Abstract
RNA trans-splicing has been explored as a therapeutic option for a variety of genetic diseases, but not for cardiac genetic disease. Hypertrophic cardiomyopathy (HCM) is an autosomal-dominant disease, characterized by left ventricular hypertrophy (LVH) and diastolic dysfunction. MYBPC3, encoding cardiac myosin-binding protein C (cMyBP-C) is frequently mutated. We evaluated the 5′-trans-splicing strategy in a mouse model of HCM carrying a Mybpc3 mutation. 5′-trans-splicing was induced between two independently transcribed molecules, the mutant endogenous Mypbc3 pre-mRNA and an engineered pre-trans-splicing molecule (PTM) carrying a FLAG-tagged wild-type (WT) Mybpc3 cDNA sequence. PTMs were packaged into adeno-associated virus (AAV) for transduction of cultured cardiac myocytes and the heart in vivo. Full-length repaired Mybpc3 mRNA represented up to 66% of total Mybpc3 transcripts in cardiac myocytes and 0.14% in the heart. Repaired cMyBP-C protein was detected by immunoprecipitation in cells and in vivo and exhibited correct incorporation into the sarcomere in cardiac myocytes. This study provides (i) the first evidence of successful 5′-trans-splicing in vivo and (ii) proof-of-concept of mRNA repair in the most prevalent cardiac genetic disease. Since current therapeutic options for HCM only alleviate symptoms, these findings open new horizons for causal therapy of the severe forms of the disease.
Keywords: hypertrophic cardiomyopathy, Mybpc3, RNA-based therapy, trans-splicing
Introduction
In the last decade repair of mRNA by spliceosome-mediated RNA trans-splicing has raised interests as a novel therapeutic intervention (for reviews, see refs. 1,2). Trans-splicing has many attractive features such as preservation of the endogenous regulation, replacement of selected portions of the target gene and, most importantly, corrections of dominant-negative mutations.3,4 The 5′-, 3′- or even internal exons of a target pre-mRNA can be replaced by trans-splicing using engineered pre-trans-splicing molecules (PTMs). PTMs carry the wild-type (WT) sequence, a binding domain complementary to the endogenous target and an appropriate set of splicing elements. After nuclear import, PTMs are transcribed and can specifically hybridize the target mutant pre-mRNA via their binding domain, giving rise to a repaired mRNA molecule (Figure 1a). As a positive side effect of trans-splicing, cis-splicing should be reduced due to competition for access to the RNA splicing machinery. So far, successful trans-splicing between PTMs and endogenous targets has been described for different genetic diseases such as hemophilia A,5 cystic fibrosis,6 spinal muscular atrophy,7,8 hyper-IgM-X-linked immunodeficiency,9 frontotemporal dementia with Parkinsonism linked to chromosome 17,10,11 epidermolysis bullosa with muscular dystrophy,12 and Huntington's disease,13 most of them being 3′-trans-splicing approaches.
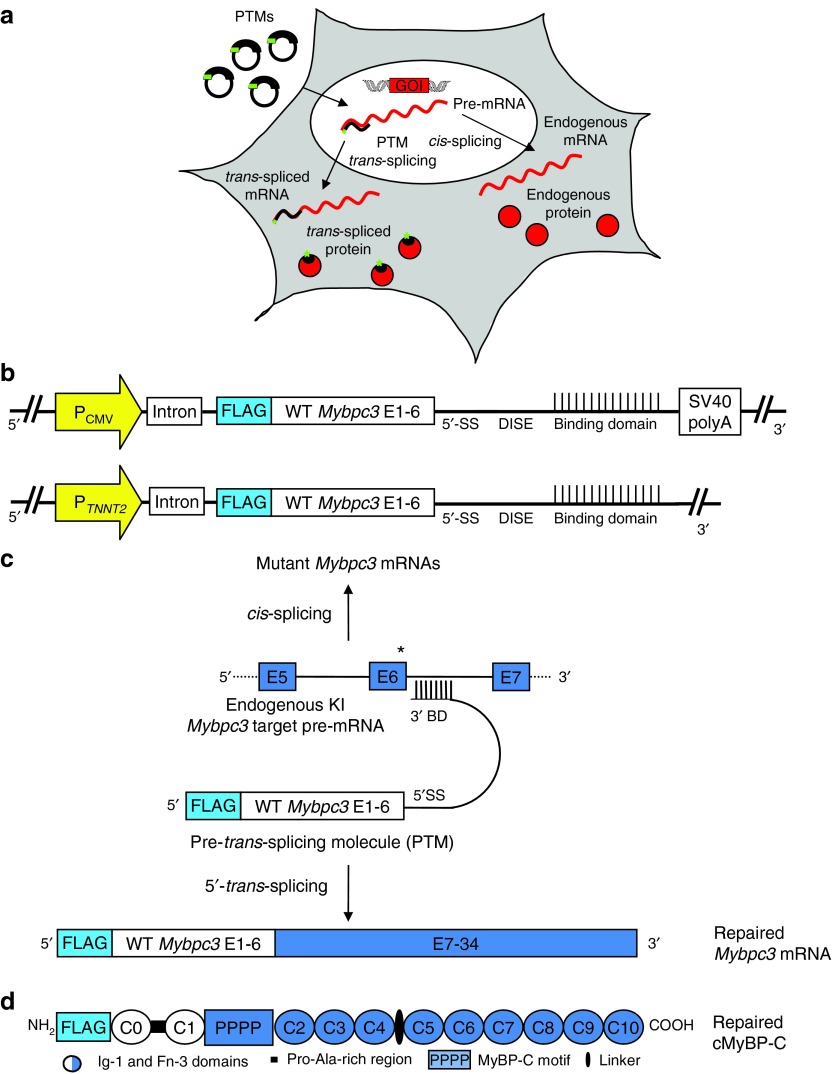
Figure 1.
RNA trans-splicing strategy. (a) Pre-trans-splicing molecules (PTMs) are transferred into cells using plasmids or virus particles, and are transcribed in the nucleus. They target the pre-mRNA of the gene of interest and produce repaired mRNA and protein. This process is competing with cis-splicing, which gives rise to endogenous mRNA and proteins. (b) Schematic illustration of PTMs. We generated several PTMs containing the same Mybpc3 wild-type sequence (exons 1–6) but different binding domains covering the Mybpc3 intron 6. PTMs also included a chimeric intron, containing sequences from the human β-globin and immunoglobulin (IgG) genes, and a DISE from the rat fibroblast growth factor receptor 2 gene. (c) Schematic representation of 5′-trans-splicing bypassing a Mybpc3 point mutation (not in scale). The endogenous KI Mybpc3 pre-mRNA target containing the mutation on the last nucleotide of exon 6 is shown together with the PTM and the resulting transcripts obtained by cis- or trans-splicing. (d) Schematic illustration of repaired FLAG-tagged cMyBP-C protein. The protein is composed of 10 Ig-1 and Fn-3 domains and has 4 phosphorylation sites (PPPP) in the MyBP-C motif. The C0–C1 domains, including the Pro-Ala-rich region derived from the PTM (white). BD, binding domain; CMV, cytomegalovirus; DISE, downstream intronic splicing enhancer; E, exon; P, promoter; TNNT2, human cardiac troponin T; 5′SS, 5′splice donor site.
