Abstract
Estimating moment to moment changes in blood oxygenation level dependent (BOLD) activation levels from functional magnetic resonance imaging (fMRI) data has applications for learned regulation of regional activation, brain state monitoring, and brain-machine interfaces. In each of these contexts, accurate estimation of the BOLD signal in as little time as possible is desired. This is a challenging problem due to the low signal-to-noise ratio of fMRI data. Previous methods for real-time fMRI analysis have either sacrificed the ability to compute moment to moment activation changes by averaging several acquisitions into a single activation estimate or have sacrificed accuracy by failing to account for prominent sources of noise in the fMRI signal. Here we present a new method for computing the amount of activation present in a single fMRI acquisition that separates moment to moment changes in the fMRI signal intensity attributable to neural sources from those due to noise, resulting in a feedback signal more reflective of neural activation. This method computes an incremental general linear model fit to the fMRI timeseries, which is used to calculate the expected signal intensity at each new acquisition. The difference between the measured intensity and the expected intensity is scaled by the variance of the estimator in order to transform this residual difference into a statistic. Both synthetic and real data were used to validate this method and compare it to the only other published real-time fMRI method.
1. Introduction
People can be taught to control their own neural activity when they are given feedback that provides information about ongoing neural activity (Rockstroh et al., 1990; Weiskopf et al., 2003). Initial neurofeedback experiments relied on electroenchephalography (EEG) to estimate neural activity, but subsequent experiments have employed functional magnetic resonance imaging (fMRI)-based neurofeedback of the blood oxygenation level dependent (BOLD) signal because its spatial specificity allows for feedback from specific brain regions known to be involved in particular mental operations or compromised in particular mental health disorders (Weiskopf et al., 2003; Yoo et al., 1999; Posse et al., 2003; deCharms et al., 2004, 2005; Caria et al., 2007).
Spatially specific neurofeedback has several important potential applications. It opens the possibility that patients with certain neurological diseases can be treated by learning to control activation in affected brain regions (deCharms et al., 2005). Also, healthy people could improve perceptual or cognitive abilities by learning to manipulate their brain state (Thompson et al., 2009). Brain-computer interfaces built around fMRI or related technologies such as functional near-infrared spectroscopy could be employed to enhance the capabilities of the human body, for example to allow locked-in (Birbaumer and Cohen, 2007) or minimally conscious (Owen and Coleman, 2008) patients to communicate.
Despite widespread interest, neurofeedback training based on fMRI has grown slowly in terms of number of publications, due at least partly to methodological challenges associated with data quality. Existing methods for real-time fMRI either do not compute moment to moment changes in activation (Cox et al., 1995; Yoo et al., 1999; Gembris et al., 2000), which is crucial in learning to control brain activation (Rockstroh et al., 1990), or provide a real-time neurofeedback signal (Goebel, 2001; deCharms et al., 2004) computed without accounting for the substantial noise corrupting fMRI data (Friston et al., 1994).
Here we present a new method for computing fMRI-based neurofeedback that separates moment to moment changes in the fMRI signal intensity attributable to neural sources from those due to the non-random fMRI noise, resulting in a feedback signal more reflective of neural activation. We accomplish this by computing at each time point an incremental general linear model (GLM) fit to the previously acquired timeseries. The GLM incorporates basis functions modeling both neural and nuisance signal contributions. As soon as a new measurement is available, the model fit is updated, and the expected fMRI signal intensity excluding neural signal components is removed from the acquired signal. The residual intensity is attributed to both neural and random noise. This residual is then scaled by the variance of the full model fit (including neural contributions) to derive an estimate of the strength of the neural signal at that timepoint. This value–in units of standard deviation from the expected baseline activation–serves as a neural activation estimate particular to that single measurement (fMRI volume). Activation estimates are computed and used independently for each measurement, thus moment to moment changes in activation reflect only fluctuations in the activation estimates themselves.
Applying an incremental GLM to fMRI data has previously been studied by several groups (Cox et al., 1995; Gembris et al., 2000; Bagarinao et al., 2003b). Our technique is novel because it 1) uses the real-time GLM parameter estimates to reconstruct an estimate of the nuisance signals from the most recently acquired measurement, which can then be removed, 2) scales activation estimates using the variance of the estimator to convert activation into units of standard deviation from baseline, 3) introduces new methods for combining activation estimates across voxels.
We validated an implementation of our novel method for moment to moment neurofeedback computation using both synthetic and real data. Synthetic data with signal and noise properties that can not be explicitly accounted for by our model were generated and used as a test set to examine the effects of non-ideal input to our algorithm. Also, real fMRI data from an experiment where subjects attempted self-regulation of regional brain activation were used to compare BOLD signal changes computed post-hoc with the neurofeedback signal computed online using our method.
2. Method
As is common practice (Friston et al., 1994; Smith, 2004), we model a measured fMRI voxel timeseries as a linear combination of basis functions:
| (1) |
where N is a set of nuisance bases, X is a set of bases representing the modeled hemodynamic response, and is a random vector where each element is drawn from a zero mean, stationary Gaussian noise process with unknown variance. The weights and represent the contribution of each basis function to the measured signal. In a standard fMRI analysis, the average contribution of each basis function over the entire timeseries would be estimated such that the mean squared error between the measured signal and the GLM reconstruction is minimized. However, in this work we aim to estimate the BOLD signal magnitude (purported neural activation convolved with the hemodynamic response) related to the task at each individual timepoint rather than the average BOLD response over a set of acquisitions.
2.1. Moment to moment neural signal estimation
At time t we estimate the BOLD (neural) signal component of the measured fMRI timecourse as
| (2) |
which is the measured signal at time t without the estimated contribution from nuisance signal. is the tth row of N, and is the estimate of the nuisance basis weights computed up to time t. Intuitively, can be though of as the expected signal intensity if there were no neural signal contribution, and therefore is the signal contributions from both neural sources and random noise. To compute we use an incremental linear least squares GLM fit (Gentleman, 1974) to all data thus far acquired, which provides an estimate of the basis weights and . This estimate is updated as each new fMRI volume is acquired.
