elite controllers, including those examining host genetic polymorphisms, should facilitate the definition of an effective HIV-specific immune response and guide HIV vaccine design.
Keywords: Acquired Immune Deficiency Syndrome (AIDS), CD8+ T cell, CD4+ T cell, Human leucocyte antigen (HLA), Cell mediated immunity, Elite controller, Human Immunodeficiency Virus (HIV), Review, Simian Immunodeficiency Virus (SIV), Vaccine, Viral load
Introduction
Every day more than 6,800 people become infected with HIV and more than 5,700 people die of AIDS. There are currently an estimated 33.2 million people living with HIV worldwide [1]. The number of individuals receiving antiretroviral (ARV) therapy in both the developed and the developing world has increased dramatically during the last few years due in part to financial commitments by organizations such as the Global Fund to fight AIDS, TB, and Malaria (GFATM) and the President's Emergency Plan for AIDS Relief (PEPFAR) [2]. However, the world has failed to meet the goals set forth in the WHO's ‘3 by 5’ plan [3] and as of the end of 2007 it was estimated that only 31% of persons in low- and middle-income countries who were in need of treatment were receiving it [2]. The low rates of treatment in resource-limited settings mean that most HIV+ individuals in these areas have not benefited from Highly-Active Antiretroviral Therapy (HAART), treatment that has led to significant survival gains in the US and other developed countries [2, 4, 5]. As a result, the vast majority of global AIDS-related deaths during 2007 occurred among people in the developing world without access to ARVs, with over three-quarters of these in Sub-Saharan Africa [1].
A prophylactic HIV vaccine that conferred sterilizing immunity, fully preventing or clearing the infection, could bring an end to the HIV epidemic, and by preventing new infections it would limit future treatment needs. Although the generation of a sterilizing vaccine remains the ultimate goal, two decades of research suggest that it will be difficult to create a vaccine that prevents infection [6]. Moreover, all effective vaccines currently in use have been shown to prevent disease, not infection or colonization per se, and HIV infection has not yet been shown to be cleared naturally in infected persons. It is unknown whether a vaccine can accomplish something that nature has been unable to do.
Vaccines traditionally rely on the induction of a strong neutralizing antibody response to prevent infection, an approach whose success for controlling HIV is extremely challenging due to a number of unique properties of the virus. Foremost among these is the enormous sequence diversity of the HIV envelope proteins, the propensity for N-linked glycans to occlude the most immunogenic and highly-conserved regions of the envelope protein, and steric constraints that make it difficult for antibodies to attach to the envelope proteins [7, 8]. Although broadly cross-reactive neutralizing antibodies have been isolated from some HIV+ individuals and thus serve as a proof-of-principle that such antibodies can be generated, people with broadly cross-reactive antibodies are extremely rare [7].
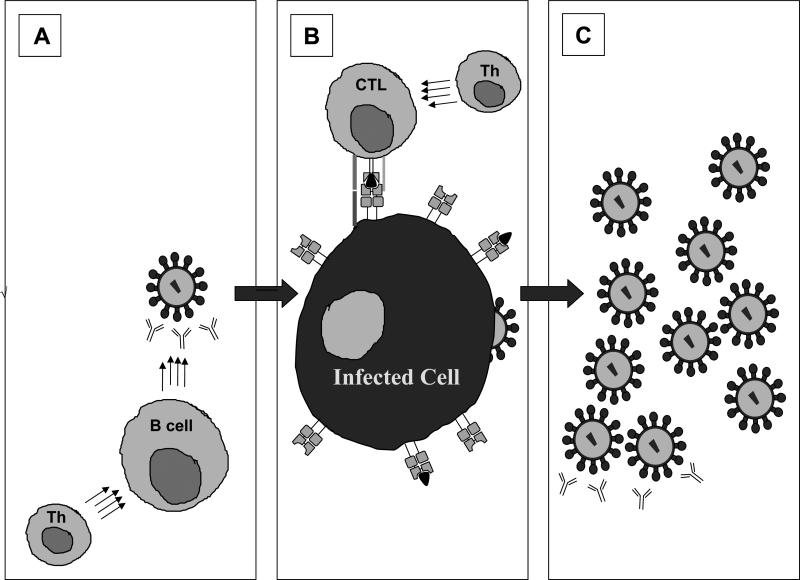
Due to the many obstacles that HIV presents to the design of a neutralizing antibody-mediated sterilizing vaccine which would have the potential to interact with free virions and provide sterilizing immunity (Figure 1A), much of the field has turned its attention towards the design of a HIV vaccine stimulating cellular immune responses that would provide durable protection from HIV disease progression and reduce the risk of transmission [6, 9]. Such a “T cell vaccine” would attenuate HIV disease progression by enabling vaccinated and subsequently infected individuals to control viral replication most likely through cytotoxic T lymphocyte (CTL) targeting of virus infected cells [9], thereby limiting progeny virus production. (FIGURE 1B).
Figure 1. Immune control of HIV infection.
A) Prior to cell entry, antibodies produced by B cells may neutralize free virus particles, preventing cells from becoming HIV infected. B) Once a cell is infected, CTL may mount effective cytolytic responses through recognition by a unique TCR of a viral peptide presented as a complex with HLA class I on the surface of the infected cell. C) If the immune response is not successful in controlling viral replication in stages A or B, progeny virions will be produced and will bud from the infected cell, creating new virus particles that can go on to infect other cells, though these may also be limited by neutralizing antibodies
This reduction of viral load would be anticipated to provide considerable benefit to the health of the individual while also reducing rates of forward-transmission and treatment demands, representing a significant public-health accomplishment [10-14].
Remarkably, there are individuals who naturally suppress HIV replication, spontaneously maintaining viral loads below the limits of detection without the assistance of ARV therapy [15, 16]. Unraveling the mechanism(s) by which these “elite controllers” limit viral replication should inform design of a vaccine that limits HIV disease progression. In this article, we review the current research on the influence of viral genetics, host genetics, and host immune responses on HIV disease progression and viral replication. We also delineate the pressing questions that remain in the field, discuss implications of a recently terminated T cell based HIV vaccine trial, and describe a new collaborative study that should help deepen our understanding of the mechanisms that enable elite controllers to suppress viral replication.
1. Viral Genetics and HIV Replication
Before sensitive HIV RNA assays became clinically available in the mid 1990's it was not possible to quantify HIV levels produced in vivo. Hence, early descriptions of individuals who did not progress to AIDS were largely based on the duration of infection and CD4 counts, not levels of viremia. “Long Term Non-Progressors” (LTNP), HIV-infected individuals with stable CD4 counts and no history of AIDS defining illnesses, were estimated to constitute 5-15% of the HIV positive population [17, 18].
With the advent of sensitive assays that quantify the number of copies of viral RNA in plasma, it became clear that the majority of LTNP have ongoing viral replication at detectable levels. In fact, some LTNP have been identified who have high levels of circulating virus [19]. However, these assays also identified a subset of LTNP, “elite controllers,” who consistently maintain undetectable viral loads, defined as <50 copies/ml, the limit of detection of standard commercial assays used clinically to monitor patients [16]. Studies of LTNP have shown that some of these individuals are infected with defective virus [20, 21]. A logical question, then, is whether elite controller status is attributable to inefficient viral replication or due to host immune control or other host-specific genetic factors.