To the best of our knowledge, no study has provided evidence for successful 5′-trans-splicing in vivo yet and this promising strategy has not been evaluated for cardiac genetic diseases. The aim of the present study was therefore to investigate this approach in hypertrophic cardiomyopathy (HCM). HCM is a myocardial disease mainly characterized by left ventricular hypertrophy (LVH) and diastolic dysfunction.14,15,16 The clinical outcome of HCM is highly variable and ranges from an asymptomatic benign course to heart failure, atrial fibrillation and sudden cardiac death caused by arrhythmias.14,15 HCM is a genetic disease transmitted as an autosomal-dominant trait and caused by mutations in genes encoding sarcomeric proteins.17 Among them, mutations in MYBPC3 encoding cardiac myosin-binding protein C (cMyBP-C) are the most frequent ones.18,19 cMyBP-C is a component of the thick filaments of the sarcomere, and plays important structural and functional roles.18,20,21
In the present study the feasibility of 5′-trans-splicing to repair HCM-mutant mRNA was assessed in isolated cardiac myocytes and in vivo in Mybpc3-targeted knock-in (KI) mice that have been generated previously.22 KI mice carry a G>A transition on the last nucleotide of exon 6, which is associated with a severe phenotype and a poor prognosis in humans19 and occurs in 13% of all HCM patients in Toscany.23 KI mice exhibit LVH with systolic and diastolic dysfunction.22,24
Results
Design of PTMs
The G>A transition mutation leads to three different mutant mRNAs in homozygous KI mice (Supplementary Figure S1). Mutant-1 contains the mutation (missense), whereas mutant-2 (nonsense) and mutant-3 (deletion/insertion) are due to skipping of exon 6.
We generated different PTMs encoding exons 1–6 of WT Mybpc3 under the control of a ubiquitous (cytomegalovirus) or cardiac myocyte-specific (TNNT2, human cardiac troponin T) promoter (Figure 1b). To specifically detect repaired Mybpc3 mRNA and protein, an N-terminal FLAG-tag was introduced in the coding sequence. In addition, an intron was inserted right after the promoter to increase mRNA stability and enhance expression of the constructs.25,26 The splicing domain included a canonical 5′ splice donor site sequence followed by a downstream intronic sequence enhancer element, which has been shown to markedly increase the trans-splicing efficiency.27 Importantly, the binding domain of the PTM is an essential part because it confers specificity to the target pre-mRNA.28 To evaluate the feasibility and efficacy of 5′-trans-splicing, we designed several constructs differing only with respect to the length of the binding domain and to the target site in Mybpc3 intron 6 (Supplementary Table S1). The binding domains are complementary to this intron, but leave out its 3′ splicing elements. Moreover, to maintain the PTM in the nucleus and reduce its translation we deleted the SV40 polyadenylation (polyA) signal, which is known to contribute to mRNA stability and nuclear export (Figure 1b).29 As negative controls we designed PTMs with reversed binding domains (PTM-R), which should not induce 5′-trans-splicing events. PTM-driven 5′-trans-splicing on the endogenous KI Mybpc3 pre-mRNA target should produce a full-length repaired Mybpc3 mRNA, in which the mutation is bypassed, resulting in a FLAG-tagged WT repaired cMyBP-C protein, and simultaneously cis-splicing should be reduced (Figure 1c,d).
Evidence for repair of Mybpc3 mRNA by 5′-trans-splicing in vitro
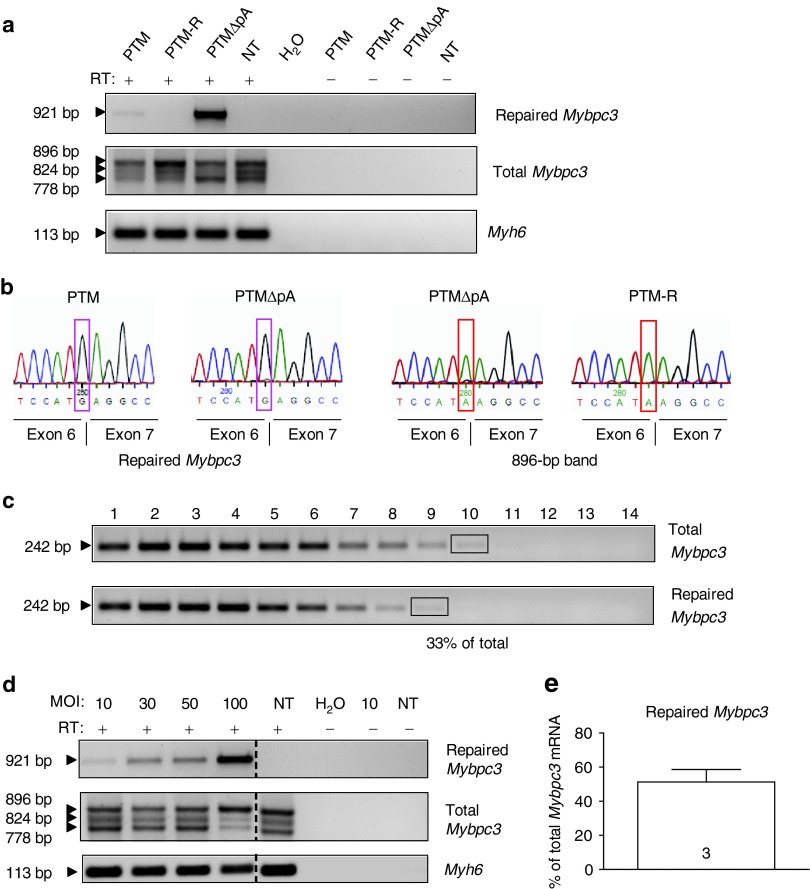
To allow gene transfer in neonatal mouse cardiac myocytes (NMCMs), PTMs were packaged into self-complementary adeno-associated virus serotype 6 (AAV6), a serotype known to efficiently transduce cardiac myocytes in culture.30,31 NMCMs were isolated from KI mice and transduced with AAV6-PTMs either with or without polyA signal (ΔpA) or AAV6-green fluorescent protein (GFP) as a control. After 4 days of transduction (multiplicity of infection (MOI): 3,000) about 80% of cells expressed GFP (Supplementary Figure S2). Using PCR primers that specifically amplify the repaired Mybpc3 mRNA (Supplementary Figure S1 and Supplementary Table S2), we obtained a specific signal in AAV6-PTM- and AAV6-PTMΔpA-transduced NMCMs, but not in untransduced or PTM-R-transduced NMCMs (Figure 2a). The absence of 5′-trans-splicing in AAV6-PTM-R-transduced NMCM excluded the possibility that recombination occurred between the highly homologous sequences of PTMs and endogenous Mybpc3. The amount of repaired Mybpc3 was higher in the absence than in the presence of the polyA signal in the PTM. To evaluate whether cis-splicing was reduced, we used Mybpc3 primers binding in exons 1 and 9, which amplify total (repaired plus mutant) Mybpc3 mRNA. Although no major difference was detected between samples, reduced signals were observed for certain Mybpc3 mRNA species in AAV6-PTM- and in AAV6-PTMΔpA-transduced NMCMs. This suggests a reduction in Mybpc3 cis-splicing when 5′-trans-splicing occurred (Figure 2a). Sequencing of repaired Mybpc3 mRNA amplicons confirmed the presence of the WT guanine (G) at the exon 6–exon 7 junction (Figure 2b). Conversely, sequencing of the upper 896-bp band of total Mybpc3 mRNA in AAV6-PTMΔpA- and AAV6-PTM-R-transduced NMCMs showed the presence of the mutant adenine (A) at the same position (Figure 2b). To estimate the amount of repaired Mybpc3 mRNA, we performed two rounds of PCR to amplify either total or only repaired Mybpc3 mRNA (Figure 2c). Comparison of amplicon intensities revealed that up to 33% of total Mybpc3 transcripts were repaired. To evaluate whether the efficiency of 5′-trans-splicing can be improved by increasing the dose of virus, we generated bicistronic recombinant adenovirus (AdV) encoding the PTMΔpA and GFP both under the control of the TNNT2 promoter. KI NMCMs were transduced with different MOI of AdV-PTMΔpA and analyzed 7 days after. Repaired Mybpc3 mRNA was detected in all transduced samples and its amount increased with increasing MOI (Figure 2d). The pattern of total Mybpc3 mRNA did not reveal major difference from one MOI to another, except at a MOI of 100 at which the intensity of the mutant-3 and mutant-2 mRNAs was lower than in untransduced cardiac myocytes (Figure 2d). Fluorescence analysis of AdV-GFP transduced cardiomyocytes confirmed a complete transduction with a MOI of 100 (Supplementary Figure S2). We further determined the efficiency of 5′-trans-splicing in several samples with AdV-PTMΔpA at a MOI of 100, and estimated by semi-quantitative analysis that 51 ± 7% of total Mybpc3 mRNA was repaired (Figure 2e).