Using this framework the projection of the fMRI timeseries onto any desired nuisance basis set can be accomplished in real-time, providing a flexible method for removal of unwanted signals. Although only is required to compute the neural contribution to the measured signal at time t, the full model fit (including ) is also updated incrementally so that signal variance associated with activation is accounted for during the model fit. Incremental estimation of the full model also facilitates voxel efficiency estimation via the variance of the model fit, as described below. Figure 1 is a pictorial description of these variables and computations.
Figure 1.
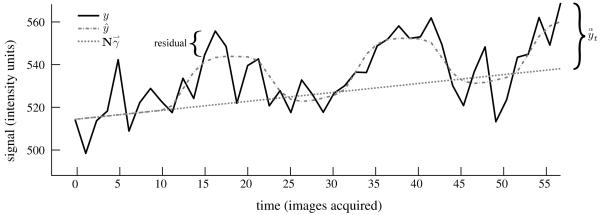
Schematic demonstrating the moment to moment activation estimation method. The activation estimate at time t is the difference between the data at t and the model reconstruction using only nuisance bases However, the full model fit is still computed 1) so that the residual error in the model fit can be used to estimate the efficiency of the voxel signal and 2) so that the timecourse variance associated with task-related neural signals is not attributed to drift, which would bias the model fit.
2.2. Regions of interest
To increase the signal-to-noise ratio (SNR, which here refers to the ratio between the magnitude of the signal changes of interest and the standard deviation of the signal changes of no interest) of the fMRI timeseries, neurofeedback is usually computed over a set of voxels comprising a brain region of interest (ROI; Weiskopf et al., 2003). This practice is based on the idea that the brain is composed of functional fields that perform distinct functions, which implies that nearby voxels will exhibit similar neural responses. Under ideal conditions, averaging data across ROI voxels reduces the white noise component while leaving the BOLD signal component unchanged. However, in practice the benefits of simple ROI averaging are compromised because the BOLD response strength varies across ROI voxels and fMRI noise is not spatially white due to physiological noise and head motion. Our method for moment-to-moment activation estimation provides a voxel-specific BOLD signal efficiency estimate, which can be used to weight activation estimates prior to ROI combination with the goal of more reliable ROI activation signals.
2.2.1. Voxel efficiency weighting
The quantity represents the amount of raw signal at time t that is attributable to either neural sources or random noise. Absolute signal change is represented in image intensity units, which have a relationship to activation strength that varies substantially among voxels. Most offline fMRI analyses transform intensities into percent signal change, which is more closely related to neural signal change. However, large signal changes due to random noise can result in large percent signal changes, and therefore deriving ROI signals via mean voxel percent signal change can result in ROI signals with large contributions from noise sources if voxel efficiency is not considered.
To overcome this issue, we center and scale activation based on a voxel-specific efficiency estimate computed from noise statistics. The incremental computation of activation described above yields an estimate of the activation present at each voxel at time t. Differences in the mean signal across voxels are implicitly accounted for by the model fit used to compute the activation. The efficiency of the neural signal is accounted for by scaling by the estimated variance of the noise (the residual of the full model fit to ). The centering and scaling results in a z-score
| (3) |
where σ1..t is the standard deviation of the full model fit given by (1) up to time t. The value zt indicates the number of standard deviations away from the expected value of the voxel signal at time t excluding neural signal contribution.
2.2.2. Approaches for ROI combination
In experiments utilizing moment to moment activation changes, a single activation measure representing the overall activation of a brain region is usually desired. To produce such a measure, zt must be combined across all the voxels in an ROI. Previous real-time fMRI methods (deCharms et al., 2004) perform ROI combination as the mean percent signal change across ROI voxels. However, direct averaging of percent signal change across ROI voxels can introduce an undesired noise weighting on the neurofeedback signal. Our activation scaling method provides several alternate choices for combining activation levels across the set of voxels in an ROI. We have explored three methods for ROI combination: mean, median, and weighted average. First, because all voxels are centered and scaled identically, direct averaging (mean combination) of zt is appropriate and does not suffer from the undesired noise weighting. However, the low SNR of fMRI data dictates a high likelihood of outlier activation levels within the ROI at any particular timepoint, and simple mean computation is sensitive to such outliers.
Computing the median of zt over voxels is one method for robustly combining ROI data. This method works well when ROI specification is accurate and the effect of interest is present over the entire ROI. However, if a substantial portion of the ROI voxels do not exhibit the effect of interest, the result of median computation can fail to reflect even a quite strong effect that happens to be present in less than half of ROI voxels. Such a situation can easily arise in practice due to differences in neural response between the localizer task for ROI delineation and the real-time experimental task when ROIs are functionally defined, variability in the relationship between brain anatomy and function when ROIs are anatomically defined, or unaccounted for subject head motion.
Combining ROI activations via voxel efficiency weighting can provide a more reliable signal than a simple mean computation while avoiding the sensitivity to ROI delineation accuracy of median computation. Additional efficiency weighting is imposed by computing a weighted average of zt over the ROI, weighting . This further reduces the contribution of voxels that are likely to be noisy without explicitly considering the voxel activations for outliers.
3. Validation
To characterize the accuracy of our neurofeedback computations we used synthetic fMRI timeseries to compare our incremental GLM fit to a post-hoc GLM fit. We also compared the only fMRI-based neurofeedback method published in sufficient detail to implement (deCharms et al., 2004) with our method by computing the ROI feedback signal for each on input data from an actual neurofeedback experiment and comparing this with activation levels from standard offline fMRI analysis.