The isolation of replication-competent virus from elite controllers suggests that control is not entirely due to deficient virus [22, 23]. Findings of discordant couples from a recent study as well as from our own cohort of elite controllers also implicate host immune responses in the control of HIV replication [24]. Several sets of discordant couples have been found in which one individual is an HIV controller while the other person experiences standard, progressive infection. Viral sequencing analysis has confirmed transmission and all viruses are replication competent. The fact that one of these individuals is viremic while the other is aviremic suggests that elite controller status is due to host immune responses or host genetics (T. Miura and BDW, unpublished) [25]. Learning precisely what host genetics and immune responses correlate with this protection is thus a topic of great importance as it can guide design of a cell-mediated vaccine.
2. Host Genetic Correlates of HIV Control
2.1. The Human Leukocyte Antigens
During viral infection, Class I Human Leukocyte Antigens (HLA) present endogenously processed viral antigens on the surface of infected cells to CD8+ T cells, which then attempt to clear the virus-infected cells by direct lysis of the infected cell and/or production of antiviral cytokines. HLA loci are the most polymorphic sites in the human genome [26], and therefore represent a good starting point for examining the impact of host genetics on HIV disease progression.
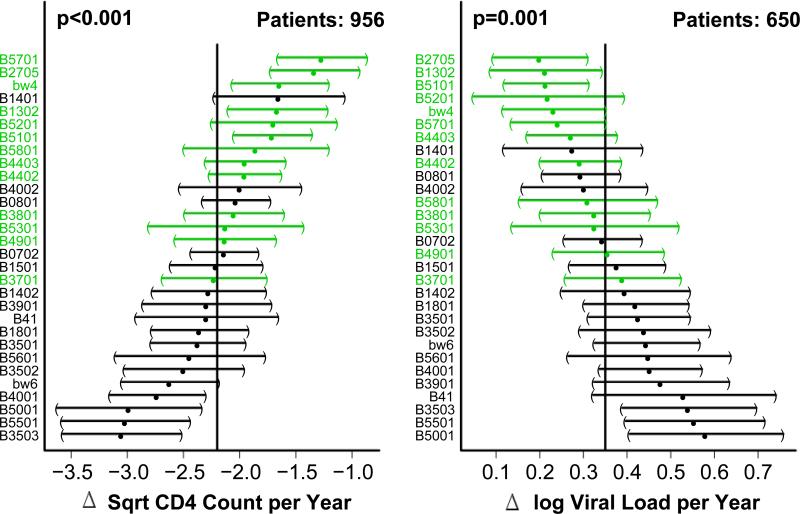
Large cohort studies have shown that certain HLA Class I alleles correlate with HIV control (FIGURE 2), providing strong evidence that the immune system plays a role in determining the rate of disease progression and implicating CTL immunity in this control. The HLA Class I allele with the strongest and most consistently recognized association with viral control is B*5701 [27-30]. HLA-B27 has also been repeatedly linked with HIV nonprogression [30, 31]. Interestingly, many individuals with this allele display an immunodominant CTL response to a single HIV-Gag p24 epitope (KK10) that is subject to viral escape. A predictable series of point mutations both within and near this epitope leads to inadequate HLA binding, loss of CTL recognition, and an increase in viral load [32]. In addition to HLA-B57 and HLA-B27, there are several other HLA-B alleles that have been associated with viral control and others that have been associated with a lack of control [28, 30, 33]. The mechanisms underlying these differences are not known.
Figure 2. Effect of HLA-B alleles on CD4 decline and viral load increase over time after HIV infection.
Average change in the square root CD4 counts (left side) and log viral load measurements (right side) per year and 95% confidence intervals were determined using a linear mixed effects regression (LMER). HLA-B alleles that are most to least protective are listed from top to bottom, respectively. The vertical line in the middle of each plot represents the sample average and confidence intervals for alleles deviating significantly from this value do not cross the line. Alleles shown in red are HLA-B Bw4+ , while those in black are HLA-B Bw6+. Data from both seroconverter and seroprevalent individuals were used in the analysis. Seroconversion date is used as the intercept in the LMER, but in seroprevalents, the first observed data point is used instead. P values were determined using the Wald test.
In addition to the identification of specific “protective” or “deleterious” alleles, several studies have shown that homozygosity at one or more HLA loci is associated with faster progression to AIDS[30, 33]. This “heterozygote advantage” model suggests that heterozygosity at each HLA Class I allele allows for presentation of a greater range of epitopes, producing a greater breadth of T cell responses and enabling more effective control of HIV replication [30, 33].
HLA-B alleles can be categorized as either Bw4 or Bw6 based on amino acid sequences at positions 77-83, which form a part of the Class I binding pocket for viral peptides. Interestingly, most of the alleles associated with HIV disease resistance are members of the HLA-Bw4 motif family, while those associated with HIV disease susceptibility are part of the Bw6 family. Given that HLA-Bw4 alleles can bind the Natural Killer (NK) cell receptor KIR3DL1 while HLA-Bw6 alleles cannot [30], it is possible that polymorphisms within NK cell receptors affect HIV disease progression. Studies to this end have demonstrated an epistatic protective effect for KIR and HLA genes, showing that the combined genotype of KIR3DS1, an activating KIR receptor with high sequence homology to KIR3DL1, and HLA-Bw4-80I, a HLA-Bw4 motif with isoleucine at position 80, is enriched in a population of slow progressors, linked to low viral load set points, and protective against AIDS-defining opportunistic infections [34, 35]. Certain KIR3DL1 alleles matched with HLA-Bw4 alleles also associate with lower mean viral loads and slower progression to AIDS [36]. The additive protective effect of certain HLA and KIR alleles implies coordination between the innate and adaptive HIV immune responses, suggesting that enhancement of innate immunity alongside stimulation of adaptive immunity could be important for HIV vaccine design. However, the precise mechanisms by which these responses mediate an effect are unknown.
2.2. HIV Co-receptors and their ligands
2.2.1. CCR5 and CCR2b
In order to attach to, fuse with, and enter a cell, HIV must bind to both the CD4 receptor and a chemokine co-receptor. M-tropic (R5) viruses utilize CCR5 as their co-receptor, while T-tropic (X4) viruses, which predominate later in the course of infection, use CXCR4. A 32-base pair deletion within the CCR5 gene (CCR5-Δ32) limits CCR5 cell-surface expression of this receptor. In persons heterozygous for this allele, this mutation correlates with lower viral load set points, and persons who are homozygous for this mutated allele experience complete protection from HIV infection, at least from viruses requiring CCR5 for entry [37-39]. A polymorphism at position −2459 within the CCR5 promoter region which leads to lower CCR5 expression on macrophages also associates with slower disease progression, lower viral propagation, and higher β-chemokine expression [40, 41].
A mutation that similarly impacts the ability of HIV to infect CD4+ cells has been identified in another chemokine co-receptor, CCR2b, which is found primarily in persons of African descent [38]. Collectively, the effects of these co-receptor polymorphisms on the ability of HIV to infect CD4 cells support blocking chemokine receptors as an effective therapeutic strategy. Indeed, CCR5 antagonists are already being used as a new class of anti-HIV drugs.
