Abstract
One of the most challenging aspects of lung cancer therapy is the rapid acquisition of multidrug-resistant (MDR) phenotype. One effective approach would be to identify and downregulate resistance-causing genes in tumors using small interfering RNAs (siRNAs) to increase the sensitivity of tumor cells to chemotherapeutic challenge. After identifying the overexpressed resistance-related antiapoptotic genes (survivin and bcl-2) in cisplatin-resistant cells, the siRNA sequences were designed and screened to select the most efficacious candidates. Modifications were introduced in them to minimize off-target effects. Subsequently, the combination of siRNA and cisplatin that gave the maximum synergy was identified in resistant cells. We then demonstrated that the combination treatment of the selected siRNAs and cisplatin encapsulated in CD44-targeting hyaluronic acid (HA)-based self-assembling nanosystems reversed the resistance to cisplatin and delayed the tumor growth significantly (growth inhibition increased from 30 to 60%) in cisplatin-resistant tumors. In addition, no abnormalities in body weights, liver enzyme levels or histopathology of liver/spleen tissues were observed in any of the treatment groups during the study period. Overall, we demonstrate that the combination of siRNA-mediated gene-silencing strategy with chemotherapeutic agents constitutes a valuable and safe approach for the treatment of MDR tumors.
Keywords: cisplatin resistance, combination therapy, lung cancer, multidrug resistance, nanoparticles, siRNA delivery
Introduction
Lung cancer is known to be the most frequent cancer worldwide and the incidence of this epidemic disease is continuing to increase at 0.5% per year globally.1 In contrast to the significant reduction of mortality from heart disease, the survival rate of lung cancer patients has been at a plateau for almost three decades, with a relative 5-year survival rate of <18% in most countries. Because of the size and distribution of lung cancer, the cytoreductive surgery is not very effective for this disease and therefore chemotherapy and/or radiation are the only treatments of choice. Despite major advances in patient management, chemotherapy and radiotherapy, nearly 80% of the patients still die within 1 year of diagnosis and long-term survival is obtained only in 5–10% of the cases.1
The major obstacle in lung cancer chemotherapy is the emergence of inherent and acquired drug resistance in cancer cells.2,3 The efficacy of chemotherapy is thus limited. To overcome this resistance, often higher doses of toxic anticancer drugs are administered to cancer patients, thus resulting in adverse side effects to healthy organs and tissues. In this regard, reversal of drug resistance is one of the most attractive ways to significantly enhance therapeutic efficacy in lung cancers.
Cancer cells become resistant to anticancer drugs by several mechanisms.4 One well-studied mechanism is the pronounced activity of efflux pumps such as ATP transporters, which efficiently pump the drugs out of tumor cells. This resistance is called pump-mediated resistance and is conferred by at least two proteins such as P-glycoprotein (P-gp, encoded by the mdr1 gene) and the multidrug resistance (MDR)-associated proteins (mrp1).2,5 In lung cancer samples, P-gp is known to be infrequently expressed,6 mdr1 mRNA expression was reported to be increased in 15–50% of tumors and the incidence of mrp1 gene expression is much higher (about 80%) in small cell lung cancer (SCLC) samples.
A growing body of evidence also suggests that the drug resistance, not mediated by pump-related genes and proteins are attributed primarily to the mechanisms responsible for the activation of antiapoptotic cellular defense. Most chemotherapeutic agents, including cisplatin, induces cell death by apoptosis.7 Cisplatin, together with a third-generation anticancer agent, is the standard regimen used in the first-line treatment of advanced non SCLC (NSCLC) in the clinic and has shown superior efficiency in multiple trials. It is believed that the DNA damage caused by the chemotherapeutic drugs induces the release of an enzyme that activates the caspases.8 Programmed cell death or apoptosis, is generally known to occur by the activation of caspase family of enzymes that cleave cellular proteins.9,10,11,12 One class of molecules that block apoptosis by directly binding to caspases is the inhibitor of apoptosis proteins such as survivin.13,14 bcl-2 is also an antiapoptotic gene and overexpressed in variety of human tumors including NSCLC and involved in tumorigenesis and chemoresistance.12 It has been shown that the overexpression of bcl-2 delays the onset of apoptosis induced by several chemotherapeutic drugs.11 Substantial evidence indicates that downregulation of antiapoptotic genes such as survivin and bcl-2 can sensitize cancer cells to anticancer drugs.5,15,16,17 Post-transcriptional silencing of MDR-related genes thus seems to be a promising strategy.18
Such an objective can be effectively achieved if the agents are safely and selectively delivered to tumors by utilizing the anatomical and pathophysiological abnormalities of solid tumors.19,20 It has been well established that nanoparticle surface decorated with tumor targeting agents, such as antibodies and peptides, can also be exploited for active targeting to tumor cells.21,22 In the previous study, we demonstrated that the hyaluronic acid (HA)-based self-assembling nanoparticle system efficiently delivered small interfering RNA (siRNA) to multiple types of tumors that express high levels of CD44.23 In the current study, we utilized the HA-based nanosystems to efficiently deliver siRNAs and chemotherapeutic agent, cisplatin, to demonstrate combination effect for reversing the multiprolonged MDR cellular defense for pronounced antitumor response. Anti-MDR strategies may thus show high levels of clinical efficacy when administered in combination with chemotherapeutic regimens.
Results
NSCLC cells show resistance to cisplatin, but not to doxorubicin
As our goal was to reverse the resistance in lung cancer, a pair of NSCLC cells that are sensitive and resistant to cisplatin (A549/A549DDP) and a SCLC pair of cells that are sensitive and resistant to doxorubicin (H69/H69AR), respectively were selected for the initial study. As previously reported, A549 and its resistant counterpart (A549DDP) demonstrated saturating levels of CD44 expression and thus exhibited efficient gene downregulation when treated with siRNA encapsulated HA nanosystem.23 However, the apparent lower level expression of CD44 on H69AR (~90%) and H69 (~60%) cells led to lower level activity in H69AR and almost no activity in H69 cells even at higher siRNA concentrations.23 Due to the above observations, the primary focus of our study was directed on resistant/sensitive NSCLC cells.
