Abstract
Hispanic women in the USA have lower breast cancer incidence than non-Hispanic white (NHW) women. Genetic factors may contribute to this difference. Breast cancer genome-wide association studies (GWAS) conducted in women of European or Asian descent have identified multiple risk variants. We tested the association between 10 previously reported single nucleotide polymorphisms (SNPs) and risk of breast cancer in a sample of 4697 Hispanic and 3077 NHW women recruited as part of three population-based case–control studies of breast cancer. We used stratified logistic regression analyses to compare the associations with different genetic variants in NHWs and Hispanics classified by their proportion of Indigenous American (IA) ancestry. Five of 10 SNPs were statistically significantly associated with breast cancer risk. Three of the five significant variants (rs17157903-RELN, rs7696175-TLR1 and rs13387042-2q35) were associated with risk among Hispanics but not in NHWs. The odds ratio (OR) for the heterozygous at 2q35 was 0.75 [95% confidence interval (CI) = 0.50–1.15] for low IA ancestry and 1.38 (95% CI = 1.04–1.82) for high IA ancestry (P interaction 0.02). The ORs for association at RELN were 0.87 (95% CI = 0.59–1.29) and 1.69 (95% CI = 1.04–2.73), respectively (P interaction 0.03). At the TLR1 locus, the ORs for women homozygous for the rare allele were 0.74 (95% CI = 0.42–1.31) and 1.73 (95% CI = 1.19–2.52) (P interaction 0.03). Our results suggest that the proportion of IA ancestry modifies the magnitude and direction of the association of 3 of the 10 previously reported variants. Genetic ancestry should be considered when assessing risk in women of mixed descent and in studies designed to discover causal mutations.
Introduction
Hispanic women in the United States (US) have a lower incidence of breast cancer compared with non-Hispanic white (NHW) women (1). In 2002–2006, age-adjusted breast cancer incidence rates in US NHWs and African Americans were 123.5 and 113.0 per 100000, respectively, compared with 90.2 per 100000 in US Hispanic women (1). Hispanics are a genetically admixed population with European, Indigenous American (IA) and African descent. We have shown previously that higher European ancestry among Hispanics in the USA and Mexico is associated with increased breast cancer risk (2–4), and we have mapped at least one locus that may explain this difference (5). The observed disparity in cancer incidence may also be the result of differences in reproductive and lifestyle factors, such as number of full-term pregnancies, alcohol consumption and menopausal hormone therapy use (2,3,6–8).
Genome-wide association studies (GWAS) of breast cancer conducted in women of European or Asian descent have identified multiple risk-associated variants (9–9). Some of these associations have been independently replicated in studies that included Hispanics (20) or African Americans (21,22). However, the previously published replication study in Hispanics did not evaluate the interaction between risk-associated variants and genetic ancestry, which is of great interest given the known heterogeneity in genetic ancestry among Hispanics (23–34).
We investigated the association between 10 previously reported and confirmed genetic variants and risk of breast cancer in a combined sample of 7774 women (3077 NHW women and 4697 Hispanics from the USA and Mexico) that were pooled for the Breast Cancer Health Disparities Study. We compared the direction and strength of the associations with the different genetic variants in NHWs and in Hispanics classified according to three levels of IA ancestry.
Methods
Study subjects
The Breast Cancer Health Disparities Study (4) utilized DNA samples and data from two population-based case–control studies conducted in the USA: the 4-Corners Breast Cancer Study (4-CBCS) (35) and the San Francisco Bay Area Breast Cancer Study (6,36); and a population-based multicenter case–control study conducted in Mexico (37). Our analyses included 3077 NHWs from the USA and 4697 women of Hispanic and Indigenous origin living in the USA or Mexico. The study was approved by the Institutional Review Board for Human Subjects at each institution. All participants signed a written informed consent.
4-Corners Breast Cancer Study.
Details about the 4-CBCS have been published previously (35). Briefly, this study recruited women residing in non-reservation areas in the states of Arizona, Colorado, New Mexico and Utah. Participants were NHW, Hispanic or IA women between 25 and 79 years of age with histological confirmed diagnosis of in situ or invasive breast cancer living in these areas at the time of diagnosis or selection between October 1999 and May 2004 (35). Population-based controls were matched to cases based on ethnicity and 5 year age distribution. Participant information was collected in English or Spanish by trained interviewers using a structured questionnaire (38–40). The present analysis included 1839 cases (603 Hispanics, 1236 NHWs) and 2059 controls (730 Hispanics, 1329 NHWs) with complete genotype data.
Mexico Breast Cancer Study.
This population-based case–control study of breast cancer included Mexican women aged 35–69 years, who resided in Mexico for at least 5 years. Details of the study have been published previously (37). Briefly, newly diagnosed cases were identified at 12 hospitals from the major healthcare systems in Mexico. The study included women with a histological confirmed new diagnosis of breast cancer (invasive or in situ) between 2004 and 2007. Controls were selected based on a probabilistic multistage sampling design that took into account the hospital’s catchment area. Data collection included the administration of a structured questionnaire at the participant’s home and collection of anthropometric measurements and a blood sample at the hospital. Part of the participant information was obtained with a questionnaire adapted from the one used in the 4-CBCS study. The present analyses included 812 Mexican cases and 989 controls with complete genotype data.
San Francisco Breast Cancer Study.
Details about this population-based case–control study have been described elsewhere (6,36). Briefly, participating women aged 35–79 years resided in the San Francisco Bay Area when diagnosed with a first primary histologically confirmed invasive breast cancer between April 1999 and April 2002. Controls identified by random-digit dialing were frequency-matched to cases based on race/ethnicity and the expected 5 year age distribution of cases. Trained interviewers administered a structured questionnaire in English or Spanish and took anthropometric measurements. The present study included 943 cases (692 Hispanics and 251 NHWs) and 1132 controls (871 Hispanics and 261 NHWs) with complete genotype data.
