Abstract
Purpose of review
To review the most promising genetic markers associated with the variability in the safety or efficacy of warfarin and clopidogrel and highlight the verification and validation initiatives for translating clopidogrel and warfarin pharmacogenetic tests to clinical practice.
Recent findings
Rapid advances in pharmacogenetics, continuous decrease in genotyping cost, development of point-of-care devices and the newly established clinical genotyping programs at several institutions hold the promise of individualizing clopidogrel and warfarin based on genotype. Guidelines have been established to assist clinicians in prescribing clopidogrel or warfarin dose based on genotype. However, the clinical utility of clopidogrel and warfarin is still limited. Accordingly, large randomized clinical trials are underway to define the role of clopidogrel and warfarin pharmacogenetics in clinical practice.
Summary
Pharmacogenetics has offered compelling evidence toward the individualization of clopidogrel and warfarin therapies. The rapid advances in technology make the clinical implementation of clopidogrel and warfarin pharmacogenetics possible. The clinical genotyping programs and the ongoing clinical trials will help in overcoming some of the barriers facing the clinical implementation of clopidogrel and warfarin pharmacogenetics.
Keywords: clinical implementation, clopidogrel, personalized medicine, pharmacogenetics, warfarin
INTRODUCTION
Cardiovascular disease (CVD) remains the primary cause of death in the United States and worldwide. In 2008, CVD deaths represented 30% of all deaths globally [1,2]. Despite the presence of numerous highly efficacious drugs for the management of CVD, however, many of these drugs exhibit large interpatient variability in their efficacy or side-effects risk. Warfarin and clopidogrel are excellent examples of widely prescribed cardiovascular medications that are highly efficacious in the treatment and prevention of CVDs and their thrombotic complications. Warfarin and clopidogrel are also considered excellent examples of medications with wide interpatient variability in the efficacy, safety or dose requirements. The variability in warfarin dosing makes it extremely challenging in the clinical setting, and influences the risk of both bleeding and thrombotic complications [3–7]. Similarly, clopidogrel exhibits wide interpatient variability in its antiplatelet effect, which affects its ability to limit cardiovascular events in some.
The completion of the Human Genome Project in 2001 led to promises of improving care through genomic medicine [8,9]. The rapid growth in pharmacogenetics research and the better understanding of the impact of genetic variation on drug response for certain drugs prompted the Food and Drug Administration (FDA) to relabel more than 100 medications with genetic information in the last decade [10]. As was projected at the time of completion of the Human Genome project, pharmacogenetics represents the first major use of genomic medicine to improve the safety and effectiveness of a variety of drugs, with the goal of delivering better, individualized medical care [11–13].
Cardiovascular medications like warfarin and clopidogrel illustrate well the role of inheritance in variability of drug safety and efficacy [3,7,14]. Candidate gene approaches and genome-wide association studies (GWASs) have shown important genetic markers significantly linked with the safety or the efficacy of warfarin and clopidogrel [14–18].
Herein we review the most promising genetic markers associated with the variability in the safety or efficacy of warfarin and clopidogrel and highlight the verification and validation initiatives for translating clopidogrel and warfarin pharmacogenetic tests to clinical practice.
CLOPIDOGREL
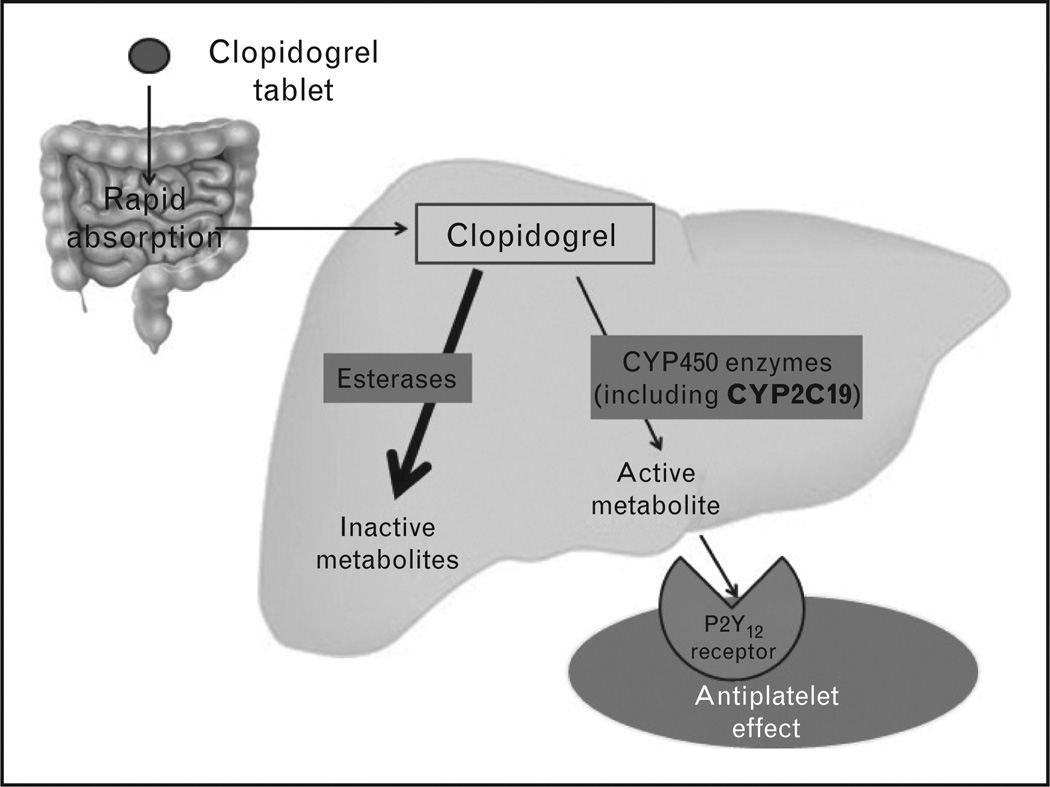
Clopidogrel is a thienopyridine antiplatelet agent widely prescribed for the prevention of ischemic events in patients with acute coronary syndrome (ACS), percutaneous coronary intervention (PCI) and myocardial infarction (MI) [19]. Despite the importance of clopidogrel in reducing the risk of adverse cardiovascular outcomes, including stent thrombosis, platelet function in response to clopidogrel varies [17,20–22]. Studies showed that about 30% of the patients taking clopidogrel do not respond effectively [17,20,21,23]. Clopidogrel is a prodrug that is rapidly absorbed from the intestine and extensively metabolized in the liver through two pathways [24–26] (Fig. 1). Several cytochrome P-450 (CYP) enzymes, including CYP2C19, work on activating clopidogrel to give an active metabolite, which irreversibly binds to the P2Y12 receptor, resulting in the inhibition of platelet aggregation [27–29].
FIGURE 1.
Clopidogrel pharmacokinetics and pharmacodynamics.
Clopidogrel pharmacogenetics discovery
Among genetic polymorphisms studied to date, the CYP2C19 polymorphisms show the strongest evidence for association with variability in the efficacy and safety of clopidogrel [30–35]. CYP2C19*2 (c.681G>A; rs4244285) creates a cryptic splice site and eventual premature stop codon. This polymorphism is the most common loss-of-function (LOF) polymorphism (Table 1) and carriers of this polymorphism have lower levels of clopidogrel active metabolite, reduced platelet inhibition, and increased risk of cardiovascular events [17,23,30, 35,36]. Conversely, the CYP2C 19*17 (c.806C>T; rs12248560) is a common gain of function polymorphism (Table 1) in which its carriers have higher levels of active metabolite, increased platelet inhibition, and increased bleeding risk [30,35,38,39]. Other variants (CYP2C19*3, CYP2C19*4, CYP2C19*5, CYP2C19*6, CYP2C19*7, and CYP2C19*8) have reduced function or LOF, but most are rare [30,40–43,44▪▪].
