Abstract
Research on regulation of cellulases and hemicellulases gene expression may be very useful for increasing the production of these enzymes in their native producers. Mechanisms of gene regulation of cellulase and hemicellulase expression in filamentous fungi have been studied, mainly in Aspergillus and Trichoderma. The production of these extracellular enzymes is an energy-consuming process, so the enzymes are produced only under conditions in which the fungus needs to use plant polymers as an energy and carbon source. Moreover, production of many of these enzymes is coordinately regulated, and induced in the presence of the substrate polymers. In addition to induction by mono- and oligo-saccharides, genes encoding hydrolytic enzymes involved in plant cell wall deconstruction in filamentous fungi can be repressed during growth in the presence of easily metabolizable carbon sources, such as glucose. Carbon catabolite repression is an important mechanism to repress the production of plant cell wall degrading enzymes during growth on preferred carbon sources. This manuscript reviews the recent advancements in elucidation of molecular mechanisms responsible for regulation of expression of cellulase and hemicellulase genes in fungi.
Keywords: Cellulase, Cellulose, CRE1, Hemicellulase, Sophorose, Xylan, XYR1.
1. INTRODUCTION
In order to enhance energy security and mitigate climate change, interest in finding renewable fuels to replace petroleum-based ones is enormously increasing. The biofuels ethanol and biodiesel represent potential options for meeting these needs in the transportation sector. The uniqueness of cellulosic ethanol as a sustainable liquid transportation fuel, which can be produced in high volumes and at low cost, and its many powerful benefits have been recognized for decades [1-5]. A recent awareness of the urgent need to advance cellulosic ethanol production is evidenced by the number of reviews reported on the theme of ethanol fuel production from lignocellulosic biomass, with great attention to ethanol production from lignocellulosic residues, such as crop and wood residues and municipal solid waste [6-15]. The key step for conversion of lignocellulosic biomass into fermentable sugars for fuel production is represented by the hydrolysis of polysaccharides, resulting from biomass pretreatment, by cellulases and hemicellulases. Filamentous fungi are the major source of cellulases and hemicellulases. As far as cellulases are concerned, three main enzymatic activities are involved in cellulose hydrolysis: 1) endoglucanases (EC 3.2.1.4); 2) exoglucanases, including d-cellodextrinases (EC 3.2.1.74) and cellobiohydrolases (EC 3.2.1.91); and 3) β-glucosidases (EC 3.2.1.21). As far as hemicellulases are concerned, endo-β-1,4-xylanases (EC 3.2.1.8) and β-xylosidases (EC 3.2.1.37) are required for degradation of the xylan backbone, while auxiliary enzymes such as α-glucuronidases (EC 3.2.1), α-arabinofuranosidases (EC 3.2.1.55), acetylesterases or acetyl xylan esterases (EC 3.1.1.6) are required to achieve the complete degradation of complex substituted xylans. Research on regulation of cellulase and hemicellulase genes’ expression may be very useful for increasing production of these enzymes in their native producers. Mechanisms of cellulase and hemicellulase genes regulation have been studied in filamentous fungi, mainly in Aspergillus [16, 17] and Trichoderma [18]. The production of these extracellular enzymes is an energy-consuming process, so the enzymes are produced only under conditions in which the fungus needs to use plant polymers as an energy and carbon source. Moreover, production of many of these enzymes is coordinately regulated, and induced in the presence of the substrate polymers. Induction mechanisms of cellulase and hemicellulase genes expression involve activation of gene expression by the respective hydrolysis and/or transglycosylation products of cellulose and/or xylan, such as gentiobiose for Penicillium [19], and sophorose for A. terreus and T. reesei [20, 21]. In addition to induction by mono- and oligo-saccharides, genes encoding hydrolytic enzymes involved in plant cell wall deconstruction in filamentous fungi can be repressed during growth in the presence of easily metabolizable carbon sources, such as glucose. Carbon catabolite repression (CCR) is an important mechanism to repress the production of plant cell wall degrading enzymes during growth on preferred carbon sources [22-25].
This manuscript reviews the recent advancements in elucidation of molecular mechanisms responsible for regulation of expression of cellulase and hemicellulase genes in fungi.
2. REGULATION OF PRODUCTION OF CELLULASES AND HEMICELLULASES IN TRICHODERMA REESEI
The cellulolytic machinery of T. reesei is one of the most widely studied [26]. T. reesei genome (http://genome.jgi psf.org/Trire2/Trire2.home.html) contains ten cellulase and sixteen hemicellulase genes [27]. The enzymes so far identified and characterized as responsible for the cellulolytic activity of T. reesei include five endoglucanases -EGI/Cel7B, EGII/Cel5A, EGIII/Cel12A [28, 29], EGIV/Cel61A [30], and EGV/Cel45A [31] and two exoglucanases -the cellobiohydrolases CBHI/Cel7A and CBHII/Cel6A [32]. These enzymes act synergistically to convert cellulose into cellobiose [28-33], whose hydrolysis into glucose involves then two β-glucosidases -BGLI/Cel3A [34] and BGLII/Cel1A [35]. An additional protein, swollenin (encoded by the gene swo1), has been described, that disrupts crystalline cellulose structures, presumably making polysaccharides more accessible to hydrolysis [36]. The cellulases CBHI/Cel7A, CBHII/ Cel6A, EGI/Cel7B, and EGII/Cel5A are the most abundantly produced by T. reesei secreting them up to 40 g/liter [37]. Due to the enormous level of cellulase production, T. reesei revealed to be a potential candidate for advancing cellulosic ethanol by I Consolidated BioProcessing, engineering it to ferment monosaccharides into ethanol in high yields [38].
The T. reesei genome also contains sixteen hemicellulases including two GH43, one GH10, four GH11, one GH74, one GH62, two GH54, one GH67 and four GH95 [27]. Among these, two major endo-β-1,4-xylanases XYNI and XYNII (EC 3.2.1.8) [39]; and one β-xylosidase, BXLI (EC 3.2.1.37) [40] have been characterized.
The presence of cellulose, xylan or mixtures of plant polymers in the fungal culture medium causes abundant production of cellulolytic and xylanolytic activities by T. reesei, as already reported by the earlier studies [41-44]. Pure (oligo)saccharides, such as sophorose [20, 21], β-cellobiono-1,5-lactone, D-xylose, xylobiose, galactose, and lactose, have been also reported to induce cellulase and hemicellulase production in T. reesei (Table 1) [24, 45-49].
Table 1.
Inducibility of Cellulases, Hemicellulases and Related Enzymes in T. reesei, N. crassa and Aspergillus spp. X: repressor; +: inductor; -: no action
| Substrate | Glycerol | Glucose | Sorbitol | Cellulose | Cellobiose | Xylose | Sophorose | Lactose | Xylobiose | Galactose | Laminaribiose | Gentiobiose | Aryl- β-glucosides | Maltose | Xylan | GalacTuronic acid | Fructose | Mannitol | Arabinose | Arabinitolo | Mannose | Sorbose | Twen 80 | C18 Fatty acids | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | |||||||||||||||||||||||||
| T. reesei | |||||||||||||||||||||||||
| bgl1 | + | + | + | + | [24] | ||||||||||||||||||||
| bgl2 | + | + | + | [48] | |||||||||||||||||||||
| bxl1 | + | + | + | [48] | |||||||||||||||||||||
| cbh1 | - | - | + | + | + | + | + | + | [51, 59] | ||||||||||||||||
| cbh2 | - | - | + | + | + | + | + | [59] | |||||||||||||||||
| cel5b | + | + | + | + | [54] | ||||||||||||||||||||
| egl1 | - | - | + | + | [51, 59] | ||||||||||||||||||||
| egl2 | - | - | + | + | [59] | ||||||||||||||||||||
| egl3 | + | + | [192] | ||||||||||||||||||||||
| egl5 | - | - | + | + | [59] | ||||||||||||||||||||
| xynI | x | + | + | + | + | + | + | [192] | |||||||||||||||||
| xynII | x | + | + | + | + | + | + | [192] | |||||||||||||||||
| xyn3 | - | + | + | [192, 193] | |||||||||||||||||||||
| a-AF | + | [192] | |||||||||||||||||||||||
| abf1 | x | + | + | [71] | |||||||||||||||||||||
| abf2 | x | + | + | [71] | |||||||||||||||||||||
| abf3 | x | + | + | [71] | |||||||||||||||||||||
| agl1 | + | + | [24] | ||||||||||||||||||||||
| agl2 | + | + | [24] | ||||||||||||||||||||||
| glr1 | + | [24] | |||||||||||||||||||||||
| axe1 | + | [24] | |||||||||||||||||||||||
| N. crassa | |||||||||||||||||||||||||
| Endocellulase | + | + | + | [121] | |||||||||||||||||||||
| β-glucosidases | x | - | [121] | ||||||||||||||||||||||
| aryl-β-glucosidase | x | + | + | + | + | + | + | [127] | |||||||||||||||||
| Cellobiase | x | + | + | + | + | [127] | |||||||||||||||||||
| Xylanase | - | + | + | [122] | |||||||||||||||||||||
| xyr-1 | + | [143] | |||||||||||||||||||||||
| xdh-1 | + | [143] | |||||||||||||||||||||||
| xyk-1 | + | [143] | |||||||||||||||||||||||
| Cellulase | + | - | + | + | [121] | ||||||||||||||||||||
| N. crassa | |||||||||||||||||||||||||
| gh 43-2 | + | + | [143] | ||||||||||||||||||||||
| gh 51-1 | + | [143] | |||||||||||||||||||||||
| gh 10-1 | + | [143] | |||||||||||||||||||||||
| gh 43-5 | + | + | [143] | ||||||||||||||||||||||
| arabinase | + | [143] | |||||||||||||||||||||||
| gh 11-2 | + | + | [143] | ||||||||||||||||||||||
| gh 10-2 | + | + | [143] | ||||||||||||||||||||||
| gh 53-1 | + | + | [143] | ||||||||||||||||||||||
| gh 1-1 | + | [143] | |||||||||||||||||||||||
| β-galactosidases | + | + | + | [199] | |||||||||||||||||||||
| gh5-1 | + | [143] | |||||||||||||||||||||||
| gh 61-7 | + | [143] | |||||||||||||||||||||||
| gh 61-4 | + | [143] | |||||||||||||||||||||||
| gh 61-1 | + | [143] | |||||||||||||||||||||||
| gh 61-3 | + | [143] | |||||||||||||||||||||||
| gh 61-6 | + | [143] | |||||||||||||||||||||||
| gh 7-1 | + | [143] | |||||||||||||||||||||||
| gh 6-3 | + | [143] | |||||||||||||||||||||||
| gh 61-13 | + | [143] | |||||||||||||||||||||||
| gh 61-5 | + | [143] | |||||||||||||||||||||||
| gh 11-1 | + | [143] | |||||||||||||||||||||||
| gh 74-1 | + | [143] | |||||||||||||||||||||||
| gh 10-1 | + | [143] | |||||||||||||||||||||||
| Aspergillus spp. | |||||||||||||||||||||||||
| Endoglucanase | x | + | + | + | [160] | ||||||||||||||||||||
| β-glucosidase | x | + | + | + | [160] | ||||||||||||||||||||
| Xylanolytic enzymes | + | + | + | + | [20, 161] | ||||||||||||||||||||
| eglA/B | + | + | [161, 200] | ||||||||||||||||||||||
| pelA/B/C/D/E/F | + | [168, 200] | |||||||||||||||||||||||
| plyA | + | [168, 200] | |||||||||||||||||||||||
| pgaA/B/C/D/E | + | [168, 200] | |||||||||||||||||||||||
| Aspergillus spp. | |||||||||||||||||||||||||
| pgaX/I/II | + | [168, 200] | |||||||||||||||||||||||
| pmeA | + | [168, 200] | |||||||||||||||||||||||
| rglA | + | [168, 200] | |||||||||||||||||||||||
| afbA | + | + | + | [24,169,170] | |||||||||||||||||||||
| afbB | + | + | + | [24,169,170] | |||||||||||||||||||||
| abnA | + | [168,169,200] | |||||||||||||||||||||||
| lacA | + | + | + | + | + | [168,169,200] | |||||||||||||||||||
| aguA | + | + | [169, 200] | ||||||||||||||||||||||
| axeA | + | [169, 200] | |||||||||||||||||||||||
| faeA | + | [169, 200] | |||||||||||||||||||||||
| axhA | + | + | [169, 200] | ||||||||||||||||||||||
| rhgA/B | + | [169, 200] | |||||||||||||||||||||||
| axhA | + | + | + | [24] | |||||||||||||||||||||
Inability of the fungal cells to incorporate insoluble polymeric compounds, such as cellulose and xylan, aroused the question on how these polymers can initiate production of hydrolytic enzymes. Several studies investigating this aspect postulated the inducer function of a low molecular weight and soluble compound derived from cellulose. One of the proposed mechanisms is that the fungus produces basal levels of cellulase (mainly CEL7A and CEL6A) and that the activity of these extracellular enzymes on cellulose produces a soluble inducer, which can enter the cell and affect induction [50, 51]. In support of this mechanism, it was shown that antibodies against CBHI, CBHII, EGI and EGII blocked the expression of cbh1/cel7a gene in the presence of cellulose but not the soluble inducer sophorose [50]. The constitutive levels of these cellulases and their role in cellulase induction were afterwards demonstrated by Carle-Urioste et al. [51]. These authors showed that the mRNAs cbh1 and egl1 are transcribed under uninduced conditions, and that induction with cellulose results in at least 1100-fold increase of both transcripts, as demonstrated by Northern blots. The basal activity of the cbh1 promoter was also examined by using a chimeric vector in which the gene encoding hygromycin B phosphotransferase [52] was placed under the control of the 59-flanking DNA sequence of the cbh1 gene. Under uninduced conditions, resistance to the antibiotic hygromycin B was observed with T. reesei cells transformed with this vector and grown on medium lacking cellulose. An antisense RNA strategy was also adopted by the same authors to gain in vivo evidence for the requirement of the basal expression of the cellulase in induction of the cellulase transcripts by cellulose [51]. The results demonstrated that the expression of this antisense RNA produced marked effects on the induction of the cbh1 transcript using cellulose (reduction of the cbh1 transcript expression between 80 and 90%) but not sophorose as an inducer. The authors also showed that the initial hydrolysis of cellulose is the rate-limiting step in the induction, as suggested by the observation that the addition of the cellulase system or its purified enzyme members to a culture of T. reesei, in the presence of cellulose, resulted in earlier detection of the cbh1 and egl1 transcripts. The time required for induction of cbh1 and egl1 transcripts using cellulose, cellulose + cellulase, or sophorose is 14, 10, and 4 h, respectively. This result supports the hypothesis that oligosaccharide( s) is(are) formed in vivo from cellulose by the activity of a low, constitutive, and extracellular cellulase activity. The relatively slow induction by sophorose could be explained by the fact that the inductive process is protein synthesis-dependent. In addition, it has recently been shown that a sophorose-inducible β-diglucoside permease is involved in the induction of the cellulase system in T. reesei [53]. Subsequently, Foreman et al. [54] identified further genes whose regulatory behavior is consistent with their role in primary inducer formation for cellulase expression. Among them, the mRNA of cel5b was moderately expressed during growth on glycerol, glucose, sophorose and lactose, and only slightly induced over this level by cellulose. It is worth noting that CEL5B contains the consensus sequence for membrane-anchoring via a glycosylphosphatidylinositol residue. All these properties make it an interesting candidate for generating the inducer of cellulase formation. Similarly, the acetyl xylan esterase Axe2, which is also predicted to contain a glycosylphosphatidylinositol anchor, may be involved in primary induction of some hemicellulases [54].