Genetic data
Genotyping was conducted as part of the Breast Cancer Health Disparities Study aimed at evaluating the association between genetic variants in genes related to inflammation, hormones and energetic factors and risk of breast cancer in Hispanic and NHW women (4). In addition, GWAS-identified single nucleotide polymorphisms (SNPs) associated with breast cancer risk in other populations (11,12,15,16,18) (published before the time during which the Breast Cancer Health Disparities Study SNP genotyping panel was designed) were genotyped for this analysis. Specifically, we genotyped rs13387042 in the 2q35 region (G/A), rs17157903 in 7q22 (C/T) within the RELN gene, rs2067980 in 5q11 (A/G) near the MRPS30 gene, rs2180341 in 6q22.1–q22.33 (A/G) within the RNF146 gene, rs2981582 in 10q26 (C/T) within the FGFR2 gene, rs3803662 in 16q12.1 (C/T) within the TOX3 gene, rs3817198 in 11p15.5 (T/C) within the LSP1 gene, rs7696175 in 4p14 (C/T) near the TLR1 gene, rs889312 in 5q11.2 (A/C) near the MAP3K1 gene and rs999737 in 14q23–q24.2 (C/T) within the RAD51L1 gene (Supplementary Material Table S1, available at Carcinogenesis Online). Additionally, 104 ancestry-informative markers were genotyped. Details about these ancestry-informative markers have been published previously (4). All markers were genotyped using a multiplexed bead array assay based on GoldenGate chemistry (Illumina, San Diego, CA) attaining a genotyping call rate of 99%.
Statistical methods
As we have described previously (4), individual genetic ancestry was estimated using the program STRUCTURE (41) and a two-source population model, which provided higher levels of repeatability and correlation among runs compared with the three-founding populations model (4). Estimates of IA ancestry were used as a continuous variable and also as a categorical three-level variable. For the latter, study participants were classified into one of three ancestry categories by level of percent IA ancestry using arbitrary cut-points based on the distribution of ancestry affiliation in the total population (low IA ancestry: ≤28%, intermediate IA ancestry: 29–70% and high IA ancestry: ≥71%) and the necessary minimum number of individuals per category to achieve sufficient statistical power, as we have reported previously (4).
Differences between cases and controls or between women in the different ancestry categories, by age at diagnosis/recruitment, menopausal status and IA genetic ancestry, were tested using Student’s t-tests (age and genetic ancestry) or Fisher’s exact test (menopausal status).
SNP association and interaction with ancestry analyses.
The association between genotypes and breast cancer risk was evaluated using logistic regression models adjusted for genetic ancestry (as a continuous variable), age at diagnosis or interview (5 year categories) and study (4-CBCS, Mexico Breast Cancer Study and San Francisco Breast Cancer Study). We also conducted stratified analyses to investigate the association of these genetic variants within each of the three ancestry categories (low IA ancestry: ≤28%, intermediate IA ancestry: 29–70% and high IA ancestry: ≥71%). We considered a statistically significant replication of a previously reported association to be any result with a P-value ≤ 0.05. In order to reduce the number of comparisons made, we only evaluated the heterogeneity of associations in Hispanics by ancestry category for those SNPs that showed significant associations in the replication analysis. For this latter analysis, we used logistic regression models that included an interaction term between genotypes and the three IA ancestry categories. When testing for interactions, we took a false discovery rate approach to define what results were considered statistically significant: We tested five hypotheses, which had their corresponding P-values (from the global test for the interaction term): P 1, P 2, …, P 5. If P (1) ≤ P (2) ≤ … ≤ P (5) are the ordered P-values and k is the largest i for which P(i) ≤ i/m × q [where m is equal to the total number of P-values (five in this analysis) and q is the specified false discovery rate (0.05)]; all i = 1, 2, …, k are considered statistically significant at a false discovery rate of 5% (42,43). We also evaluated whether the interaction with genetic ancestry was observed when ancestry was defined as a continuous variable (and therefore without the arbitrary cut-points). Finally, we tested the association between the cumulative number of at-risk alleles in NHW and Hispanics in the three ancestry categories. To create this variable, we defined the subgroup-specific risk allele based on the stratified association results. For example, for the low IA ancestry groups, the risk allele for rs7696175 is the opposite allele compared with that for the intermediate and high categories. All analyses were performed using STATA 11 (44). For all the genetic analyses that involved a distinction between NHW and Hispanics, we excluded 121 NHW women that had more than 10% IA ancestry.
Results
The study included 7774 participants from three case–control studies (3594 cases and 4180 controls). Table I shows genetic ancestry characteristics of the study participants. Among Hispanics, average IA genetic ancestry was lower among cases compared with controls in the San Francisco Breast Cancer Study and Mexico Breast Cancer Study. As expected, IA ancestry was equally low (~4%) among women who self-reported their ethnicity as NHW. Among women who self-reported being Hispanic, the proportion of postmenopausal women in the three studies was similar (~60%) and slightly smaller than the proportion in NHWs (~70%).
Table I.