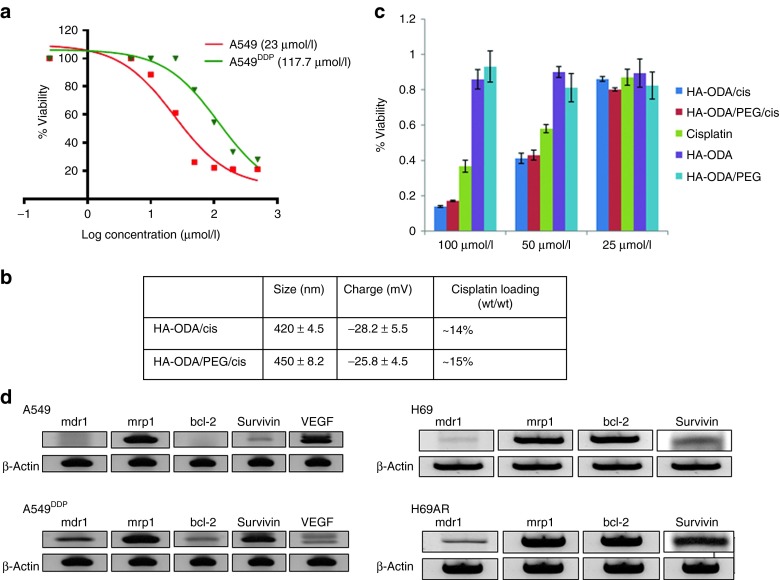
In order to assess the resistance levels of cisplatin in NSCLC cells, cisplatin drug alone was tested in sensitive and resistant A549 cells. The half maximal inhibitory concentration (IC50) (117 µmol/l) was found to be fivefold higher in resistant cells compared with the sensitive cells (Figure 1a). Likewise, the cisplatin resistance was also assessed in doxorubicin resistant/sensitive SCLC cells and found no difference in IC50's (data not shown). In order to improve the delivery efficiency of cisplatin to the resistant A549DDP cells, we attempted cisplatin encapsulation in multiple types of lipid-functionalized HA derivatives. Among them, a C8 lipid-modified derivative of HA, namely HA-1,8-diaminooctane (ODA), exhibited relatively high cisplatin encapsulation (~15% wt/wt) and was used for all future in vitro and in vivo evaluations (Figure 1b). The blank HA nanoparticles did not exhibit any toxicity in tested cells (Figure 1c). However, the cisplatin loaded in HA-ODA nanoparticles with and without polyethylene glycol (PEG) demonstrated slightly better IC50 when compared with cisplatin alone in resistant A549DDP cells (Figure 1c).
Figure 1.
Optimizing hyaluronic acid (HA)/cisplatin nanoparticles and identifying the resistant genes in drug-resistant cells. To determine the cisplatin resistance in resistant NSCLC cells, cisplatin drug was incubated with both sensitive and resistant cells (A549/A549DDP) at various concentrations for 2 days. (a) Cell viability was assessed to determine the IC50 (half maximal inhibitory concentration). (b) To improve the delivery, cisplatin was encapsulated in 1,8-diaminooctane (ODA)-modified HA nanosystems with and without poly(ethylene glycol) (PEG) and characterized. (c) The cytotoxicity of HA-ODA and HA-ODA/PEG nanoparticles were measured with and without cisplatin along with cisplatin alone as a control in resistant A549DDP cells. In order to identify the resistant genes in NSCLC and SCLC cells, RNA was extracted from both resistant and sensitive cells. (d) With appropriate primers, the reverse transcription-PCR was run to identify the expression of resistant genes. NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
Overexpression of pump and non-pump–mediated resistant genes may be responsible for resistance in NSCLC and SCLC resistance cells
Using the appropriate primers that were specific for pump and non-pump–mediated resistant genes, reverse transcription-PCR was run using the RNA extracted from both sensitive and resistant SCLC (H69/H69AR) and NSCLC (A549/A549DDP) cells (Figure 1d). Survivin is overexpressed in resistant cell lines (A549DDP and H69AR) compared with their sensitive counterparts (A549 and H69), whereas, the mrp-1 is expressed in both resistant and sensitive cell lines (A549, A549DDP, H69, H69AR) almost at the same levels. In addition to survivin and mrp1 genes, bcl-2 and mdr1 genes were also identified from this experiment. bcl-2 was slightly overexpressed in resistant A549 cells but the overall expression level was much lower than the survivin expression level in this cell line. Just like the mrp1, bcl-2 was expressed almost equally in both resistant and sensitive H69 cells. mdr1 was not expressed in sensitive cell lines, but slightly expressed in both the resistant cells. Despite the differential expression levels, all four genes were selected in this study to carry out combination experiments and to address both pump and non-pump–mediated resistance (Figure 1d). In order to downregulate the genes overexpressed in the resistance cells, highly efficacious, target specific siRNA sequences were selected by computational in silico methods using the BioPred algorithm.24
Selection of potent and specific siRNA sequences lead to efficient target knockdown in resistant cells
All possible siRNA sequences for the given target were passed through rigorous computational filtration process (involving potency and specificity filtration) using BioPred algorithm.24 Top four to five predicted most potent siRNAs (score >96%) were acquired for further in vitro analysis (Supplementary Figure S1a,b). These selected siRNA sequences were then tested in resistant A549DDP cells at series of concentrations to rank the potency (Figure 2a–d). Based on the activity in cells, two most potent sequences were selected, introduced modifications and retested in cells to confirm activity. The best-modified one was selected for in vivo evaluation. Five different unmodified survivin sequences were initially screened at four different concentrations using lipofectamine as the transfection agent in A549DDP cells to rank their potency (Figure 2a). Based on the activity, the two most potent sequences (#1 and #2) were selected for further modifications. A standard 2′-OMe modification was introduced into these two sequences and tested again in cells to confirm the activity. Since the sequence #2 did not lose any activity after introducing the modification, it was selected to test in vivo along with its unmodified version to make a choice (Figure 3a). Out of the four mrp1 siRNAs screened in cells, the best one showed only 60% target knockdown even at 50 nmol/l concentration in cells (Figure 2d). The lower values may be attributed due to issues with the siRNA potency or its target. All four mdr1 and bcl-2 sequences screened were found equally potent in cells (Figures 2b,c). The same 2′OMe modification was introduced into the best two bcl-2 sequences (#2 and #3) and tested in cells (Figure 3b). In this case, activity was slightly lost in cells when the modification was introduced in both the sequences. However, to reduce or minimize the off-target/immune stimulatory effects coming from an unmodified sequence, the best-modified version was selected for in vivo studies.