Figure 2.
Detection of repaired Mybpc3 mRNA induced by 5′-trans-splicing in vitro. (a) KI NMCMs were transduced with AAV6 (MOI: 3,000–30,000) for 7 days before harvesting. RT-PCR analysis using different primer pairs: FLAG/E9-R primers amplified a specific 921-bp fragment that correspond to the repaired Mybpc3 mRNA in KI NMCMs transduced with AAV6 encoding PTM or PTMΔpA that contain a complementary binding domain to intron 6, but not in NMCMs transduced with a PTM containing a reversed binding domain (PTM-R), in not-transduced NMCMs (NT), nor in the amplifications without RT (−). With E1-F/E9-R primers bands of 896, 824, and 778 bp were amplified, corresponding to mutant-1/repaired, mutant-3 and mutant-2, respectively. Amplification of Myh6 mRNA encoding α-myosin heavy chain was used as a loading control. (b) Sequencing of the FLAG/E9-R amplicons from AAV6-PTM and AAV6-PTMΔpA transduced NMCMs confirmed replacement of the mutant adenine with the wild-type guanine (G, pink box) at the exon 6–exon 7 junction. In addition sequencing of the gel-excised 896-bp band obtained by RT-PCR with primers E1-F/E9-R from AAV6-PTMΔpA and AAV6-PTM-R transduced NMCMs validate the presence of the endogenous mutant adenine (A, red box) on the last nucleotide of exon 6. (c) Determination of the percentage of repaired Mybpc3 mRNA by semi-quantitative RT-PCR. cDNA from AAV6-PTMΔpA-transduced KI NMCMs was used to amplify total (E1-F/E9-R primers) or only repaired (FLAG/E9-R primers) Mybpc3 by PCR (25 cycles). PCR fragments were column-purified and serially diluted (1:3). In a second round of PCR, a common primer pair (E1-F/E2-R) was used to amplify a 242-bp fragment in all Mybpc3 transcripts. The percentage of total Mybpc3 mRNA that was repaired was estimated by comparing bands of similar intensities (black rectangles), that is, lane 9 (dilution 38) of repaired Mybpc3 mRNA and lane 10 (dilution 39) of total Mybpc3 mRNA. (d) KI NMCMs were transduced with AdV-PTMΔpA (MOI: 10–100). RT-PCR analysis was performed using same primer pairs as in a. Repaired Mybpc3 mRNA was amplified in all tranduced samples but not in NT or in −RT (amplification without RT) samples. (e) KI NMCMs were transduced with AdV-PTMΔpA (MOI: 100). Bar represents repaired Mybpc3 mRNA expressed as percentage of total Mybpc3 mRNA (mean ± SEM) obtained by semi-quantitative analysis (as described above but with 1:1.5 dilutions). Number of samples is indicated in the bar. MOI, multiplicity of infection; RT-PCR, reverse transcription-PCR.
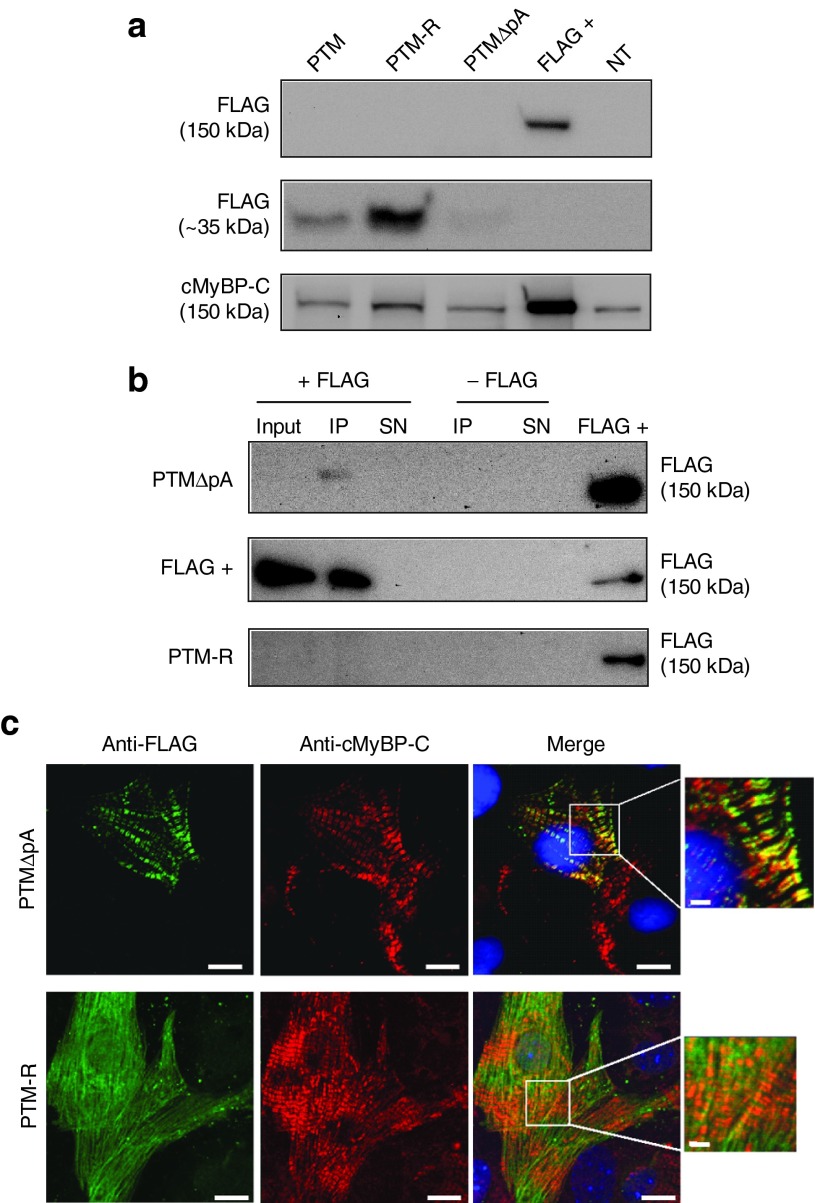
We then investigated whether the repaired Mybpc3 mRNA is translated into protein and whether the repaired cMyBP-C is properly incorporated into the sarcomere. The presence of the FLAG-tag allowed specific detection of repaired cMyBP-C. Whereas repaired cMyBP-C was not detected by standard western blot with the anti-FLAG antibody, it was detected at the correct molecular weight after FLAG-immunoprecipitation (Figure 3a,b), confirming that 5′-trans-splicing occurred in cardiac myocytes. FLAG-immunoprecipitation of AAV6-PTM-R-transduced NMCMs did not show any band at 150 kDa, while FLAG-Mybpc3 transfected HEK293 cells, used as a positive control, did show it (Figure 3b). On the other hand, we detected a major FLAG-positive band around 35 kDa in AAV6-PTM- and AAV6-PTM-R-transduced NMCMs, which corresponds to the translated PTM transcripts (Figure 3a). This band was barely detected in AAV6-PTMΔpA-transduced NMCMs, supporting the view that the absence of the polyA signal prevented translation and putative accumulation of toxic PTM proteins in cells. Endogenous and/or repaired cMyBP-C, but not translated PTMs were stained with a specific cMyBP-C antibody, which recognizes the MyBP-C motif (Figures 3a and 1d). To investigate whether the repaired cMyBP-C was incorporated into the sarcomere, we performed immunofluorescence analysis of transduced cardiac myocytes. About 9% of cMyBP-C-positive cells (= cardiac myocytes) were co-stained with the anti-FLAG antibody, and the repaired cMyBP-C showed the expected doublets in the A-band of the sarcomeres, indicating correct incorporation (Figure 3c). In contrast, the 35-kDa FLAG-PTM-R proteins showed a cellular and nuclear diffuse pattern without colocalization with endogenous cMyBP-C (Figure 3c).