2.2.2. Chemokine Receptor Ligands
The association of certain polymorphisms in HIV-1 co-receptor genes with disease progression has prompted investigation of whether differences in expression levels of the natural ligands of the co-receptors, such as RANTES, MIP-1α, MIP-1β, and SDF-1, also impact disease progression. It is currently suspected that these ligands, such as CCL3L1, can inhibit HIV entry into cells by competing for co-receptor binding, and thus retard disease progression [38, 42-45]. The extent to which these polymorphisms or combinations of these polymorphisms may contribute to durable HIV control is unclear at this time.
2.3. Other host factors that influence HIV disease progression: APOBEC3G, TRIM5α, and IL-10
APOBEC3G (CEM15, hA3G) is a human cytidine deaminase that limits infectivity of HIV-1 in strains lacking the viral protein Vif (virion infectivity factor) [46]. A study that included 8 LTNP demonstrated an association between APOBEC3G mRNA levels and lower viral loads as well as higher CD4 counts, although other studies have not been able to replicate this association [47, 48]. While evidence has shown increased levels of APOBEC3G expression in exposed-uninfected individuals, expression levels have not been found to be increased in elite controllers [49] leaving the role of APOBEC3G in attenuating progression and/or preventing infection still under debate [50].
TRIM5α is an innate immunity factor that can block HIV-1 capsid uncoating in Old World Monkeys but not in humans [51]. Using chimeric proteins produced from human and rhesus monkey TRIM5α sequences, the unique amino acid residue within the protein's SPRY domain that confers HIV-1 resistance in rhesus monkeys has been identified [52]. Mimicking this blocking mechanism found in Old World Monkeys could prove to be an effective therapeutic in humans. However, whether host polymorphisms in these genes might contribute to differences in disease outcome remains unclear.
Interleukin-10 (IL-10) is a cytokine that induces immunosuppression through inactivation of T cells and inhibition of a Th1 response [26]. Murine studies of LCMV have shown that removal of IL-10 or inhibition of the IL-10 receptor leads to increases in T cell activity and viral clearance [53]. However, IL-10 has also been shown to reduce HIV-1 replication in macrophages, but not in dendritic cells, through a blockade in an entry or post-entry stage [54]. A dominant polymorphism in the promoter region, termed IL10 5’A, which leads to a pronounced reduction in IL-10 levels, correlates with faster progression to AIDS [29, 37]. These studies highlight the delicate balance of immunosuppression in HIV infection. While cellular activation may lead to increased susceptibility to infection and disease progression [55] it is likely that an effective T cell response is needed to durably control HIV infection.
3. Innate Immunity and HIV Control
3.1. Natural Killer Cells
Natural Killer cells control infection by detecting changes in HLA Class I expression through inhibitory and activating receptors, leading to both lytic activity and cytokine production [26]. The viral control associated with this interaction has been described above through the epistatic relationship between NK KIR inhibitory receptor subtypes and HLA alleles [34-36].
Studies comparing NK cells derived from LTNP and progressors have found that LTNP possess NK cells with a higher cytotoxic capacity, despite showing similar cell surface expression of inhibitory and activating receptors [56]. Studies have also shown greater NK killing and production of IFN-γ, TNF-α, and β-chemokines in exposed uninfected individuals compared to unexposed individuals or seroconverters [56, 57]. However, it is still not clear how much of the difference in lytic ability is due to persistent HIV viremia and immune activation that causes NK dysfunction in viremic individuals [58-60]. Since very early events following acute HIV infection appear to have a major impact on subsequent disease progression, specific features of the innate immune responses may well play a role in establishing an elite controller state.
3.2. Toll-Like Receptors
Toll-Like Receptors (TLRs) are pattern recognition molecules that recognize foreign particles and trigger the production of Type-1 interferons and other cytokines [61]. TLR7/8 and TLR9, which recognize ssRNA and CpG DNA motifs respectively, have repeatedly been found to play a role in modulating HIV-1 disease progression. TLR7/8 activates monocytes and plasmacytoid dendritic cells (pDCs) through the MyD88 signaling pathway upon recognition of specific HIV-1 RNA sequences [62]. These activated pDCs go on to produce IFN-α that contributes to an apoptotic pathway, leading to CD4+ T cell decline in lymphoid tissue [63]. HIV nonprogressors were shown to produce lower levels of IFN-α and the apoptotic pathway ligands compared to progressors, which suggests a role for the TLR7/8-MyD88 activating pathway in disease progression [63]. Certain polymorphisms found within the receptor gene for TLR9 also associate with rapid HIV disease progression, providing further evidence that innate immunity and TLRs may influence HIV disease outcome [64].
4. Acquired Immunity and HIV Control
4.1. Neutralizing Antibodies
Neutralizing antibodies are crucial for preventing infection of susceptible cells and can facilitate cell killing through ADCC [9, 26]. However, it is not clear whether neutralizing antibodies help limit HIV disease progression.
Recent evidence from the rhesus macaque model has shown that the presence of a late broadly reactive neutralizing antibody response spanning two years does not associate with lower viral loads or slowed disease progression, and that these high antibody titers could merely be a result of continual exposure to high levels of viremia [65]. Although this study seems to downplay the importance of neutralizing antibodies in control of SIV disease progression, there is also evidence that post-infection passive immunization with anti-SIV neutralizing antibodies in rhesus macaques can lead to viral load suppression through dendritic cell antigen uptake and T cell stimulation [66]. A similar study has shown that passive immunization with anti-SIV IgG can lead to a decrease in the number of SIV-infected cells, most likely through ADCC [67].
Analyses of our own cohort of elite controllers have shown that very few of them have broadly cross-reacting neutralizing antibodies, suggesting that antibodies are not the main mechanism mediating control of replication, despite other studies that have found patient sera capable of broadly neutralizing a panel of different viruses [68-70]. This observation supports a previous study that demonstrated that plasma from elite controllers lacked the ability to neutralize autologous virus [71]. In contrast, a study examining neutralizing antibody responses to autologous viruses in acute and chronically infected patients found low levels of neutralizing antibody responses during chronic infection, but not during acute stages, and greater responses among controllers than other chronically infected patients [72].
5. Cellular Immunity
5.1. Cytotoxic T-lymphocytes (CTL)
Numerous lines of evidence suggest that CTL responses are critically important for controlling HIV replication, but their exact role in elite control remains unclear. It has long been known that CTL can recognize and kill HIV-infected cells [73], and that the appearance of the CTL response coincides temporally with the reduction of viremia and resolution of symptoms of acute infection [74, 75]. Furthermore, studies in rhesus macaques, including one that specifically looked at an “elite controller” animal model with rhesus macaques that controlled a pathogenic SIV infection for 1-5 years, have shown that depletion of CD8+ T cells leads to a loss of viral control that is restored upon reappearance of SIV-specific CTL [76, 77]. The correlation between certain HLA Class I alleles and HIV disease progression [30], coupled with HIV's propensity for mutating to evade CTL responses, provides more evidence for the importance of CTL in controlling HIV disease progression [78, 79].