Figure 2.
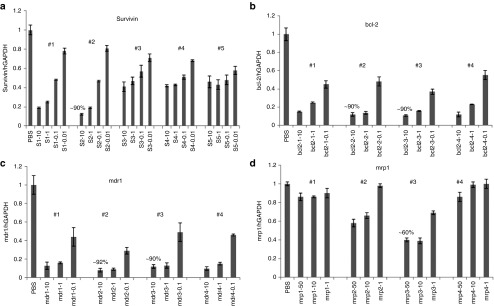
Screening small interfering RNAs (siRNAs) in resistant A549 cells to identify the most potent sequence based on the mRNA knockdown. A total of (a) five survivin, (b) four bcl-2, (c) four mdr1, and (d) four mrp1 unmodified siRNA sequences were screened at 3–4 different concentrations in A549DDP cells to rank the potency of the sequences.
Figure 3.
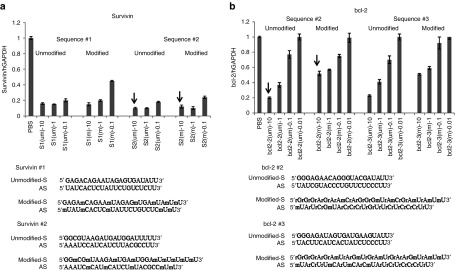
Introducing modifications to the selected small interfering RNA (siRNA) sequences and evaluating their activity. After introducing the modifications to (a) two best survivin sequences and (b) two bcl-2 siRNA sequences, they were tested again along with their unmodified counterparts in resistant A549DDP cells to compare the activity.
Combination strategies involving downregulation of non-pump–mediated genes and cisplatin treatments lead to synergy in resistant A549 cells
Based on the previous knockdown studies, the survivin siRNA (um #2) sequence that gave >90% silencing was selected for initial combination studies. The IC50 of cisplatin determined from the previous cytotoxicity data (Figure 1a) and few concentrations below IC50 were used with siRNAs in the combination study. Cisplatin treatment was given either 24 hours post-siRNA treatment or together with siRNA treatment to see if the timing of cisplatin treatment affords any benefit to the synergistic effect (Figure 4a,b). The results of this study indicated that the combination of survivin downregulation (>90%) and cisplatin treatment (at IC50) was found to be more effective when compared with cisplatin alone treatment and the activity was very similar for both co-treatment as well as for treatments that were given apart from each other. These results suggest that the downregulation of one of the resistant genes may enhance the sensitivity of the cells to cisplatin. In addition to survivin siRNA, bcl-2, mdr1, and mrp1 siRNAs (um #2, 2, and 3) were also then included in the follow-up combination studies with cisplatin to identify the best combination that gives the highest cell killing. For these experiments, bcl-2 and mdr1 siRNAs sequence that gave ~90% knockdown, mrp1 siRNA that gave the maximum knockdown (60%) were evaluated as described in the Materials and Methods. The combination of survivin and/or bcl-2 with cisplatin demonstrated combination/synergistic effect but not the mrp1 and mdr1 siRNAs. Downregulation of both survivin and bcl-2 together with cisplatin treatment showed slightly better cell-killing effect compared with the single-agent combinations (survivin + cisplatin or bcl-2 + cisplatin) suggesting a higher level of reversal of resistance with the downregulation of both antiapoptotic genes (Figure 4c).
Figure 4.
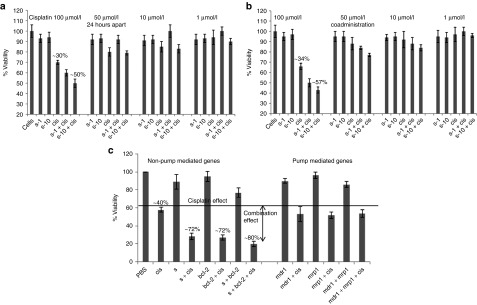
Evaluating the combination effect of small interfering RNA (siRNA) and cisplatin treatment in A549DDP cells. Cells were treated first with survivin siRNA for 24 hours followed by (a) the cisplatin treatment at different concentrations or (b) co-treated simultaneously. Forty-eight hours after the cisplatin treatment, cell viability was evaluated by MTS assay. (c) In the follow-up study, the combination effect of siRNAs against survivin, bcl-2, mdr1, and mrp1 combined with cisplatin was evaluated using the method used in a.
Combination therapy with siRNA and cisplatin yields significantly better antitumor efficacy compared with the single-agent treatment
First, the unmodified and modified survivin and bcl-2 sequences that were selected from in vitro screens (survivin: um #2 versus m #2, bcl-2: um #2 and 3 versus m#2 and 3 and control siRNA) were formulated in HA-PEI/HA-PEG nanoparticles as described previously.23 The resulting nanoparticles with siRNA had a mean hydrodynamic diameter of 85 ± 7.5 nm and zeta potential of −10 ± 4.5 mV. These forumlations were then dosed at 0.5 mg/kg every day for three consecutive days in mice bearing resistant A549 tumors for assessing target knockdown at different timepoints (24, 72, and 120 hours after last dose) to pick the best possible sequence for the combination efficacy study. In the first target knockdown study, the unmodified survivin siRNA sequence gave only 15% target knockdown in tumors 24 hours after the last injection. However, the activity was increased to ~40% at 72 hours and this activity was still maintained for at least 120 hours (Figure 5a). The non-targeting control siRNA (CTL) sequence in the same study did not show any target knockdown. The best-modified sequence selected from the in vitro screening (#2) was tested in the same tumor model in another study to compare the activity with the unmodified sequence. The activity of unmodified sequence was ~40% at 72 and 120 hours after the last injection, almost identical to the previous study. Whereas, the activity of the modified sequence was only about 25–35% at the doses tested, suggesting that there may be a slight loss of potency when the modification was introduced or it is also possible that there was some immune stimulatory effect demonstrated by the unmodified sequence (Figure 5b). In the bcl-2 screening study, the best two sequences found in the in vitro study were tested along with the corresponding modified versions in the same resistant A549 tumors. The unmodified sequences (#2 and 3) gave about 55 and 40% target knockdown, respectively at 72-hour timepoint. The modified versions, however, gave only 20–30% activity at the same timepoint (Figure 5c). Based on the above in vivo, knockdown studies, the survivin #2 (m) and bcl-2 #2 (m) sequences were selected for the combination efficacy study to minimize any off-target effects associated with the unmodified sequences.