The surface-bound cellulolytic activity displayed by conidia of T. reesei, mainly due to CEL6A/CBHII [55, 56] is also considered important for cellulase induction since its elimination by detergents hinders germination of the conidia on cellulose. These conclusions were deduced by the observation that introduction of multiple copies of the cel6a gene into T. reesei caused an enhanced secretion of CEL7A and CEL6A on cellulose and an increased cellulase activity on cellulose corresponding to enhanced level of conidial-bound CEL6A [56, 57]. Consistently, a cel6a knocked out strain showed a delay in growth and cellulase formation on cellulose [58]. In more details, comparing strains in which the corresponding genes of the main cellulases (cel6a, cel7a, cel7b, cel5a) had been deleted, Seiboth et al. [58] showed that strains knocked out for cel6 and cel5a, respectively, exhibited a significantly reduced expression of the remaining cellulase genes, while strains carrying the cel7a or cel7b deletion showed these transcripts. A strain showing both the cellobiohydrolases cel6a and cel7a deletion, was unable to initiate growth on cellulose. During growth on lactose, these strains showed no significant alteration in their ability to express the respective other cellulase genes. These data support the role of CEL6A and other conidial-bound cellulases (such as CEL5A, for which a conidial location is not yet known) in the induction of cellulases and germination on cellulose.
Ilmèn et al. [59] investigated basic features of expression regulation of the T. reesei cellobiohydrolases cbh1 and cbh2 and endoglucanases egl1, egl2 and egl5 encoding genes, at the mRNA level, showing that these cellulase genes are coordinately expressed and the steady-state mRNA levels of cbh1/Cel7A is the highest. The highest induction level was achieved with cellulose and sophorose and moderate expression was observed when cellobiose or lactose were used as the carbon source. No expression could be observed on glucose-containing medium and high glucose levels abolish the inducing effect of sophorose. However, derepression of cellulase expression occurs without apparent addition of an inducer once glucose has been depleted from the medium. This expression seems not to arise simply from starvation, since the lack of carbon or nitrogen as such is not sufficient to trigger significant expression. It was also found that glycerol and sorbitol do not promote expression but, unlike glucose, do not inhibit it either, because the addition of 1 to 2 mM sophorose to glycerol or sorbitol cultures provokes high cellulase expression levels.
The best inducer of cellulase expression so far known is sophorose ((2-O-β-glucopyranosyl-D-glucos) [60, 21, 61], whose synthesis from cellobiose involves the transglycosylation activity of β-glucosidase [62]. Induction by sophorose is affected by various parameters such as its concentration and rate of uptake [61, 63]. Two pathways of sophorose utilisation were for the first time hypothesised by Loewenberg and Chapman [64]: a catabolic pathway characterized by a high capacity but low affinity for sophorose, and a cellulase inducing pathway endowed with a lower capacity but higher affinity for sophorose. As a matter of fact, Kubicek et al. [53] showed that sophorose is transported by a cellobiose permease, characterized by low KM and Vmax for sophorose, and thus competing with the extracellular β-glucosidase, which has a much higher KM but also Vmax for it.
Most authors implied a β-glucosidase in the process of sophorose production. T. reesei produces β-glucosidases having different cellular localizations [65-69]. The gene cel3a [65, 70] encodes the major extracellular β-glucosidase identified as one of the β-glucosidases involved in inducer formation. Knock-out of the cel3a gene causes a delay in induction of the other cellulase genes by cellulose, but not by sophorose, whilst a cel3a-multicopy strain is able to produce higher levels of cellulases than the wild-type strain under nonsaturating concentrations of sophorose, but both strains were comparably efficient at saturating concentrations [48]. The observation that the β-glucosidase inhibitor nojirimycin inhibits cellulase induction also in the cel3a disrupted strain suggests that the CEL3A is not the only β-glucosidase involved in inducer formation [48]. An additional β-glucosidase-encoding gene has been cloned [35] and properties and intracellular localisation of the corresponding enzyme have been characterised [69]. However, as no multicopy or gene deletion studies have yet been carried out, ascertainment of its involvement in cellulase induction requires further investigation.
2.1. Transcriptional Factors Involved in Regulation of Cellulase and Hemicellulase Genes’ Expression in T. reesei
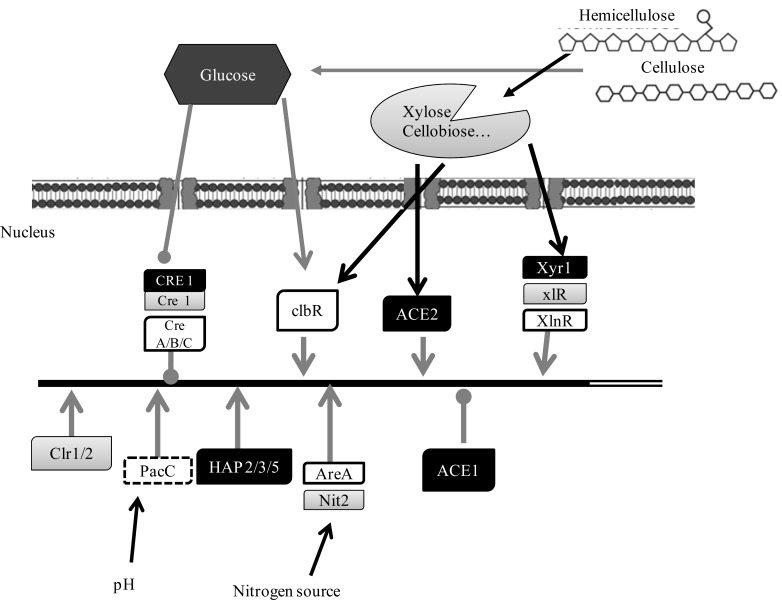
Foreman et al. [54] performed investigations on regulation of cellulase and hemicellulase genes’ expression in T. reesei by microarrays showing that most of the genes encoding known and putative biomass-degrading enzymes are transcriptionally co-regulated. This co-regulation indicates a tightly coordinated cooperation of the corresponding transcription factors, five of which have been so far identified (Fig. 1): the positive regulators XYR1, ACE2 and the HAP2/3/5 complex, the repressor ACE1 and the carbon catabolite repressor CRE1 (Table 2) [25].
Fig. (1).
Schematic representation of transcriptional factors affecting cellulases and xylanases expression in T. reseei (black box), N. crassa
(grey box) and Aspergillus spp. (white box). The carbon catabolite repressor CRE, the activators clbR, Xyr/xlR/XlnR,Clr,ACE2, the repressor
ACE1, the CCAAT binding Hap2/3/5 complex, the pH regulator PacC, and the nitrogen regulators AreA and Nit2 are shown. The repression
activity ( ), the induction activity (
), the induction activity ( ) and also promoter (
) and also promoter ( ) and coding region (
) and coding region ( ) are indicated.
) are indicated.
Table 2.
Positive and Negative Regulators of Expression of Genes Coding for (hemi)cellulolytic Enzymes and Their Binding consensus Sequences in the Target Promoters
| Trichoderma reesei | |||
|---|---|---|---|
| Positive Regulators | |||
| Name | Structure | Consensus Region | References |
| XYR1 | Zinc binuclear cluster protein | 5’-GGCTAA | [194] [25] |
| ACE2 | Zinc binuclear cluster proteins | 5ʹGGCTAATAA | |
| HAP2 | Multimeric protein complex | 5’- CCAAT | |
| HAP3 | |||
| HAP5 | |||
| Negative regulators | |||
| Name | Structure | Consensus region | References |
| ACE1 | Three Cys2His2-type zinc fingers | 5’-AGGCA | [25] [195] |
| CRE1 | Cys2His2 type transcription factor | 5’-SYGGRG | |
| Neurospora crassa | |||
| Positive regulators | |||
| Name | Structure | Consensus region | References |
| CLR-1/-2 | Two zinc binuclear cluster | - | [145] |
| PacC | Three Cys2His2 zinc fingers | 5’-GCCARG | [147] |
| Neurospora crassa | |||
| Positive regulators | |||
| Name | Structure | Consensus region | References |
| XLR1 | Zinc binuclear cluster protein | [148] | |
| NIT2 | Single zinc finger protein | 5’- TATCTA | [149] |
| Negative regulators | |||
| Name | Structure | Consensus region | References |
| CRE1 | Cys2His2 type transcription factor | 5’-SYGGRG- | [152] |
| Aspergillus spp. | |||
| Positive regulators | |||
| Name | Structure | Consensus region | References |
| AmyR | Zn(II)2Cys6-binuclear cluster DNA-binding motif | 5’-CGGN8CGG-3' | [194,-196] |
| AraR | Zn(2)Cys(6) binuclear cluster domain | [197,198] | |
| PacC | Three Cys2His2 zinc fingers | 5’- GCCARG | [22] |
| XlnR | Zinc binuclear cluster protein | 5'-GGCTAAA | [48] |
| ClbR | Zn(II)2Cys6-binuclear cluster DNA-binding motif | CGG or CCG triplets | [174] |
| AreA | Highly conserved DNA binding motif comprising a Cys(4) zinc finger followed by a basic domain | 5’-GATA (core sequence) | [182] |
| Negative regulators | |||
| Name | Structure | Consensus region | References |
| CREA | Cys2His2 type transcription factor | 5’-SYGGRG | [183-187] |
| CREB | |||
| CREC | |||
The main positive regulator of cellulase and hemicellulase gene expression is represented by XYR1 (xylanase regulator 1) [48, 18], a zinc binuclear cluster protein binding to a GGCTAA-motif arranged as an inverted repeat [48]. xyr1 deletion abolishes cellulase induction on cellulose and sophorose and impairs the induction of hemicellulase genes involved in xylan and arabinan degradation [71, 18], thus proving its essential role in the induction process. xyr1 transcription seems not to be induced during growth on cellulose [72]. Most of eukaryotic transcriptional activators are present in cells only in small amounts required to start gene expression [73], and, in many cases, they are further induced by the conditions for which they are needed and are degraded once they are no longer required [74]. On the contrary, xyr1 expression is regulated solely by CRE1-dependent CCR and by repression by the specific transcription factor ACE1, not by induction [72, 49]. Whether an increase in constitutive expression of xyr1 would increase enzyme formation is not sufficiently understood. Aigner-Mach et al. [72] fused the xyr1 gene under the regulatory signals of the nag1 (N-acetyl-β-D-glucosaminidase) promoter, which resulted in a slightly earlier beginning of xylanase formation but did not significantly enhance the final enzyme titre. However, these studies used the uninduced, basal expression level of nag1, which is not much higher than that of xyr1 itself, and studies using stronger expressed promoters (such as those for glycolytic or hydrophobin genes) must be used to clarify whether the constitutive expression of xyr1 would enhance cellulase and/or xylanase formation.
The cellulase activator ACE2 also belongs to the class of zinc binuclear cluster proteins [75]. It has so far been shown to occur only in Trichoderma spp. Deletion of ace2 lowers the transcript levels of the major cellulases and causes a decrease of cellulase activity during growth on cellulose [75, 76], whilst it does not affect cellulase induction by sophorose [75]. It is worth noting that the DNA-binding domain of ACE2 is able to bind to the promoter motif [GGC(T/A)4] present in the cbh1 promoter also recognized by XYR1 [77]. Stricker et al. [76] suggested that phosphorylation and dimerization are needed for the binding of ACE2 to the corresponding promoter element.