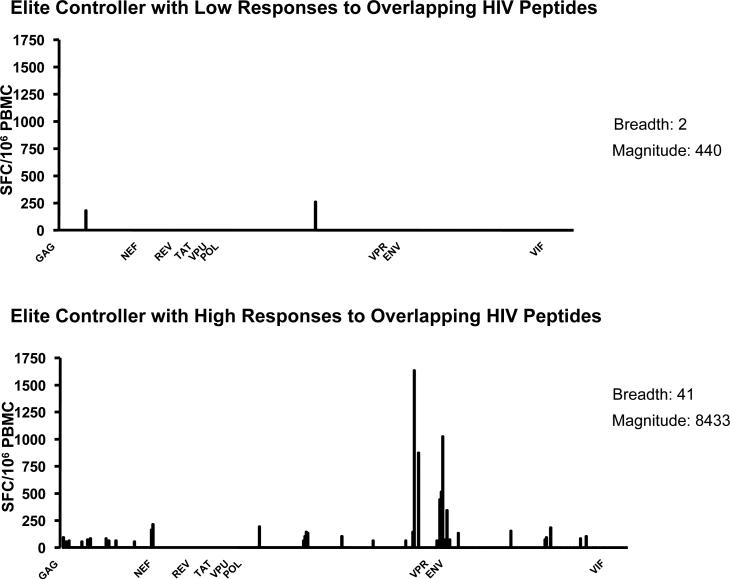
Due to CD8+ T cells’ apparent ability to impact disease progression, it was initially suspected that elite controllers would display a high breadth and magnitude of CTL responses to HIV. However, studies have failed to demonstrate a correlation between the quantity of CTL responses, as determined by IFN gamma Elispot assay, and viral load [80, 81]. In fact, the number of responses appears to be driven by viremia, with those individuals exposed to more virus mounting greater CTL responses [69]. However, even among elite controllers there is substantial heterogeneity in CD8 T cell responses, with some controllers having very few HIV-specific CTL responses, as described in a recently published study and observed within our own cohort [82] (FIGURE 3). Attention has thus shifted to the quality and specificity of CTL responses, rather than the breadth of the response alone.
Figure 3. Heterogeneity of CTL responses in elite controllers measure by IFN-γ ELISpot.
IFN- γ responses from two elite controllers measured by enzyme-linked immunosorbent spot (ELISpot) assay. While each subject was tested against. 410 peptides overlapping by 10 amino acids and spanning the whole HIV proteome, they exhibit significant differences in both the breadth and the magnitude of their responses.
5.2. Associations between CD8+ T cell phenotype and disease outcome
Recently, several groups have demonstrated an inverse correlation between the quality of CTL responses and the rate of disease progression [83-85]. T cell quality, in this case, is defined by either the presence of multi- or polyfunctional responses, or the absence of negative immunoregulatory molecules, such as Programmed Death-1 (PD-1 or CD279).
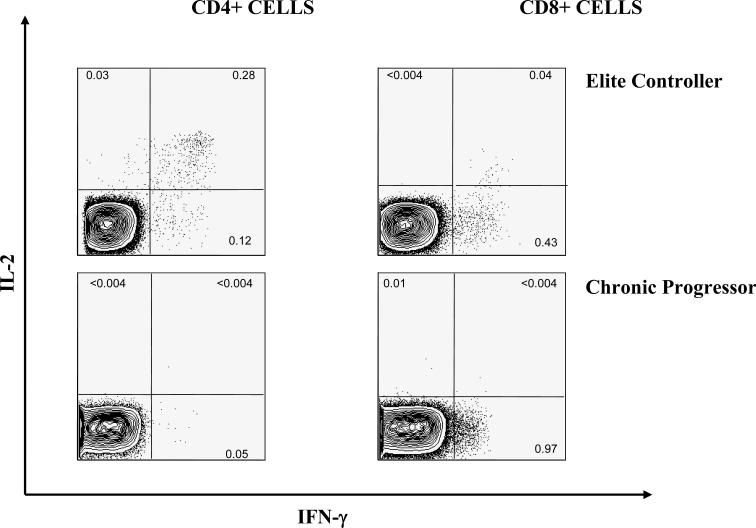
CTL are termed polyfunctional if they are able to produce multiple cytokines [84]. The most frequently assessed cytokines are interferon-gamma, MIP-1β, interleukin-2, tumor necrosis factor-alpha, and the degranulation marker CD107α. Production of multiple cytokines is thought to be indicative of a high quality response and has been correlated with viral control and a vigorous proliferative capacity in murine models [86]. In contrast, cells that are only capable of producing interferon-gamma display a diminished ability to proliferate in response to antigenic stimulation [83]. Not surprisingly, HIV controllers seem to possess a higher proportion of durable, multi-functional CD8+ T cells that persist in the systemic circulation throughout the course of infection compared to progressors [84] —suggesting that poly-functional CTL may contribute to control of HIV infection (FIGURE 4).
Figure 4. Comparison of flow cytometric-measured HIV-specific T cell responses between an elite controller and an untreated chronic progressor.
Flow analysis of Gag specific CD4+ and CD8+ T cell responses measured by intracellular cytokine staining. Dead cells are excluded from analysis by gating only on dead cell dye negative cells, CD3+ CD14/19 – cells are identified as T cells. CD4+ and CD8+ subpopulations are identified based on the presence of surface markers. The percentage of cells within the CD4+ and CD8+ subpopulations that produce Il-2, IFN-γ or both cytokines is measured for a representative elite controller and a chronic progressor.
Interestingly, these polyfunctional T cells from controllers are not only capable of rapid proliferation, but they also seem to be particularly potent killers. Recent studies have shown that the ability to proliferate is coupled to increased perforin expression. These higher levels likely facilitate cell killing and thus contribute to the efficacy of the CTL response [87]. Together, these findings indicate that control of HIV replication is marked by the presence of poly-functional, potent cellular-immune responses that persist throughout the course of infection. Unfortunately, however, it is impossible to be certain whether this is cause or effect. It may be that prolonged low-level viremia allows for accumulation of these cells, and therefore it is not certain that they contribute to control.
Further data have revealed that certain combinations of CTL surface markers associate predictably with varying levels of viral control and immunologic differentiation. Significant attention has been given to HLA-DR and CD38 molecules. HLA-DR is a marker of effector CD8+ T cells and CD38 is an activation marker. In HIV controllers, HLA-DR+/CD38− expressing cells predominate, whereas in non-controllers, CTL are more frequently HLA-DR+/CD38+ as a result of chronic stimulation [88]. HLA-DR+/CD38− CTL exist in a state of low immune activation with the capacity for extensive proliferation [88].
Other studies have shown that HIV controllers have a higher proportion of circulating effector CTL that are terminally-differentiated (CD45RA+ and CCR7−) than HIV progressors [89]. A block in the full differentiation of memory effector cells may contribute to a lack of valuable CTL responses produced by HIV progressors.
5.3. Associations between CTL specificity and disease outcome
When it comes to CTL targeting of HIV-infected cells, it appears that not all responses are created equal. In cohort studies of individuals in South Africa with chronic clade C HIV infection, increased breadth of targeting of HIV-Gag protein, taken as a percentage of total HIV-directed CTL responses, correlates with lower viral load. In contrast, individuals with higher viral loads focus more of their CTL against HIV-Env [90]. Studies of our Clade B-infected controller population have found a similar trend—individuals with lower viral loads tend to more extensively target HIV-Gag [91]. Similarly, a study in pigtail macaques showed that monkeys that generated SIV-Gag-specific CTL responses following prime/boost vaccination had lower viral loads and increased retention of CD4+ T cells after challenge with SHIVmn229 than monkeys who made similar CTL responses against SIV-Env or monkeys who made no CTL responses at all [92].
Even within a single HIV protein, there are differences in response patterns to different epitopes. A recent study found that just one out of four B*57-restricted epitopes in Nef had IFN-gamma responses that were conserved in B*57 positive LTNP but not progressors [93]. However, a recent report has suggested that in spite of significant selection pressure applied to the virus by B*57-restricted CTL responses, elite controllers have the capacity to maintain responses to wild type sequence and to generate de novo responses to viral variants [94].