Table 1.
Population prevalence of CYP2C*2, CYP2C*3, and CYP2C*17 genetic polymorphisms and their effect on clopidogrel response
| Genetic polymorphism (rs#) | Asiansa | Whitesa | African–Americansa | Effect |
|---|---|---|---|---|
| CYP2C19*2 (rs4244285) | 55% | 28% | 24% | ↓Active metabolite concentration |
| ↓ Antiplatelet effect | ||||
| ↑Risk of cardiovascular events | ||||
| CYP2C19*3b (rs4986893) | 17% | <1% | <1% | ↓ Active metabolite concentration |
| ↓ Antiplatelet effect | ||||
| ↑ Risk of cardiovascular events | ||||
| CYP2C19*17 (rs12248560) | 4% | 41% | 23% | ↑ Active metabolite concentration |
| ↑ Antiplatelet effect | ||||
| ↑ Risk of bleeding |
Population prevalence of carrying at least one of the variant alleles in this polymorphism derived from http://hapmap.ncbi.nlm.nih.gov/ and http://browser.1000genomes.org/index.html.
Clopidogrel pharmacogenetics and cardiovascular outcomes
Recently, there was an extensive focus on defining the clinical importance of CYP2C19 genotypes for clopidogrel response. Many studies verified the association between CYP2C19 genotype and its clinical impact on the efficacy and safety in patients with coronary artery disease (CAD) taking clopidogrel [17,36,38,39,45–55]. Studies consistently showed that the presence of the CYP2C19*2 allele, especially in those patients on clopidogrel post-PCI, is associated with a significantly higher risk of adverse cardiovascular events, particularly stent thrombosis [17,32,47,51,52,55,56].
A recent meta-analysis of aggressively managed CAD patients, over 90% with a PCI, addressed the association between CYP2C19*2 genotype and cardiovascular outcomes in 9685 patients from nine independent studies [30]. CYP2C19*2 hetero-zygotes and homozygotes had 55 and 76% higher risk, respectively, of a composite endpoint of cardiovascular death, MI, or stroke, compared with non-carriers. This meta-analysis found carriers of the CYP2C19*2 LOF allele had a significantly higher risk of stent thrombosis [heterozygous CYP2C19*2: hazard ratio = 2.67, 95% confidence interval (CI) 1.69–4.22, P< 0.0001; homozygous CYP2C19*2: hazard ratio = 3.97, 95% CI 1.75–9.02, P = 0.001], compared with noncarriers [30].
In contrast, meta-analyses focused on lower risk populations (e.g. those without PCI) showed a less profound effect of genotype, with only modest effect sizes and borderline statistical significance [48,50,57,58]. The most likely explanation for the differential impact of the association observed is that the magnitude of benefit from clopidogrel is smaller in the non-PCI patient populations, and thus it is more difficult to observe the effects of the LOF CYP2C19 alleles [59–63].
The CYP2C19*17 gain of function polymorphism has also been extensively studied and some studies showed a significant association between CYP2C19*17 polymorphism and higher antiplatelet response or adverse cardiovascular events [17,38,39, 47,48,58,64–66], whereas others showed no association [17,36,67]. A recent meta-analysis found that CYP2C19*17 carriers have a lower risk of cardiovascular events (hazard ratio = 0.75; 95% CI 0.66– 0.87; P< 0.001) and a higher risk of major bleeding (hazard ratio = 1.26; 95% CI 1.05–1.50; P = 0.011) compared with noncarriers [58].
Based on the early literature on this topic, in March 2010, the FDA added a boxed warning to the clopidogrel label. The warning stated the relationship between CYP2C19 genotypes and drug response and recommended an alternative therapy for clopidogrel-poor metabolizer patients, meaning those homozygous for LOF alleles. Studies have shown that prasugrel and ticagrelor are good alternatives to clopidogrel in carriers with the CYP2C19 reduced function allele [49,50], as these drugs are not impacted by CYP2C19 genotypes [67–70].
Clinical implementation of clopidogrel pharmacogenetics testing
In 2010, the American College of Cardiology (ACC) and American Heart Association (AHA) issued a consensus statement that, in the absence of prospective randomized clinical trials, ‘the evidence base is insufficient to recommend either routine genetic or platelet function testing at the present time’ [71]. There are mixed opinions in the clinical cardiology community on clopidogrel pharmacogenetics testing, with arguments both for [72] and against [73]. Collectively, a series of consensus statements, including the original ACC/AHA statement, suggest it might be reasonable to consider pharmacogenetics testing in high-risk PCI patients [30,61,73].
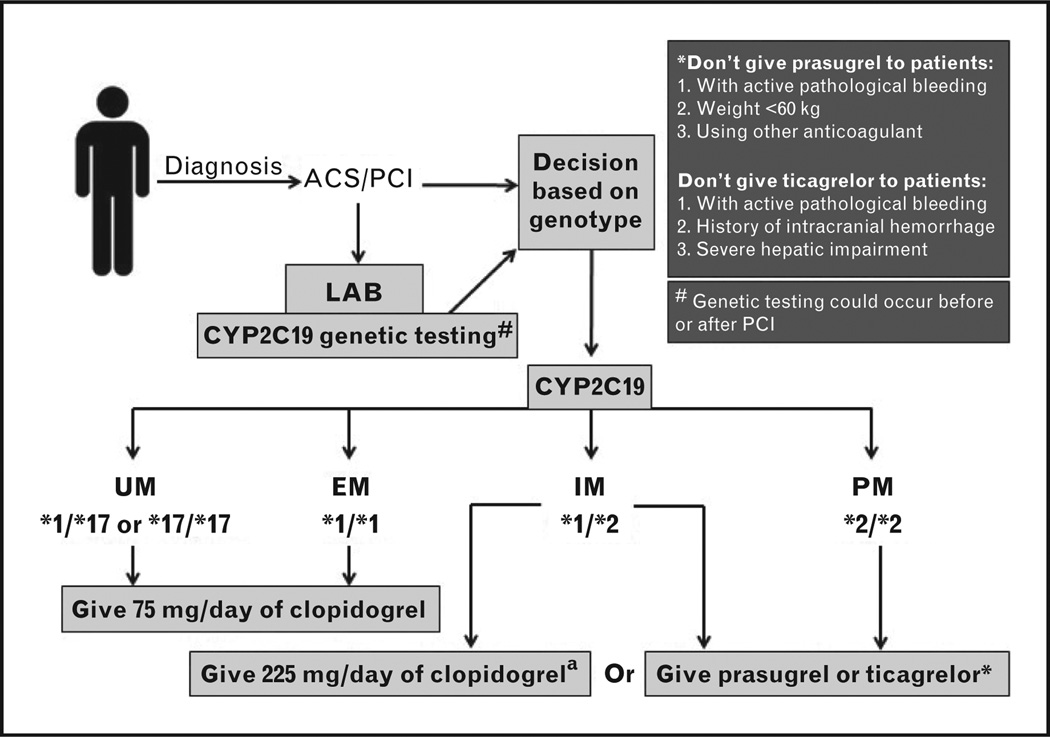
Recently, the clinical pharmacogenetics implementation consortium (CPIC) recommended guidelines for the use of CYP2C19 genetic information to guide clopidogrel therapy in ACS/PCI patients if CYP2C19 genetic testing results are available [44▪▪]. Alternative therapy is recommended in those carrying a CYP2C19*2 allele, as shown in Fig. 2. For those carrying the CYP2C19*1/CYP2C19*2 genotype, increased clopidogrel dose may be an option. Studies have shown that doubling the clopidogrel maintenance dose to 150 mg per day is not sufficient to overcome the resistance in those patients carrying the CYP2C19*2 LOF allele [75,76]. However, a recent study by Mega et al. [74▪] has shown that giving a clopidogrel dose of 225mg per day to CYP2C19*2 heterozygotes provides similar levels of platelet reactivity to those achieved by non-carriers taking 75 mg per day of clopidogrel. Doses as high as 300mg per day were not sufficient to achieve similar degrees of platelet inhibition in the homozygous carriers. Although there are no data on cardiovascular outcomes for the higher dose in heterozygotes, some would argue that the platelet reactivity data support 225 mg daily as an option for the CYP2C19*1/CYP2C19*2 genotype.