The CCAAT motif is a common cis-acting element found in either orientation in the promoter and enhancer region of a large number of eukaryotic genes. Particularly, in yeasts, as well in filamentous fungi, the CCAAT box-binding proteins identified so far all belong to the group of HAP-like factors. Site-directed mutagenesis of the promoter of one of the most abundant cellulase produced by T. reesei, cbh2, revealed the existence of an undecameric nucleotide motif which is essential for gene expression in vivo. Moreover, experiments of promoter mutation and in vivo footprinting analysis allowed to show that expression from the cel6a promoter is dependent on a CCAAT box bound by the HAP2/3/5 protein complex [78]. The CCAAT motif is found in approximately 30% of the 5'- non-coding regions of eukaryotic genes [79]. In analogy to the mammalian NF-Y complex containing NF-YA, NFYB and NF-YC orthologues of HAP2, HAP3 and HAP5, respectively, they contain a histone fold motif, a structural feature of histones suggesting that NF-Y might be involved in the organisation of the chromatin structure [80]. Thereby the action of acetyltransferases may play a role in the local disruption of nucleosomes since an association of GATA-1 and NF-Y with acetyltransferases p300/CBP has been shown [81, 82]. The corresponding hap2, hap3 and hap5 genes from T. reesei were cloned by Zeilinger et al. [83] showing that they encode proteins similar to Hap homologues from other organisms and essential for binding to the CAE (cbh2-activating element) in the T. reesei cel6a promoter. The HAP2/3/5 complex is considered needed for generating an open chromatin structure required for full transcriptional activation [84]. The hypothesis that the CCAAT sequences in the cellulase promoters could play a conserved role in the generation of an open chromatin structure necessary for full transcriptional activation is supported by the detection of a nucleosome-free region around the XYR1/ACE2/HAP2/3/5- binding area in the cel6a promoter, which is flanked by strictly positioned nucleosomes [84]. Induction by sophorose results in a loss of positioning of nucleosomes -1 and -2 downstream of the binding area, thus making the TATA box accessible. A mutation in the CCAAT box shifted this positioning, thus proving the role of the HAP2/3/5 complex in this process [84]. These data provide an experiment based explanation of the advantage for clustering of cellulases in the genome of T. reesei and illustrate that chromatin regulation is a suitable target for strain improvement. For instance, it is worth noting that Zou et al. [85] have recently demonstrated that replacement of the CREI binding sites within the cbh1 promoter of T. reesei with the binding sites of transcription activator, namely the HAP2/3/5, besides the ACEII, led to improvement of promoter efficiency. The new developped promoter was shown able to induce expression of the green fluorescent protein reporter by 5.5-fold in inducing culture medium and 7.4-fold in repressing culture medium.
ACE1 contains three Cys2His2-type zinc fingers and it was shown to bind in vitro to eight sites containing the core sequence 5'-AGGCA scattered along the 1.15-kb cel7a promoter [86]. Deletion of ace1 resulted in an increase in the expression of all the main cellulase and hemicellulase genes in sophorose- and cellulose-induced cultures, indicating that ACE1 acts as a repressor of cellulase and xylanase expression [87] and of xyr1 during growth on D-xylose [72]. A strain bearing a deletion of both the ace1 gene and ace2 gene expressed cellulases and xylanases similar to the Δace1 strain, probably due to the remaining activity of XYR1 [87].
All together the above data suggest that the substrate-unspecific activator XYR1 is fine-tuned by more specific transcriptional regulators such as ACE1 and ACE2 (Fig. 1). This working model concurs with the findings that XYR1 binds to an inverted repeat either as a homo- or a heterodimer, respectively, thereby providing the opportunity for specific regulatory proteins to interact with the accordant promoter and/or XYR1. The role of the HAP2/ 3/5 complex in this regulation may be that of a general transcriptional enhancer raising the accessibility of the other factors to the cellulase promoters.
The putative methyltransferase LaeA is a global regulator that affects the expression of multiple secondary metabolite gene clusters in several fungi, and it can modify heterochromatin structure in Aspergillus nidulans. Seiboth et al. [88] showed that the expression of genes for lignocellulose degradation are controlled by the orthologous T. reesei LAE1: the protein methyltransferase LAE1. In a lae1 deletion mutant a complete loss of expression of all seven cellulases was observed, auxiliary factors for cellulose degradation, β-glucosidases and xylanases were no longer expressed. Conversely, enhanced expression of lae1 resulted in significantly increased cellulase gene transcription. Lae1- modulated cellulase gene expression was dependent on the function of the general cellulase regulator XYR1, but also xyr1 expression was LAE1-dependent. Chromatin immunoprecipitation followed by highthroughput sequencing (‘ChIP-seq’) showed that lae1 expression was not obviously correlated with H3K4 dior trimethylation (indicative of active transcription) or H3K9 trimethylation (typical for heterochromatin regions) in CAZY (Carbohydrate-Active enZYmes) coding regions, suggesting that LAE1 does not affect CAZyme gene expression by directly modulating H3K4 or H3K9 methylation. These data demonstrate that the putative protein methyltransferase LAE1 is essential for cellulase gene expression in T. reesei through mechanisms that remain to be identified.
To learn more about the function of LAE1 in T. reesei, Karimi-Aghcheh et al. [89] further assessed the effect of deletion and overexpression of lae1 on genome-wide gene expression. They found that in addition to positively regulating 7 of 17 polyketide or nonribosomal peptide synthases, genes encoding ankyrinproteins, iron uptake, heterokaryon incompatibility proteins, PTH11-receptors, and oxidases/monoxygenases are major gene categories also regulated by LAE1. Chromatin immunoprecipitation sequencing with antibodies against histone modifications known to be associated with transcriptionally active (H3K4me2 and -me3) or silent (H3K9me3) chromatin detected 4089 genes bearing one or more of these methylation marks, of which 75 exhibited a correlation between either H3K4me2 or H3K4 me3 and regulation by LAE1.
CRE1 is the main transcription factor mediating CCR [90, 91], a mechanism promoting the assimilation of high-energy yielding carbon sources over that of sources yielding less energy, described in more details below.
2.2. Carbon Catabolite Repression of Cellulase and Hemicellulase Genes’ Expression in T. reesei
Expression of most of T. reesei cellulase and hemicellulases genes does not occur in the presence of glucose in culture medium. Two mechanisms are responsible for this phenomenon: inducer exclusion (that is, inhibition of inducer [= sophorose] uptake) by D-glucose [53] and glucose repression [59, 84, 92]. The latter specifies a transcriptional regulation controlling the preferential use of substrates such as D-glucose or other monosaccharides whose catabolism provides a high yield of ATP namely CCR.
Consequently, one of the earliest attempts for engineering cellulase production was removal of CCR. Classical mutagenesis combined with selection for 2-desoxyglucose resistance (an agent believed primarily to enrich carbon catabolite-resistant mutants) [93] has led to increased cellulase producers such as T. reesei RUT C30 [94], RL-P37 [95] and CL847 [96], thus supporting the possible importance of CCR in cellulase formation.
In Trichoderma spp., the key player in this glucose repression is the Cys2His2 type transcription factor CREI [90, 97]. cre1 is missing in the cellulase hyperproducer strain RUT C30 [90] and importance of its deletion for the increase of cellulase production has been highlighted recently [98]. The cre1 gene is located on scaffold 2: 786955-789433 (ID 120117), and the mutant is characterized by a loss of a 2478-base pair fragment, which starts downstream of the region encoding the CRE1 zinc finger and reaches into the 3'-non-coding region [99]. Le Crom et al. [100] discovered that in Rut-C30, in addition to the 29 genes deleted during the generation of NG14, the truncation of cre1 gene and the frameshift in glucosidase II, nearly 45% of the genes mutated encode transcription factors, components of nuclear import, mRNA metabolism, protein secretion, and vacuolar sorting.
The knowledge of mutations in the hyperproducer T. reesei strains was widened by Vitikainen et al. [101], reporting an aCGH (Array-Comparative Genomic Hybridization) analysis of the high-producing strains QM9123, QM9414, NG14 and Rut-C30. These authors showed that the 85 kb deletion is not responsible for the high ability of cellulase producing in Rut-C30.
In vivo functionality of the CRE1 binding sites has been shown for the cbh1 and xyn1 promoters of T. reesei where mutations in the binding sequences led to constitutive expression of these genes in the presence of D-glucose [92, 102]. Functional CREI binding sites have been shown to consist of two closely spaced 5'-SYGGRG motifs, and it has been suggested that direct CREI repression would occur only through such double binding sites. Phosphorylation of a serine in a conserved short stretch within an acidic region of T. reesei CREI has been demonstrated to regulate its DNA binding [103]. Phosphorylation of this serine may involve a casein kinase 2. Casein kinases of this class are known from various other organisms to play a role in the regulation of a large number of transcription factors [104]. However, the SNF1 kinase, which plays a central role in the regulation of CCR in yeasts [105], appears not to be involved in the phosphorylation of CRE1 in T. reesei [106].
Another gene whose product is involved in CCR in T. reesei is represented by cre2 whose disruption led to deregulation of genes normally subjected to CCR [107]. Interestingly, the E3 ubiquitin ligase LIM1 also responds to cellulase inducing conditions and binds to the cbh2-promoter [108].
The way in which the presence of glucose triggers CCR is still only poorly understood in filamentous fungi. In S. cerevisiae, the D-glucose and D-fructose phosphorylating enzymes are also involved in D-glucose and carbon catabolite sensing, due to the presence of three hexose-6-phosphorylating enzymes including two hexokinases and one glucokinase. Each of them enables S. cerevisiae to grow on D-glucose, but the hexokinase Hxk2p is responsible for the main enzymatic activity and glucose repression mediated by the carbon catabolite repressor Mig1p (whose DNA-binding domain is highly similar to that of CRE1) [109-111]. The mechanism by which Hxk2p contributes to glucose repression has not yet been fully elucidated, but its catalytic activity seems to be dispensable and thus signal transmission may rather depend on substrate binding-induced conformational changes in the Hxk2p protein or a direct regulatory role of the Hxk2p in the nucleus (discussed, for example, in Linhoff et al. [80]).
Portnoy et al. [112] investigated how xyr1, ace1 and ace2 are regulated in cellulase induction conditions and how this regulation relates to carbon catabolite repression in the low cellulase producer strain T. reesei strain QM 9414, the high-producer strain RUT C30 [94, 113] and the hyperproducer strain T. reesei CL847 [96]. They demonstrated that in QM 9414 all three genes are induced by lactose and xyr1 is also induced by D-galactose. Moreover, ace1 is carbon catabolite repressed, whereas full induction of xyr1 and ace2 requires CRE1. These regulatory patterns showed significant differences in RUT C30 and CL847 strains. Rate of cellulase production by strain CL847 on lactose was around 15-fold higher than that for strain QM 9414, consistently with the 15-fold-increase of the cbh1 transcript level. These data indicate that gene expression is a major limiting step for cellulase biosynthesis. Consistent with its role as the major transcriptional regulator of cellulase gene expression, a strongly increased basal expression of xyr1 was observed in strain CL847, which was further induced by lactose. This increase indicates an improved function of the transcriptional machinery required for xyr1 expression in strain CL847. The basal expression of ace2 was not significantly altered in strain CL847, and the inducible level was the same as that in strain QM 9414. This indicates that the lack of CRE1 function, which seems to be required for ace2 gene expression, as indicated by the lower expression levels in the Δcre1 mutant, has been overcome during the breeding of CL847. While these data suggest that ace2 expression is not limiting for cellulase induction on lactose, they nevertheless show that wild-type expression levels appear to be necessary for the formation of high levels of cellulase. Expression of ace1—even though it is a repressor of cellulase formation—was also increased in the mutant strain CL847. However, ace1 is subject to CRE1-dependent CCR. The comparison reveals that the basal expression level of ace1 in CL847 is lower than that in the Δcre1 strain and decreases during the glucose feed. The approximate doubling of this level during the lactose feed is conserved, however. It has therefore been concluded that carbon catabolite derepression of ace1 has partially reverted in CL847, leading to a lower concentration of this repressor under cellulase-producing conditions. The present findings of reduced xyr1 but increased cbh1 transcription in the Δcre1 strain would be consistent with the operation of post-translational modification of XYR1. Nevertheless, these data show clearly that the expression of xyr1, ace1, and ace2 has been significantly altered in the hyperproducer CL847, suggesting that their wild-type expression was insufficient for hyperproduction. Identification of the proteins and genes responsible for the mechanisms observed may result in a major breakthrough in the understanding of cellulase formation and may offer a straightforward means for its improvement. These observations suggest that a strongly elevated basal transcription level of xyr1 and reduced upregulation of ace1 by lactose may have been important for generating the hyperproducer strain and that thus, these genes are major control elements of cellulose production.
3. REGULATION OF PRODUCTION OF CELLULASES AND HEMICELLULASES IN NEUROSPOSRA CRASSA
Neurospora crassa, a non pathogenic filamentous fungus of the class ascomycetes, is a well-known model organism that has been used for 90 years to study genetics, biochemistry, and fungal biology [114]. It was genetically characterized [115] and it has been shown to be able to degrade cellulose since 30 years ago [116]. N. crassa is able to synthesize and secrete high levels of all three enzyme types involved in cellulose degradation [117-121], as well as endoxylanase and β-xylosidase activities [122, 123]. There are 23 predicted cellulase genes and 19 predicted hemicellulase genes in the genome of N. crassa (http://www.broadinstitute.org/ annotation/genome/neurospora/MultiHome.html). In addition, N. crassa is a well-known ethanol producing microorganism that has been used for fermentation of agricultural residues [124].