Subject characteristics by study, ethnicity and IA ancestry
| Study | Hispanic | NHWs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | P a | Low IA ancestry | Intermediate IA ancestry | High IA ancestry | P a | Controls | Cases | P a | |
| 4-CBCS | ||||||||||
| n | 730 | 603 | 232 | 1046 | 55 | 1329 | 1236 | |||
| Age at diagnosis, mean (SD) | 54 (12) | 53 (11) | 0.04 | 54 (12) | 53 (12) | 53 (11) | 0.64 | 56 (12) | 56 (11) | 0.13 |
| IA ancestry, mean (SD) | 0.41 (0.17) | 0.42 (0.17) | 0.17 | 0.17 (0.09) | 0.45 (0.10) | 0.83 (0.10) | 0.04 (0.04) | 0.04 (0.06) | 0.18 | |
| Postmenopausal (%) | 65 | 61 | 0.11 | 61 | 64 | 55 | 0.34 | 69 | 65 | 0.08 |
| San Francisco Breast Cancer Study | ||||||||||
| n | 871 | 692 | 285 | 1134 | 144 | 261 | 251 | |||
| Age at diagnosis, mean (SD) | 53 (11) | 54 (11) | 0.03 | 54 (11) | 54 (11) | 52 (11) | 0.04 | 59 (13) | 58 (12) | 0.55 |
| IA ancestry, mean (SD) | 0.48 (0.20) | 0.42 (0.20) | <0.01 | 0.16 (0.08) | 0.48 (0.10) | 0.81 (0.09) | 0.04 (0.04) | 0.05 (0.06) | 0.32 | |
| Postmenopausal (%) | 59 | 60 | 0.59 | 64 | 59 | 52 | 0.09 | 67 | 71 | 0.38 |
| Mexico Breast Cancer Study | ||||||||||
| n | 989 | 812 | 37 | 892 | 872 | N/A | N/A | |||
| Age at diagnosis, mean (SD) | 50 (9) | 52 (10) | 0.01 | 52 (9) | 51 (10) | 51 (9) | 0.67 | |||
| IA ancestry, mean (SD) | 0.72 (0.18) | 0.67 (0.19) | <0.01 | 0.20 (0.07) | 0.56 (0.10) | 0.85 (0.10) | ||||
| Postmenopausal (%) | 56 | 58 | 0.38 | 59 | 57 | 56 | 0.92 | |||
N/A, not available.
a P-value of t-test for age at diagnosis/interview and ancestry and Fisher exact test for % postmenopausal.
The genotype analysis included a total of 7653 women (2956 NHW and 4697 Hispanic). Of the 10 SNPs, 2 were associated with breast cancer risk in both Hispanics and NHWs (rs2981582 in FGFR2 and rs3803662 in TOX3) (Table II). The odds ratio (OR) estimates for the variants in the FGFR2 and TOX3 genes were homogeneous across populations and ancestry groups and the magnitude of associations among all women combined was similar to those previously reported [FGFR2 OR for rare allele heterozygous: 1.17 (95% CI = 1.06–1.30) and TOX3 OR: 1.24 (95% CI = 1.12–1.36)] (Table II). However, the Mantel–Haenzel test of homogeneity between studies was statistically significant for the TOX3 variant (OR for trend test in 4-Corners Breast Cancer Study 1.01, Mexico Breast Cancer Study 1.08 and San Francisco Breast Cancer Study 1.72, P = 0.0008). Three other variants were not statistically significantly associated in NHWs, but were associated among Hispanics, with heterogeneous associations across the three genetic ancestry categories and no statistically significant heterogeneity observed between studies (Table II). Specifically, rs13387042 (2q35) was positively associated among all Hispanic women combined (P trend < 0.01) and among those with intermediate (P trend < 0.01) or high (P trend < 0.01) IA ancestry, but no association was observed among Hispanic women with low IA ancestry (P trend = 0.45, test of heterogeneity P = 0.02). Similarly, rs7696175 (4p14, TLR1) was positively associated among all Hispanic women combined (P trend < 0.01) and among those with intermediate (P trend < 0.01) or high (P trend < 0.01) IA ancestry, but not among Hispanic women of low IA ancestry or among NHW women (test of heterogeneity P = 0.03). The rs17157903 (7q22, RELN) variant was inversely associated with breast cancer risk among all Hispanic women combined (P trend = 0.02) and among Hispanic women of low IA ancestry (P trend = 0.04), but positively associated among Hispanic women with high IA ancestry (test of heterogeneity P = 0.03) (Table II). The test of interaction between this SNP and IA ancestry was statistically significant at a false discovery rate of 5%. Interaction tests that defined genetic ancestry as a continuous variable were consistent with those observed for the categorical variable (Table II). Given that the magnitude and direction of the associations for Hispanic women with low IA ancestry and NHW women were generally similar, we tested the associations by ancestry categories pooling NHWs with Hispanics of low IA. Results were similar to those for NHW women only (Supplementary Table S2, available at Carcinogenesis Online). Results of analyses stratifying by menopausal status were not statistically significant after adjustment for multiple comparisons (Supplementary Table S3, available at Carcinogenesis Online).
Table II.