FIGURE 2.
Evidence-based clopidogrel dosing based on CYP2C19 genotype in acute coronary syndrome / percutaneous coronary intervention patients (ACS/PCI), adapted from CPIC guidelines [44▪▪] with revisions based on more recent literature. UM, ultra metabolizer; EM, extensive metabolizer; IM, intensive metabolizer; PM, poor metabolizer. aIt is preferable to use the alternative antiplatelet (prasugrel or ticagrelor) as a first option unless there are any contraindications to its use. Giving a clopidogrel dose of 225 mg per day to CYP2C19*2 heterozygote carriers was reported to be sufficient for getting similar levels of platelet reactivity achieved by noncarriers taking 75 mg per day of clopidogrel [74▪]. However, further studies are still needed to confirm this finding.
Despite the concrete evidence of the association between CYP2C19*2 genotype and variability in clopidogrel response, the clinical implementation of clopidogrel genetic testing is still challenging. Several institutions have established preemptive genotyping programs and tried to overcome the barriers that hinder the implementation of pharmacogenetic tests through practice [77–79]. Schildcrout et al. [80▪] suggested an improvement in the safety and efficacy of six drugs, including clopidogrel, based on this preemptive genotyping approach.
In an attempt to overcome some of the barriers facing the implementation of clopidogrel pharmacogenetics, a novel bedside CYP2C19*2 genetic test with a buccal swab has recently been developed (Spartan RX CYP2C19, Spartan Biosciences, Ottawa, Ontario, Canada) [81]. The rapidity of this test (results are obtained within an hour) facilitated the use of pharmacogenetics for guiding clopidogrel therapy after PCI. The verification of this test was done in a prospective, randomized proof of concept (POC) study that included 200 patients undergoing PCI [82▪▪]. Patients were randomized to either CYP2C19*2 rapid point-of-care genotyping or to standard treatment. Patients in the rapid genotyping arm were screened for the CYP2C19*2 allele. CYP2C19*2 carriers were given 10mg per day of prasugrel, whereas noncarriers and patients in the standard treatment group were given 75 mg per day of clopidogrel. After randomization and a week of follow-up, higher risk of stent thrombosis and cardiovascular events was present in patients in the standard treatment arm than the rapid genotyping arm. This study is considered a valuable step toward clopidogrel individualization based on genetics and provides insight into the potential clinical benefits of genotype-guided clopidogrel therapy [83].
In summary, there is a solid literature suggesting the risk of reduced clopidogrel efficacy in the presence of a CYP2C19 LOF allele. Furthermore, the recent technical advances in genotyping have made the individualization of clopidogrel based on genotype possible. Large prospective randomized trials (NCT01742117, NCT01761786 and NCT01452152) are underway to further evaluate the safety, efficacy and cost-effectiveness of CYP2C19*2 genetic testing and its clinical utility in PCI patients. However, the data suggest the potential benefit to guide clopidogrel therapy now for those patients undergoing PCI, particularly those at high risk for adverse outcomes. Increasing numbers of centers are implementing this approach, and it may become increasingly common in clinical practice.
WARFARIN
Warfarin is a highly effective therapy, and the cheapest and the most prescribed oral anticoagulant worldwide [5,84,85]. However, warfarin’s narrow therapeutic index and wide interindividual variability present challenges in its clinical utilization [3,86–88]. Many studies have shown the important role of both genetic and nongenetic factors in explaining the wide interindividual variability in warfarin dose [16,89–92]. Warfarin is a racemic mixture of R-warfarin and S-warfarin, in which the latter is three to five times more potent in its pharmacodynamic effect than R-warfarin [93]. S-warfarin is mainly metabolized by the CYP2C9 enzyme, whereas the R-warfarin is metabolized by other CYP-450 enzymes [94–97]. Warfarin works by inhibiting the vitamin K epoxide reductase complex 1 (VKORC1) [98,99]. This inhibition antagonizes the conversion of oxidized vitamin K to functional reduced vitamin K, hindering the conversion of premature clotting factors to active clotting factors, causing an anticoagulation effect [100] (Supplementary Fig. 1, http://links.lww.com/HCO/A16).
Warfarin pharmacogenetics discovery
Over the last decade, many of the several hundred publications on warfarin pharmacogenetics have shown that both clinical and demographic factors explain part of the interindividual variability in warfarin dose [14,16,18,101–104]. Countless studies have consistently documented that CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) carriers require lower warfarin dose as they have 40–70% reduction in S-warfarin clearance, respectively [105–108], which increases bleeding risk, particularly early in the course of therapy [88,105,106]. In 2004, the gene encoding the VKORC1 enzyme was identified [98,99] and numerous studies have since documented the impact of VKORC1 polymorphisms, particularly −1639 G>A (rs9923231), on warfarin dose requirements [14,16,18,90,92,109– 120]. Other polymorphisms in VKORC1, CYP2C9, CYP4F2, gamma glutamyl carboxylase (GGCX), epoxide hydrolase 1 (EPHX1), and apolipoprotein E (APOE) were identified, but their associations with warfarin dose requirements among African-Americans, whites and Asians were inconsistent [16,109,111,113,120–143].
Verifying the clinical utility of genotype-guided warfarin dosing
In 2007, and again in 2010, the FDA updated the warfarin label with genetic information [144]. The 2010 label includes a table to facilitate the dosing of warfarin based on CYP2C9*2, CYP2C9*3 and VKORC1 (−1639 G>A) genotypes (Supplementary Table 1, http://links.lww.com/HCO/A16). Furthermore, several dosing algorithms were published to assist clinicians with genotype-guided dosing [14,18,92,104,145,146]. Among those, the Gage algorithm [18] and the International Warfarin Pharmacogenetics Consortium (IWPC) algorithm, which included more than 5000 patients from four continents, were considered the best validated and most accurate [14,147]. Furthermore, studies have shown a better prediction of warfarin dose requirements by using the IWPC algorithm compared with a clinical algorithm, and with the FDA genotype dosing table [14,144].