The cellulase complex in N. crassa is composed by four endoglucanases, three exoglucanases and one β-glucosidase [125]. A summary of inducibility of cellulases, hemicellulases and related enzymes in N. crassa is reported in the Table 1. In 1964, Eberhart et al. [126] showed the presence of two β-glucosidases, including an aryl-β-glucosidase and a cellobiase, acting complementarily in N. crassa. Based on their production in response to specific inducers or various conditions of growth, these enzymes represent two fundamentally different classes of disaccharidases. Results of Eberhart et al. [127] on the induction of β-glucosidases (EC 3.2.1.21) in N. crassa, showed that the aryl-β-glucosidase can be induced either by disaccharides, that are usually used as substrates by this class of enzyme, or by monosaccharides that are not their usual substrates [120, 128-132]. Induction in the presence of monosaccharides and spontaneous production associated with conidiation is reversal of catabolite repression [133-135] because enzymes are not produced when a significant level of glucose is present in the induction medium. Both cellobiase and aryl-β-glucosidase seem to be exceptions to the general situation that disaccharide substrates are not the best inducers of specific disaccharidases in Neurospora [120, 130, 132], being production of both enzymes induced by cellobiose. Aryl-β-glucosidase production is semiconstitutive at late stages of culture growth prior to conidiation. At early stages, aryl-β-glucosidase is induced by cellobiose, laminaribiose, and gentiobiose, and in small part induced by galactose, amino sugars, and aryl-β-glucosides. Among the monosaccharides, xylose and galactose induce aryl-β-glucosidase. Cellobiase is induced by cellobiose, but other inducers have little effect on this enzyme such as galactose and maltose. Cellobiase activity is very low in all stages of the vegetative life cycle in the absence of β-glucoside inducer. Experimental results showed that a mixture of xylose and cellobiose induced increase of cellobiase, while added xylose did not change significantly the induction of aryl-β-glucosidase. Cellobiose is clearly the best inducer, with an optimum effect from 0.05 to 1 mM. The induction of β-glucosidases was inhibited by glucose, 2-deoxy-D-glucose, and sodium acetate. Sodium phosphate concentrations between 0.01 and 0.1 M stimulated induction of both enzymes, while concentrations above 0.1 M were inhibitory. The optimal condition for induction of both β-glucosidases was pH 6.0. Cellobiase induction was relatively more inhibited than aryl-β-glucosidase in the range of pH 6.0 to 8.0. The time required for the induction of these enzymes by cellobiose is 6 hours. As described below, fatty acids and surfactants have positive effects on the cellulases production [121], however, experimental results showed that oleic acid had no effect on production of β-glucosidase, while Tween 80 decreased its production [121]. This is probably due to some difference in the cellulase and β-glucosidase released. In fact in most organisms studied, β-glucosidase is an intracellular enzyme, released only by autolysis [136]. A recent study [137] demonstrated that a N. crassa mutant carrying deletions of two genes encoding extracellular β-glucosidase enzymes and one intracellular β-glucosidase lacks β-glucosidase activity, but its cellulase gene expression is efficiently induced in the presence of cellobiose, cellotriose, or cellotetraose as a sole carbon source while sophorose does not act as an inducer. Furthermore, the inclusion of a deletion of the catabolite repressor gene, cre-1, in the triple β-glucosidase mutant resulted in a strain that produces higher concentrations of secreted active cellulases on cellobiose. So cellobiose is an inducer of β-glucosidases but carbon catabolite repression (CCR, see the following paragraph) masks this inducing activity.
Eberhart et al. [116] studied the extracellular endocellulase (EC 3.2.1.4) production in mycelia and ungerminated conidia of N. crassa. They demonstrated a simple induction system of cellobiose and potassium phosphate buffer (pH 6.0) of extracellular cellulase to provide energy and substrates for protein synthesis. Yazdi et al. [125, 121, 138] have shown for the first time that N. crassa is capable of synthesizing and secreting high levels of the cellulase complex enzymes growing on microcrystalline cellulose, and other carbon sources, as an inducer for the enzymes. They also studied the role of surfactants and fatty acids on the production of the cellulases, since in other species it has been reported that surfactants and fatty acids stimulate production of the cellulase complex [139-141]. Yazdi et al. [121] demonstrated that the presence of C18 fatty acids and surfactants, such as Tween 80, increases production of both endoglucanase and exoglucanase in the medium. It is probably due to an increase in the permeability of the cell membrane, thus permitting more of the enzymes to be secreted, as postulated for other species by Reese & Maguire [139] and Demain & Birnbaum [142].
Analysis of the effects of different carbon sources, such as glucose, xylan and cellulose, on the production of extracellular cellulases and xylanases by N. crassa, reported by Mishra et al. [122], showed that the extracellular activities were very poor when the fermentation was carried out with glucose, while the maximum xylanase production was observed when N. crassa was grown on commercial xylan. However, significant amounts of xylanase were also produced when cellulose powder was used as a carbon source.
In 2012, Sun et al. [143] showed that N. crassa responds to the presence of cellulose (Avicel) by inducing both cellulase and hemicellulase gene expression, while the exposure to xylan only induces hemicellulase gene expression. In addition, exposure to Avicel induces some hemicellulase genes to a much higher expression level than exposure to xylan. These data suggest crosstalk between inducer molecules and regulatory pathways that are involved in deconstruction of plant cell walls in filamentous fungi. In this work, 353 genes have been identified that were significantly induced by xylan; in particular three genes, gh51-1 (arabinofuranosidase), gh10-2 (endoxylanase) and gh43-5 (β-xylosidase) showed increased expression levels of over 200 fold. Among 353 identified genes only 30 genes were induced by exposure to xylose, although none of the xylanolytic-related genes was essential for growth of N. crassa on xylan, in fact mutations in a number of them affected xylanase activity. These observations indicate some redundancy among enzymes associated with hemicellulose degradation, similar to those identified with cellulose degradation [48].
3.1. Transcriptional Factors Involved in Regulation of Cellulase and Hemicellulase Genes Expression in N. crassa
The knowledge of N. crassa genome sequence [144] has allowed the identification of the proteins involved in regulation of cellulase ad hemicellulase genes expression (Table 2, Fig. 1).
Two zinc binuclear cluster transcription factors (CLR-1 and CLR-2) are important regulators of genes encoding both cellulases and hemicellulases in the presence of cellulose as carbon source, but they are not required for growth or hemicellulase activity production in the presence of xylan as reported by Coradetti et al. [145]. In particular, Coradetti et al. [145] demonstrated that CLR-1 is a crucial element in cellobiose sensing mechanism of N. crassa during its growth on avicel. CLR-1 promotes expression of several genes necessary for cellobiose utilization, as well as that of clr-2. CLR-2, maybe in a complex with CLR-1 directly induces cellulase and hemicellulase gene expression, when N. crassa is grown on avicel. Phylogenetic analyses of CLR-1 and CLR-2 protein sequences performed by the same group, showed that these factors are conserved in the genomes of most filamentous ascomycete fungi degrading cellulose suggesting that homologs of CLR-1 and CLR-2 play an important role in plant cell-wall degradation.
In nature, the enzymatic breakdown of plant cell wall polymers can occur in different surrounding pHs and there is a regulatory mechanism controlling pH-dependent transcriptional regulation. pacc gene in N. crassa (ORF NCU00090) is the pacc/RIM101 orthologue, extensively studied in A. nidulans and S. cerevisiae [146]. The transcription factor PacC responds to changes in extracellular pH by activating specific alkaline genes and repressing specific acid genes [147, 148].
A xylan degradation regulator-1 (xlr-1 NCU06971) is essential for hemicellulose degradation in N. crassa. xlr-1 encodes a member of a TF family containing conserved fungal Zn(2)-Cys(6) binuclear cluster domain with significant amino acid homology to xyr1 in Trichoderma species [148], sharing 57.6% identity with homolog in T. reesei. Recently, Sun et al. [143] demonstrated that a deletion of xlr-1gene abolishes growth of N. crassa in both xylan and xylose containing media, but it slightly affects the growth on Avicel and the production of cellulase activity in the presence of this substrate. To determine regulatory mechanisms for hemicellulose degradation, the authors explored the transcriptional regulation of XLR-1 under xylose, xylanolytic and cellulolytic conditions. Their results showed that XLR-1 regulates only some predicted hemicellulase genes in N. crassa and was required for a full induction of several cellulase genes. Moreover, among the genes induced by xylan there are 19 permease/transporter genes and their full induction requires a functional xlr-1. Of these 19 transporters, five have been functionally tested for transport of D-glucose and D-xylose. Another transcription factor identified in N. crassa was NIT2 protein (AreA in A. nidulans), a member of the GATA factors family, characterized in N. crassa as a positive regulator of genes encoding enzymes for nitrogen source catabolism under nitrogen limiting conditions [149, 150]. Goncalves et al. [148] suggested that nit-2 also acts as a repressor of carbon metabolism. They analyzed cis elements present in the gene encoding glycogen synthase (gsn) promoter and showed that nit-2 is able to bind these cis elements. Moreover, the knocked-out nit-2 strain showed loss of glycogen accumulation despite having low gsn gene expression as compared to the wild-type strain, suggesting they may have a role in glycogen metabolism regulation. A link between carbon and nitrogen regulation was already reported by Lockington et al. [151] in A. nidulans. Although the result was preliminary, the authors suggested the existence of a link in the regulation of the carbon and nitrogen utilization pathways in filamentous fungi.
3.2. Carbon Catabolite Repression of Cellulase and Hemicellulase Genes Expression in N. crassa
Sun et al. [152] investigated CCR of cellulase expression in N. crassa and they showed that, under cellulolytic conditions, CRE-1 regulates genes involved in plant cell wall utilization by directly binding to adjacent motifs in promoter regions and also may compete for binding with positive regulatory factors. They demonstrated that deletion of cre-1 caused constant expression of cellulase genes, resulting in higher cellulolytic enzyme activity. Moreover, cre-1 caused the repression of cellulolytic genes during growth on Avicel. Some genes known to be directly regulated by CRE-1 homologs in other systems (such as cbh-1of T.reesei and XlnA of A. nidulans) and also a large number of other target genes of predicted or unknown function in N. crassa were identified. These genes may be regulated directly or indirectly by CRE-1. For example, CRE-1 binds to the promoter region of cbh-1 in N. crassa and may compete for binding with pathway specific cellulolytic regulator required for induction; the identity of cellulolytic regulators in N. crassa is currently unknown. Among CRE-1 targets identified in N. crassa there are a hypothetical protein of unknown function (NCU 03181), an additional xylanase (NCU07225) and gh6-3 (NCU07190). It is worth of note that a MFS monosaccharide transporter (NCU04963) was identified as a direct target of CRE-1 in Neurospora. These results suggest that CRE-1 may directly regulate genes involved in sugar transport, in addition to regulating genes encoding regulatory/enzymes associated with utilization of alternative carbon sources. In summary, CRE-1 functions as a global transcription factor in N. crassa and affects both gene repression and activation, both directly and indirectly.
The same authors recently [143] studied the cre-1 regulation in hemicellulase expression. As described above, in N. crassa, transcription of most hemicellulase genes is via induction by xylanolytic molecules and is regulated via xlr-1 and/or other transcription factors. However, the hemicellulolytic system is also responsive to CCR. CRE-1 mediated CCR regulates the expression level of some, but not all, hemicellulase genes in N. crassa under Avicel conditions. xlr-1 is regulated by a combination of induction and derepression and it is also subjected to non-CRE-1 mediated CCR. These observations imply that other mechanisms regulate CCR in filamentous fungi in addition to CRE-1, similar to what has been described for S. cerevisiae [153].
4. REGULATION OF PRODUCTION OF CELLULASES AND HEMICELLULASES IN ASPERGILLUS SPP
The genomes of four Aspergillus spp., A. nidulans, A. oryzae, A. niger and A. fumigatus, have been recently sequenced (http://www.aspergillusgenome.org/), and shown to contain around 200 genes -out of 14,600- involved in polysaccharides’ degradation [154]. Aspergillus spp. have been so far described as high cellulases’ producers and many genes coding for cellulase, endoxylanases, β-xylosidases and pectinases have been cloned and characterized from Aspergillus spp. strains [24]. A summary of inducibility of cellulases, hemicellulases and related enzymes in Aspergillus spp. is reported in the Table 1.
The influence of carbon and nitrogen sources on the production of cellulases has been so far investigated showing that the enzyme production is strongly variable according to the carbon source.
For instance, Hanif et al. [155] showed that even low concentrations of glucose negatively affect β-cellobiohyd rolase (CBH) production in Aspergillus niger, whilst cellulose and wheat bran stimulate β-cellobiohydrolase and filter paperase (FPase) activities, respectively. It was shown that addition of glucose inhibited cellulase production, even in cultures of A. niger gowing on wheat bran, shown to be a good inducer.
In several manuscripts, lactose has been defined as the best inducer for cellulase production in Aspergillus spp. Mrudula and Murugammal [156] confirmed that lactose is the best inducer of cellulase activity production by Aspergillus niger. In fact, lactose was shown the best carbon source to obtain high level of both CMCase and FPase activities, in both liquid and solid state fermentation.
As shown by Ali and Sayed [157], xylose is the best carbon source for induction of both endo- and exo-cellulase activity production in A. terreus, whilst the production of β-glucosidase is positively affected by both glucose and xylose. Lignocellulosic substrates, like agroindustrial wastes, have been so far described as good substrate for cellulase and xylanase production by filamentous fungi, as Aspergillus spp. Among the several examples, Ghori et al. [158] recently demonstated the properties of corn-stover as an inducer of cellulase activity production by A. niger. Moreover, they demonstated that addition of cane molasses and yeast sludge to the fermentation medium leads to an increase of cellulase production. However, the induction mechanism involved in solid state fermentation have been shown to be more complex [159].