Figure 5.
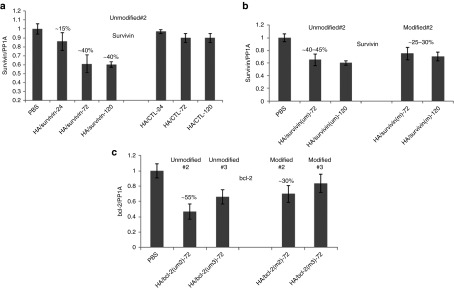
Survivin/bcl-2 knockdown in A549DDP tumors at different timepoints. Tumor-bearing mice were injected with (a) survivin siRNA (um) and (b) um versus m encapsulated hyaluronic acid-poly(ethyleneimine) (HA-PEI)/HA-poly(ethylene glycol) (HA-PEG) self-assembled nanoparticles at 0.5 mg/kg for 3 days. Tumors were harvested at different timepoints (24, 72, and 120 hours for a and 72 and 120 hours for b) to look at the target specific knockdown. (c) Similarly, two bcl-2 siRNA sequences (um and m) were encapsulated in the same HA system and treated the mice bearing the same tumors and monitored the target knockdown at 72 hours. CTL, non-targeting control siRNA.
To determine the optimum cisplatin doses that are needed for the efficacy study, a pilot cisplatin efficacy study was run before performing the actual combination study (Figure 6a). To check the activity of cisplatin encapsulated HA-ODA nanoparticles with and without PEG, mice bearing A549DDP tumors were grown and sorted out to accommodate four groups with five mice in each group. Mice were injected with either phosphate-buffered saline (PBS), cisplatin alone, HA-ODA/cisplatin and HA-ODA/HA-PEG/cisplatin nanoparticles at 1 mg/kg, twice at 4 days apart. Mice treated with either cisplatin or HA-ODA/cisplatin or HA-ODA/PEG/cisplatin nanoparticels showed tumor growth inhibition compared with the PBS-treated group (treatment-to-control ratio (T/C) of 0.6–0.65). Out of these three groups, the HA-ODA/cisplatin nanoparticles tend to be slightly better than the other two in the efficacy curve at the early timepoints. Given this, the HA-ODA/cisplatin nanoparticle system was selected for the combination study.
Figure 6.
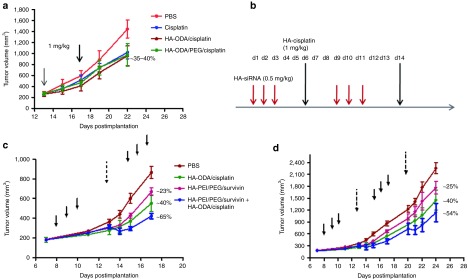
Effect of the combination of cisplatin treatment and survivin silencing on growth of resistant A549DDP tumors. (a) To find the optimum cisplatin dose and timing for the combination efficacy study, a pilot study was initially run with cisplatin or 1,8-diaminooctane-modified hyaluronic acid (HA-ODA)/cisplatin or HA-ODA/poly(ethylene-glycol) (PEG)/cisplatin at 1 mg/kg doses and monitored the tumor growth. Based on this study results and the previous knockdown study data, (b) an efficacy study was designed with synchronized dosing of small-interfering RNA (siRNA) and cisplatin. (c) As planned, A549DDP tumor-bearing mice were grouped and injected with either siRNA alone at 3 × 0.5 mg/kg or with cisplatin at 1 mg/kg or with siRNA and cisplatin (3 × 0.5 and 1 mg/kg) together and monitored the tumor growth after the first round of treatment. (d) The mice were injected with the second set of doses and monitored the tumor growth for another week.
Next, to run a pilot efficacy study to combine the survivin downregulation and cisplatin treatment, mice bearing A549DDP tumors were treated with PBS, cisplatin, HA-PEI/PEG/ survivin or combination of HA-PEI/PEG/survivin + cisplatin. As the activity of three consecutive siRNA doses sustained for 5 days, the cisplatin treatment was given 72 hours after the third siRNA dose to accommodate the activity of both treatments (Figure 6b). The second round of siRNA treatment started 5 days after the last siRNA dose of the first round. There was tumor growth inhibition observed in mice that had survivin alone (~23%) or cisplatin alone (~46%) treatments. But the combination group showed significantly better growth inhibition (~65%) compared with PBS or either of the single-agent treatment on day 17 (Figure 6c). The corresponding T/C values were 0.77, 0.60, and 0.35 suggesting that there was combination or synergistic effect to some extent with the combination treatment. It suggests that the cell death induction by an anticancer drug in combination with the suppression of non-pump resistance is required for effective killing of drug-resistant cancer cells. However, the rate of tumor growth inhibition was slightly reduced after the second round of treatment (Figure 6d). After two rounds of treatment, the growth inhibition was only 25% (T/C of 0.75) for survivin, 40% (T/C 0.6) for cisplatin, and 54% (T/C 0.46) for combination treatment. This may be due to the development of further resistance for treatments or doses that were not sufficiently high enough to kill all the cells. Based on the results, further modifications were incorporated in the study plan for the next efficacy study.