In an attempt to investigate the clinical utility and the accuracy of pharmacogenetic-guided dose prediction algorithms, several case–control studies and prospective randomized clinical trials have been conducted [117,118,145,148,149]. Most of those studies provided some evidence for the clinical utility of a pharmacogenetics-based approach in determining loading and maintenance doses. Nevertheless, the results from those trials were limited by small sample size, limitations in design and using surrogate outcomes [5,150]. In 2010, a prospective study compared the incidence of hospitalization during 6 months in 896 patients receiving warfarin genotyping for CYP2C9*2, CYP2C9*3, and VKORC1 (−1693 G>A) versus 2688 in a matched historical control group with standard care [151]. The results of this study showed a one-third reduction in the incidence of hospitalization due to bleeding or thromboembolism in the genotyping-guided approach group compared with the standard care group (hazard ratio = 0.72). However, the results of this nonrandomized trial have been criticized because of flaws in study design, susceptibility to physician treatment bias and lack of temporal plausibility [85,152].
Most recently, CoumaGen-II, a well-powered clinical trial, provided additional support for the potential benefits of warfarin therapy initiation using warfarin pharmacogenetics-based algorithms versus warfarin standard care [153▪▪]. The first part of this study was a randomized double-blind trial comparing a one-step pharmacogenetics algorithm, derived from a modified IWPC algorithm, with a three-step algorithm. The three-step pharmacogenetics algorithm was neither superior nor inferior to the one-step algorithm, suggesting the simpler one-step algorithm might be preferable in clinical practice. Furthermore, the second part of this study was a nonrandom trial comparing the clinical effectiveness of patients dosed via the pharmacogenetics algorithm versus standard of care. The pharmacogenetic algorithm arm showed 11% absolute and 26% relative reduction in the percentage of out-of-range international normalized ratio (%OOR INR) at 1 month compared with the standard dosing care. Additionally, a similar difference in %OOR INR was also shown between the two groups after 3 months. Moreover, patients with pharmacogenetic dosing also had a higher percentage of time in the therapeutic range at 1 and 3 months (68 and 71%, respectively) compared with standard dosing (58 and 59%; P< 0.001). Furthermore, there were also statistical differences in clinical outcomes that favored pharmacogenetic dosing. Despite the concern of confounding and bias due to the non-randomization of the standard care management group [5,154], this study provided insight into the promise of pharmacogenetics in controlling warfarin therapy, suggesting the broader use of warfarin pharmacogenetic dosing in clinical practice.
Clinical implementation of warfarin pharmacogenetics testing
The warfarin CPIC guidelines recommended the Gage or the IWPC dosing algorithms as the preferred approaches for estimating the stable warfarin dose [155▪▪]. They secondarily recommended the FDA dosing table as an alternative approach in case of absence of electronic access to these algorithms. Moreover, they have suggested that inclusion of polymorphisms confined to a particular population, such as CYP2C9*5, CYP2C9*6, CYP2C9*8, and CYP2C9*11 in African–Americans, might help in better estimating the stable warfarin dose required. On the basis of these findings, more studies are underway to detect other genetic and nongenetic factors associated with warfarin dose variability in underrepresented populations [156,157].
Rapid and affordable genotyping is a commonly cited barrier facing the implementation of warfarin pharmacogenetics. In an attempt to facilitate the clinical utilization of warfarin pharmacogenetic testing, a POC warfarin-based genetic test was recently validated. This test provides precise results in less than 2h [158]. Additionally, one of the largest ongoing randomized trials, European Pharmacogenetics of Anticoagulant Therapy, is using a genotyping test that can be done at the bedside or in the clinic using whole blood and provides results within 1.5h [159]. Other scientists are working on designing and evaluating faster and simpler methods for warfarin pharmacogenetic testing [160]. Although the genotyping cost for one patient might range from less than US$ 25 to about US$ 200 for commercial platforms, the rapid advances in genotyping and sequencing technologies promise rapid genotyping with lower cost.
Despite the strong evidence for the clinical and analytical validity of warfarin pharmacogenetic testing, at least for whites, its clinical use is still limited. Accordingly, large randomized controlled trials are underway to assess the use of warfarin pharmacogenetics in clinical practice (Table 2). Furthermore, scientists worldwide are intensifying their efforts to identify other genetic and nongenetic factors that might be associated with warfarin dose variability in nonwhites.
Table 2.
Ongoing randomized controlled trials evaluating pharmacogenetic-guided warfarin dosing
| Study | Targeted enrolment |
Design | Primary endpoint |
|---|---|---|---|
| COAG (NCT00839657)b [161] | 1238 | Multicenter, DB, two arms PGx vs clinical algorithma | PTTR during the first 4 weeks of Therapy |
| EU-PACT (NCT01119300)b [159] | 970 | Multicenter, SB, two arms PGx vs clinical algorithm | PTTR during the first 12 weeks of therapy |
| GIFT (NCT01006733)b [162] | 1600 | Multicenter, DB, two arms PGx vs clinical algorithma | Composite of: nonfatal VTE, nonfatal major hemorrhage, death from any cause, and INR ≥4.0 |
| WARFARIN (NCT01305148)b | 4300 | Multicenter, DB, two arms GenoSTAT test (PGx)+clinical factors vs clinical factors alone | Incidence of major hemorrhage and thromboembolic events during the first 30 days of therapy |
| Pharmacogenetic dosing of warfarin: a controlled randomized trial | 600 | Three arms study IWPC PGx vs Taiwanese algorithm vs standard care | Time to target INR and PTTR |
COAG, Clarification of Optimal Anticoagulation through Genetics; DB, double blind; EU-PACT, European Pharmacogenetics of Anticoagulant Therapy; GIFT, Genetics Informatics Trial; INR, international normalized ratio; IWPC, International Warfarin Pharmacogenetic Consortium; PGx, pharmacogenetics; PTTR, percentage of time within therapeutic range; SB, single blind; VTE, venous thromboembolic event; WARFARIN, Warfarin Adverse Event Reduction For Adults Receiving Genetic Testing at Therapy Initiation.
Each study arm includes a baseline dose initiation algorithm and a dose revision algorithm applied over the first four to five doses of warfarin therapy.
Further information can be found at www.clinicaltrials.gov.