Figure 3.
Detection of repaired cMyBP-C induced by 5′-trans-splicing in vitro. KI NMCMs were transduced with AAV6 at a multiplicity of infection of 3,000 for 7 days before analysis. (a) Western blots were stained with antibodies directed against the FLAG epitope and cMyBP-C. The anti-FLAG antibody revealed a 150-kDa FLAG-cMyBP-C band only in the protein sample derived from HEK293 cells transiently transfected (48 hours) with a FLAG-cMyBP-C plasmid (FLAG +), but not in AAV6-transduced NMCMs (upper panel). On the other hand, the anti-FLAG antibody revealed 35-kDa proteins (middle panel). The anti-cMyBP-C-antibody (directed against the MyBP-C motif) detected only full-length cMyBP-C in all samples (lower panel). However, it was not possible to distinguish between endogenous and potentially repaired proteins. (b) FLAG-immunoprecipitation (IP), followed by FLAG-western blot revealed a specific band in the IP fraction of AAV6-PTMΔpA-transduced KI NMCMs as well as in the positive control (FLAG +), but not in the IP fraction of AAV6-PTM-R-transduced KI NMCMs and of samples without antibody. (c) Immunofluorescence analysis of AAV6-PTMΔpA- and AAV6-PTM-R-transduced KI NMCMs. Cells were double-stained with anti-FLAG (green) and anti-cMyBP-C (red) antibodies. Nuclei were stained with DRAQ5 (blue). Bars 10 and 2 µm in the higher magnifications. SN, supernatant.
Evidence for repair of Mybpc3 mRNA by 5′-trans-splicing in vivo
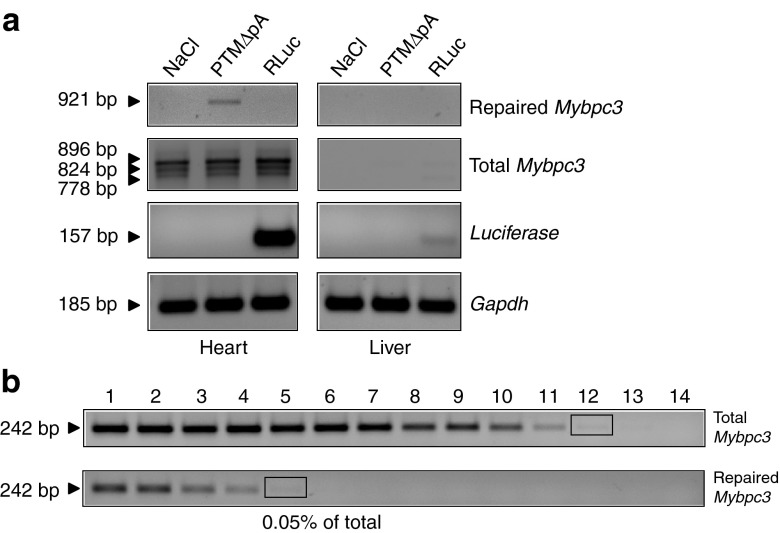
We next assessed the feasibility of PTM-driven 5′-trans-splicing in KI mice in vivo. The PTMΔpA and Renilla luciferase (RLuc) were inserted in the pdsAAV transfer vector under the control of the TNNT2 promoter and were packaged in AAV serotype 9 (AAV9), which has proven efficient cardiac transduction in mice in vivo.32 AAV9 (mean dose 5.2 × 1012 vg/kg of body weight (BW)) was administered systemically into 7-week-old animals. Echocardiographic analysis performed during one month after injection did not display major differences in cardiac function between mice that received either AAV9 or NaCl (Supplementary Table S3). After 28 days, luciferase expression was evaluated by in vivo bioluminescence imaging and luminescence was recorded only in the heart of the AAV9-RLuc–injected mouse (Supplementary Figure S3). Accordingly, luciferase mRNA level was high in the heart and very low in the liver of the mouse that received AAV9-RLuc (Figure 4a), validating efficient and preferential cardiac transduction with AAV9. Importantly, the repaired Mybpc3 mRNA was detected in the heart of the mouse that received AAV9-PTMΔpA, but not in the others (Figure 4a). No effect on cis-splicing was discernible (Figure 4a). Semi-quantitative analysis showed that 0.05% of total Mybpc3 mRNA was repaired (Figure 4b).
Figure 4.
Evidence for Mybpc3 mRNA and cMyBP-C protein repair by 5′-trans-splicing in the heart of 7-week-old KI mice. (a) Adult mice were sacrificed 4 weeks after AAV9 administration and RT-PCR analysis was performed using specific Mybpc3 primers. The 921-bp fragment (= repaired Mybpc3) was amplified only in the heart of the mouse that received AAV9-PTMΔpA, whereas total Mybpc3 mRNA amplification showed the expected bands corresponding to the different mutants in all hearts. Luciferase mRNA was markedly accumulated in heart and to a lower extent in liver of the mouse that received AAV9-RLuc. Gapdh mRNA was used as a loading control. (b) Determination of the percentage of repaired Mybpc3 mRNA by semi-quantitative RT-PCR. After two rounds of PCR, bands with similar intensity (black rectangles) allowed the estimation of the percentage of total Mybpc3 mRNA that is repaired, that is, lane 5 (dilution 34) of repaired Mybpc3 and lane 12 (dilution 311) of total Mybpc3. RT-PCR, reverse transcription-PCR.