The discordant inhibitory capacity of cells targeting epitopes in different HIV proteins is likely due to a combination of viral and host factors. The 9 HIV proteins vary in their ability to accommodate escape mutations without inducing a fitness cost. In the case of Gag and Env, it is likely that Gag is subject to more stringent sequence constraints than Env, making it more difficult for HIV to mutate away from Gag-specific CTL responses [95]. More generally, eliciting immune responses against the most highly-conserved regions of HIV appears to be a good strategy [96], given HIV's high mutation rate and evidence that escape mutations can result in a loss of suppressive ability in HIV controllers [32]. In the case of elite controllers, a recent publication has highlighted the ability of elite controllers to maintain CTL responses in spite of the rise of viral mutants, which may be one of the key characteristics separating controllers from non-controllers and allowing for prolonged resistance to progressive infection [97].
Host factors may also contribute to the discordant efficacy of CTL responses against different HIV proteins. It is possible that CD8+ T cells specific for certain HIV proteins bind with a higher avidity, proliferate more extensively and thus inhibit more efficiently. Recent data from our cohort indicates that even clonal populations that are specific for the same epitope can have varying anti-viral effects due to the specific T cell receptor (TCR) that is engaged (H. Chen and B. Walker, unpublished). Further studies into antigen processing, presentation, and CD8 recognition of the HLA-peptide complex are needed to elucidate the mechanisms for the observed differences in order to gain a better understanding of how to optimize the cytolytic functions of CD8+ T cells.
5.4. CD4+ T cell responses
Preservation of CD4+ T cell responses throughout HIV infection also appears to be crucial for immune control of HIV replication. Studies have demonstrated statistically significant correlations between disease progression and the proportion of poly-functional CD4+ T cell responses that include interleukin-2 production, although it is unclear whether or not viremia levels are driving CD4+ T cell dysfunction [85, 98-101]. As was observed for CD8+ T Cells, interleukin-2 production by CD4+ T cells is positively correlated with the ability to proliferate following exposure to antigen [102]. A recent study suggests that inactivation of the FOXO3a pathway, which regulates Fas-mediated apoptosis, may contribute to the preservation of CD4+ T cells observed in elite controllers[103].
Interestingly, while classical immunology characterizes CD4+ T cells as helper cells, CD4+ cells in controllers can also carry out cytolytic activity and thus directly contribute to viral suppression. This killing is likely perforin-dependent, as is evidenced by a statistically significant difference in levels of perforin expression between controllers and uninfected controls [104].
5.5. Immune Activation
HIV viral load is widely regarded as a good indicator for risk of disease progression [10, 13, 14]. However, it is becoming apparent that the level of immune activation in an individual is one of the strongest predictors of disease progression [105], even among elite controllers [106].
Low viral loads generally correlate with low levels of immune activation, making it difficult to determine whether HIV disease progression is driven by HIV viremia, immune activation, or both. A recent study describes several HIV-infected persons experiencing non-progressive infection despite having very high levels of viremia [19]. These individuals have similar immune patterns to Sooty Mangabees, the reservoir species for HIV-2 and the source of many lab strains of SIV, in that they live with high levels of virus but do not mount a vigorous anti-viral cellular immune response to it and do not experience disease [107]. The lack of disease progression despite chronically high viremia in these individuals suggests that immune activation, rather than HIV itself, may cause disease progression.
Further evidence that immune activation could be deleterious is available in both the macaque model and in human studies. A recent report in the SIV literature described extensive depletion of CD4+ T cells with a regulatory phenotype during acute infection [108], suggesting that the removal of a barrier to T cell activation may be partly responsible for the rapid infection and depletion of CD4+ T cells during acute infection. In humans, studies examining the level of the inhibitory cytokine IL-10 have found that decreased levels of IL-10 correlate with increased immune activation and faster HIV disease progression [29, 37]. Finally, studies of elite controllers have shown that among individuals with undetectable viremia, those with higher levels of T cell activation, marked by the HLA-DR+CD38+ phenotype, have lower CD4 counts [106].
5.6. Negative Regulators of Immune Activation
5.6.1. PD-1 and CTLA-4
Inhibitory immunoregulatory markers are elevated on both CD4+ and CD8+ T cells in individuals experiencing progressive HIV-infection [109-111]. Binding of Programmed Death-1 (PD-1 or CD279) with either of its ligands, PD-L1 or PD-L2, on CD8+ T cells delivers an immunosuppressive signal presumably aimed at preventing autoimmunity. PD-1 binding leads to decreased cytokine production by CD8+ cells, and in turn, a reduction in the proliferative capacity of these cells [111]. PD-1 expression is increased on the surface of HIV-specific T lymphocytes, suggesting that these cells are somehow being selectively blocked from carrying out their cytolytic activity [110]. In fact, PD-1 expression levels on both CD4+ and CD8+ lymphocytes correlates positively with viral load and negatively with CD4 count [109, 111].
HIV controllers express significantly lower levels of PD-1 than people with progressive disease [112]. It has been hypothesized that the lower levels of PD-1 in controllers enable them to maintain competent, polyfunctional HIV-specific CTL. Conversely, T cells in individuals with high levels of PD-1 are thought to be “exhausted” because they are unable to efficiently carry out their cytolytic activity, and this exhaustion could permit HIV to replicate virtually unchecked. However, the theory that PD-1 expression levels on CTL determine their ability to mount an effective immune response may have the causal relationship reversed. A newly published report offers evidence that the high levels of circulating antigen in persons failing to effectively control HIV viremia may in fact cause T cell exhaustion, rather than being a result of T cell exhaustion [113]. This finding would seem to corroborate the report of another recent publication which found that CD8+ cells specific for the HLA B*27 KK10 epitope, which one would expect to be highly effective CTL because this epitope is highly-immunodominant and seems to be a critical CTL target, express just as much PD-1 as those CTL that do not target this epitope [114].
In sum, there does seem to be a strong correlation between PD-1 expression and disease progression, but the directionality of this relationship is unclear. Further studies, in particular those looking at elite controllers, are thus needed to clarify the relationship between PD-1 expression and control of HIV infection.
Like PD-1, CD152 (a.k.a CTLA-4) is often upregulated during HIV infection [115]. CTLA-4 is an immunoregulatory molecule that is primarily expressed on CD4+ T lymphocytes. It has been implicated in the loss of CTL responses observed in some individuals during chronic HIV infection [116]. Currently, the leading hypothesis is that CTLA-4high T cells decrease interleukin-2 secretion, thus preventing the initiation of an immune response [115].
Recent studies looking at both HIV and CMV infection have found that CTLA-4 expression correlates inversely with CD4 count in both acutely and chronically infected individuals [115, 116]. Interestingly, HIV controllers do not demonstrate an upregulation of CTLA-4 on CD4+ T cells and do not develop an exhausted phenotype [115].
Learning why the PD-1 and CTLA-4 pathways are not upregulated on T lymphocytes in elite controllers will help to develop strategies for circumventing upregulation of these immunoregulatory pathways in recipients of a cell-mediated HIV vaccine.