CONCLUSION
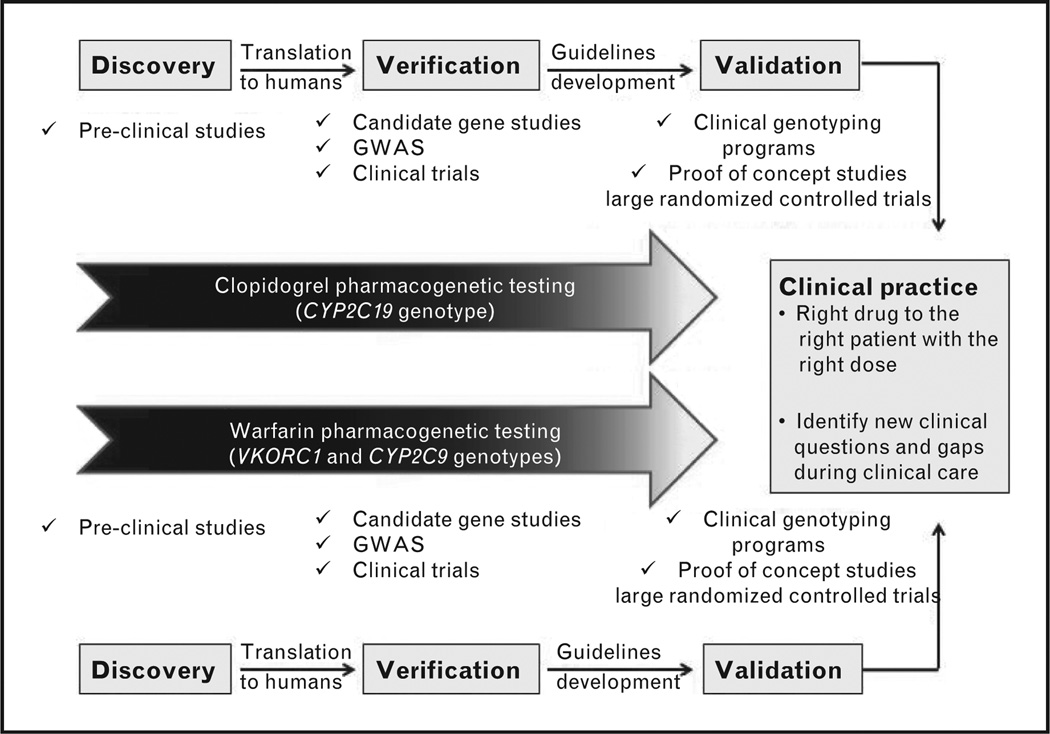
Pharmacogenetics holds the promise of giving the right drug to the right patient with the right dose. The rapid advances in pharmacogenetics have offered compelling evidence toward the individualization of clopidogrel and warfarin therapies (Fig. 3). Despite the concrete literature supporting the importance of pharmacogenetics in tailoring both therapies, several barriers, like turnaround time of genotyping, cost, and lack of large randomized clinical data to confirm beneficial outcomes of pharmacogenetic testing, still limit its clinical implementation. The rapid advances in genotyping and the recent POC devices hold promise to overcome the problem of turnaround time. Furthermore, the approach of preemptive genotyping might also be helpful in overcoming some of those barriers, as, in the future, it is envisioned that large amounts of genetic information will be generated on a person at some point in his life and then stored in his medical record for future use. This would facilitate the clinical implementation of clopidogrel and warfarin pharmacogenetics, where a delay in therapy initiation is not typically possible. Moreover, results from the ongoing large randomized trials will further define the role of clopidogrel and warfarin pharmacogenetics in clinical practice.
FIGURE 3.
Status of clopidogrel and warfarin pharmacogenetics: from identification toward clinical implementation. GWAS, genome-wide association studies.
Supplementary Material
KEY POINTS.
A large number of studies have shown important genetic markers significantly associate with the safety or the efficacy of clopidogrel and warfarin.
The FDA has updated clopidogrel and warfarin label with genetic information and guidelines have been established to assist clinicians in prescribing clopidogrel or warfarin dose based on genotype.
Despite the strong evidence for the clinical and analytical validity of clopidogrel and warfarin pharmacogenetic testing, however, its clinical utility is still limited.
Large randomized clinical trials are underway to define the role of clopidogrel and warfarin pharmacogenetics in clinical practice.
Acknowledgements
This work was supported in part by NIH grants U01 HL074492, RO1 NS073346 and UL1 TR000064.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (p. 371).
- 1.Roger VL, Go AS, Lloyd-Jones DM, et al. Heart disease and stroke statistics: 2012 update - a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global status report on noncommunicable diseases 2010. Geneva, Switzerland: WHO; 2010. [Google Scholar]
- 3.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28:1084–1097. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JA. Warfarin pharmacogenetics: a rising tide for its clinical value. Circulation. 2012;125:1964–1966. doi: 10.1161/CIRCULATIONAHA.112.100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JA. Warfarin: an old drug but still interesting. Pharmacotherapy. 2008;28:1081–1083. doi: 10.1592/phco.28.9.1081. [DOI] [PubMed] [Google Scholar]
- 7.Freedman JE, Hylek EM. Clopidogrel, genetics, and drug responsiveness. N Engl J Med. 2009;360:411–413. doi: 10.1056/NEJMe0810513. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 9.Khoury MJ. The case for a global human genome epidemiology initiative. Nat Genet. 2004;36:1027–1028. doi: 10.1038/ng1004-1027. [DOI] [PubMed] [Google Scholar]
- 10.Wei CY, Michael Lee MT, Chen YT. Pharmacogenomics of adverse drug reactions: implementing personalized medicine. Hum Mol Genet. 2012;21(R1):R58–R65. doi: 10.1093/hmg/dds341. [DOI] [PubMed] [Google Scholar]
- 11.Lesko LJ, Schmidt S. Individualization of drug therapy: history, present state, and opportunities for the future. Clin Pharmacol Ther. 2012;92:458–466. doi: 10.1038/clpt.2012.113. [DOI] [PubMed] [Google Scholar]
- 12.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JA, Cavallari LH, Beitelshees AL, et al. Pharmacogenomics: application to the management of cardiovascular disease. Clin Pharmacol Ther. 2011;90:519–531. doi: 10.1038/clpt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahin MH, Khalifa SI, Gong Y, et al. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21:130–135. doi: 10.1097/FPC.0b013e3283436b86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright RS, Anderson JL, Adams CD, et al. 2011 ACCF/AHA Focused Update of the Guidelines for the Management of Patients With Unstable Angina/Non-ST-Elevation Myocardial Infarction (Updating the 2007 Guideline): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;123:2022–2060. doi: 10.1161/CIR.0b013e31820f2f3e. [DOI] [PubMed] [Google Scholar]
- 20.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49:1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 21.Gurbel PA, Bliden KP, Guyer K, et al. Platelet reactivity in patients and recurrent events poststenting: results of the PREPARE POST-STENTING Study. J Am Coll Cardiol. 2005;46:1820–1826. doi: 10.1016/j.jacc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 22.Breet NJ, van Werkum JW, Bouman HJ, et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA. 2010;303:754–762. doi: 10.1001/jama.2010.181. [DOI] [PubMed] [Google Scholar]
- 23.Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 24.Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenet Genomics. 2010;20:463–465. doi: 10.1097/FPC.0b013e3283385420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caplain H, Donat F, Gaud C, Necciari J. Pharmacokinetics of clopidogrel. Semin Thromb Hemost. 1999;25(Suppl 2):25–28. [PubMed] [Google Scholar]
- 26.Lins R, Broekhuysen J, Necciari J, Deroubaix X. Pharmacokinetic profile of 14C–labeled clopidogrel. Semin Thromb Hemost. 1999;25(Suppl 2):29–33. [PubMed] [Google Scholar]
- 27.Savi P, Pereillo JM, Uzabiaga MF, et al. Identification and biological activity of the active metabolite of clopidogrel. Thromb Haemost. 2000;84:891–896. [PubMed] [Google Scholar]
- 28.Hollopeter G, Jantzen HM, Vincent D, et al. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. 2001;409:202–207. doi: 10.1038/35051599. [DOI] [PubMed] [Google Scholar]
- 29.Kazui M, Nishiya Y, Ishizuka T, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38:92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 30.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 32.Sofi F, Giusti B, Marcucci R, et al. Cytochrome P450 2C19*2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a metaanalysis. Pharmacogenomics J. 2011;11:199–206. doi: 10.1038/tpj.2010.21. [DOI] [PubMed] [Google Scholar]
- 33.Xie HG, Zou JJ, Hu ZY, et al. Individual variability in the disposition of and response to clopidogrel: pharmacogenomics and beyond. Pharmacol Ther. 2011;129:267–289. doi: 10.1016/j.pharmthera.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Gladding P, Webster M, Zeng I, et al. The pharmacogenetics and pharmacodynamics of clopidogrel response: an analysis from the PRINC (Plavix Response in Coronary Intervention) trial. JACC Cardiovasc Interv. 2008;1:620–627. doi: 10.1016/j.jcin.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Simon T, Bhatt DL, Bergougnan L, et al. Genetic polymorphisms and the impact of a higher clopidogrel dose regimen on active metabolite exposure and antiplatelet response in healthy subjects. Clin Pharmacol Ther. 2011;90:287–295. doi: 10.1038/clpt.2011.127. [DOI] [PubMed] [Google Scholar]
- 36.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 37.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 38.Sibbing D, Koch W, Gebhard D, et al. Cytochrome 2C1 9*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 39.Tiroch KA, Sibbing D, Koch W, et al. Protective effect of the CYP2C1 9*17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160:506–512. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 40.de Morais SM, Wilkinson GR, Blaisdell J, et al. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 41.De Morais SM, Wilkinson GR, Blaisdell J, et al. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 42.Xiao ZS, Goldstein JA, Xie HG, et al. Differences in the incidence of the CYP2C1 9 polymorphism affecting the S-mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther. 1997;281:604–609. [PubMed] [Google Scholar]
- 43.Ibeanu GC, Goldstein JA, Meyer U, et al. Identification of new human CYP2C1 9 alleles (CYP2C19*6 and CYP2C19*2B) in a Caucasian poor metabolizer of mephenytoin. J Pharmacol Exp Ther. 1998;286:1490–1495. [PubMed] [Google Scholar]
- 44. Scott SA, Sangkuhl K, Gardner EE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450–2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132.A comprehensive review on clopidogrel pharmacogenetics that provides guidelines to assist clinicians in prescribing clopidogrel based on genetic information