Association between 10 previously reported breast cancer risk variants by global genetic ancestry in US NHWs, US Hispanics and Mexicans
| SNP | NHW (n = 2956a) | Hispanics (n = 4697) | P interactionb | P homogeneity, M–H test by studyc | Hispanics (n = 4697) | P interactionb | All women (n = 7653) | ||
|---|---|---|---|---|---|---|---|---|---|
| Low IA ancestry (n = 554) | Intermediate IA ancestry (n = 3072) | High IA ancestry (n = 1071) | |||||||
| rs13387042 (2q35) | |||||||||
| MAF | 0.53 | 0.33 | 0.50 | 0.36 | 0.18 | 0.41 | |||
| GA | 1.06 (0.88–1.28) | 1.18 (1.04–1.34) | 0.75 (0.50–1.15) | 1.19 (1.02–1.39) | 1.38 (1.04–1.82) | 1.15 (1.04–1.28) | |||
| AA | 1.16 (0.94–1.42) | 1.49 (1.23–1.80) | 1.20 (0.75–1.94) | 1.37 (1.10–1.72) | 2.64 (1.38–5.05) | 1.34 (1.17–1.53) | |||
| P for trend | 0.17 | <0.01 | 0.15 | 0.74 | 0.45 | <0.01 | <0.01 | 0.02 | <0.01 |
| P for trend ancestry continuous | 0.04 d | ||||||||
| rs17157903 (7q22) RELN | |||||||||
| MAF | 0.13 | 0.09 | 0.15 | 0.10 | 0.04 | 0.11 | |||
| CT | 1.06 (0.89–1.27) | 0.93 (0.80–1.09) | 0.87 (0.59–1.29) | 0.86 (0.72–1.04) | 1.69 (1.04–2.73) | 1.00 (0.89–1.12) | |||
| TT | 1.33 (0.77–2.28) | 0.44 (0.23–0.87) | 0.20 (0.04–0.96) | 0.57 (0.27–1.24) | N/A | 0.86 (0.58–1.29) | |||
| P for trend | 0.30 | 0.02 | 0.04 | 0.30 | 0.04 | 0.16 | 0.03 | 0.47 | |
| P for trend ancestry continuous | 0.31d | ||||||||
| rs2067980 (5q11) MRPS30 | |||||||||
| MAF | 0.15 | 0.16 | 0.15 | 0.16 | 0.18 | 0.16 | |||
| AG | 1.17 (0.99–1.38) | 1.00 (0.87–1.13) | 1.14 (0.79–1.68) | 0.91 (0.77–1.07) | 1.16 (0.88–1.53) | 1.07 (0.97–1.19) | |||
| GG | 1.33 (0.81–2.18) | 0.87 (0.60–1.26) | 0.89 (0.18–4.30) | 0.92 (0.57–1.48) | 0.79 (0.41–1.51) | 1.00 (0.74–1.33) | |||
| P for trend | 0.26 | 0.45 | 0.57 | 0.88 | 0.72 | 0.48 | 0.98 | ||
| rs2180341 (6q22) RNF146 | |||||||||
| MAF | 0.24 | 0.23 | 0.26 | 0.23 | 0.21 | 0.24 | |||
| AG | 0.97 (0.83–1.13) | 0.96 (0.85–1.08) | 0.98 (0.68–1.41) | 0.93 (0.80–1.08) | 1.05 (0.81–1.37) | 0.96 (0.87–1.06) | |||
| GG | 1.17 (0.85–1.62) | 1.02 (0.79–1.32) | 0.90 (0.47–1.75) | 1.04 (0.75–1.43) | 1.07 (0.58–2.00) | 1.08 (0.88–1.32) | |||
| P for trend | 0.34 | 0.88 | 0.84 | 0.76 | 0.82 | 0.82 | 0.46 | ||
| rs2981582 (10q26) FGFR2 | |||||||||
| MAF | 0.42 | 0.42 | 0.44 | 0.42 | 0.41 | 0.42 | |||
| CT | 1.16 (0.98–1.36) | 1.19 (1.05–1.36) | 1.33 (0.90–1.98) | 1.22 (1.04–1.43) | 1.07 (0.81–1.41) | 1.17 (1.06–1.30) | |||
| TT | 1.43 (1.15–1.77) | 1.49 (1.26–1.77) | 1.78 (1.08–2.91) | 1.51 (1.23–1.86) | 1.27 (0.88–1.83) | 1.48 (1.29–1.68) | |||
| P for trend | <0.01 | <0.01 | 0.94 | 0.16 | 0.02 | <0.01 | 0.2 | 0.79 | <0.01 |
| P for trend ancestry continuous | 0.58 | ||||||||
| rs3803662 (16q12) TOX3 | |||||||||
| MAF | 0.29 | 0.41 | 0.35 | 0.40 | 0.46 | 0.36 | |||
| CT | 1.15 (0.99–1.34) | 1.27 (1.12–1.45) | 1.12 (0.77–1.62) | 1.27 (1.08–1.49) | 1.41 (1.05–1.89) | 1.24 (1.12–1.36) | |||
| TT | 1.54 (1.16–2.04) | 1.25 (1.05–1.49) | 1.67 (0.96–2.90) | 1.21 (0.98–1.50) | 1.32 (0.92–1.88) | 1.30 (1.12–1.50) | |||
| P for trend | <0.01 | 0.01 | 0.25 | 0.0008 | 0.07 | 0.08 | 0.13 | 0.57 | <0.01 |
| P for trend ancestry continuous | 0.30 | ||||||||
| rs3817198 (11p15) LSP1 | |||||||||
| MAF | 0.34 | 0.20 | 0.27 | 0.21 | 0.13 | 0.25 | |||
| TC | 1.10 (0.94–1.28) | 1.02 (0.90–1.16) | 0.96 (0.67–1.38) | 1.01 (0.87–1.18) | 1.07 (0.80–1.44) | 1.06 (0.96–1.17) | |||
| CC | 0.98 (0.77–1.25) | 1.20 (0.90–1.60) | 0.93 (0.46–1.85) | 1.24 (0.89–1.74) | 1.35 (0.44–4.14) | 1.03 (0.86–1.24) | |||
| P for trend | 0.89 | 0.22 | 0.76 | 0.83 | 0.20 | 0.60 | 0.72 | ||