3.1. Incremental versus post-hoc model fit
The incremental estimate of the GLM parameters and changes with each newly acquired image. Changes in the model parameters are associated with changes in the model reconstruction of the data, which changes the residual error at each timepoint. This dictates that the activation estimate will differ from a similar activation estimate at time t computed post-hoc using all acquired data. To measure the error in model reconstruction due to incremental computations we take the post-hoc GLM fit as truth data and use a Monte Carlo simulation to compare it to the incrementally computed fit to synthetically generated fMRI data. The goal of this characterization is to provide a qualitative sense of the difference in moment to moment BOLD estimates between the incremental and offline GLM approaches.
We generated several types of synthetic fMRI data for comparing incremental and post-hoc model fits. Each fMRI time-course was generated starting from a four-cycle, 1% signal change boxcar with 30 TR period beginning after 20 TRs and convolved with a two Gamma function estimate of a standard hemodynamic response (Friston and Ashburner, 1994; Boynton et al., 1996). Such timecourses are meant to resemble idealized data from neurofeedback studies like the one considered in the comparison between feedback methods below. To investigate behavior of the incremental computation, these idealized time-courses were corrupted with various types of noise:
Gaussian random noise
Gaussian noise and linear drift
Gaussian noise and non-linear drift derived from actual resting-state fMRI scans
Colored noise combined with non-linear drift derived from the resting scans
The fMRI signals resulting from cases 1 and 2 are designed to probe model differences when SNR is varied but the noise conforms to the assumptions of the GLM, which always included only 0th and 1st order drift terms here. Cases 3 and 4 add noise that was unable to be modeled given this particular GLM design matrix to investigate behavior under non-ideal conditions.
In case 1, SNR was varied from 0.25 to 4.0 by multiples of 2.0 by changing the variance of the Gaussian noise. For each SNR level 1000 fMRI timeseries were generated, each from a different random noise vector. Gaussian noise was generated in the same way for cases 2-4. The strength of the drift was manipulated by changing its percent signal change over the same range as the SNR.
Cases 3 and 4 rely on nonlinear drift and colored noise signals taken from an fMRI scan of a single subject at rest with eyes open. EPI parameters were: TE=30ms, TR=3000ms, matrix size=64×64, field of view=192×192mm2, 32 3.6mm thick slices, bandwidth=2298Hz/px, flip angle=90°, and 200 measurements. A set of gray matter voxels were identified by thresholding the first EPI volume using a threshold value obtained by manual adjustment and inspection. Six non-linear drift signals were extracted from six individual random gray matter voxel timecourses, which were low-pass filtered at 128s cutoff. An example drift timecourse is shown in the second panel (nuisance signal) of Figure 2.
Figure 2.
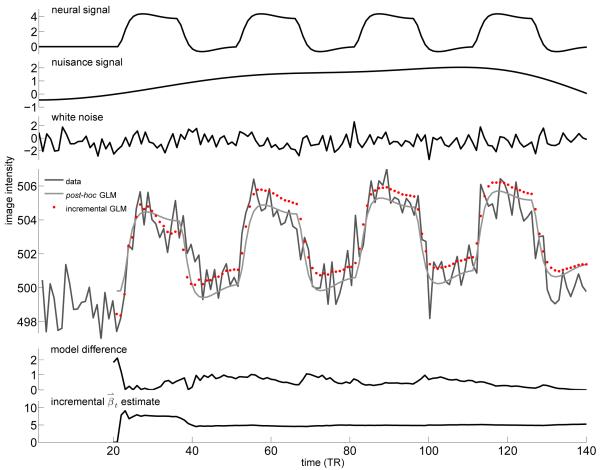
Example of synthetic fMRI timecourse generation. The top three panels show individual components of the resulting signal. The top panel shows the contribution to the final signal from hemodynamic response to activation (1% signal change). The second panel shows the contribution from scanner drift or physiological noise, measured from real resting fMRI scans (in this case 0.5% signal change). The third panel shows the Gaussian white noise vector generated for this example (SNR of 2). The large panel shows the resulting fMRI timecourse data (the sum of the first three panels and a constant 500 unit offset), the post-hoc GLM reconstruction of this timecourse, and the incremental GLM reconstruction from a fit performed only on data up to that time. Below this, the model difference panel shows the reconstruction error, which is the squared difference between the post-hoc and incremental reconstructions. The bottom panel shows the evolution of the model parameter estimate of the neural basis, which stabilizes near the correct value of 4 after the first stimulus block.
In case 4, realistic noise autocorrelations were added to synthetic timecourses by using actual timecourses from the resting scans. For each of the 1000 timecourses, a colored noise signal was generated by selecting a gray matter voxel at random, calculating the Fourier transform of the timecourse, randomizing the phase, and computing an inverse Fourier transform. These colored noise signals were combined with the convolved neural response and non-linear drift signals to derive a synthetic fMRI timecourse.
For each synthetic fMRI timeseries both an incremental GLM fit using our method and a post-hoc GLM fit using the regress function in MATLAB 7.5 (The Mathworks, Natick, MA) were performed. At each timepoint after the initial 20 volume window the difference between the model reconstruction of the signal computed using the post hoc and incremental GLMs was measured. For all 1000 timeseries in each noise type, the mean squared error (MSE) between the incremental and post hoc estimates was measured across time. The mean percent error for each noise type was then computed as mean over timeseries of the root MSE scaled by the mean signal value. Figure 2 shows a schematic demonstrating fMRI timecourse generation and post-hoc/incremental error computation.
We assessed the errors associated with computing a GLM fit incrementally by comparing the discrepancy in model reconstruction at each timepoint of synthetic fMRI timeseries using a Monte Carlo simulation. Figure 3 shows the effect of varying the SNR of the fMRI timeseries on the error associated with the incremental GLM fit. Average fit error is low for all SNRs (below 0.5%) and decreases sharply with increasing SNR. Figure 4 suggests that, when linear trends are modeled in the design matrix, the strength of linear trends present in the timeseries have little effect on incremental computation error. This implies that when the model bases span the signal space incremental computation is effective.