- 45.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51:1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 46.Giusti B, Gori AM, Marcucci R, et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am J Cardiol. 2009;103:806–811. doi: 10.1016/j.amjcard.2008.11.048. [DOI] [PubMed] [Google Scholar]
- 47.Zou JJ, Xie HG, Chen SL, et al. Influence of CYP2C19 loss-of-function variants on the antiplatelet effects and cardiovascular events in clopidogrel-treated Chinese patients undergoing percutaneous coronary intervention. Eur J Clin Pharmacol. 2012 doi: 10.1007/s00228-012-1392-5. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 48.Pare G, Mehta SR, Yusuf S, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 49.Sorich MJ, Vitry A, Ward MB, Prasugrel vs, et al. clopidogrel for cytochrome P450 2C19-genotyped subgroups: integration of the TRITON-TIMI 38 trial data. J Thromb Haemost. 2010;8:1678–1684. doi: 10.1111/j.1538-7836.2010.03923.x. [DOI] [PubMed] [Google Scholar]
- 50.Wallentin L, James S, Storey RF, et al. Effect of CYP2C1 9 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 51.Sibbing D, Stegherr J, Latz W, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J. 2009;30:916–922. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 52.Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto K, Hokimoto S, Chitose T, et al. Impact of CYP2C19 polymorphism on residual platelet reactivity in patients with coronary heart disease during antiplatelet therapy. J Cardiol. 2011;57:194–201. doi: 10.1016/j.jjcc.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Małek LA, Przyłuski J, Spiewak M, et al. Cytochrome P450 2C19 polymorphism, suboptimal reperfusion and all-cause mortality in patients with acute myocardial infarction. Cardiology. 2010;117:81–87. doi: 10.1159/000320093. [DOI] [PubMed] [Google Scholar]
- 55.Harmsze AM, van Werkum JW, Ten Berg JM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J. 2010;31:3046–3053. doi: 10.1093/eurheartj/ehq321. [DOI] [PubMed] [Google Scholar]
- 56.Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012;91:257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmes MV, Perel P, Shah T, et al. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306:2704–2714. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 58.Zabalza M, Subirana I, Sala J, et al. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012;98:100–108. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 59.Shuldiner AR, Vesely MR, Fisch A. CYP2C1 9 genotype and cardiovascular events. JAMA. 2012;307:1482. doi: 10.1001/jama.2012.443. author reply 1484–1485. [DOI] [PubMed] [Google Scholar]
- 60.Mega JL, Topol EJ, Sabatine MS. CYP2C1 9 genotype and cardiovascular events. JAMA. 2012;307:1482–1483. doi: 10.1001/jama.2012.444. author reply 1484–1485. [DOI] [PubMed] [Google Scholar]
- 61.Johnson JA, Roden DM, Lesko LJ, et al. Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012;91:774–776. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ten Berg JM, Deneer VH. Antiplatelet therapy: does CYP2C19 genotype affect clinical outcome? Nat Rev Cardiol. 2012;9:192–194. doi: 10.1038/nrcardio.2012.29. [DOI] [PubMed] [Google Scholar]
- 63.Fuster V, Sweeny JM. Clopidogrel and the reduced-function CYP2C19 genetic variant: a limited piece of the overall therapeutic puzzle. JAMA. 2010;304:1839–1840. doi: 10.1001/jama.2010.1566. [DOI] [PubMed] [Google Scholar]
- 64.Frere C, Cuisset T, Gaborit B, et al. The CYP2C19*17 allele is associated with better platelet response to clopidogrel in patients admitted for non-ST acute coronary syndrome. J Thromb Haemost. 2009;7:1409–1411. doi: 10.1111/j.1538-7836.2009.03500.x. [DOI] [PubMed] [Google Scholar]
- 65.Sibbing D, Gebhard D, Koch W, et al. Isolated and interactive impact of common CYP2C1 9 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost. 2010;8:1685–1693. doi: 10.1111/j.1538-7836.2010.03921.x. [DOI] [PubMed] [Google Scholar]
- 66.Pareé G, Mehta SR, Yusuf S, et al. Effects of CYP2C1 9 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 67.Tantry US, Bliden KP, Wei C, et al. First analysis of the relation between CYP2C19 genotype and pharmacodynamics in patients treated with ticagrelor versus clopidogrel: the ONSET/OFFSET and RESPOND genotype studies. Circ Cardiovasc Genet. 2010;3:556–566. doi: 10.1161/CIRCGENETICS.110.958561. [DOI] [PubMed] [Google Scholar]
- 68.Varenhorst C, James S, Erlinge D, et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J. 2009;30:1744–1752. doi: 10.1093/eurheartj/ehp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maeda A, Ando H, Asai T, et al. Differential impacts of CYP2C19 gene polymorphisms on the antiplatelet effects of clopidogrel and ticlopidine. Clin Pharmacol Ther. 2011;89:229–233. doi: 10.1038/clpt.2010.268. [DOI] [PubMed] [Google Scholar]
- 70.Kelly RP, Close SL, Farid NA, et al. Pharmacokinetics and pharmacodynamics following maintenance doses of prasugrel and clopidogrel in Chinese carriers of CYP2C19 variants. Br J Clin Pharmacol. 2012;73:93–105. doi: 10.1111/j.1365-2125.2011.04049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmes DR, Dehmer GJ, Kaul S, et al. ACCF/AHA clopidogrel clinical alert: approaches to the FDA ‘boxed warning’: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010;56:321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 72.Sibbing D, Bernlochner I, Kastrati A, et al. Current evidence for genetic testing in clopidogrel-treated patients undergoing coronary stenting. Circ Cardiovasc Interv. 2011;4:505–513. doi: 10.1161/CIRCINTERVENTIONS.111.962183. discussion 513. [DOI] [PubMed] [Google Scholar]
- 73.Pare G, Eikelboom JW, Sibbing D, et al. Testing should not be done in all patients treated with clopidogrel who are undergoing percutaneous coronary intervention. Circ Cardiovasc Interv. 2011;4:521. doi: 10.1161/CIRCINTERVENTIONS.111.962142. discussion 521. [DOI] [PubMed] [Google Scholar]
- 74. Mega JL, Hochholzer W, Frelinger AL, 3rd, et al. Dosing clopidogrel based on CYP2C1 9 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA. 2011;306:2221–2228. doi: 10.1001/jama.2011.1703.This study aimed to evaluate whether higher doses can overcome the resistance in those patients carrying the CYP2C19*2 LOF allele. They reported that a clopidogrel dose of 225 mg/day in CYP2C19*2 heterozygous carriers provides similar levels of platelet reactivity to those achieved by noncarriers taking 75 mg per day of clopidogrel
- 75.Price MJ, Berger PB, Teirstein PS, et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA. 2011;305:1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 76.Barker CM, Murray SS, Teirstein PS, et al. Pilot study of the antiplatelet effect of increased clopidogrel maintenance dosing and its relationship to CYP2C19 genotype in patients with high on-treatment reactivity. JACC Cardiovasc Interv. 2010;3:1001–1007. doi: 10.1016/j.jcin.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 77.Lieb W, Volzke H, Pulley JM, et al. Strategies for personalized medicine-based research and implementation in the clinical workflow. Clin Pharmacol Ther. 2012;92:443–445. doi: 10.1038/clpt.2012.119. [DOI] [PubMed] [Google Scholar]
- 78.O’Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson JA, Burkley BM, Langaee TY, et al. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012;92:437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schildcrout JS, Denny JC, Bowton E, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92:235–242. doi: 10.1038/clpt.2012.66.They examined the frequency with which 56 medications with known outcomes influenced by variant alleles were prescribed in a cohort of 52 942 medical home patients at Vanderbilt University Medical Center. They suggested a significant improvement in the safety and efficacy of six drugs, including clopidogrel and warfarin, based on effective preemptive genotyping programs
- 81.Crews KR, Hicks JK, Pui CH, et al. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther. 2012;92:467–475. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roberts JD, Wells GA, Le May MR, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379:1705–1711. doi: 10.1016/S0140-6736(12)60161-5. A prospective randomized proof-of-concept study that included 200 patients undergoing PCI. They used a novel point-of-care genetic test to identify carriers of the CYP2C19*2 allele and aimed to assess a pharmacogenetic approach to dual antiplatelet treatment after PCI. Higher risk of stent thrombosis and cardiovascular events was documented with the standard care compared with the rapid genotyping arm.