| rs7696175 (4p14) TLR1 | |||||||||
| MAF | 0.44 | 0.38 | 0.38 | 0.37 | 0.40 | 0.40 | |||
| CT | 0.98 (0.82–1.15) | 1.14 (1.00–1.29) | 1.19 (0.82–1.72) | 1.19 (1.01–1.38) | 0.98 (0.74–1.29) | 1.06 (0.96–1.17) | |||
| TT | 0.83 (0.68–1.03) | 1.36 (1.14–1.63) | 0.74 (0.42–1.31) | 1.39 (1.11–1.74) | 1.73 (1.19–2.52) | 1.06 (0.93–1.22) | |||
| P for trend | 0.09 | <0.01 | <0.01 | 0.30 | 0.31 | <0.01 | <0.01 | 0.03 | 0.37 |
| P for trend ancestry continuous | 0.005 d | ||||||||
| rs889312 (5q11) MAP3K1 | |||||||||
| MAF | 0.29 | 0.43 | 0.34 | 0.42 | 0.50 | 0.38 | |||
| AC | 1.04 (0.89–1.21) | 1.04 (0.91–1.19) | 1.35 (0.94–1.95) | 1.01 (0.86–1.19) | 0.95 (0.71–1.28) | 1.04 (0.94–1.15) | |||
| CC | 1.03 (0.79–1.34) | 1.09 (0.92–1.29) | 1.48 (0.84–2.60) | 1.07 (0.87–1.32) | 0.93 (0.66–1.31) | 1.07 (0.93–1.23) | |||
| P for trend | 0.82 | 0.30 | 0.80 | 0.18 | 0.50 | 0.68 | 0.33 | ||
| rs999737 (14q23) RAD51L1 | |||||||||
| MAF | 0.23 | 0.17 | 0.21 | 0.17 | 0.15 | 0.19 | |||
| CT | 0.94 (0.80–1.10) | 1.01 (0.88–1.15) | 1.00 (0.69–1.43) | 0.94 (0.80–1.11) | 1.20 (0.90–1.59) | 0.97 (0.88–1.07) | |||
| TT | 0.84 (0.60–1.17) | 1.17 (0.83–1.66) | 0.76 (0.31–1.89) | 1.20 (0.78–1.84) | 1.61 (0.70–3.71) | 0.97 (0.76–1.22) | |||
| P for trend | 0.30 | 0.38 | 0.24 | 0.56 | 0.40 | 0.27 | 0.77 | ||
All estimates with an associated P-value ≤ 0.05 are in bold.
aNHW women with more than 10% IA ancestry were excluded from the genetic analysis, n = 121.
bGlobal test for interaction, 2 and 4 degrees of freedom according to number of categories compared.
c P-value of Mantel–Haenzel test of homogeneity between studies.
dThese are the P-values for a global test for the interaction term with ancestry defined as a continuous variable. OR for the interaction term: for rs13387042, the OR for heterozygous was 2.02 (95% CI = 1.13–3.60); for rs17157903, the OR for heterozygous was 1.74 (95% CI = 0.79–3.81); for rs7696175, the OR for rare allele homozygous was 2.82 (95% CI = 1.27–6.28).
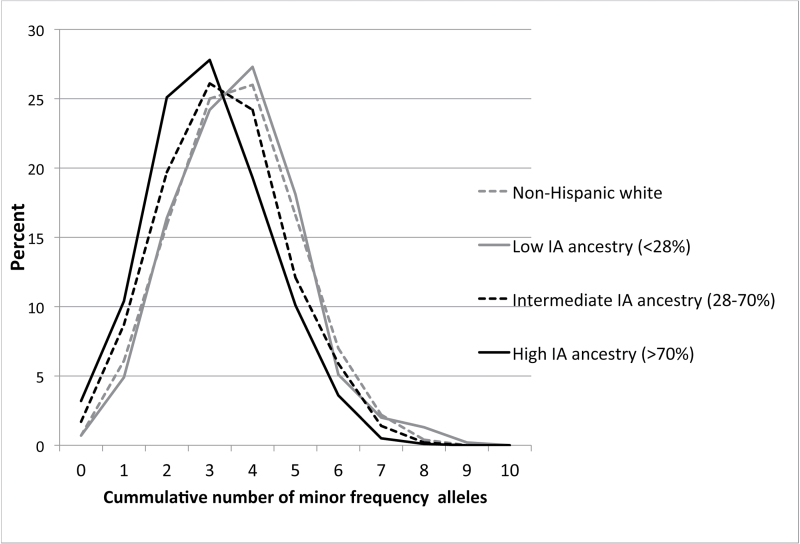
Finally, we compared the cumulative effect of risk-associated alleles between NHW and Hispanic women in the three ancestry categories. To make these analyses more precise, we only included the five replicated SNPs. As expected, we observed increased odds of breast cancer among women who carried a larger number of risk-associated variants compared with those with fewer risk variants (Table III). The association appeared to be stronger among women who had either intermediate or high IA ancestry compared with women with low IA ancestry (Table III). However, it should be noted that associations for the self-reported Hispanic low IA ancestry group were imprecise given few women were in this category. The distribution of the cumulative number of risk alleles varied by ancestry group (Figure 1), and therefore, in the case of women with high IA ancestry, there were not enough individuals with a large number of risk alleles (>6) to adequately estimate the magnitude of the association at the upper level of at-risk alleles. As we did for the genotype analyses, we also tested the associations by ancestry categories combining NHW women with Hispanics of low IA ancestry; results for the low IA category of this latter analysis were similar to those previously obtained for the NHW and low IA self-reported Hispanics, but achieved higher statistical significance due to the larger sample size (Supplementary Table S4, available at Carcinogenesis Online).
Table III.