Figure 3.
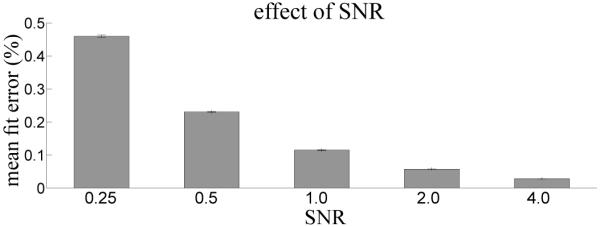
Effect of manipulating SNR of the synthetic fMRI timeseries on the error in model reconstruction between post-hoc and incremental GLM fits. Error bars indicate standard error over the 1000 synthetic timeseries.
Figure 4.
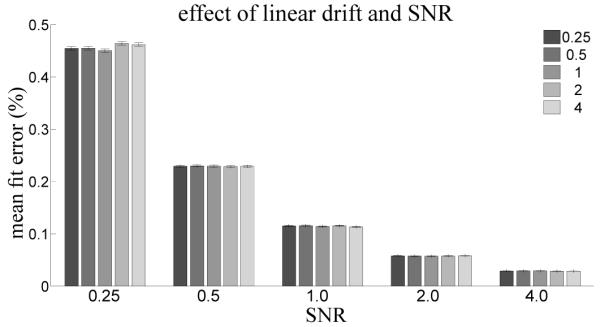
Effect of manipulating the strength of white noise and linear drift in the synthetic fMRI timeseries on the error in model reconstruction between post-hoc and incremental GLM fits. The strength of linear drift in % signal change is indicated by bar color, as shown in the legend. Error bars indicate standard error over the 1000 synthetic timeseries.
Figure 5 and Figure 6 explore scenarios where the model bases do not span the signal space. They demonstrate the degree of breakdown of incremental computation under non-ideal conditions. Figure 5 shows the average error between model fits when the signal is corrupted with unmodeled non-linear drift. It appears that there is an interaction between white noise strength and drift % signal change, with trend strength exhibiting a greater effect when there is less white noise. For each signal type the average error is below 1%, which is low.
Figure 5.
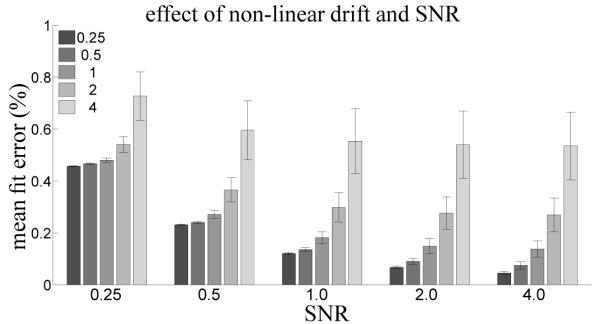
Effect of manipulating the strength of non-linear drift in the synthetic fMRI timeseries. Bar color indicates % signal change of the drift. Error bars indicate standard error over the mean error from each of the 6 different drift signals.
Figure 6.
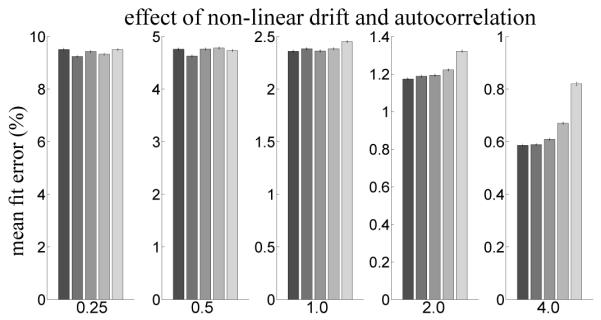
Effect on GLM reconstruction error of adding temporal autocorrelation to the synthetic fMRI data. The strength of non-linear drift is indicated by bar color using the same color and error bar scheme as in Figure 4 and Figure 5.
Figure 6 shows the average error between model fits when temporal correlation and non-linear drift are added to the signal. When white noise strength is low (SNRs 2 and 4), the model fit error is low (less than 1.5%) and drift strength affects the error. However, the figure indicates that as white noise strength increases, error increases rapidly and drift strength has no apparent effect on the degree of error, suggesting that autocorrelations dominate.
3.2. Feedback quality
We assessed the quality of real-time neurofeedback computed using our method and compared it to the neurofeedback computation method of deCharms et al. (2004) (henceforth referred to as the average percent signal change method; APSC) using data collected during a self-regulation experiment. The results of the self regulation experiment have been presented in abstract form (Thompson et al., 2009) and a manuscript is in preparation, but for completeness we briefly describe the methods used to collect this data.
3.2.1. Neurofeedback data collection
16 subjects underwent neurofeedback training to gain control of activation in cortical regions involved in auditory attention. All scans were performed on a Trio 3T MR system with a 32-channel head coil (Siemens Healthcare, Erlangen, Germany). To define an ROI from which to provide neurofeedback, each subject performed an initial functional localizer EPI scan while performing a behavioral task that required that they alternately attend to or ignore the acoustic noise produced by the EPI pulse sequence in 30s blocks separated by 16s of passive fixation. EPI parameters for this scan were: TE=30ms, TR=1500ms, matrix size=64×64, field of view=200×200mm2, 24 4.5mm thick slices, bandwidth=2298Hz/px, flip angle=90°, and 160 measurements. Online subject head motion compensation was accomplished using the Siemens PACE/MoCo system (Thesen et al., 2000).