- 83.Barrett PM, Topol EJ. Pharmacogenetics: point-of-care genetic testing - a new frontier explored. Nat Rev Cardiol. 2012;9:315–316. doi: 10.1038/nrcardio.2012.63. [DOI] [PubMed] [Google Scholar]
- 84.Cavallari LH, Shin J, Perera MA. Role of pharmacogenomics in the management of traditional and novel oral anticoagulants. Pharmacotherapy. 2011;31:1192–1207. doi: 10.1592/phco.31.12.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ginsburg GS, Voora D. The long and winding road to warfarin pharmacogenetic testing. J Am Coll Cardiol. 2010;55:2813–2815. doi: 10.1016/j.jacc.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 86.Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med. 2007;147:755–765. doi: 10.7326/0003-4819-147-11-200712040-00006. [DOI] [PubMed] [Google Scholar]
- 87.Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007;167:1414–1419. doi: 10.1001/archinte.167.13.1414. [DOI] [PubMed] [Google Scholar]
- 88.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 89.Li C, Schwarz UI, Ritchie MD, et al. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009;113:3925–3930. doi: 10.1182/blood-2008-09-176859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 91.Jonas DE, McLeod HL. Genetic and clinical factors relating to warfarin dosing. Trends Pharmacol Sci. 2009;30:375–386. doi: 10.1016/j.tips.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 92.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 93.Choonara IA, Haynes BP, Cholerton S, et al. Enantiomers of warfarin and vitamin K1 metabolism. Br J Clin Pharmacol. 1986;22:729–732. doi: 10.1111/j.1365-2125.1986.tb02966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rettie AE, Korzekwa KR, Kunze KL, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5:54–59. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Z, Fasco MJ, Huang Z, et al. Human cytochromes P4501A1 and P4501A2: R-warfarin metabolism as a probe. Drug Metab Dispos. 1995;23:1339–1346. [PubMed] [Google Scholar]
- 96.Ngui JS, Chen Q, Shou M, et al. In vitro stimulation of warfarin metabolism by quinidine: increases in the formation of 4’- and 10-hydroxywarfarin. Drug Metab Dispos. 2001;29:877–886. [PubMed] [Google Scholar]
- 97.Wienkers LC, Wurden CJ, Storch E, et al. Formation of (R)-8-hydroxywarfarin in human liver microsomesA new metabolic marker for the (S)-mephenytoin hydroxylase, P4502C19. Drug Metab Dispos. 1996;24:610–614. [PubMed] [Google Scholar]
- 98.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 99.Li T, Chang CY, Jin DY, et al. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 100.Wajih N, Hutson SM, Owen J, Wallin R. Increased production of functional recombinant human clotting factor IX by baby hamster kidney cells engineered to overexpress VKORC1, the vitamin K 2,3-epoxide-reducing enzyme of the vitamin K cycle. J Biol Chem. 2005;280:31603–31607. doi: 10.1074/jbc.M505373200. [DOI] [PubMed] [Google Scholar]
- 101.Limdi NA, Beasley TM, Baird MF, et al. Kidney function influences warfarin responsiveness and hemorrhagic complications. J Am Soc Nephrol. 2009;20:912–921. doi: 10.1681/ASN.2008070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Limdi NA, Limdi MA, Cavallari L, et al. Warfarin dosing in patients with impaired kidney function. Am J Kidney Dis. 2010;56:823–831. doi: 10.1053/j.ajkd.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ansell J, Hirsh J, Hylek E, et al. Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. (8th) 2008;133:160S–198S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 104.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005;7:97–104. doi: 10.1097/01.gim.0000153664.65759.cf. [DOI] [PubMed] [Google Scholar]
- 107.Scordo MG, Pengo V, Spina E, et al. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther. 2002;72:702–710. doi: 10.1067/mcp.2002.129321. [DOI] [PubMed] [Google Scholar]
- 108.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 109.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7:99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 110.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 111.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 112.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 113.Caldwell MD, Berg RL, Zhang KQ, et al. Evaluation of genetic factors for warfarin dose prediction. Clin Med Res. 2007;5:8–16. doi: 10.3121/cmr.2007.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schalekamp T, Brasse BP, Roijers JF, et al. VKORC1 and CYP2C9 genotypes and acenocoumarol anticoagulation status: interaction between both genotypes affects overanticoagulation. Clin Pharmacol Ther. 2006;80:13–22. doi: 10.1016/j.clpt.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 115.Lee SC, Ng SS, Oldenburg J, et al. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther. 2006;79:197–205. doi: 10.1016/j.clpt.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 116.D’Andrea G, D’Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 117.Millican EA, Lenzini PA, Milligan PE, et al. Genetic-based dosing in orthopedic patients beginning warfarin therapy. Blood. 2007;110:1511–1515. doi: 10.1182/blood-2007-01-069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Voora D, Eby C, Linder MW, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005;93:700–705. doi: 10.1160/TH04-08-0542. [DOI] [PubMed] [Google Scholar]
- 119.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 121.Dickmann LJ, Rettie AE, Kneller MB, et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60:382–387. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 122.Allabi AC, Gala JL, Horsmans Y. CYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a Black Beninese population. Pharmacogenet Genomics. 2005;15:779–786. doi: 10.1097/01.fpc.0000174787.92861.91. [DOI] [PubMed] [Google Scholar]
- 123.Blaisdell J, Jorge-Nebert LF, Coulter S, et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–537. doi: 10.1097/01.fpc.0000114759.08559.51. [DOI] [PubMed] [Google Scholar]
- 124.Cavallari LH, Langaee TY, Momary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 125.Cavallari LH, Perera M, Wadelius M, et al. Association of the GGCX (CAA)1 6/17 repeat polymorphism with higher warfarin dose requirements in African Americans. Pharmacogenet Genomics. 2012;22:152–158. doi: 10.1097/FPC.0b013e32834f288f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perera MA, Gamazon E, Cavallari LH, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scott SA, Jaremko M, Lubitz SA, et al. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10:1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McDonald MG, Rieder MJ, Nakano M, et al. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol. 2009;75:1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Danese E, Montagnana M, Johnson JA, et al. Impact of the CYP4F2 p.V433M polymorphism on coumarin dose requirement: systematic review and metaanalysis. Clin Pharmacol Ther. 2012;92:746–756. doi: 10.1038/clpt.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liang R, Li L, Li C, et al. Impact of CYP2C9*3, VKORC1–1639, CYP4F2rs2108622 genetic polymorphism and clinical factors on warfarin maintenance dose in Han-Chinese patients. J Thromb Thrombolysis. 2012;34:120–125. doi: 10.1007/s11239-012-0725-7. [DOI] [PubMed] [Google Scholar]