In the next efficacy study, two siRNAs (survivin and bcl-2) were used in combination with cisplatin to see if the efficacy could be improved by knocking down both gene expression levels before cisplatin treatment. In this study, in addition to two therapeutic siRNAs, a control siRNA was also used in the presence and absence of cisplatin treatment to eliminate any nonspecific activity. The dose regimen was also adjusted slightly from the previous efficacy study to accommodate the peak activity of both treatments in a shorter window (Figure 7a). As expected, the untreated tumors and mice that had control siRNA treatments displayed rapidly progressive tumor growth, whereas the tumors in animals that had therapeutic siRNA alone, cisplatin alone or combination treatments grew relatively slowly. After two rounds of cisplatin and siRNA treatments, (six siRNA doses and two cisplatin doses), the groups that had combination treatment (survivin + cisplatin, bcl-2 + cisplatin or survivin + bcl-2 + cisplatin) showed significantly better tumor growth inhibition compared with PBS or CTL group (Figure 7b) and the groups that had single-agent treatment (survivin, bcl-2, survivin + bcl-2 or cisplatin) with growth inhibition of 62% (T/C of 0.38) for survivin + bcl-2 + cisplatin group, 58% (T/C of 0.42) for bcl-2+cisplatin group, and 52% (T/C 0.48) for survivin + cisplatin group. Knocking down both survivin and bcl-2 together with cisplatin treatment, although did not show significant difference in efficacy compared to the ones with single siRNA + cisplatin (survivin + cisplatin or bcl-2 + cisplatin) groups, the combination did however show much greater significant difference from its control groups. (survivin + bcl-2 + cisplatin versus survivin + bcl-2; P = 0.02, survivin + cisplatin versus survivin; P = 0.07 or bcl-2 + cisplatin versus bcl-2; P = 0.03 or cisplatin versus survivin + cisplatin; P = 0.07, cisplatin versus bcl-2 + cisplatin; P = 0.05, cisplatin versus survivin + bcl-2 + cisplatin; P = 0.01 or PBS versus survivin + cisplatin; P = 0.01, PBS versus bcl-2 + cisplatin; P = 0.002, PBS versus survivin + bcl-2 + cisplatin; P = 0.0001). There was a slightly added benefit of knocking down both survivin and bcl-2 expression levels together with cisplatin treatment over either of the siRNA + cisplatin treatment in this study (62% growth inhibition versus 58 or 52% growth inhibition) supporting the pattern that was found in in vitro study results (80 versus ~72% killing).
Figure 7.
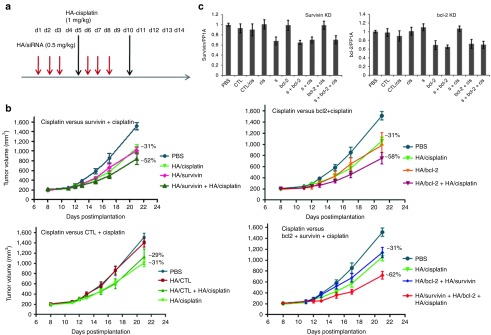
Effect of combination of downregulation of two antiapoptotic genes (survivin and bcl-2) and cisplatin treatment. (a) Based on the previous study, the dose regimen was changed slightly to improve the outcome. Mice bearing A549DDP tumors were treated with nine different combinations as described (n = 5) for over 2 weeks and monitored the tumor growth and other safety measurements. (b) Comparing the antitumor efficacy of cisplatin-treated mice with survivin + cisplatin-treated mice, cisplatin-treated mice with bcl-2 + cisplatin-treated mice, cisplatin-treated mice with CTL siRNA + cisplatin-treated mice, cisplatin-treated mice with survivin + cisplatin and survivin + bcl-2 + cisplatin-treated mice. (c) The corresponding target knockdown (survivin and bcl-2) were also determined in the tumors collected at the end of the study. Tumor samples were collected 72 hours after last siRNA dose. CTL, non-targeting control siRNA; siRNA, small interfering RNA.
The tumor growth inhibition in mice that had only cisplatin treatment was 31% (T/C of 0.69) and it was 29% (T/C of 0.71) for mice that had CTL siRNA + cisplatin treatment. The siRNA treatment groups such as survivin alone, bcl-2 alone, and survivin+bcl-2 groups had 31, 31, and 30% growth inhibition (T/C values of 0.69, 0.69, and 0.70), respectively. The control group with no cisplatin had almost no difference in tumor growth from PBS-treated group. To see if the tumor growth inhibition observed correlates with the knockdown levels, tumor samples collected at the end of the study were analyzed for survivin and bcl-2 specific knockdown (Figure 7c). There was about 30–40% survivin knockdown observed in tumors that had survivin or survivin + cisplatin or survivin + bcl-2 + cisplatin treatment and a similar level bcl-2 specific knockdown seen in mice that had bcl-2 alone or bcl-2 + cisplatin or bcl-2 + survivin + cisplatin treatment. All the tumors that had survivin (or bcl-2) treatment showed similar downregulation of either survivin or bcl-2; however, the ones that had the combination of survivin (or bcl-2) and cisplatin demonstrated the knockdown along with significant tumor growth inhibition compared with the single-agent treatment. Similarly, the tumors that had survivin + bcl-2 or survivin + bcl-2 + cisplatin treatment showed about the same levels of both survivin and bcl-2 downregulation, but the group that had cisplatin showed the highest tumor growth inhibition compared with all the other groups in the study supports the effective combination strategy (Figure 7c). This result suggests that the combination of survivin or bcl-2 or survivin + bcl-2 downregulation and cisplatin treatment together demonstrated combination or synergistic effect. Again, the study results suggest that the combination of anticancer drug with suppression of non-pump resistance seems to be required for effective killing of MDR cells. Alternatively, one could also say that by downregulating the overexpressed resistant gene/genes in this study, the resistance to cisplatin could be overcome to some extent. More exploration should be carried out to identify any other genes that could also contribute to the resistance, the right set of dosing schedules and frequency of doses to improve the synergistic effect. Taken together, our findings provide strong evidence that by silencing resistance-related genes, higher levels of efficacy can be achieved with chemotherapeutic agents.
Cisplatin-resistant tumor-bearing mice well tolerated the combination treatments of siRNA and cisplatin with no adverse effects
During this study period, to monitor the safety of the formulations, the body weights of the mice used in the study was measured. Following two rounds of treatments, there was no obvious weight loss (Figure 8a) seen in any of the groups suggesting that the formulations/nanoparticles that were used for treatment are reasonably safe. Mice tolerated the single as well as the combination treatments quite well. In addition, there was no elevation in liver enzyme levels observed during the study period (Figure 8d). Aspartate transaminase (AST) and alanine transaminase (ALT) levels were at the background levels (as same as the levels noted for PBS-treated mice) on day 14, 48 hours after the first round of siRNA/cisplatin treatment. Similar results were also found at the end of the study point (day 21). Lactate dehydrogenase levels were also unchanged at both timepoints indicating a lack of damage to the liver. Histopathology of liver and spleen tissues of mice from each group in this study on day 21 was found to be consistent with what is regarded as within normal limits (Figures 8b,c). Taken together, these results suggest that these treatments with HA/siRNA or HA/cisplatin nanoparticles were well tolerated by the mice with resistant tumors. Systemic administration of siRNA/cisplatin encapsulated HA nanoparticles thus provide safe and sequence specific inhibition of tumor growth in resistant tumor model.