During the scan the EPI volumes were transferred directly to an external computer via TCP/IP connection just after reconstruction. After the scan a standard fMRI analysis using FSL (http://www.fmrib.ox.ac.uk/fsl/)was conducted using these volumes to locate brain regions more activated when attending toward the noise than away from it. Three-dimensional spatial smoothing with a Gaussian kernel of 6.25mm3 and intensity normalization respecting differences in the mean intensity between timepoints were performed as preprocessing before performing a GLM fit of a design matrix made up of bases representing the condition blocks themselves and their temporal derivatives. A contrast attend toward minus attend away was computed and t-statistic maps were generated. These t-statistic maps were used to locate a cluster of activation in the posterior and superior portion of the left temporal lobe of each subject, which was taken as the ROI for neurofeedback. In addition, 9 subjects exhibited a cluster in the right hemisphere that was also included.
Neurofeedback took place over six functional runs, each having the same EPI parameters as the functional localizer except that scans lasted 210 measurements. Each scan consisted of five 45s blocks of either up or down regulation separated by 12.5s of passive fixation and the whole scan was preceded by 30s of initial fixation. During each block subjects were instructed to either increase or decrease their activation in response to the scanner acoustic noise using whatever strategy they found to be effective. They were told to judge effectiveness by observing the neurofeedback that was provided via a vertical thermometer display after the 30s initial fixation period. The first and the last scans served as pre- and post-training scans where no feedback was provided but subjects were instructed to attempt to control activation as best as they could.
Neurofeedback was computed using an implementation of our newly proposed method. EPI volumes were sent from the scanner via TCP/IP connection to an external analysis computer just after image reconstruction and motion correction (Siemens MoCo). Incoming images were included in the incremental GLM fit, then linear trend nuisance signals were discounted, and scaled activation estimates were computed for each voxel within a precomputed brain mask. Next, neurofeedback was computed by combining activation across ROI voxels using the weighted average method we describe here. The resulting activation estimate (in units of standard deviations from baseline) was immediately sent via TCP/IP connection to the computer that presented the stimulus to the subject, where the height of the thermometer was changed to reflect ROI activation.
3.2.3. Feedback quality comparison
To compare feedback quality between our method and previously proposed methods, we estimated the moment to moment activation over the auditory attention ROI incrementally using both our new method and APSC for comparison with a truth data neurofeedback signal computed post-hoc. To compute the truth data signal, we used the same data collected during the neurofeedback experiment, but used data from the entire run to determine activation deviations frombaseline, rather than only the data collected up to the neurofeedback timepoint.
Preprocessing was accomplished using the SPM5 software package (www.fil.ion.ucl.ac.uk/spm/software/spm5). Each functional run was motion corrected and high-pass filtered (cutoff 114s), then a GLM with a design matrix consisting only of subject head motion parameters was fit to each voxel timeseries. Next, timeseries from voxels within the ROI were extracted, and the GLM reconstruction of the nuisance signal was subtracted prior to conversion to percent signal change. The average percent signal change timecourse over the ROI was taken to be the truth data for the moment to moment activation. As discussed above, percent signal change is suboptimal for combining voxels, however using z-scores would have unfairly biased our comparison. This truth data was correlated with the moment to moment ROI activation level computed by our algorithm using mean, median, and weighted average ROI combination methods separately, as well as with the neurofeedback signal computed by a custom implementation of APSC.
The only difference between our implementation of APSC and that described in (deCharms et al., 2004) is that we compute feedback using only the main ROI and ignore the signal from the background ROI, which they subtract from the main ROI. This is appropriate as the truth data has been constructed to validate methods for combining voxels across a single ROI. Therefore, subtracting the background ROI signal would likely reduce the correlations with truth data and unfairly bias the comparison toward ourmethod, which only contains signal fromthe main ROI. In practice, ROI subtraction can be computed with either our method or APSC after ROI combination has been performed.
3.2.3. Feedback quality comparison
Figure 7 shows the results of comparing post-hoc, ground truth moment to moment activation estimates to those computed incrementally using our method and APSC. Strong correlations were observed for all neurofeedback methods. However, the feedback signal produced by the our method shows significantly higher correlation with the ground truth activation signal than APSC, regardless of ROI combination method (p < 0.0001 for each pairwise comparison with APSC). This demonstrates that explicitly modeling non-neural portions of the signal provides substantial improvement in feedback quality.
Figure 7.
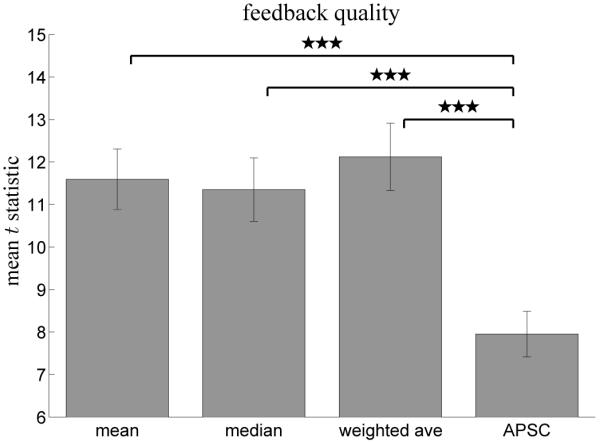
Comparison of ground truth neurofeedback with feedback computed using our method with either mean, median, or weighted average ROI combination as well as neurofeedback computed using APSC. The mean t-statistic shown here is the mean regression statistic over all subjects and functional runs of the self-regulation experiment. Methods were compared using a paired t-test, and all differences are significant to p < 0.0001. Error bars represent standard error from the mean.
These results also allow comparison of different ROI combination methods. Weighted average ROI combination was significantly higher than both mean and median feedback (p < 0.001), whereas mean and median feedback did not differ significantly. This suggests that the additional efficiency weighting imposed by weighted average ROI combination increases the quality of neurofeedback.
4. Discussion
We have developed a novel method for estimating moment to moment changes in BOLD signal using real-time analysis of fMRI data. Development of this algorithm was motivated by the need to account for the substantial contribution to the fMRI signal arising from non-neural sources. We validated an implementation of this method against traditional post-hoc analyses of both synthetic and real fMRI data to probe its characteristics in a range of practical contexts. In addition, we have compared the quality of neurofeedback provided by our algorithm against the only other published method for computing fMRI-based neurofeedback, demonstrating a significant improvement.