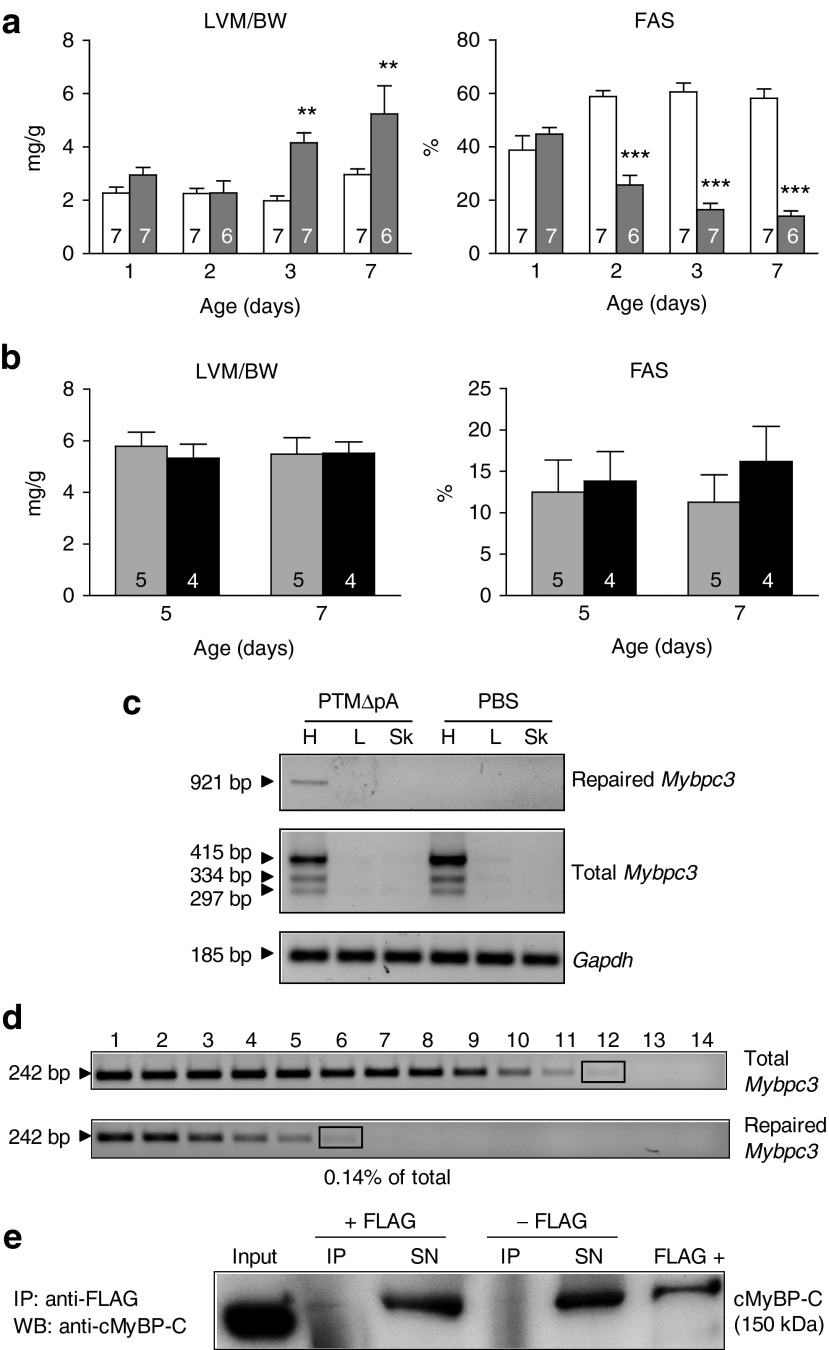
To augment the dose of virus and thus the 5′-trans-splicing efficiency, we performed experiments in neonates (Figure 5). Longitudinal echo analysis in neonatal mice revealed that KI mice developed first systolic dysfunction, as shown by lower fractional area shortening than WT mice at day 2, followed by LVH, as shown by higher left-ventricular-mass-to-BW than WT mice at day 3 (Figure 5a). We then systemically administered AAV9-PTMΔpA into 1-day-old KI mice (3.4 × 1014 vg/kg BW). This dose of AAV9 resulted in an almost complete transduction of cardiac tissue at postnatal day 7 (Supplementary Figure S4). Although the dose per BW was ~65-fold higher than in the adult mouse, no beneficial effect on left ventricular mass/BW and on fractional area shortening were observed at day 4 and 7 (Figure 5b) as well as 7 weeks after injection (Supplementary Table S3). Despite the absence of rescue, we evaluated the 5′-trans-splicing efficiency in one mouse 7 weeks after injection. The full-length repaired Mybpc3 mRNA was detected by reverse transcription-PCR only in the heart of the AAV9-PTMΔpA-injected mouse and represented 0.14% of total Mybpc3 transcripts (Figure 5c,d and Supplementary Figure S5), which thus showed 2.8-fold higher 5′-trans-splicing event in the newborn than in the adult mouse. In addition, the repaired cMyBP-C was detected, although faintly after FLAG-immunoprecipitation (Figure 5e).
Figure 5.
Evidence for Mybpc3 mRNA and cMyBP-C protein repair by 5′-trans-splicing in the heart of newborn KI mice. (a) Echocardiographic analysis of 1- to 7-day-old Mybpc3-targeted KI (gray bars) and WT mice (white bars). Left ventricular mass (LVM) to body weight (BW) ratio and fractional area shortening (FAS) are shown. Values are expressed as mean ± SEM. **P < 0.01, ***P < 0.001, two-way ANOVA and Bonferroni post-hoc test. Number of animals is indicated in the bars. (b) One-day-old KI mice received PBS (gray bars) or AAV9-PTMΔpA (black bars) by systemic administration into the temporal vein and were analyzed by echocardiography at 5 and 7 days of age. Values are expressed as mean ± SEM. (c) Heart (H), liver (L), and skeletal muscle (Sk) were extracted 7 weeks after administration of AAV9-PTMΔpA or PBS in newborn mice. RT-PCR analysis showed the repaired Mybpc3 mRNA band only in the heart of the mouse that received AAV9-PTMΔpA, whereas total Mybpc3 mRNA was also amplified in the control mouse. Gapdh mRNA was used as a loading control. (d) Determination of the percentage of repaired Mybpc3 mRNA by semi-quantitative RT-PCR. After two rounds of PCR the percentage of total Mybpc3 mRNA that was repaired was estimated by using bands of similar intensities (black rectangles), that is, lane 6 (dilution 35) of repaired Mybpc3 and lane 12 (dilution 311) of total Mybpc3. (e) Ventricular tissue of AAV9-PTMΔpA-injected mouse was used for immunoprecipitation (IP) with or without anti-FLAG antibody, followed by anti-cMyBP-C western blot. This revealed a faint signal in the IP fraction, corresponding to repaired cMyBP-C protein, a clear signal in the input and in the supernatants, corresponding to endogenous cMyBP-C as well as in the positive control (FLAG +). ANOVA, analysis of variance; PBS, phosphate-buffered saline; RT-PCR, reverse transcription-PCR; SN, supernatant.
Discussion
RNA trans-splicing as a potential therapeutic technology has been applied to several diseases both in cell systems and in mouse models (for reviews, see refs. 1,2). The present study provides the first evidence of successful 5′-trans-splicing both in cardiac myocytes and in the heart in vivo for the most prevalent cardiac genetic disease.