5.6.2. Regulatory T cells
T Regulatory Cells, or Tregs, are T cells that inhibit the effector functions of other T cells. Tregs are characterized by the presence of the transcription factor FOXP3, which is critical for Treg development, and surface expression of the inhibitory ligand CTLA-4, which binds to the CD80 and CD86 receptors on T lymphocytes [117]. The vast majority of Tregs are CD4+, although some CD8+ T cells also express FOXP3 [118].
Tregs constitute approximately 5% of overall CD4+ T cells and are likely involved in the prevention of autoimmunity, as their depletion in healthy hosts leads to autoimmune complications [118]. Tregs have been the subject of extensive research in many cancers as they are thought to prevent anti-tumor immune activity. For example, studies of ovarian cancer have shown that higher concentrations of circulating Tregs correlate with poorer prognosis, and that higher CTL:Treg ratios predict increased survival [119, 120].
In the case of HIV, it remains unclear whether Tregs are protective or deleterious, and it may be that Tregs exert different effects in different anatomical locations and/or stages of infection [121]. For instance, Tregs capable of suppressing lymphoproliferative responses against HIV have been identified in the blood of HIV-infected persons, and cohort studies have shown that the percentage of Tregs correlates with viral load [122] and is elevated in patients with CD4<200/ul [123]. These findings seem to indicate that Treg levels during the chronic phase correlate with disease progression. However, research in the SIV model has shown that Tregs are severely depleted in the intestinal lamina propria during acute SIV infection [108]. This depletion of Tregs is coincident with the initial burst of HIV viremia, which suggests that depletion of Tregs increases cell-susceptibility to HIV infection and that Tregs may actually be protective during acute infection.
6. Conclusions
More than a quarter century after the start of the AIDS epidemic, HIV continues to kill millions worldwide while affecting countless more. Vast improvements in treatment have transformed HIV into a chronic, manageable disease for some [4], however many people in need of treatment are still not receiving it [3]. With tens of millions of people infected and several million new infections occurring each year, an HIV vaccine remains the best hope for ending the epidemic.
The ideal HIV vaccine would be 100% effective at preventing new infections. Unfortunately, it is unlikely that a first generation HIV vaccine will be able to confer sterilizing immunity [6]. Vaccines that achieve protective immunity rely on the induction of a strong neutralizing antibody response, which is proving extremely difficult to achieve in HIV vaccine design. Neutralizing antibodies against HIV are rarely detected in HIV+ individuals [7], and vaccines that have elicited neutralizing antibodies in vitro have failed to confer any degree of protection [124].
Given the difficulty of eliciting an effective neutralizing antibody response, the focus of vaccine design has turned to a cell-mediated approach. Although it would permit infection, a T cell vaccine would ideally attenuate disease progression and reduce the risk of transmission. In order to create a successful HIV vaccine, we must first dissect the correlates of immune protection. Specifically, we must learn: 1) How can we generate highly functional CTL? 2) What are the most potent CTL targets? 3) How can we make effective immune responses durable? Elite controllers likely hold the key to answering some of these questions and learning what factors are needed to create a T cell based vaccine that enables individuals to mount an immune response that is both effective and durable. While the answers to these questions are unlikely to surmount the current obstacles to designing a sterilizing vaccine, they will hopefully guide us towards an effective T cell vaccine.
7. Expert Opinion
HIV vaccine design efforts were recently paused by the termination of Merck's Phase II trial of its recombinant adenovirus vaccine. This vaccine consisted of HIV Gag, Pol and Nef genes delivered by an adenovirus vector. Early results of the trial showed that the vaccine failed to demonstrate any protection from acquisition, an endpoint that is not expected for a T cell based vaccine, or a lowered viral set point, a marker of viral control and a more realistic endpoint of a T cell based vaccine. In fact, the vaccine may have actually increased susceptibility to HIV in persons seropositive for adenovirus prior to immunization [125]. This unexpected finding raises questions whether induction of strong cellular immune responses alone, at least with this particular vaccine vector and protocol, guarantees vaccine efficacy or even safety. Because of these safety concerns and the fact that no HIV vaccine candidates so far have produced beneficial results in Phase II/III efficacy trials, we are in agreement with others [126] who suggest that more basic research into what defines an effective HIV-specific immune response is necessary before proceeding with human clinical trials. Studying elite controllers, individuals who by definition are effectively suppressing HIV replication, can help us answer this question.
Though much is now known about elite controllers, no single genetic characteristic has been identified that is shared by a majority of elite controllers; however, it is clear that there are several genetic components that account for at least part of their viral control. Similarly, while there is strong evidence for cellular immunity contributing to control and we are beginning to develop an understanding of which HIV proteins make the best CTL targets, we have yet to identify any cellular responses that are either necessary or sufficient for control. Therefore, we should continue to pursue avenues of basic research, identifying differences between the immune responses observed in HIV controllers and progressors, and also seeking to define new genes that are implicated in protection from disease progression. As we learn more about elite controllers, our understanding of precisely what genetic factors and immune responses naturally correlate with HIV control will guide vaccine design.
New technologies that allow for rapid and affordable sequencing of the human genome may be useful in evaluating genetic associations with HIV disease outcome [127]. Indeed, this technique has already enabled researchers to scan across the entire human genome for polymorphisms that correlate with outcomes in other disease settings. As part of the International HIV Controllers Study [128] we are performing a genome wide association scan looking at 650,000 Single-Nucleotide-Polymorphisms (SNPs) in HIV controllers and progressors, in order to find previously-unidentified genetic associations with HIV disease outcome. 1,500 subjects have already been scanned by this genome-wide association scan (GWAS) method, and preliminary analyses are underway. A similar study has already yielded genetic associations with lower level viremia following acute infection [129]. Two of the polymorphisms discovered, however, were not found in a majority of a cohort of elite controllers that was subsequently studied [130]. This disparity underscores the need for more and larger studies like the International HIV Controllers Study, which take a systematic, focused, and unbiased approach to trying to learn how elite controllers mediate their suppression of HIV replication. Indeed, it is our hope that by focusing specifically on comparing elite controllers, who are by definition exceptional, with patients who have high viral loads, the International HIV Controllers Study will be powered to uncover novel mechanisms of immune control of HIV infection and could thus have major implications for generation of HIV vaccines.
Acknowledgements
The authors would like to acknowledge Mary Carrington, PhD, Laboratory of Genomic Diversity, NCI-Frederick, and Florencia Pereyra, MD, Partners AIDS Research Center, for providing figures #2 and #4 respectively. We would also like to acknowledge the following members of the HIV Elite Controller Study for their input and guidance: Marylyn Addo, MD, PhD, Boris Julg, MD, and Alicja Piechocka-Trocha, DVM.