4.1. Previous methods for computing neurofeedback
Unfortunately, many neurofeedback fMRI experiments do not describe their methods in sufficient detail to compare them with the proposed method (Weiskopf et al., 2003; Bray et al., 2007; Caria et al., 2007; Pedersen et al., 2008; Johnston et al., 2009; Rota et al., 2009; Sitaram et al., 2009; Sorger et al., 2009). This section is devoted to examining those real-time fMRI methods that have been adequately described in published work.
The first method for real-time fMRI was presented by Cox et al. (1995), who were the first to apply the incremental GLM to fMRI data. This method and later optimizations for accuracy and speed (Bagarinao et al., 2003b,a, 2005) aim both to enable online assessment of scan quality and to increase the speed with which standard offline fMRI analyses could be performed. At any point in the scan, the result would be the same as if an offline analysis was computed on all the data collected up to that point. Real-time processing is applied to incoming fMRI data, however over time the meaning of real-time fMRI has evolved away from the meaning proposed in their work and toward the concept of computing the amount of BOLD signal present in a subset of recently acquired data during the scans.
Yoo and Jolesz (2002) provided the first demonstration that fMRI-based neurofeedback could be used to train subjects to modify their own activation patterns. They applied the method of (Cox et al., 1995) to 30s blocks of fMRI data, providing feed-back only after each block was collected and analyzed. This block-wise analysis is a simple version of a sliding-window neurofeedback computation proposed by Gembris et al. (2000); Posse et al. (2001) and used by Posse et al. (2003) in a neurofeedback study of amygdala activation due to self-induced sadness. In this sliding window neurofeedback computation, an incremental GLM is used to compute parameter estimates restricted to a short sliding window (22s in these studies) of data behind the current time. To accomplish this the data falling off the end of the window is discounted at the same time that incoming data is incorporated into the incremental GLM. Neurofeedback based on sliding window approaches has better temporal resolution than that based on approaches that rely on collection of all the data within a block before providing feedback, but worse than methods that compute feedback based only on the most recently acquired data.
Although Weiskopf et al. (2003) described the first study using neurofeedback reflecting activation localized to the most recently acquired measurement, the algorithms they used to compute neurofeedback were not adequately described. deCharms et al. (2004) presented a similar study where subjects were trained to control activation in motor cortex using neurofeedback computed using the APSC method described in Section 3.2. Our method can be seen as an improvement over APSC in that arbitrary nuisance signals can be removed from the feedback signal, the activation in each voxel is scaled by the estimator variance particular to that voxel, and that a more flexible ROI combination scheme has been introduced. We have provided quantitative demonstration that these improvements can lead to higher quality feedback.
A promising new approach to neurofeedback computation using multivariate statistics was introduced by LaConte et al. (2007). In their work a support vector machine (SVM) classification algorithm was applied to the volumes of an fMRI run that serve as a training set–conceptually similar to a functional localizer–to determine brain states which are later queried for similarity to the current brain state using the trained classifier. This similarity can be interpreted as neurofeedback not of BOLD signal itself but instead indicating the match with some template brain state represented by a target activation pattern. This approach has been applied in a self-regulation training experiment where smokers were asked to modulate their activation response to cues designed to induce nicotine craving (La Conte et al., 2009).
This approach is conceptually quite different from the traditional univariate approach where the BOLD activation levels of at most a small set of brain regions are considered in computing neurofeedback. However, the scheme that we introduce here for the removal of nuisance signals and scaling voxel activation estimates can be used as preprocessing steps for multivariate approaches.
4.2. Contributions
Unlike other methods for real-time fMRI analysis, our method allows explicit modeling and discounting of nuisance signals in fMRI data, which are numerous. There were significant improvements to the quality of neurofeedback when modeling subject head motion as a nuisance signal. Because the linear modeling framework allows addition of arbitrary signal bases, many additional nuisance signals beyond head motion could potentially be discounted. For example, outlier timepoints due to large amounts of brief equipment noise that are occasionally observed in fMRI can be explicitly accounted for by using outlier indicator regressors (Mazaika et al., 2007). Also, BOLD signal confounds such as respiration, heart-rate variation, and eye-movements, as well as physiological noise signals from white matter or cerebrospinal fluid can be accounted for in real-time, potentially increasing estimate quality.
The ability to account for arbitrary signal shapes in real-time makes new experiments possible. When measuring baseline BOLD fluctuations, the evoked response to stimuli can be removed prior to baseline estimation, allowing more rapid and robust estimation. Baseline BOLD measurements correlate with performance ability (Boly et al., 2008), so measuring them in real-time allows manipulation of task performance (Hinds et al., 2009; Yoo et al., 2009).
The addition of several new methods for ROI voxel combination can provide neurofeedback that is significantly more similar to that which would have been estimated offline than previously proposed methods. We hope that this increase in the efficiency of moment to moment activation computations will increase the likelihood success for future experiments that rely on neurofeedback.
4.3. Limitations
Although this method represents an advance in real-time fMRI analysis, it can not account for the many limitations of fMRI itself. As with any BOLD-based neuroimaging method, the accuracy of fMRI is dependent on the coupling between blood oxygenation and neural activity, which is not always reliable (Sirotin and Das, 2008). Also, GLM fits to fMRI data assume that the fMRI signal can be modeled as a linear combination of the model bases. This assumption is easily violated if the specified model does not include bases that can account for non-noise signal variation. In addition, the GLM fit assumes that noise in the signal is temporally uncorrelated, which is not the case for fMRI data (Friston et al., 1994). It is important to note that although we do not explicitly model non-linear drifts or perform temporal autocorrelation correction in our validations, these factors could be accounted for to increase the accuracy of activation estimation.