The percentage of total Mybpc3 mRNA that was repaired was estimated to be between 33 and 66% in transduced KI NMCMs. This is much higher than what has been reported in previous studies using endogenous targets.5,33 However, despite the high efficiency of Mybpc3 5′-trans-splicing at the mRNA level, the amount of repaired cMyBP-C protein was rather low. This suggests a low efficiency of translation and underlines that mRNA copy number and protein levels do not need to be correlated.34 On the other hand, the amount of total Mybpc3 mRNAs is 80% and the level of cMyBP-C protein 90% lower in KI than in WT mice.22 Therefore, when 33% of total Mybpc3 mRNA is repaired, it represents less than 7% of the Mybpc3 mRNA amount and less than 4% of the cMyBP-C protein amount found in WT mice. This may well be under the limit of detection by western blot.
Our study provides additional evidence for removing the polyA signal in the PTM construct to prevent translation and therefore accumulation of PTM proteins that could exert a dominant-negative effect on the structure and/or function of cardiac myocytes.
Recently, in vivo 3′-trans-splicing has been shown to improve the phenotype of a mouse model of spinal muscular atrophy.8 The present study provides the first evidence for successful mRNA repair by 5′-trans-splicing in vivo. The combination of AAV9 and TNNT2 promoter allowed efficient cardiac transduction in vivo. Although this resulted in detectable levels of repaired cMyBP-C, the amount was still too low to ameliorate the cardiac phenotype. Thus, further optimization of the technique is needed to increase the amount of “therapeutic” protein.
Among strategies that aim at specifically targeting mutant mRNA in dominant genetic disease, such as mutant-specific RNA interference,35 trans-splicing has potential advantages for the therapy of HCM. First, it allows the repair of even complex consequences on RNA splicing that, as exemplified in the present HCM mouse model,22 can result from a single point mutation and will be difficult/impossible to target with siRNA without affecting WT mRNA. Second, and in contrast to RNA interference therapies targeting a specific mutation, two different PTMs would be enough to treat the 40–60% of HCM patients who carry a MYBPC3 mutation18,19,36,37,38—one targeting the 5′ mutations and the other the 3′ mutations. Therefore, trans-splicing represents a promising, potentially causal therapy of severe forms of HCM.
Materials and methods
Animals. The investigation conforms to the guidelines for the care and use of laboratory animals published by the National Institutes of Health (Publication no. 85-23, revised 1985). The experimental procedures were in accordance with the German Law for the Protection of Animals and accepted by the Ministry of Science and Public Health of the City State of Hamburg, Germany (Nr. 69/10). Mybpc3-targeted KI mice were generated previously,22 and maintained on a Black Swiss background.
Design of PTMs. The sequences of primers used are listed in Supplementary Table S2. The coding sequence of the PTMs was generated by PCR from WT Mybpc3 cDNA with a forward primer (PTM F) containing an XhoI restriction site, the ATG followed by the FLAG sequence and the first 20 nucleotides of Mybpc3 exon 1. The reverse primer (PTM R) contained a BamHI restriction site and the 5′ canonical splice donor site sequence followed by a downstream intronic splicing enhancer element/sequence from the rat fibroblast growth factor receptor 2 gene and last 23 nucleotides of Mybpc3 exon 6. The binding domains were obtained by PCR on genomic KI DNA using a forward primer (BD F) containing a BamHI restriction site, and 21 nucleotides of Mybpc3 intron 6. The reverse primer (BD R) contained a NotI restriction site and 28 nucleotides complementary to Mybpc3 intron 6. The reverse binding domain was amplified in the same way (primers BD-R F and BD-R R) but reverse complemented. PCR products were sequentially cloned into pdsAAV6-TNNT2 vector (human TNNT2 promoter) and accuracy of the insertion was verified by DNA sequencing analysis. The SV40 polyA signal was removed in one of the PTM plasmids by digestion with NotI and Mva1269I followed by religation of the plasmid.
Production and titration of AAV particles. AAV6 pseudotyped vectors were generated by cotransfection of HEK293-AAV cells (Biocat, Heidelberg, Germany) with the pdsAAV-TNNT2 transfer plasmid and the AAV packaging plasmid pDP6rs, which provides the AAV2 rep and AAV6 cap genes and adenoviral helper functions.39 AAV9 pseudotyped vectors were generated by triple-transfection of pdsAAV-TNNT2 transfer plasmid with pAAV2/9 and pHelper encoding adenoviral helper functions (Biocat). Generation of recombinant AAV6 and AAV9 particles was carried out as described previously,40 with some modifications. HEK293-AAV cells were cultivated in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum, 0.1 mmol/l MEM non-essential amino acids, 2 mmol/l l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Tissue culture reagents were obtained from Life technologies. Briefly, 1.5 × 107 HEK293-AAV cells were seeded on 15-cm plates and transfected with polyethylenimine. After 72 hours, cells were harvested, washed three-times with phosphate-buffered saline (PBS) and resuspended in PBS. After three freeze–thaw cycles, benzonase (Merck, Darmstadt, Germany; final concentration 250 U/ml) was added and the lysates incubated for 1 hour at 37 °C. Cell debris was pelleted and vector-containing lysates were purified using iodixanol step gradients.
The genomic titers of DNase-resistant recombinant AAV particles were determined by quantitative PCR using the SYBR Green qPCR Master MIX 2 (Fermentas, Darmstadt, Germany) and an ABI PRISM 7900HT cycler (Applied Biosystems, Foster City, CA). Vectors were quantified using primers specific for the TNNT2 promoter sequence. Real-time PCR was performed in a total volume of 10 μl with 0.3 μmol/l for each primer. pdsAAV-GFP plasmid was used as a copy number standard. A standard curve for quantification was generated by serial dilutions of the respective plasmid DNA. The cycling conditions were as follows: 50 °C for 2 minutes, 95 °C for 10 minutes, followed by 35 cycles of 95 °C for 15 seconds and 60 °C for 60 seconds. Calculations were done using the SDS 2.4 software (Applied Biosystems).
Generation of recombinant AdV. To generate the AdV-PTMΔpA under the control of TNNT2 promoter, we used the In-fusion kit (Clontech, St Germain-en-Laye, France) to fuse together the two cassettes into pShuttle85706. The pShuttle containing the TNNT2-PTMΔpA insert as well as the TNNT2-GFP in a bicistronic manner was electroporated into Escherichia coli BJ5183-D1 (Stratagene, Darmstadt, Germany) to produce adenoviral DNA through recombination. This DNA was used to transfect HEK293 cells and recombinant AdV was amplified using standard techniques.
Isolation, culture, and transduction of neonatal mouse cardiac myocytes. We isolated and cultured NMCMs using a well-established protocol.22 AAV6-mediated transductions of cardiac myocytes were performed for 30 minutes at 37 °C in suspension before plating (4.4 × 105 cells/well) at a MOI of 3,000 (AAV6-PTMΔpA, both RNA and protein analysis, AAV6-PTM and AAV6-PTM-R for protein analysis) or 30,000 (AAV6-PTM and AAV6-PTM-R for RNA analysis). Cardiac myocytes were kept in culture for 7 days at 37 °C and 10% CO2 before harvesting.
In vivo AAV9 administrations. Seven-week-old KI mice received AAV9-PTMΔpA (1.04 × 1011 vg), AAV9-RLuc (1.36 × 1011 vg) or NaCl via systemic administration into the tail vein with a 29-G needle. Intravenous injections of neonatal KI mice (postnatal day 1) with AAV9-PTMΔpA (4.7 × 1011 vg) or PBS were performed into the temporal vein using a 30-G needle.41 All mice recovered quickly from the injection.