The effect of several carbon sources on glycosyl hydrolases gene expression has been studied by Nazir et al. [160] who reported differential expression of endoglucanase and beta-glucosidase isoforms of A. terreus, in both solid and liquid cultures. Maximal expression of four endoglucanase isoforms was observed in presence of rice straw and corn cobs, in solid state and liquid fermentation, respectively. Addition of fructose and cellobiose to corn cobs containing medium caused the up-regulation of endoglucanase activity, whereas addition of mannitol, ethanol and glycerol selectively repressed the expression of at least three endoglucanase isoforms. As far as the beta-glucosidase profiling is concerned, addition of glucose, fructose, sucrose, cellobiose, mannitol and glycerol resulted in down-regulation of most of the isoforms.
Many manuscripts have been reported concerning induction of xylanase production in Aspergillus spp.. It is well known that xylose, xylan and crude xylan-containing substrates mainly induce xylanolytic enzymes production in Aspergilli spp.. There are rare cases where other monomeric or polymeric substrates, such as glucose and cellulose, induce xylanolytic expression. For instance, Hrmova et al. [161] observed the induction of xylanolytic enzymes by cellulose, cellobiose and even by a heterodysaccharide consisting of glucose and xylose in A. terreus. The regulation of xyalonlytic enzymes is not identical in all Aspergillus spp.. As a matter of fact, Kimura et al. [162] cloned a xylose-inducible, glucose repressed endoxylanase gene from A. oryzae in A. nidulans, where its expression was instead increased by adding glucose.
Pinaga et al. [163] studied the effect of several compounds on xylanase production by A. nidulans. Xylooligosaccharides such as xylobiose, xylotriose and xylotetraose induced xylanase activity production, their efficiency being directly related to their chain length. However, xylans such as wheat arabinoxylan, oat spelt xylan, birchwood xylan and 4-O-methyl-D-glucorono-D-xylan were found to be the most powerful inducers. Xylose, on the contrary, was not shown to be a good inducer.
Xylanases production by A. phoenicis was shown positively affected by xylan, xylose and b-methylxyloside, similarly to the cases of other fungi belonging to the Aspergillus genus such as Aspergillus sydowii and A. tubingensis, as studied and discussed by Rizzatti et al. [164]. This study also demonstrated that the levels of production of xylanase by A. phoenics, decreased when glucose was added to the inducers xylan or xylose, similarly to A. sydowii whose xylanase production is inhibited by glucose [165].
Methyl β-d-xyloside was shown a more effective inducer than xylan, for both extracellular xylanase and intracellular β-xylosidase by Simao et al. [166]. The same group also demonstated that both glucose and cycloheximide inhibit the positive effect of methyl β-d-xyloside on xylanase production. However, not much is known about the uptake system for the inducers xylose and xylobiose in Aspergillus spp,. but more is known about the formation of the inducing compounds. For instance, the A.niger β-xylosidase, encoded by xlnD, has been shown to have an important role in xylanolytic inducer formation, being active towards xylan and xylooligosaccharides for the formation of D-xylose [167].
Galacturonic acid is the main inducer of several pectinolytic enzymes encoding genes such as pelA, plyA, pgaX, rglA and pmeA [168]. As reported by de Vries et al. [169], galacturonic acid positively affects even the expression of several genes encoding enzymes which act on the pectin side chains such as arabinofuranosidases (abfA and abfB), endoarabinase (abnA), endogalactanase (galA) and galactosidase (lacA).
As far as the induction of extracellular arabinases is concernd, pentose sugars and polyols generated by the metabolic pathway of L-arabinose and D-xylose catabolism were shown to be invooved in Aspergillus niger arabinases production. Particularly, induction occurred with L-arabinose and L-arabitol but not with D-xylose or xylitol, L-arabitol being the best inducer for a-L-arabinofuranosidase and endo-arabinase activities [170].
4.1. Transcriptional Factors Involved in Regulation of Cellulase and Hemicellulase Genes Expression in Aspergillus spp
Transcription factors involved in the regulation of Aspergillus spp. (hemi)cellulolytic enzymes encoding genes, mostly belong to Zn(II)2Cys6 binuclear cluster DNA-binding motif family (Table 2, Fig. 1).
XlnR is the main transcriptional activator which has been largely studied for its involvement in the regulation of cellulases, hemicellulases and accessory enzyme genes for xylan degradation in Aspergillus spp.. It is an orthologue of the xyR1 gene of T. reesei [18].
van Peiji et al. [171] finely described the role of XlnR transcriptional activator. It has been demonstrated that XlnR regulates the transcription of the xlnB, xlnC and xlnD genes encoding endoxylanases B, endoxylanase C and β-xylosi dase, respectively. It is also involved in the activation of cellulase genes transcription, such as those coding for the two endoglucanases eglA and eglB. In addition, XlnR has been shown to positively affect the transcription of several accessory enzymes gene involved in hemicelluloses degradation, including glucuronidase A, acetylxylan esterase A, arabinoxylan arabinofuranohydrolase A and feruloyl esterase A.
Several northern blot analyses have been performed on A. niger strains, in order to demonstrate the important role of XlnR on the activation of different glycosyl hydrolases genes transcription. These analyses allowed the comparison of level of expression of genes of interest in an A. niger wild-type strain, a xlnR loss-of-function mutated strain and a multiple-copy strain [171, 172].
More recently, similar studies have been performed by Tani et al. [173] who demonstrated that cellulose affects positively both cellulase and hemicellulase activities production in A. aculeatus, through two different pathways, namely XlnR-dependent and XlnR-independent pathways. Real-time PCR (Polymerase Chain Reaction) experiments have been performed to identify the genes controlled by the XlnR-independent pathway. Particularly, both cellobiose and cellulose were shown to induce the expression of the gene regulated by XlnR-independent signaling pathway, the latter stimulating expression of FIII-avicelase (cbhI), FII-carboxymethyl cellulase (cmc2), and FIa-xylanase (xynIa).
Recently, further analyses on (hemi)cellulase genes regulation in A. aculeatus have been performed by Kunitake et al. [174]. ClbR, a new activator with a Zn(II)2Cys6 binuclear cluster DNA-binding motif specific for fungi, has been identified. It has been shown to control the cellobiose and cellulose responsive induction of cellulase and xylanase genes which are regulated by both XlnR-dependent and XlnR-independent signaling pathways. For instance, disruption of clbR gene caused the decrease of the cellobiose- and cellulose-responsive induction of the cbhI, cmc2, and xynIa genes and the cellulose-responsive induction of the cmc1 and xynIb genes Kunitake et al. [174].
Differently from T. reesei, fine-tuning transcription factors like Ace1 and Ace2 cannot be found in Aspergillus spp. The putative ACEI proteins of A. nidulans, stzA (AF202995), is deposited into the database as a gene encoding a protein that alleviates sensitivity to salt and DNA damaging agents. Interestingly, stzA has been identified as an orthologue of the T. Reesei ACE1 gene [175]. The authors provided evidence of competition, or interaction, between the ACE1/StzA and AreA binding sites in promoters of stzA and its orthologs, and in genes involved in the metabolism of amino acids. The A. nidulans and A. fumigatus cpcA (cross pathway control regulator of amino acid biosynthesis) promoters have seven potential ACE1/StzA binding sites, six of which are highly conserved in position. The presence of potential CPC1 binding sites (5'-TGAC/GTCA) in the stzA and ace1 promoters suggests an intriguing link between intracellular amino acid availability and cellulase gene expression. In accordance with these findings, a recent study by Gremel et al. [108] indeed revealed that cellulase gene expression can be enhanced by the addition of methionine.
PacC is the major factor involved in pH-dependent expression in Aspergillus spp.. pH regulation of genes encoding cell wall-degrading enzymes has not been studied in detail in Aspergillus. However, indications for pH-dependent expression of xylanolytic and pectinolytic genes have been obtained [176]. Kojma et al. [176] demonstrated that A. kawachii produces different polygalacturonases using culture media with different pHs whilst A. nidulans PacC mutant strain does not produce arabinofuranosidase activity [177] and two endoxylanase, xlnA and xlnB [178]. Even cellulase production in A. fumigatus is affected by pH [179]. Indeed, two and one PacC consensus sites have been revealed in the promoter regions of xlnA, xlnB [180] and xlnD [181], respectively. The role of the transcriptional activator AreA on total cellulase production has been studied by Lockington et al. [182]. areA gene product is known to control the expression of genes encoding the enzymes involved in nitrogen metabolism in ammonium derepressing conditions [183]. The homologous of AreA was identified in N. crassa as NIT2 protein, [149, 150].
It has been shown that the amount of total secreted cellulase activity increased in a strain containing the constitutively activating areA allele, xprD1, and decreased in a strain containing the loss of function allele, areA217. To deepen AreA role in cellulase genes regulation, two genes encoding exocellulases, and one gene encoding an endocellulase were cloned. The putative regulatory regions of all the genes contain potential binding sites for the global carbon and nitrogen regulatory proteins, CreA and AreA, potential consensus binding sites for XlnR, whilst the AceII DNA binding consensus sequence involved in induction in T. reesei, misses in all the genes [182]. Real-time PCR techniques were used to assess the relative expression levels of genes encoding hydrolase activities and of the genes encoding regulatory elements such as AreA, PacC and CreA in an effort to identify possible transcriptional regulation mechanisms in A. oryzae solid state fermentation [159]. This study showed the complexity of the regulation of genes coding for hydrolytic enzymes under solid state fermentation, as other factors such as post-transcriptional regulation appeared to be involved.
4.2. Carbon Catabolite Repression of Cellulase and Hemicellulase Genes Expression in Aspergillus spp
It has been reported that creA, cre B and creC genes products are involved in the regulatory mechanism of carbon catabolite repression in Aspergillus spp. [183-187]. CreA-mediated repression in Aspergillus has been demonstrated for genes encoding cellulase, arabinases, several endoxylanases and other xylanolytic activities such as xylosidase, feruloyl esterase and some pectinases [22]. The binding consensus motif for A. nidulans CreA was determined to be 5'-SYGGRG [188]. Besides glucose, other monomeric carbon sources result in CreA-mediated repression of gene expression, such as xylose. For instance, high concentrations of xylose have been shown to activate the CreA-mediated repression, by down-regulating the expression levels of several xylanolytic and cellulolytic genes in A. niger [169] and A. terreus [157].
Interestingly, Flipphi et al. [189] showed that in A. nidulans, mutations in both the single glucokinase and the single hexokinase genes belonging to the fungus, lead to a CreA-mediated carbon catabolite derepression, similarly to T. reesei which also features only one glucokinase and one hexokinase.
CreB encodes a deubiquitinating enzyme and it is a functional member of a novel subfamily of the ubp family defined by the human homolog UBH1 [186]. It forms a complex with a WD40-repeat protein encoded by creC [182], which is required to prevent the proteolysis of CreB in the absence of CCR [187]. Interestingly, the E3 ubiquitin ligase LIM1 also responds to cellulase inducing conditions and binds to the cbh2-promoter [108].
In addition, CreD has been reported to be involved in CCR of Aspergillus sp.. Mutations in creD suppress the phenotypic effects of mutations in creC and creB [190]. CreD contains arrestin domains and PY motifs and is highly similar to S. cerevisiae Rod1p and Rog3p, which interact with the ubiquitin ligase Rsp5p [191].
5. CONCLUSIONS
The regulation of (hemi)cellulolytic genes appears to be basically the same among filamentous fungi such as T. reesei, N. crassa, Asperigillus spp., although their regulatory mechanisms are quite complex and present some differences. In particular, cross talks between expression of cellulolytic and hemicellulolytic genes make the regulatory mechanisms more complicated. Xyr homologs (Xyr1 of T.reesei, XlR of N. crassa, and XlnR of Aspergillus spp) mediate expression of both xylanolytic and cellulolytic genes in response to xylan. Moreover, Xyr homologs mediate cellulose-inductive expression of the xylanolytic genes as well as the cellulolytic genes. In addition to Xyr homologues, AceI and AceII in T. reesei, and ClbR in A. aculeatus, are suggested to be involved in the regulation of the expression of these genes. This further makes the regulatory mechanisms of (hemi) cellulolytic genes complicated. Unfortunately, the roles of AceI and AceII in the regulation of cellulolytic genes remain ambiguous and need to be investigated further. Carbon catabolite repression of cellulase expression appears to be essentially the same among fungi. As described above, major inductive signals for fungi to degrade plant cell wall are derived from cellulose and xylan. Xyr homologs receive inducing signals from these two different polysaccharides and activate transcription of many (hemi)cellulolytic genes. Xyr homologues could recognize two distinct inductive signals, in response to cellulose and xylan, prior to induction of the target genes due to their differential conformational changes.
As a main difference among the cellulase and hemicellulase regulatory systems from different fungi, these systems appear to be more specialized in T. reesei than in the other fungi, considering that T. reesei fine-tuning transcription factors like Ace1 and Ace2 cannot be found in Aspergillus and Neurospora spp.
However, more research is needed to completely disclose the molecular mechanisms of regulation of cellulase and hemicellulase gene expression in T. reesei, Aspergillus and Neurospora spp.. Their elucidation could provide a basis for the rational application of transcriptional regulators for biotechnological processes in filamentous fungi, leading to efficient bioethanol production from lignocellulosic biomass.