Association between cumulative number of risk alleles from five replicated GWAS hits and breast cancer risk in US NHWs, US Hispanics and Mexicans, by genetic ancestry
| Number of rare allelesa | Frequency (%) | ORb | 95% CI | P | |
|---|---|---|---|---|---|
| NHW (n = 2956c) | 0 | 22 (0.7) | |||
| 1 | 179 (6.1) | ||||
| 2 | 470 (15.9) | Ref. (≤2) | |||
| 3 | 739 (25.0) | 1.47 | 0.86–2.50 | 0.16 | |
| 4 | 769 (26.0) | 1.64 | 1.00–2.69 | 0.05 | |
| 5 | 491 (16.6) | 1.72 | 1.06–2.79 | 0.03 | |
| 6 | 208 (7.0) | 1.93 | 1.18–3.13 | <0.01 | |
| 7 | 65 (2.2) | 1.95 | 1.18–3.22 | <0.01 | |
| 8 | 13 (0.4) | 2.46 | 1.44–4.22 | <0.01 | |
| 9 | 1 (0.0) | ||||
| 10 | 0 | ||||
| P trend | <0.01 | ||||
| Low IA ancestry (n = 554) | 0 | 4 (0.7) | |||
| 1 | 27 (4.9) | ||||
| 2 | 91 (16.4) | Ref. (≤2) | |||
| 3 | 134 (24.2) | 1.27 | 0.18–9.15 | 0.81 | |
| 4 | 151 (27.3) | 1.44 | 0.22–9.47 | 0.71 | |
| 5 | 100 (18.1) | 1.53 | 0.23–10.09 | 0.66 | |
| 6 | 28 (5.1) | 1.85 | 0.28–12.18 | 0.52 | |
| 7 | 11 (2.0) | 2.52 | 0.38–16.69 | 0.34 | |
| 8 | 7 (1.3) | 3.01 | 0.43–21.12 | 0.27 | |
| 9 | 1 (0.2) | ||||
| 10 | 0 | ||||
| P trend | 0.11 | ||||
| Intermediate IA ancestry (n = 3072) | 0 | 53 (1.7) | |||
| 1 | 267 (8.7) | ||||
| 2 | 606 (19.7) | Ref. (≤2) | |||
| 3 | 801 (26.1) | 1.59 | 1.03–2.46 | 0.04 | |
| 4 | 744 (24.2) | 1.89 | 1.25–2.85 | <0.01 | |
| 5 | 371 (12.1) | 2.01 | 1.34–3.02 | <0.01 | |
| 6 | 182 (5.9) | 2.32 | 1.54–3.51 | <0.01 | |
| 7 | 43 (1.4) | 3.22 | 2.04–5.09 | <0.01 | |
| 8 | 5 (0.2) | 3.55 | 2.11–5.97 | <0.01 | |
| 9 | 0 | ||||
| 10 | 0 | ||||
| P trend | <0.01 | ||||
| High IA ancestry (n = 1071) | 0 | 34 (3.2) | |||
| 1 | 111 (10.4) | ||||
| 2 | 269 (25.1) | Ref. (≤2) | |||
| 3 | 298 (27.8) | 1.37 | 1.00–1.87 | 0.05 | |
| 4 | 207 (19.3) | 1.38 | 0.97–1.95 | 0.07 | |
| 5 | 108 (10.1) | 1.89 | 1.22–2.92 | <0.01 | |
| 6 | 38 (3.6) | 2.43 | 1.23–4.81 | 0.01 | |
| 7 | 5 (0.5) | 7.11 | 0.78–64.86 | 0.08 | |
| 8 | 1 (0.1) | ||||
| 9 | 0 | ||||
| 10 | 0 | ||||
| P trend | 0.09 |
ars13387042, rs17157903, rs2981582, rs3803662 and rs7696175.
bThe allele that is considered the risk allele differs by group, depending on the OR of the SNP association for that group: rs7696175 is inverted for the NHWs and for rs17157903, the rare allele is the risk allele in the IA group.
cNHW women with more than 10% IA ancestry were excluded from the genetic analysis (n = 121).
Fig. 1.
Distribution of cumulative number of minor frequency alleles among NHWs (gray solid line) and Hispanics with low (gray dotted line), intermediate (black dotted line) and high (black solid line) levels of IA genetic ancestry.
Discussion
We evaluated the association between breast cancer case–control status and 10 SNPs previously reported to be associated with the disease, in a large sample of US NHW, US Hispanic and Mexican women. Five of the SNPs replicated among Hispanics (SNPs on or near genes: FGFR2, TOX3, TLR1, RELN and one in region 2q35) and two of those five replicated in NHWs (SNPs on FGFR2 and TOX3). We also found that among Hispanics, the SNPs within TLR1, RELN and the 2q35 region showed evidence of heterogeneity by level of IA ancestry. In general, associations for Hispanic women with low IA genetic ancestry were similar in magnitude and direction to those in NHW women. For SNPs within the 2q35 and TLR1 regions, stronger associations were observed among women with high IA ancestry compared with those with low IA ancestry. For the SNP within the RELN region, we observed a more complicated pattern: an inverse association in Hispanic women, but when stratifying into three ancestry categories, the inverse association was limited to women with low IA ancestry, whereas a positive association was found in women with high IA ancestry. When we considered the cumulative effect of multiple risk alleles, overall, the associations were stronger among women with intermediate to high levels of IA ancestry and similar in Hispanic women with low IA ancestry and NHW women. To our knowledge, this is the first study with sufficient variation in IA ancestry to evaluate the associations between breast cancer risk and GWAS-identified risk alleles considering heterogeneity by genetic ancestry.