- 132.Singh O, Sandanaraj E, Subramanian K, et al. Influence of CYP4F2 rs2108622 (V433M) on warfarin dose requirement in Asian patients. Drug Metab Pharmacokinet. 2011;26:130–136. doi: 10.2133/dmpk.dmpk-10-rg-080. [DOI] [PubMed] [Google Scholar]
- 133.Borgiani P, Ciccacci C, Forte V, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10:261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 134.Sagreiya H, Berube C, Wen A, et al. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenet Genomics. 2010;20:407–413. doi: 10.1097/FPC.0b013e328338bac2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pautas E, Moreau C, Gouin-Thibault I, et al. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 136.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kringen MK, Haug KB, Grimholt RM, et al. Genetic variation of VKORC1 and CYP4F2 genes related to warfarin maintenance dose in patients with myocardial infarction. J Biomed Biotechnol. 2011;2011:739751. doi: 10.1155/2011/739751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perini JA, Struchiner CJ, Silva-Assunção E, Suarez-Kurtz G. Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin Pharmacol Ther. 2010;87:417–420. doi: 10.1038/clpt.2009.307. [DOI] [PubMed] [Google Scholar]
- 139.Lubitz SA, Scott SA, Rothlauf EB, et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J Thromb Haemost. 2010;8:1018–1026. doi: 10.1111/j.1538-7836.2010.03792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.King CR, Deych E, Milligan P, et al. Gamma-glutamyl carboxylase and its influence on warfarin dose. Thromb Haemost. 2010;104:750–754. doi: 10.1160/TH09-11-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cha PC, Mushiroda T, Takahashi A, et al. High-resolution SNP and haplotype maps of the human gamma-glutamyl carboxylase gene (GGCX) and association study between polymorphisms in GGCX and the warfarin maintenance dose requirement of the Japanese population. J Hum Genet. 2007;52:856–864. doi: 10.1007/s10038-007-0183-9. [DOI] [PubMed] [Google Scholar]
- 142.Voora D, Koboldt DC, King CR, et al. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:445–451. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vecsler M, Loebstein R, Almog S, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 144.Finkelman BS, Gage BF, Johnson JA, et al. Genetic warfarin dosing: tables versus algorithms. J Am Coll Cardiol. 2011;57:612–618. doi: 10.1016/j.jacc.2010.08.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anti-coagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 146.Gong IY, Tirona RG, Schwarz UI, et al. Prospective evaluation of a pharmacogenetics-guided warfarin loading and maintenance dose regimen for initiation of therapy. Blood. 2011;118:3163–3171. doi: 10.1182/blood-2011-03-345173. [DOI] [PubMed] [Google Scholar]
- 147.Shin J, Cao D. Comparison of warfarin pharmacogenetic dosing algorithms in a racially diverse large cohort. Pharmacogenomics. 2011;12:125–134. doi: 10.2217/pgs.10.168. [DOI] [PubMed] [Google Scholar]
- 148.Hillman MA, Wilke RA, Yale SH, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res. 2005;3:137–145. doi: 10.3121/cmr.3.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lenzini PA, Grice GR, Milligan PE, et al. Laboratory clinical outcomes of pharmacogenetic vs clinical protocols for warfarin initiation in orthopedic patients. J Thromb Haemost. 2008;6:1655–1662. doi: 10.1111/j.1538-7836.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Johnson EG, Horne BD, Carlquist JF, Anderson JL. Genotype-based dosing algorithms for warfarin therapy: data review and recommendations. Mol Diagn Ther. 2011;15:255–264. doi: 10.1007/BF03256417. [DOI] [PubMed] [Google Scholar]
- 151.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 152.Lefevre F, Goodman SN, Piper MA. Pharmacogenetic testing for warfarin dosing still awaits validation. J Am Coll Cardiol. 2011;57:756. doi: 10.1016/j.jacc.2010.07.057. author reply 756–757. [DOI] [PubMed] [Google Scholar]
- 153. Anderson JL, Horne BD, Stevens SM, et al. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II) Circulation. 2012;125:1997–2005. doi: 10.1161/CIRCULATIONAHA.111.070920. Randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II). Results from this study provided insights into the promise of pharmacogenetics in controlling warfarin therapy
- 154.Motsinger-Reif AA, Wagner MJ. Clinical trial evidence of the promise of pharmacogenomics warfarin dosing algorithms. Pharmacogenomics. 2012;13:861–863. doi: 10.2217/pgs.12.65. [DOI] [PubMed] [Google Scholar]
- 155. Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. A comprehensive review on warfarin pharmacogenetics that provides guidelines to assist clinicians in prescribing warfarin dose based on genetic information
- 156.Cavallari LH, Perera MA. The future of warfarin pharmacogenetics in under-represented minority groups. Future Cardiol. 2012;8:563–576. doi: 10.2217/fca.12.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Limdi NA. Warfarin pharmacogenetics: challenges and opportunities for clinical translation. Front Pharmacol. 2012;3:183. doi: 10.3389/fphar.2012.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Howard R, Leathart JB, French DJ, et al. Genotyping for CYP2C9 and VKORC1 alleles by a novel point of care assay with HyBeacon® probes. Clin Chim Acta. 2011;412:2063–2069. doi: 10.1016/j.cca.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 159.van Schie RM, Wadelius MI, Kamali F, et al. Genotype-guided dosing of coumarin derivatives: the European pharmacogenetics of anticoagulant therapy (EU-PACT) trial design. Pharmacogenomics. 2009;10:1687–1695. doi: 10.2217/pgs.09.125. [DOI] [PubMed] [Google Scholar]
- 160.Poe BL, Haverstick DM, Landers JP. Warfarin genotyping in a single PCR reaction for microchip electrophoresis. Clin Chem. 2012;58:725–731. doi: 10.1373/clinchem.2011.180356. [DOI] [PubMed] [Google Scholar]
- 161.French B, Joo J, Geller NL, et al. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Do EJ, Lenzini P, Eby CS, et al. Genetics informatics trial (GIFT) of warfarin to prevent deep vein thrombosis (DVT): rationale and study design. Pharma-cogenomics J. 2012;12:417–424. doi: 10.1038/tpj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.