Abbreviations used in this manuscript
- HIV
Human Immunodeficiency Virus
- AIDS
Acquired Immune Deficiency Syndrome
- ARV
Antiretroviral
- GFATM
Global Fund to fight AIDS, TB and Malaria
- PEPFAR
President's Emergency Plan for AIDS Relief
- WHO
World Health Organization
- HAART
Highly-Active Antiretroviral Therapy
- RNA
Ribonucleic Acid
- LTNP
Long Term Non-Progressor
- HLA
Human Leukocyte Antigen
- CTL
Cytotoxic T Lymphocyte
- mRNA
messenger Ribonucleic Acid
- LCMV
Lymphocytic Choriomeningitis Virus
- TLR
Toll-Linked Receptors
- ssRNA
Single-stranded Ribonucleic Acid
- PDC
Plasmacytoid Dendritic Cell
- ADCC
Antibody-Dependent Cell-Cytotoxicity
- SIV
Simian Immunodeficiency Virus
- SHIV
SIV/HIV Chimeric Virus
- TCR
T cell receptor
- CMV
Cytomegalovirus
- Treg
Regulatory T Cell
- SNP
Single Nucleotide Polymorphism
- LMER
Linear mixed effects regression
- ELISpot
Enzyme-Linked Immunosorbent Spot
References
- 1.UNAIDS AIDS epidemic update: a special report on HIV/AIDS. 2007 [Google Scholar]
- 2.WHO . Towards universal access: Scaling up priority HIV/AIDS interventions in the health sector: Progress Report 2008. WHO Press; Geneva: [Google Scholar]
- 3.WHO Coverage and need for antiretroviral treatment. 2007 cited; Available from: http://www.who.int/hiv/facts/cov0605/en/
- 4.Walensky RP, et al. The survival benefits of AIDS treatment in the United States. J Infect Dis. 2006;194(1):11–9. doi: 10.1086/505147. [DOI] [PubMed] [Google Scholar]
- 5.Palella FJ, Jr., et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–60. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 6.Johnston MI, Fauci AS. An HIV vaccine--evolving concepts. N Engl J Med. 2007;356(20):2073–81. doi: 10.1056/NEJMra066267. [DOI] [PubMed] [Google Scholar]
- 7.Burton DR, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 9.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat Med. 2004;10(8):806–10. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez B, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. Jama. 2006;296(12):1498–506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 11.Walensky RP, et al. A therapeutic HIV vaccine: how good is good enough? Vaccine. 2004;22(29-30):4044–53. doi: 10.1016/j.vaccine.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 12.Gupta SB, et al. Estimating the benefit of an HIV-1 vaccine that reduces viral load set point. J Infect Dis. 2007;195(4):546–50. doi: 10.1086/510909. [DOI] [PubMed] [Google Scholar]
- 13.Mellors JW, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126(12):946–54. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 14.Lyles RH, et al. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J Infect Dis. 2000;181(3):872–80. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 15.Lambotte O, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41(7):1053–6. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 16.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27(3):406–16. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Munoz A, et al. Long-term survivors with HIV-1 infection: incubation period and longitudinal patterns of CD4+ lymphocytes. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8(5):496–505. doi: 10.1097/00042560-199504120-00010. [DOI] [PubMed] [Google Scholar]
- 18.Cao Y, et al. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332(4):201–8. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 19.Choudhary SK, et al. Low immune activation despite high levels of pathogenic human immunodeficiency virus type 1 results in long-term asymptomatic disease. J Virol. 2007;81(16):8838–42. doi: 10.1128/JVI.02663-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander L, et al. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74(9):4361–76. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Learmont JC, et al. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340(22):1715–22. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 22.Blankson JN, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81(5):2508–18. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura T, et al. Genetic Characterization of Human Immunodeficiency Virus type 1 in Elite Controllers: Lack of gross genetic defects or common amino acid changes. J Virol. 2008 doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey JR, et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008;82(15):7395–410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey JR, et al. Transmission of HIV-1 from a Patient Who Developed AIDS to an Elite Suppressor. J Virol. 2008 doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janeway C TP, Walport M, Shlomchik M. Immunobiology: the immune system in health and disease. 6 ed. Garland Science Publishing; New York: 2005. [Google Scholar]
- 27.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97(6):2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432(7018):769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36(6):565–74. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 30.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 31.Magierowska M, et al. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 1999;93(3):936–41. [PubMed] [Google Scholar]
- 32.Kelleher AD, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193(3):375–86. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrington M, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283(5408):1748–52. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 34.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 35.Qi Y, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2(8):e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrington M, Nelson G, O'Brien SJ. Considering genetic profiles in functional studies of immune responsiveness to HIV-1. Immunol Lett. 2001;79(1-2):131–40. doi: 10.1016/s0165-2478(01)00275-9. [DOI] [PubMed] [Google Scholar]
- 38.Ioannidis JP, et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann Intern Med. 2001;135(9):782–95. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 39.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 40.Salkowitz JR, et al. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin Immunol. 2003;108(3):234–40. doi: 10.1016/s1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hladik F, et al. Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism -2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79(18):11677–84. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci U S A. 1999;96(8):4581–5. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal F, et al. Polymorphism of RANTES chemokine gene promoter is not associated with long-term nonprogressive HIV-1 infection of more than 16 years. J Acquir Immune Defic Syndr. 2006;41(1):17–22. doi: 10.1097/01.qai.0000188335.86466.ea. [DOI] [PubMed] [Google Scholar]
- 44.Modi WS, et al. Genetic variation in the CCL18-CCL3-CCL4 chemokine gene cluster influences HIV Type 1 transmission and AIDS disease progression. Am J Hum Genet. 2006;79(1):120–8. doi: 10.1086/505331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott DH, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. Aids. 2000;14(17):2671–8. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- 46.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 47.Jin X, et al. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79(17):11513–6. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho SJ, et al. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80(4):2069–72. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandhi SK, et al. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82(6):3125–30. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biasin M, et al. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J Infect Dis. 2007;195(7):960–4. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- 51.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 52.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–8. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 53.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12(11):1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ancuta P, et al. Opposite effects of IL-10 on the ability of dendritic cells and macrophages to replicate primary CXCR4-dependent HIV-1 strains. J Immunol. 2001;166(6):4244–53. doi: 10.4049/jimmunol.166.6.4244. [DOI] [PubMed] [Google Scholar]
- 55.Castelli JC, et al. Relationship of CD8(+) T cell noncytotoxic anti-HIV response to CD4(+) T cell number in untreated asymptomatic HIV-infected individuals. Blood. 2002;99(11):4225–7. doi: 10.1182/blood-2001-11-0078. [DOI] [PubMed] [Google Scholar]
- 56.O'Connor G M, et al. Natural Killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin Immunol. 2007;124(3):277–83. doi: 10.1016/j.clim.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 57.Scott-Algara D, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol. 2003;171(11):5663–7. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 58.Fauci AS, Mavilio D, Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat Rev Immunol. 2005;5(11):835–43. doi: 10.1038/nri1711. [DOI] [PubMed] [Google Scholar]
- 59.Kottilil S, et al. Expression of chemokine and inhibitory receptors on natural killer cells: effect of immune activation and HIV viremia. J Infect Dis. 2004;189(7):1193–8. doi: 10.1086/382090. [DOI] [PubMed] [Google Scholar]
- 60.Mavilio D, et al. Characterization of CD56−/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A. 2005;102(8):2886–91. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131–7. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 62.Meier A, et al. MyD88-dependent immune activation mediated by human immunodeficiency virus type 1-encoded toll-like receptor ligands. J Virol. 2007;81(15):8180–91. doi: 10.1128/JVI.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herbeuval JP, et al. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc Natl Acad Sci U S A. 2006;103(18):7000–5. doi: 10.1073/pnas.0600363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bochud PY, et al. Polymorphisms in Toll-like receptor 9 influence the clinical course of HIV-1 infection. Aids. 2007;21(4):441–6. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 65.Kraft Z, et al. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J Virol. 2007;81(12):6402–11. doi: 10.1128/JVI.00424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto H, et al. Post-infection immunodeficiency virus control by neutralizing antibodies. PLoS ONE. 2007;2:e540. doi: 10.1371/journal.pone.0000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Binley JM, et al. Passive infusion of immune serum into simian immunodeficiency virus-infected rhesus macaques undergoing a rapid disease course has minimal effect on plasma viremia. Virology. 2000;270(1):237–49. doi: 10.1006/viro.2000.0254. [DOI] [PubMed] [Google Scholar]