Only 2 out of 10 SNPs replicated in NHW women despite the fact that the original GWAS were conducted in women of European ancestry. A possible reason might be lack of power in our study, since most GWAS in Europeans included larger sample sizes to compensate for adjustment for multiple comparisons. However, some of the SNPs that we analyzed have been reported to show ORs of 1.2 and our study was powered to detect associations of that magnitude. Another possibility is heterogeneity among the different studies in terms of the proportion of women included with a family history of breast cancer (18), premenopausal versus postmenopausal status (12) and tumor hormone receptor status (45,46). If these and other demographic and lifestyle factors are important effect modifiers of reported SNP associations, the ability to replicate associations in different studies could be compromised. It should be noted that most studies have identified different SNPs as important predictors of risk, highlighting the difficulty in identifying a common set of SNPs that might be useful to predict individual risk at the population level.
One possible explanation for the observed heterogeneity of the association between GWAS-identified SNPs and breast cancer risk by genetic ancestry is that estimates of genetic ancestry might be acting as a proxy for non-genetic risk factors that we did not consider in our models. If this were the case, then the observed heterogeneity might be reflecting gene by environment interactions. Studies done to date among women of European descent do not seem to support this possibility (47,48), but the environmental exposures may be different in Hispanic populations and future research among Hispanics is required to evaluate this explanation further. Alternatively, the heterogeneity we found may be due to variation in the linkage disequilibrium (LD) patterns among Hispanics of different genetic ancestry. The majority of the SNPs discovered through GWAS were identified in European or Asian populations and have not been confirmed as risk alleles among Hispanic women. In particular, some of the reported SNPs are in intergenic regions, such as is the case of the SNP on 2q35. Their association with breast cancer risk could be due to LD between that variant and the true causal locus. Different LD patterns in populations with different genetic background could influence our ability to detect these associations. This could lead to heterogeneous associations because LD might be tighter in one group compared with another, or the two alleles could be differently linked with the causal variant in the two groups (49). This suggests that additional genotyping and/or sequencing within the three regions that showed evidence of heterogeneity may help to identify the causal variants using an approach such as the ancestry-shift refinement mapping (49). Finally, it is also possible that the differences in the magnitude and direction of the observed associations are due to real heterogeneity in the effect of risk variants between different populations because of differential genetic and epigenetic interactions that influence susceptibility. Regardless of the interpretation, our results suggest that common variants associated with breast cancer risk in Europeans or Asians might have different effect sizes in other population groups. Moreover, our analyses suggest that among admixed populations, such as Hispanics, consideration of ancestry proportions might be relevant to understand the true associations between risk variants and disease risk.
Even though this combined sample of Hispanic breast cancer cases and controls with genotype data is the largest compiled to date, there are limitations that need to be acknowledged. Due to the lack of information on tumor characteristics for cases from Mexico, we were unable to consider factors such as stage and hormone receptor status with the broader range of admixture. Additionally, we used arbitrary cut-points to define the three ancestry categories on the basis of the minimum necessary number of individuals per category that would provide enough power for the analyses (4). However, gene by ancestry interaction analyses done using genetic ancestry defined as a continuous variable (therefore freed from the arbitrary cut-points of the ancestry categories) showed results that were consistent with those observed with the categorical ancestry variable. Therefore, our choice of arbitrary cut-points is unlikely to have introduced a bias.
In conclusion, our results suggest that the degree of IA genetic ancestry modifies the magnitude and direction of associations with currently known breast cancer risk variants among Hispanic women. Thus, it is important to consider genetic ancestry to elucidate the observed ethnic disparities in breast cancer risk.
Supplementary material
Supplementary Tables S1–S4 can be found at http://carcin.oxfordjournals.org/
Funding
The Breast Cancer Health Disparities Study was funded by the National Cancer Institute (CA14002 to M.L.S.). The San Francisco Bay Area Breast Cancer Study was supported by the National Cancer Institute (CA63446 and CA77305), the U.S. Department of Defense (DAMD17-96-1-6071) and the California Breast Cancer Research Program (7PB-0068). The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000036C awarded to the Cancer Prevention Institute of California; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute. The 4-Corners Breast Cancer Study was funded by the National Cancer Institute (CA078682, CA078762, CA078552 and CA078802; N01-PC-67000 to Utah Cancer Registry), the State of Utah Department of Health, the New Mexico Tumor Registry, the Arizona and Colorado Cancer Registries, the Centers for Disease Control and Prevention National Program of Cancer Registries and additional state support. The Mexico Breast Cancer Study was funded by Consejo Nacional de Ciencia y Tecnología (SALUD-2002-C01-7462). Work for this study was also supported by the National Cancer Institute (CA 160607 to L.F. and CA120120 to E.Z.), the Center for Aging in Diverse Communities (CADC) under the Resource Centers for Minority Aging Research program by the National Institute on Aging (P30-AG15272) and the National Cancer Institute grant from the Special Population Network program to University of Texas, San Antonio (Redes En Acción # U01CA86117).
Supplementary Material
Acknowledgements
We would also like to acknowledge the contributions of the following individuals to the study: Jennifer Herrick and Sandra Edwards for data harmonization and data management, Erica Wolff and Michael Hoffman for laboratory support, Carolina Ortega for her assistance with data management for the Mexico Breast Cancer Study, and Jocelyn Koo for data management for the San Francisco Bay Area Breast Cancer Study. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute or endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 4-CBCS
4-Corners Breast Cancer Study
- CI
confidence interval
- GWAS
genome-wide association study
- IA
Indigenous American
- LD
linkage disequilibrium
- NHW
non-Hispanic white
- OR
odds ratio
- SNP
single nucleotide polymorphism.