We have demonstrated that, on average, the incremental GLM fit is accurate based on the available data up to a given timepoint. However, when analyzing a given timeseries post-hoc the discrepancy between the incremental fit and offline fit can be substantial at early timepoints. Future work will characterize this effect and investigate methods for correcting potential errors in activation estimation. Similarly, the estimated mean squared error of the model fit varies over the course of a single acquisition. Because the computation of zt includes a scaling by the residual, the magnitude of zt will be affected. One method for alleviating this issue is to build up an estimate of the residual at the beginning of the timeseries, then fix this estimate so that zt is scaled in the same way for the remainder of the timeseries. We have found through experience that estimating the residual of the model fit over the first 20 timepoints provides an effective scaling factor, however more or fewer measurements could be required based on the particular experimental conditions.
In this work we have shown that the proposed method can provide estimates of moment to moment BOLD signal change that are more faithful to post hoc estimates than those computed using previously proposed methods. While it is highly likely that more accurate neurofeedback leads directly to more effective training of activation control, we have not tested this fact here. Comparison of the effectiveness of neurofeedback training with various feedback accuracy levels is left as future work.
4.4. Implementation
The moment to moment activation algorithm described here was implemented in C++ as part of a system for performing real-time neuroimaging data analysis. For the incremental GLM fit to the fMRI timeseries we employ a custom C++ translation of the Algol60 implementation of (Gentleman, 1974). Direct TCP/IP communication between the scanner and our software was accomplished with the help of Siemens Medical Solutions. Using our implementation on a standard desktop computer, incremental GLM fitting, nuisance signal removal, activation scaling, and weighted average ROI combination was measured to take 10ms for an ROI consisting of 364 voxels (1600mm3). This was measured from the time that the reconstructed EPI volume was received from the scanner, and thus does not include time for collection or reconstruction of the EPI data.
This method will be made publicly available as part of a real-time functional imaging software package. Our hope is that a published, validated, and freely available neurofeedback system will increase the number of neurofeedback studies and advance understanding in the field.
5. Acknowledgments
The authors would like to thank the Athinoula A. Martinos Imaging Center, McGovern Institute for Brain Research at MIT for supporting this research, and Michael Hamm from Siemens Healthcare for assistance with implementing the the custom data sender used in our real-time fMRI analysis system.
References
- Bagarinao E, Matsuo K, Nakai T. Real-time functional MRI using a PC cluster. Concepts in Magnetic Resonance Part B: Magnetic Resonance Engineering. 2003a;19(1):14–25. [Google Scholar]
- Bagarinao E, Matsuo K, Nakai T, Sato S. Estimation of general linear model coefficients for real-time application. Neuroimage. 2003b;19(2):422–429. doi: 10.1016/s1053-8119(03)00081-8. [DOI] [PubMed] [Google Scholar]
- Bagarinao E, Matsuo K, Tanaka Y, Sarmenta L, Nakai T. Enabling on-demand real-time functional MRI analysis using grid technology. Methods of Information in Medicine. 2005;44(5):665. [PubMed] [Google Scholar]
- Birbaumer N, Cohen L. Brain-computer interfaces: communication and restoration of movement in paralysis. The Journal of Physiology. 2007;579(3):621. doi: 10.1113/jphysiol.2006.125633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly M, Phillips C, Balteau E, Schnakers C, Degueldre C, Moonen G, Luxen A, Peigneux P, Faymonville M, Maquet P, et al. Consciousness and cerebral baseline activity fluctuations. Hum Brain Mapp. 2008;29:868–874. doi: 10.1002/hbm.20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton G, Engel S, Glover G, Heeger D. Linear systems analysis of functional magnetic resonance imaging in human V1. Journal of Neuroscience. 1996;16(13):4207. doi: 10.1523/JNEUROSCI.16-13-04207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Shimojo S, O’Doherty J. Direct instrumental conditioning of neural activity using functional magnetic resonance imaging-derived reward feedback. Journal of Neuroscience. 2007;27(28):7498. doi: 10.1523/JNEUROSCI.2118-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caria A, Veit R, Sitaram R, Lotze M, Weiskopf N, Grodd W, Birbaumer N. Regulation of anterior insular cortex activity using real-time fMRI. Neuroimage. 2007;35(3):1238–1246. doi: 10.1016/j.neuroimage.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Cox R, Jesmanowicz A, Hyde J. Real-time functional magnetic resonance imaging. Magnetic Resonance in Medicine. 1995;33(2):230–236. doi: 10.1002/mrm.1910330213. [DOI] [PubMed] [Google Scholar]
- deCharms R, Christoff K, Glover G, Pauly J, Whitfield S, Gabrieli J. Learned regulation of spatially localized brain activation using real-time fMRI. Neuroimage. 2004;21(1):436–443. doi: 10.1016/j.neuroimage.2003.08.041. [DOI] [PubMed] [Google Scholar]
- deCharms R, Maeda F, Glover G, Ludlow D, Pauly J, Soneji D, Gabrieli J, Mackey S. Control over brain activation and pain learned by using real-time functional MRI. Proceedings of the National Academy of Sciences. 2005;102(51):18626–18631. doi: 10.1073/pnas.0505210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K, Ashburner J. Statistical parametric mapping. Functional neuroimaging: Technical foundations. 1994;8:79–93. [Google Scholar]