- 68.Harrer T, et al. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12(7):585–92. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 69.Pereyra F, et al. Genetic and Immunologic Heterogeneity among Persons Who Control HIV Infection in the Absence of Therapy. J Infect Dis. 2008;197(4):563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–4. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bailey JR, et al. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol. 2006;80(10):4758–70. doi: 10.1128/JVI.80.10.4758-4770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deeks SG, et al. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol. 2006;80(12):6155–64. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walker BD, et al. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature. 1987;328(6128):345–8. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 74.Koup RA, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68(7):4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borrow P, et al. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68(9):6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmitz JE, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283(5403):857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 77.Friedrich TC, et al. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol. 2007;81(7):3465–76. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allen TM, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79(21):13239–49. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneidewind A, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81(22):12382–93. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Betts MR, et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75(24):11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Addo MM, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77(3):2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emu B, et al. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82(11):5398–407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peretz Y, et al. Functional T cell subsets contribute differentially to HIV peptide-specific responses within infected individuals: correlation of these functional T cell subsets with markers of disease progression. Clin Immunol. 2007;124(1):57–68. doi: 10.1016/j.clim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 84.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Emu B, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79(22):14169–78. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wherry EJ, et al. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77(8):4911–27. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3(11):1061–8. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 88.Saez-Cirion A, et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A. 2007;104(16):6776–81. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Addo MM, et al. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS ONE. 2007;2(3):e321. doi: 10.1371/journal.pone.0000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiepiela P, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 91.Florencia Pereyra BB, Rothchild Alissa, Baker Brett, Rathod Almas, Rosenberg Rachel, Padilla Priscilla, Moss Kristin, Beattie Warren, Birch Christopher, Carrington Mary, Piechocka-Trocha1 Alicja, Walker Bruce. Preferential targeting of HIV-Gag epitopes in elite controllers in HIV Vaccines: From Basic Research to Clinical Trials. Whistler; British Columbia: 2007. [Google Scholar]
- 92.Peut V, Kent SJ. Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen. J Virol. 2007;81(23):13125–34. doi: 10.1128/JVI.01408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navis M, et al. A nonprogressive clinical course in HIV-infected individuals expressing human leukocyte antigen B57/5801 is associated with preserved CD8+ T lymphocyte responsiveness to the HW9 epitope in Nef. J Infect Dis. 2008;197(6):871–9. doi: 10.1086/528695. [DOI] [PubMed] [Google Scholar]
- 94.Bailey JR, et al. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203(5):1357–69. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.LANL HIV Sequence Database. 2008 [Google Scholar]
- 96.Altfeld M, Allen TM. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 2006;27(11):504–10. doi: 10.1016/j.it.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 97.Migueles SA, et al. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J Virol. 2003;77(12):6889–98. doi: 10.1128/JVI.77.12.6889-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boaz MJ, et al. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J Immunol. 2002;169(11):6376–85. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- 99.Harari A, et al. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J Immunol. 2005;174(2):1037–45. doi: 10.4049/jimmunol.174.2.1037. [DOI] [PubMed] [Google Scholar]
- 100.Younes SA, et al. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198(12):1909–22. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tilton JC, et al. Changes in paracrine interleukin-2 requirement, CCR7 expression, frequency, and cytokine secretion of human immunodeficiency virus-specific CD4+ T cells are a consequence of antigen load. J Virol. 2007;81(6):2713–25. doi: 10.1128/JVI.01830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boaz MJ, et al. CD4 responses to conserved HIV-1 T helper epitopes show both negative and positive associations with virus load in chronically infected subjects. Clin Exp Immunol. 2003;134(3):454–63. doi: 10.1111/j.1365-2249.2003.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Grevenynghe J, et al. Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection. Nat Med. 2008 doi: 10.1038/nm1728. [DOI] [PubMed] [Google Scholar]
- 104.Norris PJ, et al. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol. 2004;78(16):8844–51. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deeks SG, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104(4):942–7. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 106.Hunt PW, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197(1):126–33. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rey-Cuille MA, et al. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol. 1998;72(5):3872–86. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chase AJ, et al. Severe Depletion of CD4+CD25+ Regulatory T Cells from the Intestinal Lamina Propria but not Peripheral Blood or Lymph Nodes During Acute SIV Infection. J Virol. 2007 doi: 10.1128/JVI.00841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 110.Petrovas C, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203(10):2281–92. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trautmann L, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12(10):1198–202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 112.Zhang JY, et al. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood. 2007;109(11):4671–8. doi: 10.1182/blood-2006-09-044826. [DOI] [PubMed] [Google Scholar]
- 113.Streeck H, et al. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 2008;5(5):e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204(10):2473–85. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaufmann DE, et al. Upregulation of CTLA-4 by HIV-specific CD4(+) T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007;8(11):1246–54. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 116.Zaunders JJ, et al. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J Virol. 2006;80(20):10162–72. doi: 10.1128/JVI.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat Immunol. 2005;6(4):338–44. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 118.Wang HY, Wang RF. Regulatory T cells and cancer. Curr Opin Immunol. 2007;19(2):217–23. doi: 10.1016/j.coi.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 119.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–9. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 120.Sato E, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(51):18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.de St Groth BF, Landay AL. Regulatory T cells in HIV infection: pathogenic or protective participants in the immune response? Aids. 2008;22(6):671–83. doi: 10.1097/QAD.0b013e3282f466da. [DOI] [PubMed] [Google Scholar]
- 122.Andersson J, et al. The prevalence of regulatory T cells in lymphoid tissue is correlated with viral load in HIV-infected patients. J Immunol. 2005;174(6):3143–7. doi: 10.4049/jimmunol.174.6.3143. [DOI] [PubMed] [Google Scholar]
- 123.Montes M, et al. Foxp3+ regulatory T cells in antiretroviral-naive HIV patients. Aids. 2006;20(12):1669–71. doi: 10.1097/01.aids.0000238415.98194.38. [DOI] [PubMed] [Google Scholar]
- 124.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 125.HIV vaccine failure prompts Merck to halt trial. Nature. 2007;449(7161):390. doi: 10.1038/449390c. [DOI] [PubMed] [Google Scholar]
- 126.Cohen J. Retrovirus meeting. Back-to-basics push as HIV prevention struggles. Science. 2008;319(5865):888. doi: 10.1126/science.319.5865.888. [DOI] [PubMed] [Google Scholar]
- 127.Telenti A, Goldstein DB. Genomics meets HIV-1. Nat Rev Microbiol. 2006;4(11):865–73. doi: 10.1038/nrmicro1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Study IHC. International HIV Controllers Study website. 2007 cited; Available from: www.hivcontrollers.org.
- 129.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han Y, et al. The role of protective HCP5 and HLA-C associated polymorphisms in the control of HIV-1 replication in a subset of elite suppressors. Aids. 2008;22(4):541–4. doi: 10.1097/QAD.0b013e3282f470e4. [DOI] [PubMed] [Google Scholar]