ACKNOWLEDGEMENTS
This work was supported by grant from the Ministero dell’Università e della Ricerca Scientifica -Industrial Research Project “Integrated agro-industrial chains with high energy efficiency for the development of eco-compatible processes of energy and biochemicals production from renewable sources and for the land valorization (Enerbio-Chem)” PON01_01966, funded in the frame of Operative National Programme Research and Competitiveness 2007–2013 D. D. Prot. n. 01/Ric. 18.1.2010.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Lynd L R, Cushman J H, Nichols R J, Wyman C E. Fuel ethanol from cellulosic biomass. Science. 1991;251:1318–23. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- 2.Lynd L R. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu. Rev. Energy. Environ. 1996;21:403–65. [Google Scholar]
- 3.Wyman C E. Ethanol production from lignocellulosic biomass: overview. In: Wyman C E, editor. Handbook on bioethanol, production and utilization. Taylor and Francis; 1996. pp. 1–18. [Google Scholar]
- 4.Wyman C E. Biomass ethanol: technical progress, opportunities, and commercial challenges. Annu. Rev. Energy. Environ. 1999;24:189–226. [Google Scholar]
- 5.Wyman C E. Twenty years of trials, tribulations, and research progress in bioethanol technology—selected key events along the way. Appl. Biochem. Biotechnol. 2001:91–93. doi: 10.1385/abab:91-93:1-9:5. 5–21. [DOI] [PubMed] [Google Scholar]
- 6.Antizar-Ladislao B, Turrion-Gomez J L. Second-generation biofuels and local bioenergy systems. Bioprod. Bioref. 2008;2:455–69. [Google Scholar]
- 7.Champagne P. Bioethanol from agricultural waste residues. Environ. Progr. 2008;27(1):51–7. [Google Scholar]
- 8.Farrell A E, Plevin R, Turner B, Jones A, O’Hare M, Kammen D. Ethanol can contribute to energy and environmental goals. Science. 2006;311:506–8. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 9.Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund M F, Lidén G, Zacchi G. Bioethanol— the fuel of tomorrow from the residues of today. Trends. Biotechnol. 2006;24(12):549–56. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. USA. 2006;103(30):11206–10. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y, Tanaka S. Ethanol fermentation from biomass resources: current state and prospects. Appl. Microbiol. Biotechnol. 2006;69:627–42. doi: 10.1007/s00253-005-0229-x. [DOI] [PubMed] [Google Scholar]
- 12.Tollefson J. Not your father’s biofuels. Nature. 2008;451(21):880–3. doi: 10.1038/451880a. [DOI] [PubMed] [Google Scholar]
- 13.Wyman C E. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol. 2007;25(4):153–7. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Solomon B D. Biofuels and sustainability. Ann. N. Y. Acad. Sci. 2010;1185:119–34. doi: 10.1111/j.1749-6632.2009.05279.x. [DOI] [PubMed] [Google Scholar]
- 15.Faraco V, Hadar Y. The Potential of Lignocellulosic Ethanol Production in the Mediterranean Basin. Renewable and Sustainable Energy Reviews. 2011;15:252–266. [Google Scholar]
- 16.Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biothechnol. 2009;85:141–154. doi: 10.1007/s00253-009-2236-9. [DOI] [PubMed] [Google Scholar]
- 17.van Peij N N, Visser J, de Graaf L H. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 1998a;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 18.Stricker A R, Mach R L, de Graff L H. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl. Microbiol. Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 19.Karasawa T, Yachi M, Suto M, Kamagata Y, Takao S, Tomita F. Induction of cellulase by gentiobiose and its sulfurcontaining analog in Penicillium purpurogenum. Appl. Environ. Microbiol. 1992;58:106–110. doi: 10.1128/aem.58.1.106-110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrmová M, Petraková E, Biely P. Induction of cellulose- and xylan-degrading enzyme systems in Aspergillus terreus by homo- and heterodisaccharides composed of glucose and xylose. J. Gen. Microbiol. 1991;137:541–547. doi: 10.1099/00221287-137-3-541. [DOI] [PubMed] [Google Scholar]
- 21.Mandels M, Parrish F W, Reese E T. Sophorose as an inducer of cellulase in Trichoderma reesei. J. Bacteriol. 1962;83:400–408. doi: 10.1128/jb.83.2.400-408.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries R P, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruijter G J, Visser J. Carbon repression in Aspergilli. FEMS Microbiol. Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 24.Aro N, Pakula T, Penttilä M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 2005;29:719–739. doi: 10.1016/j.femsre.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Kubicek C P, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol. Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubicek C P. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J. Biotechnol. 2012 doi: 10.1016/j.jbiotec.2012.05.020. http://dx.doi.org/10.1016/j.jbiotec.2012.05.020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez D, Berka R M, Henrissat B, Saloheimo M, Arvas M, Baker S E, Chapman J, Chertkov O, Coutinho P M, Cullen D, Danchin E G, Grigoriev I V, Harris P, Jackson M, Kubicek C P, Han C S, Ho I, Larrondo L F, de Leon A L, Magnuson J K, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch C L, Yao J, Barabote R, Nelson M A, Detter C, Bruce D, Kuske C R, Xie G, Richardson P, Rokhsar D S, Lucas S M, Rubin E M, Dunn-Coleman N, Ward M, Brettin T S. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat. Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 28.Okada H, Tada K, Sekiya T, Yokoyama K, Takahashi A, Tohda H, Kumagai H, Morikawa Y. Molecular characterization and heterologous expression of the gene encoding a low-molecular-mass endoglucanase from Trichoderma reesei QM9414. Appl. Environ. Microbiol. 1998;64:555–563. doi: 10.1128/aem.64.2.555-563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saloheimo M, Lehtovaara P, Penttila M, Teeri T T, Stahlberg J, Johansson G, Pettersson G, Claeyssens M, Tomme P, and Knowles J K. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene (Amst.) 1988;63:11–22. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- 30.Saloheimo M, Nakari-Setala T, Tenkanen M, Penttila M. cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur. J. Biochem. 1997;249:584–591. doi: 10.1111/j.1432-1033.1997.00584.x. [DOI] [PubMed] [Google Scholar]
- 31.Saloheimo A, Henrissat B, Hoffren A M, Teleman O, Penttilä M. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 1994;13:219–228. doi: 10.1111/j.1365-2958.1994.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 32.Teeri T T, Lehtovaara P, Kauppinen S, Salovuori I, Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene (Amst.) 1987;51:43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- 33.Ward M, Wu S, Dauberman J, Weiss G, Larenas E, Bower B, Rey M, Clarkson K, Bott R. In: Biochemistry and Genetics of Cellulose Degradation. Aubert J P, Beguin P, and Millet J, editors. New York: Academic Press; 1993. pp. 53–70. [Google Scholar]
- 34.Barnett C C, Berka R M, and Fowler T. Cloning and amplification of the gene encoding an extracellular beta-glucosidase from Trichoderma reesei: evidence for improved rates of saccharification of cellulosic substrates. Bio/Technology. 1991;9:562–567. doi: 10.1038/nbt0691-562. [DOI] [PubMed] [Google Scholar]
- 35.Takashima S, Nakamura A, Hidaka M, Masaki H, Uozumi T. Molecular cloning and expression of the novel fungal beta-glucosidase genes from Humicola grisea and Trichoderma reesei. J. Biochem. (Tokyo) 1999;125:728–736. doi: 10.1093/oxfordjournals.jbchem.a022343. [DOI] [PubMed] [Google Scholar]
- 36.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 37.Durand H, Baron M, Calmels T, and Tiraby G. Classical and molecular genetics applied to Trichoderma reesei for the selection of improved cellulolytic industrial strains. FEMS Symp. 1998;43:135–152. [Google Scholar]
- 38.Amore A, and Faraco V. Potential of fungi as category I Consolidated BioProcessing organisms for cellulosic ethanol production. Renewable & Sustainable Energy Reviews. 2012;16:3286–3301. [Google Scholar]
- 39.Törrönen A, Harkki A, Rouvinen J. Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 1994;13(11):2493–2501. doi: 10.1002/j.1460-2075.1994.tb06536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrmann M C, Vrsanska M, Jurickova M, Hirsch J, Biely P, and Kubicek C P. The beta-D-xylosidase of Trichoderma reesei is a multifunctional beta-D-xylan xylohydrolase. Biochem. J. 1997;321:375–381. doi: 10.1042/bj3210375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bisaria V S, Mishra S. Regulatory aspects of cellulase biosynthesis and secretion. CRC Crit. Rev. Biotechnol. 1989;9:61–113. doi: 10.3109/07388558909040616. [DOI] [PubMed] [Google Scholar]
- 42.Kubicek CP. From cellulose to cellulase inducers: facts and fiction. In: Suominen P, Reinikainen T, editors. Proceedings of the 2nd Tricel Symposium on Trichoderma reesei cellulases and other hydrolases. Espoo, Finland: Foundation for biotechnical and industrial fermentation research; 1993. pp. 181–188. [Google Scholar]
- 43.Zeilinger S, Mach R L. Xylanolytic enzymes of Trichoderma reesei: properties and regulation of expression. Cer. Chem. Res. Trends. 1998;1:27–35. [Google Scholar]
- 44.Mach R L, Zeilinger S. Regulation of gene expression in industrial fungi: Trichoderma. Appl. Microbiol. Biotechnol. 2003;60(5):515–522. doi: 10.1007/s00253-002-1162-x. [DOI] [PubMed] [Google Scholar]
- 45.Karaffa L, Fekete E, Gamauf C, Szentirmai A, Kubicek CP, Seiboth B. D-galactose induces cellulase gene expression in Hypocrea jecorina at low growth rate. Microbiology. 2006;152:1507–1514. doi: 10.1099/mic.0.28719-0. [DOI] [PubMed] [Google Scholar]
- 46.Kubicek CP, Penttilä ME. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocaldium. Enzymes biological control and commercial applications. London: Taylor and Francis; 1998. pp. 49–72. [Google Scholar]
- 47.Morikawa Y, Ohashi T, Mantani O, Okada H. Cellulase induction by lactose in Trichoderma reesei PC-3-7. Appl. Microbiol. Biotechnol. 1995;44:106–111. [Google Scholar]
- 48.Stricker A R, Grosstessner-Hain K, Wu¨rleitner E, Mach RL. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell. 2006;5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stricker A R, Steiger M G, Mach R L. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 2007;581:3915–3920. doi: 10.1016/j.febslet.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 50.El-Gogary S, Leite A, Crivellaro O, Eveleigh D E, El-Dorry H. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc. Natl. Acad. Sci. USA. 1989;86:6138–6141. doi: 10.1073/pnas.86.16.6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carle-Urioste J C, Escobar-Vera J, El-Gogary S, Henrique-Silva F, Torigoi E, Crivellaro O, Herrera-Estrella A, El-Dorry H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 1997;272:10169–10174. doi: 10.1074/jbc.272.15.10169. [DOI] [PubMed] [Google Scholar]
- 52.Gritz L, and Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene (Amst.) 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 53.Kubicek C P, Messner R, Gruber F, Mandels M, Kubicek-Pranz E M. Triggering of cellulase biosynthesis in Trichoderma reesei: involvement of a constitutive, sophorose-inducible, glucoseinhibited β-diglucoside permease. J. Biol. Chem. 1993;268:19364–19368. [PubMed] [Google Scholar]
- 54.Foreman P K, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman N S, Goedegebuur F, Houfek T D, England G J, Kelley A S, Meerman H J, Mitchell T, Mitchinson C, Olivares H A, Teunissen P J, Yao J, Ward M. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 55.Kubicek C P, Mühlbauer G, Grotz M, John E, Kubicek-Pranz E M. Properties of a conidial bound cellulase enzyme system from Trichoderma reesei. J. Gen. Microbiol. 1988;134:1215–1222. [Google Scholar]
- 56.Messner R, Kubicek-Pranz E M, Gsur A, Kubicek C P. Cellobiohydrolase II is the main conidial bound cellulase in Trichoderma reesei and other Trichoderma strains. Arch. Microbiol. 1991;155:601–606. doi: 10.1007/BF00245356. [DOI] [PubMed] [Google Scholar]
- 57.Kubicek-Pranz E M, Gruber F, Kubicek C P. Transformation of Trichoderma reesei with the cellobiohydrolase II gene as a means for obtaining strains with increased cellulase production and specific activity. J. Biotechnol. 1991;20:83–94. [Google Scholar]
- 58.Seiboth B, Messner R, Gruber F, Kubicek C P. Disruption of the Trichoderma reesei cbh2 gene coding for cellobiohydrolase II leads to a delay in the triggering of cellulase formation by cellulose. J. Gen. Microbiol. 1992;138:1259–1264. [Google Scholar]
- 59.Ilmén M, Saloheimo A, Onnela M L, Penttilä ME. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 1997;63:1298–1306. doi: 10.1128/aem.63.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandels M, Reese E T. Induction of cellulase in fungi by cellobiose. J. Bacteriol. 1960;79:816–826. doi: 10.1128/jb.79.6.816-826.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sternberg D, Mandels G R. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J. Bacteriol. 1979;139:761–769. doi: 10.1128/jb.139.3.761-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaheri M, Leisola M, Kauppinen V. Transglycosylation products of cellulase system of Trichoderma reesei. Biotechnol. Lett. 1979;1:41–46. [Google Scholar]
- 63.Sternberg D, Mandels G R. Regulation of the cellulolytic system in Trichoderma reesei by sophorose: induction of cellulase and repression of beta-glucosidase. J. Bacteriol. 1980;144:1197–1199. doi: 10.1128/jb.144.3.1197-1199.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loewenberg J R, Chapman C M. Sophorose metabolism and cellulase induction in Trichoderma. Arch. Microbiol. 1977;113:61–64. doi: 10.1007/BF00428581. [DOI] [PubMed] [Google Scholar]