In vivo bioluminescence imaging. Luciferase activity in the mouse heart was non-invasively assessed by in vivo bioluminescence imaging 4 weeks after AAV9 injection. AAV9-Rluc–injected mouse and the NaCl-injected mouse were anesthetized with 1.8% isofluorane. Thereafter the substrate coelenterazine (Biosynth, Staad, Switzerland) dissolved in methanol and further diluted in sodium phosphate buffer pH 7, was injected intraperitoneally (i.p.) at a dose of 2.5 mg/kg body weight in both mice. The mice were then placed in the chamber of a Xenogen in vivo Imaging System under continuous anesthesia. The oxidation of coelenterazine by Renilla luciferase releases coelentarimide and blue light at 480 nm. This bioluminescence was recorded in a manually-selected region of interest centered over the mouse heart, using 3-minute scans.
Echocardiographic analysis. Transthoracic echocardiography was performed using the Vevo 2100 System (VisualSonics, Toronto, Ontario, Canada). KI mice were anesthetized with isofluorane (1–2%) and fixed to a warming platform in a supine position. B-mode images were obtained using a MS400 transducer for adult mice and a MS550 transducer for neonatal mice. Images were obtained in a parasternal short and long-axis view and dimensions of the left ventricle were measured in a short-axis view in diastole and systole.
Reverse transcription-PCR analysis. Total RNA was isolated from cultured NMCMs or ventricular tissue (30 mg) using the SV Total RNA Isolation System Kit (Promega, Madison, WI) according to the manufacturer's instructions. RNA concentration, purity and quality were determined using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Darmstadt, Germany). Reverse transcription was performed from 150 to 200 ng RNA using oligo-dT primers (SuperScript-III kit; Life Technologies, Darmstadt, Germany). As a control for genomic contamination, a reaction without reverse transcription was performed in parallel. Touchdown PCR amplifications (65–60 °C) with different primer pairs (Supplementary Table S2) were performed using AmpliTaq Gold Polymerase (Applied Biosystems) in a total volume of 20 μl for 35 cycles. PCR products were visualized on 1.5% agarose gels. The full-length repaired Mybpc3 mRNA was amplified by touchdown PCR (67–62 °C) with Phusion Hot StartII High-Fidelity DNA polymerase (Biozym, Hessisch Oldendorf, Germany) for 31 cycles. For semi-quantitative analysis a touchdown PCR (65–60 °C) for 25 cycles was used to amplify either total (primers E1-F, E9-R) or repaired (primers FLAG, E9-R) Mybpc3 mRNA. PCR products of the first PCR round were purified on a column (QIAquick PCR Purification Kit; QIAGEN, Valencia, CA) prior a second touchdown PCR (65–60 °C, primers E1-F, E2-R) for 35 cycles.
Western blot and immunoprecipitation analyses. Crude protein extract from cultured NMCMs or HEK293 cells were extracted in lysis buffer (30 mmol/l Tris base pH 8.8, 5 mmol/l EDTA, 30 mmol/l NaF, 3% SDS, 10% glycerol) and protein concentration was determined by Bradford protein assay (Bio-Rad, Hercules, CA). Total proteins (NMCMs 30 μg/lane, HEK293 2.5 μg/lane) were separated on 10% SDS-polyacrylamide (29:1) mini-gels (Bio-Rad) and transferred on polyvinylidene fluoride membranes by electroblotting. Membranes were stained overnight with primary antibodies directed against the FLAG epitope (1:5,000; Sigma, St Louis, MO) in 5% milk in TBS-T or against the MyBP-C motif (1:1,000). After incubation with anti-mouse (1:10,000; Sigma) or anti-rabbit (1:6,000; Sigma) peroxidase-conjugated secondary antibodies, proteins were visualized using Super Signal West Dura detection reagent (Thermo Scientific) and signals were detected with the ChemiGenius2 Bio Imaging System.
For immunoprecipitation AAV6-PTMΔpA-transduced NMCMs or ventricular tissue of AAV9-PTMΔpA-injected mouse were lysed in modified RIPA buffer (500 mmol/l NaCl, 1 mmol/l EDTA, 50 mmol/l Tris-HCl pH 7.4, 1% Triton X-100, protease inhibitors Complete Mini; Roche, Indianapolis, IN), sonicated 2× for 30 seconds and centrifuged for 10 minutes at full speed at 4 °C. The supernatant containing soluble proteins was diluted in 500 μl modified RIPA buffer and gently rolled overnight at 4 °C with or without 10 μg anti-FLAG antibody (Sigma). The immunocomplexes were recovered by incubation with 50 μl of protein A/G plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) for 4 hours at 4 °C. After three washing steps in RIPA buffer and one additional wash step in 1× PBS (200g, 3 minutes, 4 °C) the FLAG-tagged proteins were eluted in 1× Laemmli buffer (20 mmol/l Tris-HCl pH 6.8, 200 mmol/l DTT, 4% SDS, 0.02% bromophenol blue, 20% glycerol) and used for western blot as described above. Anti-mouse Exacta Cruz (1:2,000; Santa Cruz Biotechnology), which does not recognize the heavy and light chains of the immunoprecipitation antibody, was used as secondary antibody. The same immunoprecipitation protocol was applied to HEK293 cells (40 μg) transiently transfected with FLAG-WT-cMyBP-C used as control.
Immunofluorescence analysis. For immunofluorescence analysis, AAV6-transduced KI NMCMs were cultured for 7 days on coverslips. Cells were rinsed once with ice-cold 1× PBS and fixed 10 minutes at −20 °C in methanol/acetone (20/80). After two short washing steps in 1× PBS, cells were permeabilized 1 hour at room temperature in solution A (10% FCS, 1% BSA, 0.5% Triton X-100 in 1× PBS). Incubation with primary antibodies (anti-FLAG, 1:800; anti-MyBP-C motif, 1:500) was done in solution B (1% BSA, 0.5% Triton X-100 in 1× PBS) for 1 hour at room temperature. Cells were then rinsed twice in solution B and incubated for 1 hour at room temperature with secondary antibodies (anti-mouse IgG Alexa 488-conjugated, 1:800 and anti-rabbit IgG Alexa 546-conjugated 1:800; Molecular Probes, Darmstadt, Germany) diluted in solution B together with DRAQ5 (1:1,000; Biostatus, Leicestershire, UK) for nuclear staining. Coverslips were embedded in Mowiol and confocal images were acquired with a Zeiss LSM 710 system using a Zeiss Axiovert microscope (Zeiss, Jena, Germany) and a 40×-oil objective.
SUPPLEMENTARY MATERIAL Figure S1. Schematic representation of the repaired and mutant Mybpc3 mRNA. Figure S2. Efficiency of AAV6- and AdV-mediated transduction in Mybpc3-targeted knock-in NMCMs. Figure S3. In vivo bioluminescence imaging. Figure S4. Efficiency of transduction after systemic administration of AAV9-GFP in a Mybpc3-targeted knock-in mouse. Figure S5. Detection of full-length repaired Mybpc3 mRNA. Table S1. Sequences of the binding domains. Table S2. Sequences of the PCR primers used in the present study. Table S3. Echocardiographic analysis.