Figure 8.
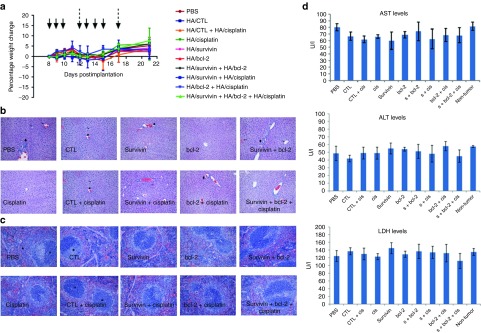
Monitoring toxicity in mice that had single and combination treatments as described in Figure 7. (a) Percentage weight change during the study period, the histopathology of (b) livers and (c) spleens of mice from each group at the end of the study were monitored. (d) Liver enzyme levels such as AST, ALT, and LDH levels were also monitored at the end of study. ALT, alanine transaminase; AST, aspartate transaminase; CTL, non-targeting control siRNA; LDH, lactate dehydrogenase.
Discussion
As discussed previously, the mechanisms of MDR is very complex and it is usually the synergistic result of a combination of several mechanisms. Overexpression of pump-mediated genes or antiapoptotic molecules are some of the examples discussed earlier.16,17 Downregulating the genes that are overexpressed and known to be responsible for resistance in those cells using siRNAs is potentially a powerful way of reversing the resistance.18 After downregulating the overexpressed genes, one would hope that the resistance will be reversed and the tumors will become sensitive to the anticancer drug treatment. Since it is possible that the inhibition of only one contribution to cellular resistance may not be sufficient for overcoming all mechanisms of cancer cell resistance to chemotherapy, combination of more than one mechanism or attacking more than one gene might be an effective strategy.17
Given the differential expression levels of pump and non-pump–mediated resistant genes in A549 and A549DDP pair, all were evaluated in the combination studies with cisplatin to understand the pump and non-pump–mediated resistance mechanism. Only, the combination of survivin and/bcl-2 with cisplatin demonstrated combination/synergistic effect but not the mrp1 and mdr1 siRNAs. As these mdr1 and mrp1 genes are known to contribute to pump-mediated resistance, it makes sense that they are not associated with the antiapoptotic pathway regulation. Downregulation of both survivin and bcl-2 together showed slightly better tumor cell-killing effect compared with the single-agent combinations suggesting a possible reversal of resistance with downregulation of more than one gene expression.
Before evaluating these combinations in tumor-bearing mice, multiple pilot studies were conducted to pick the optimum dose of cisplatin and siRNA to be used in the combination studies. Dosing schedules were carefully chosen based on those pilot studies to accommodate the maximum combination effect achievable. Since the siRNA-mediated knockdown was much higher at 72 hours than earlier timepoints, the cisplatin dose was given at 72 hours after last siRNA dose to attain maximum possible synergy. Also, the knockdown activity lasted for 5 days according to the previous studies. As such, the second set of siRNA treatments started 5 days after the last siRNA dose. Although the results suggest that there is therapeutic benefit in the combination group compared with the single-agent group, the resistance was not completely reversed by these treatments. The treatment schedules and frequency of treatments together with the data in this study suggest that a shorter treatment period to include both treatments may have a better outcome. As such, the second study was planned to incorporate this so that the siRNA-mediated knockdown coming from the first round of siRNA treatment will prevail until the effect coming from the second set of siRNA treatments commenced. Combining siRNA (either survivin or bcl-2) and cisplatin treatments clearly gave a significantly improved activity compared to single-agent treatment in the combination efficacy studies as noticed in the in vitro settings. Also, when both siRNAs are used in combination with cisplatin, the growth inhibition was the highest compared to single siRNA + cisplatin treatment. However, the results indicate that the resistance was still not completely reversed (tumor growth inhibition was increased from 30 to 60%). This may be due to the involvement of genes other than the ones that we tested. Exploring other gene involvement in drug resistance is thus critical. Despite the fact that, this combination strategy was tested in only one resistant cell line and there are other possible genes involved in the resistance mechanism other than the genes that we evaluated, our data suggest that sustained therapeutic benefits can be obtained by combining siRNA treatments along with cisplatin treatment using nanoparticle-based delivery systems. Given the clinical experience suggesting very poor efficacy or no efficacy after development of resistance, combination protocols predicted on these individual treatment modalities would be anticipated to provide superior clinical benefits with reduced toxicity burden to patients.
In addition to evaluating delivery and efficacy, it is also important to monitor the safety and tolerability of the nanoparticles that are being used to deliver both siRNA and cisplatin in the efficacy studies. To address that, the parameters such as change in body weight, plasma levels of the liver enzymes (ALT and AST), and lactate dehydrogenase were measured and compared between the treatment groups. Both ALT and AST are aminotransferases. AST is found in a variety of tissues including liver, heart, kidney, and brain. It is released into the serum when any of these tissues is damaged. It is therefore not a highly specific indicator of liver injury. Whereas the ALT is exclusively found in liver and it is released as a result of liver injury. Thus, it serves as a fairly specific indicator of liver status. To further characterize the efficacy and safety of this therapy, the histopathology of liver and spleen was also carried out and compared between the treatment groups. The results clearly suggested that the mice with resistant tumors well tolerated the current delivery system with both siRNA and cisplatin.
In contrast to other delivery systems available, the HA-based targeted systems we used here is efficacious at low siRNA and cisplatin doses and can be used to deliver multiple siRNA sequences and multiple small molecule drugs. These systems can thus serve as potential therapeutics for the treatment of multiple diseases.