- Friston K, Jezzard P, Turner R. Analysis of functional MRI time-series. Human Brain Mapping. 1994;1(2):153–171. [Google Scholar]
- Gembris D, Taylor J, Schor S, Frings W, Suter D, Posse S. Functional magnetic resonance imaging in real time (FIRE): Sliding-window correlation analysis and reference-vector optimization. Magnetic Resonance in Medicine. 2000;43:259–268. doi: 10.1002/(sici)1522-2594(200002)43:2<259::aid-mrm13>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Gentleman WM. Algorithm as 75: Basic procedures for large, sparse or weighted linear least problems. Applied Statistics. 1974;23(3):448–454. [Google Scholar]
- Goebel R. Cortex-based real-time fMRI. NeuroImage [Abstract, Organization for Human Brain Mapping] 2001;S13(6):129. [Google Scholar]
- Hinds O, Thompson TW, Gabrieli S, Triantafyllou C, Gabrieli J. Behavioral task enhancement by brain state monitoring using real-time fmri [Abstract] Soc Neurocience Abst. 2009 Sun 11:15AM, room N426. [Google Scholar]
- Johnston S, Boehm S, Healy D, Goebel R, Linden D. Neurofeedback: a promising tool for the self-regulation of emotion networks. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.07.056. [DOI] [PubMed] [Google Scholar]
- LaConte S, King-Casas B, Cinciripini P, Eagleman D, Versace F, Chiu P. Modulating rt-fMRI neurofeedback interfaces via craving and control in chronic smokers. NeuroImage, [Abstract] Organization for Human Brain Mapping. 2009 [Google Scholar]
- LaConte S, Peltier S, Hu X. Real-time fMRI using brain-state classification. Human Brain Mapping. 2007;28(10):1033–1044. doi: 10.1002/hbm.20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaika P, Whitfield-Gabrieli S, Reiss A, Glover G. Artifact repair for fMRI data from high motion clinical subjects. NeuroImage [Abstract, Organization for Human Brain Mapping] 2007 [Google Scholar]
- Owen A, Coleman M. Functional neuroimaging of the vegetative state. Nature Reviews Neuroscience. 2008;9(3):235–243. doi: 10.1038/nrn2330. [DOI] [PubMed] [Google Scholar]
- Pedersen A, Bauer J, Koelkebeck K, Kugel H, Rothermundt M, Suslow T, Arolt V, Ohrmann P. Implicit sequence learning in schizophrenia investigated with functional MRI. Schizophrenia Research. 2008;102:91–92. [Google Scholar]
- Posse S, Binkofski F, Schneider F, Gembris D, Frings W, Habel U, Salloum J, Mathiak K, Wiese S, Kiselev V, et al. A new approach to measure single-event related brain activity using real-time fMRI: feasibility of sensory, motor, and higher cognitive tasks. Human brain mapping. 2001;12(1):25–41. doi: 10.1002/1097-0193(200101)12:1<25::AID-HBM30>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse S, Fitzgerald D, Gao K, Habel U, Rosenberg D, Moore G, Schneider F. Real-time fMRI of temporolimbic regions detects amygdala activation during single-trial self-induced sadness. Neuroimage. 2003;18(3):760–768. doi: 10.1016/s1053-8119(03)00004-1. [DOI] [PubMed] [Google Scholar]
- Rockstroh B, Elbert T, Birbaumer N, Lutzenberger W. Biofeedback-produced hemispheric asymmetry of slow cortical potentials and its behavioural effects. International Journal of Psychophysiology. 1990;9(2):151–65. doi: 10.1016/0167-8760(90)90069-p. [DOI] [PubMed] [Google Scholar]
- Rota G, Gentili C, Sitaram R, Veit R, Guazzelli M, Birbaumer N, Dogil G. The default-mode network during learning of volitional control over regional brain activity. NeuroImage, [Abstract], Organization for Human Brain Mapping. 2009 [Google Scholar]
- Sirotin Y, Das A. Anticipatory haemodynamic signals in sensory cortex not predicted by local neuronal activity. Nature. 2008;457(7228):475–479. doi: 10.1038/nature07664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram R, Caria A, Birbaumer N. Hemodynamic brain-computer interfaces for communication and rehabilitation. Neural Networks. 2009;22:1320–8. doi: 10.1016/j.neunet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Smith S. Overview of fMRI analysis. British Journal of Radiology. 2004;77(Special Issue 2):S167. doi: 10.1259/bjr/33553595. [DOI] [PubMed] [Google Scholar]
- Sorger B, Dahmen B, Reithler J, Gosseries O, Maudoux A, Laureys S, Goebel R. Another kind of [] BOLD Response’: answering multiple-choice questions via online decoded single-trial brain signals. Progress in Brain Research. 2009:275–292. doi: 10.1016/S0079-6123(09)17719-1. [DOI] [PubMed] [Google Scholar]
- Thesen S, Heid O, Mueller E, Schad L. Prospective acquisition correction for head motion with image-based tracking for real-time fMRI. Magnetic Resonance in Medicine. 2000;44(3):457–465. doi: 10.1002/1522-2594(200009)44:3<457::aid-mrm17>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Thompson TW, Hinds O, Ghosh S, Lala N, Triantafyllou C, Whitfield-Gabrieli S, Gabrieli JD. Training selective auditory attention with real-time fMRI feedback. NeuroImage [Abstract, Organization for Human Brain Mapping] 2009 [Google Scholar]
- Weiskopf N, Veit R, Erb M, Mathiak K, Grodd W, Goebel R, Birbaumer N. Physiological self-regulation of regional brain activity using real-time functional magnetic resonance imaging (fMRI): methodology and exemplary data. Neuroimage. 2003;19(3):577–586. doi: 10.1016/s1053-8119(03)00145-9. [DOI] [PubMed] [Google Scholar]
- Yoo JJ, Hinds O, Thompson TW, Gabrieli S, Triantafyllou C, Gabrieli J. Enhancing memory acquisition using real-time fmri [Abstract] Soc Neurocience Abst. 2009 Sun 10:45AM, room N426. [Google Scholar]
- Yoo S, Guttmann C, Zhao L, Panych L. Real-time adaptive functional MRI. Neuroimage. 1999;10(5):596–606. doi: 10.1006/nimg.1999.0494. [DOI] [PubMed] [Google Scholar]
- Yoo S, Jolesz F. Functional MRI for neurofeedback: feasibility studyon a hand motor task. Neuroreport. 2002;13(11):1377. doi: 10.1097/00001756-200208070-00005. [DOI] [PubMed] [Google Scholar]