- 65.Fowler T, Brown R D., Jr The bgl1 gene encoding extracellular β- glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 1992;6:3225–3235. doi: 10.1111/j.1365-2958.1992.tb01777.x. [DOI] [PubMed] [Google Scholar]
- 66.Messner R, Kubicek C P. Evidence for a single, specific β-glucosidase in cell walls from Trichoderma reesei QM 9414. Enzyme Microb. Technol. 1990;12:685–690. [Google Scholar]
- 67.Umile C, Kubicek C P. A constitutive, plasma-membrane bound β-glucosidase in Trichoderma reesei. FEMS Microbiol. Lett. 1986;34:291–295. [Google Scholar]
- 68.Inglin M, Feinberg B A, Loewenberg J R. Partial purification and characterization of a new intracellular beta-glucosidase of Trichoderma reesei. Biochem. J. 1980;185:515–519. doi: 10.1042/bj1850515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saloheimo M, Kuja-Panula J, Ylösmäki E, Ward M, Penttilä M. Enzymatic properties and intracellular localization of the novel Trichoderma reesei beta-glucosidase BGLII (CEL1A) Appl. Environ. Microbiol. 2002;68:4546–4553. doi: 10.1128/AEM.68.9.4546-4553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mach R L, Seiboth B, Myasnikov A, Gonzalez R, Strauss J, Harkki A M, Kubicek C P. The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellulose-inducible β-glucosidase involved in cellulase induction by sophorose. Mol. Microbiol. 1995;16:687–697. doi: 10.1111/j.1365-2958.1995.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 71.Akel E, Metz B, Seiboth B, Kubicek C P. Molecular regulation of arabinan and L-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei) Eukaryot. Cell. 2009;8:1837–1844. doi: 10.1128/EC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mach-Aigner A R, Pucher M E, Steiger M G, Bauer G E, Preis S J, Mach R L. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 2008;74:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1998;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 74.Tansey W P. Transcriptional activation: risky business. Genes Dev. 2001;15:1045–1050. doi: 10.1101/gad.896501. [DOI] [PubMed] [Google Scholar]
- 75.Aro N, Saloheimo A, Ilmén M, Penttilä M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 76.Stricker A R, Trefflinger P, Aro N, Penttilä M, Mach R L. Role of Ace2 (activator of cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 2008;45:436–445. doi: 10.1016/j.fgb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Furukawa T, Shida Y, Kitagami N, Mori K, Kato M, Kobayashi T, Okada H, Ogasawara W, Morikawa Y. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 2009;46:564–574. doi: 10.1016/j.fgb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 78.Zeilinger S, Mach R L, Kubicek C P. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 1998;273:34463–24471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]
- 79.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Linhoff M W, Wright K L, Ting J P. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol. Cell. Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 83.Zeilinger S, Ebner A, Marosits T, Mach R, Kubicek C P. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase IIgene) activating element. Mol. Genet. Genomics. 2001;266:56–63. doi: 10.1007/s004380100518. [DOI] [PubMed] [Google Scholar]
- 84.Zeilinger S, Schmoll M, Pail M, Mach R L, Kubicek C P. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics. 2003;270:46–55. doi: 10.1007/s00438-003-0895-2. [DOI] [PubMed] [Google Scholar]
- 85.Zou G, Shi Shaohua, Jiang Y, vanden Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang C, and Zhou Z. Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microbial Cell Factories. 2012;11:21. doi: 10.1186/1475-2859-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saloheimo A, Aro N, Ilmén M, Penttilä M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 87.Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, and Kubicek C P. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Molecular Microbiology. 2012;84(6):1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, Lichius A, Omann M, Zeilinger S, Seiboth B, Rhee C, Keller NP, Freitag M, Kubicek CP. Functional Analyses of Trichoderma reesei LAE1 Reveal Conserved and Contrasting Roles of This Regulator. G3 (Bethesda) 2013;3(2):369–78. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ilmén M, Thrane C, Penttila M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 91.Strauss J, Mach R L, Zeilinger S, Hartler G, Stöffler G, Wolschek M, Kubicek C P. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 1995;376:103–107. doi: 10.1016/0014-5793(95)01255-5. [DOI] [PubMed] [Google Scholar]
- 92.Ilmén M, Onnela M L, Klemsdal S, Keränen S, Penttilä M. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 1996;253:303–314. doi: 10.1007/pl00008597. [DOI] [PubMed] [Google Scholar]
- 93.Zimmermann F K, Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol. Gen. Genet. 1977;154:75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]
- 94.Eveleigh D E, Montenecourt B S. Increasing yields of extracellular enzymes. Adv. Appl. Microbiol. 1979;25:57–74. doi: 10.1016/s0065-2164(08)70146-1. [DOI] [PubMed] [Google Scholar]
- 95.Sheir-Neiss G, Montenecourt B S. Characterization of the secreted cellulases of Trichoderma reesei wild type mutants during controlled fermentations. Appl. Microbiol. Biotechnol. 1984;20:46–53. [Google Scholar]
- 96.Durand H, Clanet H, Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb. Technol. 1988;10:341–346. [Google Scholar]
- 97.Dowzer C E, Kelly J M. Analysis of the creA gene; a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakari-Setälä T, Paloheimo M, Kallio J, Vehmaanperä J, Penttilä M, Saloheimo M. Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl. Environ. Microbiol. 2009;75:4853–4860. doi: 10.1128/AEM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seidl V, Gamauf C, Druzhinina I S, Hartl L, Seiboth B, Kubicek C P. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Le Crom S, Schackwitz W, Pennacchio L, Magnuson J K, Culley D E, Collett J R, Martin J, Druzhinina IS, Mathis H, Monot F, Seiboth B, Cherry B, Rey M, Berka R, Kubicek CP, Baker S E, Margeot A. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. PNAS. 2009;106(38):16151–1615. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vitikainen M, Arvas M, Pakula T, Oja M, Penttilä M, and Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mach R L, Strauss J, Zeilinger S, Schindler M, Kubicek C P. Carbon catabolite repression of xyn1 (xylanase I-encoding) gene expression in Trichoderma reesei. Mol. Microbiol. 1996;21:1273–1281. doi: 10.1046/j.1365-2958.1996.00094.x. [DOI] [PubMed] [Google Scholar]
- 103.Cziferszky A, Mach R L, Kubicek C P. Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei) J. Biol. Chem. 2002;277:14688–14694. doi: 10.1074/jbc.M200744200. [DOI] [PubMed] [Google Scholar]
- 104.Meggio F, Pinna L A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 105.Carlson M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 106.Czifersky A, Seiboth B, Kubicek CP. The Snf1 kinase of the filamentous fungus Hypocrea jecorina phosphorylates regulation-relevant serine residues in the yeast carbon catabolite repressor Mig1 but not in the filamentous fungal counterpart Cre1. Fungal Genet. Biol. 2003;40:166–175. doi: 10.1016/s1087-1845(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 107.Denton J, Kelly J M. Identification and characterization of the Trichoderma reesei homologue of Aspergillus nidualns creB. Fungal Genet. Newsl. 54(Suppl):98. [Google Scholar]
- 108.Gremel G, Dorrer M, Schmoll M. Sulphur metabolism and cellulase gene expression are connected processes in the filamentous fungus Hypocrea jecorina (anamorph Trichoderma reesei) BMC Microbiol. 2008;8:174. doi: 10.1186/1471-2180-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rolland F, Winderickx J, Thevelein J M. Glucose-sensing and –signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 110.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 111.Santangelo G M. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Portnoy T, Margeot A, Seidl-Seiboth V, Le Crom S, Ben Chaabane F, Linke R, Seiboth B, Kubicek C P. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell. 2011;10(2):262–71. doi: 10.1128/EC.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Montenecourt B S, Eveleigh D E. Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl. Environ. Microbiol. 1977;34:777–782. doi: 10.1128/aem.34.6.777-782.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Davis R H, Perkins D D. Neurospora: A model of model microbes. Nat. Rev. Genet. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- 115.Perkins D, Radford A, Newmeyer D, Bjorkman M. Chromosomal Loci of Neurospora crassa. Microbiological Reviews. 1982:426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eberhart B M, Beck R S, Goolsby K M. Cellulase of Neurospora crassa. J. Bacteriol. 1977;130:181–186. doi: 10.1128/jb.130.1.181-186.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marsh P B, Bollenbacher K, Butler M L, Raper K B. The fungi concerned in fiber deterioration. II. Their ability to decompose cellulose. Textile Res. J. 1949;19:461–484. [Google Scholar]
- 118.Hirsh H M. Temperature dependent cellulase production by Neurospora crassa and its ecological implications. Experientia. 1954;10:180–182. doi: 10.1007/BF02157201. [DOI] [PubMed] [Google Scholar]
- 119.Eberhart B M. Exogenous enzymes of Neurospora conidia and mycelia. J. Cell. Comp. Physiol. 1961;58:11–16. doi: 10.1002/jcp.1030580103. [DOI] [PubMed] [Google Scholar]
- 120.Bates W K, Woodward W O. Neurospora beta-galactosidase: Evidence for a second enzyme. Science. 1964;146:777–778. doi: 10.1126/science.146.3645.777. [DOI] [PubMed] [Google Scholar]
- 121.Yazdi M T, Woodwarda J R, Radford A. The cellulase complex of Neurospora crassa: activity, stability and release. Journal of General Microbiology. 1990c;136:1313–1319. doi: 10.1099/00221287-136-7-1313. [DOI] [PubMed] [Google Scholar]
- 122.Mishra C, Keskar S, Rao M. Production and properties of extracellular endoxylanase from Neurospora crassa. Appl. Environ. Microb. 1984;48:224–228. doi: 10.1128/aem.48.1.224-228.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deshpande V, Lachke A, Mishra C, Keskar S, and Rao M. Mode of action and properties of xylanase and β-Xylosidase from Neurospora crassa. Biotechnol. Bioeng. 1986;28:1832–1837. doi: 10.1002/bit.260281210. [DOI] [PubMed] [Google Scholar]
- 124.Rao M, Mishra C, Keskar S, Srinivasan MC. Production of ethanol from wood and agricultural residues by Neurospora crassa. Enzyme Microb. Technol. 1985;7:625–628. [Google Scholar]
- 125.Yazdi M T, Woodward J R, Radford A. Cellulase production by Neurospora crassa: induction and optimisation of the enzyme complex. Enzyme and Microbial Technology. 1990a;12:116–119. doi: 10.1016/0141-0229(90)90084-4. [DOI] [PubMed] [Google Scholar]
- 126.Eberhart B M, Cross D F, Chase L R. beta-Glucosidase system of Neurospora crassa I. beta-Glucosidase and cellulase activities of mutant and wild-type strains. J. Bacteriol. 1964;87:761–770. doi: 10.1128/jb.87.4.761-770.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eberhart B M, Beck R S. Induction of beta-glucosidase in Neurospora crassa. J. Bacteriol. 1973;116:295–303. doi: 10.1128/jb.116.1.295-303.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanks D L, Sussman A S. The relation between growth conidiation and trehalase activity in Neurospora crassa. Amer. J. Bot. 1969a;56:1152–1159. [PubMed] [Google Scholar]
- 129.Hanks D L, Sussman A S. Control of trehalase synthesis in Neurospora crassa. Amer. J. Bot. 1969b;56:1160–1166. [PubMed] [Google Scholar]
- 130.Landman O E. Neurospora lactase II enzyme formation in the standard strain. Arch. Biochem.Biophys. 1954;52:93–109. doi: 10.1016/0003-9861(54)90092-2. [DOI] [PubMed] [Google Scholar]
- 131.Lester G, Byers A. Properties of two beta-galactosidases of Neurospora crassa. Biochem. Biophys. Res. Commun. 1965;18:725–734. [Google Scholar]
- 132.Metzenberg R L. A gene affecting the repression of invertase and trehalase in Neurospora. Arch. Biochem. Biophys. 1962;96:468–474. doi: 10.1016/0003-9861(62)90322-3. [DOI] [PubMed] [Google Scholar]
- 133.Flavell R B, Woodward D O. The regulation of synthesis of Krebs cycle enzymes in Neurospora by catabolite end production repression. Eur. J. Biochem. 1970;13:548–553. doi: 10.1111/j.1432-1033.1970.tb00959.x. [DOI] [PubMed] [Google Scholar]
- 134.Gratzner H, Sheehan D N. Neurospora mutant exhibiting hyperproduction of amylase and invertase. J. Bacteriol. 1969;97:544–549. doi: 10.1128/jb.97.2.544-549.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Magasanik B. Catabolite repression. Cold Spring Harbor Symp. Quant. Biol. 1961;26:249–256. doi: 10.1101/sqb.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- 136.Gong C S, Tsao G T. Cellulase biosynthesis and regulation. Annual Reports on Fermentation Processes. 1979;3, 11:1–140. [Google Scholar]
- 137.Znameroski E A, Coradetti S T, Roche C M, Tsai J C, Iavarone A T, Cate J, Glass N L. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. PNAS. 2012 doi: 10.1073/pnas.1118440109. www.pnas.org/cgi/doi/10.1073/pnas.1118440109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yazdi M T, Radford A, Keen J N, Woodwarjd J R. Cellulase production by Neurospora crassa. Purification and characterization of cellulolytic enzymes. Enzyme and Microbial Technology. 1990b;12:120–123. doi: 10.1016/0141-0229(90)90084-4. [DOI] [PubMed] [Google Scholar]