Materials and methods
Tumor cell lines and tumor establishment. Human non-small lung cancer cell line A549 and SCLC cell line H69 were obtained from ATCC (Manassas, VA). The corresponding resistant cell lines (A549DDP and H69AR) were obtained from MGH (Boston, MA) and ATCC, respectively. Cells were grown in RPMI medium supplemented with 10% FBS. Animal procedures were performed according to a protocol approved by Northeastern University, Institutional Animal Care and Use Committee (NU-IACUC). Tumor models were developed in nude mice. Five to six weeks old nude mice were injected subcutaneously with tumor cells A549 (5 × 106 cells + matrigel), A549DDP (1 × 107 cells) under the right shoulder. Tumor volume was measured at least once or twice a week to monitor the tumor growth/suppression.
Baseline cisplatin resistance in lung cancer cells. To measure the cytotoxicity of cisplatin, A549/A549DDP cells were incubated with cisplatin at concentrations ranging from 1,000 up to 0.1 µmol/l for 2 days. The cellular cytotoxicity was assessed using MTS assay and expressed as % viable cells.
siRNA and chemotherapy combination. A range of cisplatin concentration at IC50 and below the IC50 was used together with the siRNAs in the first round of combination study. Out of the four sets of cells plated, two sets were transfected with survivin siRNA using lipofectamine at two different doses (1 and 10 nmol/l). Twenty-four hours after the transfection, cisplatin was added to one set of designated cells that had siRNA, at its IC50 concentration and few other concentrations below IC50. In parallel, the third set of cells was incubated with cisplatin alone at the same doses. All samples were kept at 37 °C for 2 days after the cisplatin addition. To the last set of cells, siRNA and the drug were simultaneously added and incubated for 48 hours as previously described. In the follow-up study, the other siRNAs (siRNAs for other targets such as bcl-2, mdr1, and mrp1) were also included in the combination evaluation. In these studies, the cisplatin concentration was kept at 100 µmol/l and the siRNA concentration was kept at 10 nmol/l. As described before, the siRNA treatment was given first and incubated for 24 hours. Following this, cisplatin was added to the cells that contained siRNA and incubated for another 48 hours.
Target knockdown with therapeutic siRNAs. As described previously,23 the self-assembled nanoparticles were made with HA-PEI, HA-PEG, and siRNAs (survivin, bcl-2, and control siRNA). The siRNA-loaded nanoparticles were characterized using the dynamic light scatting instrument by measuring the size and charge. For target knockdown studies, A549DDP tumor-bearing mice were treated with survivin siRNA or CTL siRNA encapsulated in HA nanoparticles at 0.5 mg/kg for 3 days. Tumors were harvested 24, 72, and 120 hours after the third dose. RNA was extracted from the tumors to analyze the mRNA knockdown by reverse transcription-quantitative PCR method. In another study, the above tested unmodified survivin siRNA sequence was injected into tumor-bearing mice along with the corresponding modified sequence. Knockdown was monitored 72 and 120 hours after the third dose. In a different study, 2 bcl-2 (um) and 2 bcl-2 (m) siRNA encapsulated HA nanoparticles were tested in the same tumor model to pick the best possible sequence for the combination efficacy study.
Cisplatin efficacy in A549-resistant tumors. To find the best HA-lipid derivative that can encapsulate and release cisplatin effectively, multiple HA derivatives (with lipids tail of chain length such as C4, C6, C8, C18, choline and PEI-modified versions) were initially tried and the HA conjugated to 1,8-diaminooctane (designated as HA-ODA) was selected for further studies; 10 mg/ml of HA-ODA solution was made in water. Likewise, 10 mg/ml cisplatin solution was also made in DMSO; 90 µl of the HA-ODA and 10 µl of the cisplatin were mixed well to form HA-ODA/cisplatin self-assembled nanoparticles. Along with this, HA-ODA solution was mixed with equal volume of HA-PEG (at 10 mg/ml) and then with cisplatin solution in the following volume ratio (0.9:0.9:0.2). All the nanoparticle formulations were kept at room temperature for 15–20 minutes for nanoparticle stabilization. Four groups of mice (n = 5) with A549DDP tumors received total of two doses of either free cisplatin or HA-ODA/cisplatin or HA-ODA/PEG/cisplatin at 1 mg/kg at 4 days apart. Tumor volumes were measured using the following formula to monitor the tumor growth inhibition: Tumor volume = (length × width/width)/2. The growth inhibitory effect was estimated using the T/C ratio.
Combination efficacy studies with survivin/bcl-2 knockdown and cisplatin treatment. In the first study, five groups of mice (n = 5) with A549DDP tumors received doses of PBS (control) or cisplatin or HA-PEI/PEG/survivin siRNA or HA-PEI/PEG/survivin siRNA + cisplatin at the doses described. siRNA doses were given at 0.5 mg/kg for 3 days. Seventy-two hours after the last siRNA dose, cisplatin dose was given at 1 mg/kg. The second round of treatments was initiated 72 hours after the cisplatin dose and repeated with the same pattern. Tumor volumes were monitored during the study period at least twice a week. In the next efficacy study, 10 groups of mice (n = 5) with A549DDP tumors received doses of different derivatives as described in the study design. In this study, both survivin and bcl-2 siRNAs in HA nanoparticles were used as single agent with and without cisplatin in HA nanoparticles and also used together to downregulate both genes at once with and without cisplatin. Unlike the first study, here the first cisplatin treatment was initiated 48 hours after the last siRNA dose and the second round of treatments were initiated 24 hours after cisplatin treatment. Tumor volume measurements were taken throughout the whole study to monitor the tumor growth/suppression. Tumors were collected 72 hours after last siRNA treatment and analyzed the survivin and bcl-2 levels as described before.
Measuring body weight changes, liver enzyme levels and histopathology. In the second efficacy study, in addition to the treatment groups (n = 5), three additional mice with tumors were used in each group to monitor acute toxicity/safety. These mice were given the same treatment as the mice in the efficacy study groups. Mice were weighed the day the treatments commenced and every day during the dosing period. Body weights were taken continuously throughout the whole study period.
To measure the liver enzyme levels, the blood was collected, 48 hours after the first round of treatment (three doses of HA/siRNA and one dose of cisplatin) from all 10 groups (n = 3/group). Also, at the end of the efficacy study, a terminal bleed was done to collect blood from all ten groups (n = 5) to look at the liver enzyme levels (both ALT and AST) and lactate dehydrogenase levels after two rounds of treatment using the manufacturer's instructions. Liver and spleen samples from mice were also collected for histopathological analysis at the end of the study (n = 5). The tissue samples analysis was performed at Tufts University Veterinary School (Grafton, MA).