References
- 1. Jemal A., et al. (2010). Cancer statistics, 2010. CA Cancer J. Clin., 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Fejerman L., et al. (2008). Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res., 68, 9723–9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fejerman L., et al. (2010). European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol. Biomarkers Prev., 19, 1074–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slattery M.L., et al. (2012). Genetic variation in genes involved in hormones, inflammation and energetic factors and breast cancer risk in an admixed population. Carcinogenesis, 33, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fejerman L., et al. (2012). Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Hum. Mol. Genet., 21, 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. John E.M., et al. (2005). Migration history, acculturation, and breast cancer risk in Hispanic women. Cancer Epidemiol. Biomarkers Prev., 14, 2905–2913 [DOI] [PubMed] [Google Scholar]
- 7. Chlebowski R.T., et al. (2005). Ethnicity and breast cancer: factors influencing differences in incidence and outcome. J. Natl Cancer Inst., 97, 439–448 [DOI] [PubMed] [Google Scholar]
- 8. Fejerman L., et al. (2008). Population differences in breast cancer severity. Pharmacogenomics, 9, 323–333 [DOI] [PubMed] [Google Scholar]
- 9. Ahmed S., et al. (2009). Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet., 41, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gold B., et al. (2008). Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl Acad. Sci. USA., 105, 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter D.J., et al. (2007). A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet., 39, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas G., et al. (2009). A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet., 41, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Easton D.F., et al. (2007). Genome-wide association study identifies novel breast cancer susceptibility loci. Nature, 447, 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng W., et al. (2009). Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet., 41, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stacey S.N., et al. (2008). Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet., 40, 703–706 [DOI] [PubMed] [Google Scholar]
- 16. Stacey S.N., et al. (2007). Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet., 39, 865–869 [DOI] [PubMed] [Google Scholar]
- 17. Long J., et al. (2010). Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet., 6, e1001002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turnbull C., et al. (2010). Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet., 42, 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fletcher O., et al. (2011). Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl Cancer Inst., 103, 425–435 [DOI] [PubMed] [Google Scholar]
- 20. Slattery M.L., et al. (2011). Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res. Treat., 129, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barnholtz-Sloan J.S., et al. (2011). Replication of GWAS “Hits” by race for breast and prostate cancers in European Americans and African Americans. Front. Genet., 2, 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hutter C.M., et al. (2011). Replication of breast cancer GWAS susceptibility loci in the Women’s Health Initiative African American SHARe Study. Cancer Epidemiol. Biomarkers Prev., 20, 1950–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. González Burchard E., et al. (2005). Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am. J. Public Health., 95, 2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price A.L., et al. (2007). A genomewide admixture map for Latino populations. Am. J. Hum. Genet., 80, 1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonilla C., et al. (2004). Substantial Native American female contribution to the population of Tacuarembó, Uruguay, reveals past episodes of sex-biased gene flow. Am. J. Hum. Biol., 16, 289–297 [DOI] [PubMed] [Google Scholar]
- 26. Sans M., et al. (2006). Population structure and admixture in Cerro Largo, Uruguay, based on blood markers and mitochondrial DNA polymorphisms. Am. J. Hum. Biol., 18, 513–524 [DOI] [PubMed] [Google Scholar]
- 27. Bertoni B., et al. (2003). Admixture in Hispanics: distribution of ancestral population contributions in the Continental United States. Hum. Biol., 75, 1–11 [DOI] [PubMed] [Google Scholar]
- 28. Santos N.P., et al. (2010). Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum. Mutat., 31, 184–190 [DOI] [PubMed] [Google Scholar]
- 29. Via M., et al. (2011). History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS One., 6, e16513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Avena S., et al. (2012). Heterogeneity in genetic admixture across different regions of Argentina. PLoS One, 7, e34695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seldin M.F., et al. (2007). Argentine population genetic structure: large variance in Amerindian contribution. Am. J. Phys. Anthropol., 132, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galanter J.M., et al. (2012). Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet., 8, e1002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang S., et al. (2008). Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet., 4, e1000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang S., et al. (2007). Genetic variation and population structure in native Americans. PLoS Genet., 3, e185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slattery M.L., et al. (2007). Body size, weight change, fat distribution and breast cancer risk in Hispanic and non-Hispanic white women. Breast Cancer Res. Treat., 102, 85–101 [DOI] [PubMed] [Google Scholar]
- 36. John E.M., et al. (2003). Lifetime physical activity and breast cancer risk in a multiethnic population: the San Francisco Bay area breast cancer study. Cancer Epidemiol. Biomarkers Prev., 12, 1143–1152 [PubMed] [Google Scholar]
- 37. Angeles-Llerenas A., et al. (2010). Moderate physical activity and breast cancer risk: the effect of menopausal status. Cancer Causes Control, 21, 577–586 [DOI] [PubMed] [Google Scholar]
- 38. Edwards S., et al. (1994). Objective system for interviewer performance evaluation for use in epidemiologic studies. Am. J. Epidemiol., 140, 1020–1028 [DOI] [PubMed] [Google Scholar]
- 39. Liu K., et al. (1994). A study of the reliability and comparative validity of the cardia dietary history. Ethn. Dis., 4, 15–27 [PubMed] [Google Scholar]
- 40. Slattery M.L., et al. (1994). A computerized diet history questionnaire for epidemiologic studies. J. Am. Diet. Assoc., 94, 761–766 [DOI] [PubMed] [Google Scholar]
- 41. Pritchard J.K., et al. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benjamini Y., et al. (2005). Quantitative trait loci analysis using the false discovery rate. Genetics, 171, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benjamini Y., et al. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res., 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 44. StataCorp (2009). Stata Statistical Software: Release 11. StataCorp LP: College Station, TX: [Google Scholar]
- 45. Garcia-Closas M., et al. (2008). Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin. Cancer Res., 14, 8000–8009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garcia-Closas M., et al. (2008). Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet., 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milne R.L., et al. (2010). Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res., 12, R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campa D., et al. (2011). Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J. Natl Cancer Inst., 103, 1252–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stacey S.N., et al. (2010). Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet., 6, e1001029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.