Acknowledgments
We thank Evelyn Bendrat and Michaela Miehe (UKE-HEXT, Hamburg, Germany), and Mareen Welzel and Andreas Jungmann (University Hospital Heidelberg, Heidelberg, Germany) for their excellent technical support in adeno-associated virus production. We thank Christiane Pharmann and Sonja Schrepfer (UKE-Transplant and Stem cell immunobiology lab, Hamburg, Germany) for in vivo bioluminescence assays. We also thank June Uebeler and Verena Behrens-Gawlik (UKE-Pharmacology, Hamburg, Germany) for help in cardiac myocytes preparation and confocal imaging. We also thank Jürgen Kleinschmidt (DKFZ Heidelberg, Heidelberg, Germany) and Julie Johnston (Penn Vector Core, Pennsylvania University, Philadelphia, PA, USA) for supplying the pDP6rs and the pAAV2/9 plasmids, respectively, and Christian Witt (University of Heidelberg, Heidelberg, Germany) for the anti-cMyBP-C antibody. This work was supported by the Fritz Thyssen Stiftung (Az. 10.09.1.139), the seventh Framework Program of the European Union (Health-F2-2009–241577; Big-Heart project), the DZHK (German Center for Cardiovascular Research), and the German Federal Ministry of Research and Education (BMBF). The authors declared no conflict of interest.
Supplementary Material
References
- Wally V, Murauer EM, Bauer JW. Spliceosome-mediated trans-splicing: the therapeutic cut and paste. J Invest Dermatol. 2012;132:1959–1966. doi: 10.1038/jid.2012.101. [DOI] [PubMed] [Google Scholar]
- Mitchell LG, McGarrity GJ. Gene therapy progress and prospects: reprograming gene expression by trans-splicing. Gene Ther. 2005;12:1477–1485. doi: 10.1038/sj.gt.3302596. [DOI] [PubMed] [Google Scholar]
- Le Roy F, Charton K, Lorson CL, Richard I. RNA-targeting approaches for neuromuscular diseases. Trends Mol Med. 2009;15:580–591. doi: 10.1016/j.molmed.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Chao H, Walsh CE. RNA repair using spliceosome-mediated RNA trans-splicing. Trends Mol Med. 2004;10:263–268. doi: 10.1016/j.molmed.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Chao H, Mansfield SG, Bartel RC, Hiriyanna S, Mitchell LG, Garcia-Blanco MA, et al. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang Q, Mansfield SG, Puttaraju M, Zhang Y, Zhou W, et al. Partial correction of endogenous DeltaF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- Coady TH, Baughan TD, Shababi M, Passini MA, Lorson CL. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady TH, Lorson CL. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J Neurosci. 2010;30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara M, Pergolizzi RG, Kobayashi H, Krause A, Luettich K, Lesser ML, et al. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin T, Anthony K, Garcia-Blanco MA, Mansfield SG, Anderton BH, Gallo JM. Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing. Hum Mol Genet. 2009;18:3266–3273. doi: 10.1093/hmg/ddp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avale ME, Rodríguez-Martín T, Gallo JM. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum Mol Genet. 2013;22:2603–2611. doi: 10.1093/hmg/ddt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wally V, Klausegger A, Koller U, Lochmüller H, Krause S, Wiche G, et al. 5′ trans-splicing repair of the PLEC1 gene. J Invest Dermatol. 2008;128:568–574. doi: 10.1038/sj.jid.5701152. [DOI] [PubMed] [Google Scholar]
- Rindt H, Yen PF, Thebeau CN, Peterson TS, Weisman GA, Lorson CL. Replacement of huntingtin exon 1 by trans-splicing. Cell Mol Life Sci. 2012;69:4191–4204. doi: 10.1007/s00018-012-1083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines et al. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58:e212–e260. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- Ehlermann P, Weichenhan D, Zehelein J, Steen H, Pribe R, Zeller R, et al. Adverse events in families with hypertrophic or dilated cardiomyopathy and mutations in the MYBPC3 gene. BMC Med Genet. 2008;9:95. doi: 10.1186/1471-2350-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron P, Carrier L, Dubourg O, Tesson F, Desnos M, Richard P, et al. Penetrance of familial hypertrophic cardiomyopathy. Genet Couns. 1997;8:107–114. [PubMed] [Google Scholar]
- Schlossarek S, Mearini G, Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–620. doi: 10.1016/j.yjmcc.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, EUROGENE Heart Failure Project et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- Pohlmann L, Kröger I, Vignier N, Schlossarek S, Krämer E, Coirault C, et al. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ Res. 2007;101:928–938. doi: 10.1161/CIRCRESAHA.107.158774. [DOI] [PubMed] [Google Scholar]
- Cazorla O, Szilagyi S, Vignier N, Salazar G, Krämer E, Vassort G, et al. Length and protein kinase A modulations of myocytes in cardiac myosin binding protein C-deficient mice. Cardiovasc Res. 2006;69:370–380. doi: 10.1016/j.cardiores.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Vignier N, Schlossarek S, Fraysse B, Mearini G, Krämer E, Pointu H, et al. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res. 2009;105:239–248. doi: 10.1161/CIRCRESAHA.109.201251. [DOI] [PubMed] [Google Scholar]
- Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- Fraysse B, Weinberger F, Bardswell SC, Cuello F, Vignier N, Geertz B, et al. Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol. 2012;52:1299–1307. doi: 10.1016/j.yjmcc.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88:478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorain S, Peccate C, Le Hir M, Garcia L. Exon exchange approach to repair Duchenne dystrophin transcripts. PLoS ONE. 2010;5:e10894. doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- Fuke H, Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2008;36:1037–1049. doi: 10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller OJ, Leuchs B, Pleger ST, Grimm D, Franz WM, Katus HA, et al. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res. 2006;70:70–78. doi: 10.1016/j.cardiores.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Friedrich FW, Wilding BR, Reischmann S, Crocini C, Lang P, Charron P, et al. Evidence for FHL1 as a novel disease gene for isolated hypertrophic cardiomyopathy. Hum Mol Genet. 2012;21:3237–3254. doi: 10.1093/hmg/dds157. [DOI] [PubMed] [Google Scholar]
- Pacak CA, Mah CS, Thattaliyath BD, Conlon TJ, Lewis MA, Cloutier DE, et al. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res. 2006;99:e3–e9. doi: 10.1161/01.RES.0000237661.18885.f6. [DOI] [PubMed] [Google Scholar]
- Chen HY, Kathirvel P, Yee WC, Lai PS. Correction of dystrophia myotonica type 1 pre-mRNA transcripts by artificial trans-splicing. Gene Ther. 2009;16:211–217. doi: 10.1038/gt.2008.150. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Friedlander RM. Using non-coding small RNAs to develop therapies for Huntington's disease. Gene Ther. 2011;18:1139–1149. doi: 10.1038/gt.2011.170. [DOI] [PubMed] [Google Scholar]
- Friedrich FW, Carrier L. Genetics of hypertrophic and dilated cardiomyopathy. Curr Pharm Biotechnol. 2012;13:2467–2476. doi: 10.2174/138920112804583041. [DOI] [PubMed] [Google Scholar]
- Brito D, Miltenberger-Miltenyi G, Vale Pereira S, Silva D, Diogo AN, Madeira H. Sarcomeric hypertrophic cardiomyopathy: genetic profile in a Portuguese population. Rev Port Cardiol. 2012;31:577–587. doi: 10.1016/j.repc.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Curila K, Benesova L, Penicka M, Minarik M, Zemanek D, Veselka J, et al. Spectrum and clinical manifestations of mutations in genes responsible for hypertrophic cardiomyopathy. Acta Cardiol. 2012;67:23–29. doi: 10.1080/ac.67.1.2146562. [DOI] [PubMed] [Google Scholar]
- Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- Sands MS, Barker JE. Percutaneous intravenous injection in neonatal mice. Lab Anim Sci. 1999;49:328–330. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.