SUPPLEMENTARY MATERIAL Figure S1. Selection of siRNA sequences using computational in silico methods.
Acknowledgments
The authors wish to gratefully acknowledge the funding support from the National Cancer Institute Alliance for Nanotechnology in Cancer, Center for Cancer Nanotechnology Excellence grant U54-CA151881, and the Cancer Nanotechnology Platform Partnership grant U01- CA151452. The authors declared no conflict of interest.
Supplementary Material
Selection of siRNA sequences using computational in silico methods.
References
- Fridman E, Skarda J, Pinthus JH, Ramon J, Mor Y. Expression of multidrug resistance-related protein (MRP-1), lung resistance-related protein (LRP) and topoisomerase-II (TOPO-II) in Wilms' tumor: immunohistochemical study using TMA methodology. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2008;152:47–51. doi: 10.5507/bp.2008.007. [DOI] [PubMed] [Google Scholar]
- Scagliotti GV, Novello S, Selvaggi G. Multidrug resistance in non-small-cell lung cancer. Ann Oncol. 1999;10 suppl. 5:S83–S86. doi: 10.1093/annonc/10.suppl_5.s83. [DOI] [PubMed] [Google Scholar]
- Takara K, Sakaeda T, Okumura K. An update on overcoming MDR1-mediated multidrug resistance in cancer chemotherapy. Curr Pharm Des. 2006;12:273–286. doi: 10.2174/138161206775201965. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Yagüe E, Higgins CF, Raguz S. Complete reversal of multidrug resistance by stable expression of small interfering RNAs targeting MDR1. Gene Ther. 2004;11:1170–1174. doi: 10.1038/sj.gt.3302269. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Minko T, Dharap SS, Pakunlu RI, Wang Y. Molecular targeting of drug delivery systems to cancer. Curr Drug Targets. 2004;5:389–406. doi: 10.2174/1389450043345443. [DOI] [PubMed] [Google Scholar]
- Debatin KM. Apoptosis pathways in cancer and cancer therapy. Cancer Immunol Immunother. 2004;53:153–159. doi: 10.1007/s00262-003-0474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Liu J, Long Y, Miao Y, Su M, Zhang Q, et al. Knockdown of survivin expression by siRNAs enhances chemosensitivity of prostate cancer cells and attenuates its tumorigenicity. Acta Biochim Biophys Sin (Shanghai) 2009;41:223–230. doi: 10.1093/abbs/gmp005. [DOI] [PubMed] [Google Scholar]
- Ocker M, Neureiter D, Lueders M, Zopf S, Ganslmayer M, Hahn EG, et al. Variants of bcl-2 specific siRNA for silencing antiapoptotic bcl-2 in pancreatic cancer. Gut. 2005;54:1298–1308. doi: 10.1136/gut.2004.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei XY, Zhong M, Feng LF, Zhu BY, Tang SS, Liao DF. siRNA-mediated Bcl-2 and Bcl-xl gene silencing sensitizes human hepatoblastoma cells to chemotherapeutic drugs. Clin Exp Pharmacol Physiol. 2007;34:450–456. doi: 10.1111/j.1440-1681.2007.04593.x. [DOI] [PubMed] [Google Scholar]
- Huang Z, Lei X, Zhong M, Zhu B, Tang S, Liao D. Bcl-2 small interfering RNA sensitizes cisplatin-resistant human lung adenocarcinoma A549/DDP cell to cisplatin and diallyl disulfide. Acta Biochim Biophys Sin (Shanghai) 2007;39:835–843. doi: 10.1111/j.1745-7270.2007.00356.x. [DOI] [PubMed] [Google Scholar]
- Trabulo S, Cardoso AM, Santos-Ferreira T, Cardoso AL, Simões S, Pedroso de Lima MC. Survivin silencing as a promising strategy to enhance the sensitivity of cancer cells to chemotherapeutic agents. Mol Pharm. 2011;8:1120–1131. doi: 10.1021/mp100426e. [DOI] [PubMed] [Google Scholar]
- Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R, et al. RNAi-based therapeutics targeting survivin and PLK1 for treatment of bladder cancer. Mol Ther. 2011;19:928–935. doi: 10.1038/mt.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Zelcer N, Prior JL, Kuil AJ, Piwnica-Worms D. In vivo RNA interference-mediated ablation of MDR1 P-glycoprotein. Clin Cancer Res. 2005;11:4487–4494. doi: 10.1158/1078-0432.CCR-05-0038. [DOI] [PubMed] [Google Scholar]
- Pakunlu RI, Cook TJ, Minko T. Simultaneous modulation of multidrug resistance and antiapoptotic cellular defense by MDR1 and BCL-2 targeted antisense oligonucleotides enhances the anticancer efficacy of doxorubicin. Pharm Res. 2003;20:351–359. doi: 10.1023/a:1022687617318. [DOI] [PubMed] [Google Scholar]
- Pakunlu RI, Wang Y, Tsao W, Pozharov V, Cook TJ, Minko T. Enhancement of the efficacy of chemotherapy for lung cancer by simultaneous suppression of multidrug resistance and antiapoptotic cellular defense: novel multicomponent delivery system. Cancer Res. 2004;64:6214–6224. doi: 10.1158/0008-5472.CAN-04-0001. [DOI] [PubMed] [Google Scholar]
- Saad M, Garbuzenko OB, Minko T. Co-delivery of siRNA and an anticancer drug for treatment of multidrug-resistant cancer. Nanomedicine (Lond) 2008;3:761–776. doi: 10.2217/17435889.3.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim JH, Park H, Kim YS, Park K, Nam H, et al. Tumor-homing multifunctional nanoparticles for cancer theragnosis: Simultaneous diagnosis, drug delivery, and therapeutic monitoring. J Control Release. 2010;146:219–227. doi: 10.1016/j.jconrel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- Ganesh S, Iyer AK, Morrissey DV, Amiji MM. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials. 2013;34:3489–3502. doi: 10.1016/j.biomaterials.2013.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selection of siRNA sequences using computational in silico methods.