- 139.Reese E, Maguire A. Surfactants as stimulants of enzyme production by microorganisms. Applied Microbiology. 1969;17:242–245. doi: 10.1128/am.17.2.242-245.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shewale G, Sadanaj C. Cellulase and b-glucosidase production by basidiomycete species. Canadian Journal of Microbiology. 1978;24:1204–1216. doi: 10.1139/m78-195. [DOI] [PubMed] [Google Scholar]
- 141.Deshpandem V, Srinavasamn C, and Deshmakh S S. Effect of fatty acids on cellulase production by Penicillium funiculosum and its mutants. Biotechnology Letters. 1987;9:301–304. [Google Scholar]
- 142.Demain A L, Birnbaum J. Alteration of permeability for the release of metabolites from microbial cells. Current Topics in Microbiology and Immunology. 1968;46:1–25. doi: 10.1007/978-3-642-46121-7_1. [DOI] [PubMed] [Google Scholar]
- 143.Sun J, Tian C, Diamond S, Glass N L. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryotic Cell. 2012 doi: 10.1128/EC.05327-11. doi:10.1128/ EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Galagan J E, Calvo S E, Borkovich K A, Selker E U, Read N D, Jaffe D, FitzHugh W, Ma L J, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen C B, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson M A, Werner-Washburne M, Selitrennikoff C P, Kinsey J A, Braun E L, Zelter A, Schulte U, Kothe G O, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg R L, Perkins D D, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt R J, Osmani S A, DeSouza C P, Glass L, Orbach M J, Berglund J A, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig D O, Alex L A, Mannhaupt G, Ebbole D J, Freitag M, Paulsen I, Sachs M S, Lander E S, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 145.Coradetti S T, Craig J P, Xiong Y, Shock T, Tian C. Glass,http://www.pnas.org/content/early/2012/04/23/1200785109. - corresp-1 N. L. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. PNAS. 2012 doi: 10.1073/pnas.1200785109. www.pnas.org/cgi/doi/10.1073/pnas.1200785109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Penalva M A, Arst H N. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 2002;66:426–646. doi: 10.1128/MMBR.66.3.426-446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Penalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goncalves R D, Cupertino F B, Freitas F Z, Luchessi A D, Bertolini M C. A genome-wide screen for Neurospora crassa transcription factors regulating glycogen metabolism. 2001 doi: 10.1074/mcp.M111.007963. Manuscript M111.007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fu Y H, Marzluf G A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 1990;10:1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fu Y H, Marzluf G A. Characterization of nit-2, the Major Nitrogen Regulatory Gene of Neurospora crassa. Mol. Cell. Biol. 1987;7:1691–1696. doi: 10.1128/mcb.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lockington RA, Rodbourn L, Barnett S, Carter CJ, Kelly JM. Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Gen. Biol. 2002;37:190–196. doi: 10.1016/s1087-1845(02)00504-2. [DOI] [PubMed] [Google Scholar]
- 152.Sun J, Glass N L. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS ONE. 2011;6:e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gancedo J M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.de Souza W, de Gouvea P F, Savoldi M, Malavazi I, de Souza Bernardes L A, Goldman M H S, de Vries R P, de Castro Oliveira J V, and Goldman GH. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnology for Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hanif A, Yasmeen A, Rajoka M I. Induction, production, repression, and de-repression of exoglucanase synthesis in Aspergillus niger. Biores Techn. 2004;94(3):311–319. doi: 10.1016/j.biortech.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 156.Mrudula S, Murugammal R. Production of cellulase by Aspergillus niger under submerged and solid state fermentation using coir waste as a substrate. Braz J Microbiol. 2011;42:1119–1127. doi: 10.1590/S1517-838220110003000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ali S, Sayed A. Regulation of cellulase biosynthesis in Aspergillus terreus. World J Microbiol Biotech. 1992;8:73–75. doi: 10.1007/BF01200691. [DOI] [PubMed] [Google Scholar]
- 158.Ghori MI, Ahmed S, Malana A, Jamil A. Corn stover-enhanced cellulase production by Aspergillus niger NRRL 567. Afr J Biotech. 2011;10(31):5878–5886. [Google Scholar]
- 159.McKelvey SM, Murphy RA. Analysis of wide-domain transcriptional regulation in solid-state cultures of Aspergillus oryzae. J. Ind. Microbiol. Biotechnol. 2010;37(5):455–69. doi: 10.1007/s10295-010-0691-z. [DOI] [PubMed] [Google Scholar]
- 160.Nazir A, Soni R, Saini HS, Kaur A, Chadha BS. Profiling differential expression of cellulases and metabolite footprints in Aspergillus terreus. Appl. Biochem. Biotechnol. 2010;162(2):538–47. doi: 10.1007/s12010-009-8775-9. [DOI] [PubMed] [Google Scholar]
- 161.Hrmova M, Biely P, Vrsanska M. Cellulose- and xylan-degrading enzymes of Aspergillus terreus and Aspergillus niger. Enzyme Microb.Technol. 1989;11:610–616. [Google Scholar]
- 162.Kimura T, Kitamoto N, Kito Y, Karita S, Sakka K, Ohmiya K. Molecular cloning of xylanase gene xynG1 from Aspergillus oryzae KBN 616, a Shoyu Koji mold, and analysis of its expression. J. Ferment. Bioeng. 1998;85:10–16. [Google Scholar]
- 163.Piñaga F, Fernández-Espinar M T, Vallés S, Ramón D. Xylanase production in Aspergillus nidulans: induction and carbon catabolite repression. FEMS Microb Lett. 1994;115(2–3):319–323. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 164.Rizzatti ACS, Zanolli F, Bertolini MC, Peixoto-Nogueira S C, Terenzi H F, Jorge J A, de Moraes Polizeli M. Regulation of xylanase in Aspergillus phoenicis: a physiological and molecular approach. J Ind Microbiol Biotechnol. 2008;35:237–244. doi: 10.1007/s10295-007-0290-9. [DOI] [PubMed] [Google Scholar]
- 165.Ghosh M, Nanda G. Physiological studies on xylose induction and glucose repression of xylanolytic enzymes in Aspergillus sydowii MG49. FEBS Microbiol Lett. 1994;117:151–156. [Google Scholar]
- 166.Simao RC, Souza C G M, Peralta R M. Induction of xylanase in Aspergillus tamarii by methyl β-d-xyloside. Appl Microbiol Biotech. 1997;47(3):267–271. [Google Scholar]
- 167.van Peij NNME, Brinkmann J, Vrsanska´ M, Visser J, de Graaff L H. b-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 168.Parenicova L. Pectinases of Aspergillus niger: a molecular and biochemical characterisation. Wageningen, The Netherlands: PhD Thesis, Wageningen University; 2000. [Google Scholar]
- 169.de Vries RP, Visser J, de Graaff LH. CreA modulates the XlnR induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 1999b;150:281–285. doi: 10.1016/s0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 170.Veen P, Flipphi M J A, Voragen A. G. J.2, Visser J. Induction of extracellular arabinases on monomeric substrates in Aspergillus niger. Arch Microbiol. 1993;159:66–71. doi: 10.1007/BF00244266. [DOI] [PubMed] [Google Scholar]
- 171.Van Peij N, Gielkens MMC, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 1998a;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tani S, Kanamasa S, Sumitani J, Arai M, Kawaguchi T. XlnR-independent signaling pathway regulates both cellulase and xylanase genes in response to cellobiose in Aspergillus aculeatus. Curr. Genet. 2012;58:93–104. doi: 10.1007/s00294-012-0367-5. [DOI] [PubMed] [Google Scholar]
- 174.Kunitake E, Tani S, Sumitani J, Kawaguchi T. A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulos-eresponsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Appl. Microbiol. Biotechnol. 2012 doi: 10.1007/s00253-012-4305-8. DOI 10.1007/s00253-012-4305-8. [DOI] [PubMed] [Google Scholar]
- 175.Chilton I J, Delaney C E, Barham-Morris J, Fincham D A, Hooley P, Whitehead M P. The Aspergillus nidulans stress response transcription factor StzA is ascomycete-specific and shows species- specific polymorphisms in the C-terminal region. Mycol. Res. 2008;112:1435–1446. doi: 10.1016/j.mycres.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 176.Kojima Y, Sakamoto T, Kishida M, Sakai T, Kawasaki H. Acidic condition-inducible polygalacturonase of Aspergillus kawachii. J. Mol. Catal. Ser. B. 1999;6:351–357. [Google Scholar]
- 177.Gielkens M M C, Gonzales-Candelas L, Sanchez-Torres P, van de Vondervoort P J I, de Graaf L H, Visser J. The abfB gene encoding the major α-L-arabinofuranosidase of Aspergillus nidulans: nucleotide sequence, regulation and construction of a disrupted strain. Microbiology. 1999;145:735–741. doi: 10.1099/13500872-145-3-735. [DOI] [PubMed] [Google Scholar]
- 178.MacCabe A P, Orejas M, Perez-Gonzalez J A, Ramon D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J. Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stewart JC, Parry JB. Factors influencing the production of celulase by Aspergillus fumigatus (Fresenius) J. Gen. Microbiol. 1981;125:33–39. doi: 10.1099/00221287-125-1-33. [DOI] [PubMed] [Google Scholar]
- 180.MacCabe A P, Fernandez-Espinar M T, de Graaff L H, Visser J, Ramon D. Identification, isolation and sequence of the Aspergillus nidulans xlnC gene encoding the 34-kDa xylanase. Gene. 1996;175:29–33. doi: 10.1016/0378-1119(96)00116-3. [DOI] [PubMed] [Google Scholar]
- 181.Perez-Gonzalez J A, van Peij N N M E, Bezoen A, MacCabe A P, Ramon D, de Graaff L H. Molecular cloning and transcriptional regulation of the Aspergillus nidulans xlnD gene encoding b-xylosidase. Appl. Environ. Microbiol. 1998;64:1412–1419. doi: 10.1128/aem.64.4.1412-1419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lockington R A, Kelly J M. The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol. Microbiol. 2002;43:1173–1182. doi: 10.1046/j.1365-2958.2002.02811.x. [DOI] [PubMed] [Google Scholar]
- 183.Arst HN, Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- 184.Dowzer C E, Kelly J M. Analysis of the creA gene; a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hynes MJ, Kelly JM. Pleiotropic mutants of Aspergillus nidulans altered in carbon metabolism. Mol. Gen. Genet. 1977;150:193–204. doi: 10.1007/BF00695399. [DOI] [PubMed] [Google Scholar]
- 186.Lockington R A, Kelly J M. Carbon catabolite repression in Aspergillus nidulans involves deubiquitination. Mol. Microbiol. 2001;40:1311–1321. doi: 10.1046/j.1365-2958.2001.02474.x. [DOI] [PubMed] [Google Scholar]
- 187.Todd R B, Lockington R A, Kelly J M. The Aspergillus nidulans creC gene involved in carbon catabolite repression encodes a WD40 repeat protein. Mol. Gen. Genet. 2000;263:561–570. doi: 10.1007/s004380051202. [DOI] [PubMed] [Google Scholar]
- 188.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol. Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 189.Flipphi M, van de Vondervoort P J, Ruijter G J, Visser J, Arst H N, Jr, Felenbok B. Onset of carbon catabolite repression in Aspergillus nidulans. Parallel involvement of hexokinase and glucokinase in sugar signalling. J. Biol. Chem. 2003;278:11849–11857. doi: 10.1074/jbc.M209443200. [DOI] [PubMed] [Google Scholar]
- 190.Boase N A, Kelly J M. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol. Microbiol. 2004;53:929–940. doi: 10.1111/j.1365-2958.2004.04172.x. [DOI] [PubMed] [Google Scholar]
- 191.Polo S, Di Fiore P P. Finding the right partner: science or ART? Cell. 2008;135:590–592. doi: 10.1016/j.cell.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 192.Nogawa M, Goto M, Okada H, Morikawa Y. L-Sorbose induce cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr. Genet. 2001;38:329–334. doi: 10.1007/s002940000165. [DOI] [PubMed] [Google Scholar]
- 193.Xu J, Nogawa M, Okada H, Morikawa Y. Regulation of xyn3 gene expression in Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol. 2000;54(3):370–5. doi: 10.1007/s002530000410. [DOI] [PubMed] [Google Scholar]
- 194.Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, Kobayashi T, Tsukagoshi N. Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr. Genet. 2001;39:10–15. doi: 10.1007/s002940000175. [DOI] [PubMed] [Google Scholar]
- 195.Suto M, Tomita F. Induction and catabolite repression mechanisms of cellulase in fungi. J Biosci Bioeng. 2001;92(4):305–11. doi: 10.1263/jbb.92.305. [DOI] [PubMed] [Google Scholar]
- 196.Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2000;64:816–827. doi: 10.1271/bbb.64.816. [DOI] [PubMed] [Google Scholar]
- 197.Battaglia E, Hansen S, Leendertse A, Madrid S, Mulder H, Nikolaev I, de Vries R. Regulation of pentose utilisation by AraR, but not XlnR, differs in Aspergillus nidulans and Aspergillus niger. Appl. Microbiol. Biotechnol. 2011a;91:387–397. doi: 10.1007/s00253-011-3242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Battaglia E, Visser L, Nijssen A, van Veluw GJ, Wösten HAB, de Vries RP. Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutinary adaptations in Eurotiales. Stud. Mycol. 2011b;69:31–38. doi: 10.3114/sim.2011.69.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bates W K, Hedman S C, Woodward D O. Comparative inductive responses of two beta-galactosidases of Neurospora. J. Bacteriol. 1967;93(5):1631. doi: 10.1128/jb.93.5.1631-1637.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.de Vries RP. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl. Microbiol. Biotechnol. 2003;61:10–20. doi: 10.1007/s00253-002-1171-9. [DOI] [PubMed] [Google Scholar]