Abstract
Genetically engineered mouse models (GEMMs) of human cancer were first created nearly 30 years ago. These early transgenic models demonstrated that mouse cells could be transformed in vivo by expression of an oncogene. A new field emerged, dedicated to generating and using mouse models of human cancer to address a wide variety of questions in cancer biology. The aim of this review is to highlight the contributions of mouse models to the diagnosis and treatment of human cancers. Because of the breadth of the topic, we have selected representative examples of how GEMMs are clinically relevant rather than provided an exhaustive list of experiments. Today, as detailed here, sophisticated mouse models are being created to study many aspects of cancer biology, including but not limited to mechanisms of sensitivity and resistance to drug treatment, oncogene cooperation, early detection, and metastasis. Alternatives to GEMMs, such as chemically induced or spontaneous tumor models, are not discussed in this review.
GENETICALLY ENGINEERED MOUSE MODELS OF HUMAN CANCER: AN OVERVIEW
To most practicing oncologists, genetically engineered mouse models (GEMMs) may represent an interesting field of cancer biology with little immediate clinical relevance. However, animal experiments and clinical observations in the late 1990s led to a convergence of the mouse modeling and oncology communities. Around the same time that the tyrosine kinase inhibitor (TKI) imatinib was starting to show effectiveness in patients with BCR-ABL–driven chronic myelogenous leukemia (CML), several research groups showed in inducible transgenic mouse models that sustained expression of a tumor-inducing oncogene (eg, MYC in hematopoietic cells, HrasG12V in the skin) was required for tumor survival.1–3 By using mice in which an oncogene could be turned on and turned off in specific tissues at specific times (by addition or removal of an inducer in the animal diet), investigators showed that, once tumors formed, turning off expression (or de-induction) of the oncogene led to rapid tumor regression. Mechanisms underlying tumor regression included apoptosis, cell cycle arrest, and differentiation of tumor and/or vasculature cells. These observations revealed that cells that previously were viable in the absence of the oncogene could somehow become dependent on its expression for survival. These first in vivo experimental demonstrations of oncogene addiction implied that therapy directed against a specific target driving tumor growth could be effective at treating the disease.4 The results directly paralleled the early clinical trial results with imatinib, in which the drug turned off ABL in patients with CML, leading to dramatic responses to therapy.5 Importantly, in mouse models with oncogene-induced tumors, the loss of tumor suppressor genes did not prevent tumor regression when expression of the driving oncogene was turned off. This observation was initially made in HrasG12V-induced melanomas arising in the absence of Ink4a/Arf.1 Analogous results were obtained in KrasG12D-induced lung adenocarcinomas and Wnt1-induced mammary tumors arising on Trp53 (mouse p53)- or Ink4A/Arf-deficient backgrounds.6–8 Collectively, these data implied that targeted therapy to inhibit a driving oncogene should work even in genetically complex human tumors.1,6 Today, multiple other human examples of oncogene addiction exist, including mutant epidermal growth factor receptor (EGFR)–driven lung adenocarcinomas, KIT-driven gastrointestinal stromal tumors, and BRAF-driven melanomas, targeted by EGFR-, KIT-, and BRAF-specific kinase inhibitors, respectively.9–13
As opposed to inactivating an oncogene, recent work has explored the effect of restoring the function of a deleted tumor suppressor gene.14–16 For example, lymphomas and sarcomas arising as a result of Trp53 loss alone or in combination with MYC overexpression regressed when Trp53 was reinstated, confirming that tumor cells also become dependent on the changes occurring within a cell when a tumor suppressor gene is lost. Thus, restoration of tumor suppressor genes in human tumors appears to be a promising therapeutic strategy.
MAKING MORE CLINICALLY RELEVANT MOUSE TUMOR MODELS USING NEW GENETIC TOOLS
The GEMMs that revealed oncogene dependence were generated by using sophisticated mouse modeling techniques developed throughout the years. Ideal GEMMs of human cancer feature tumors that are initiated in a small subset of cells within an organ by a genetic lesion found in the human counterpart and for which steps in tumor progression (eg, angiogenesis, acquisition of secondary mutations, ability to metastasize) also resemble the human disease. Moreover, to additionally mimic most human sporadic cancers, the genetic lesion being studied should be induced in adult tissues rather than during embryonic development. As a result of extensive refinement of techniques for generating mouse models, GEMMs available today have many of these features.
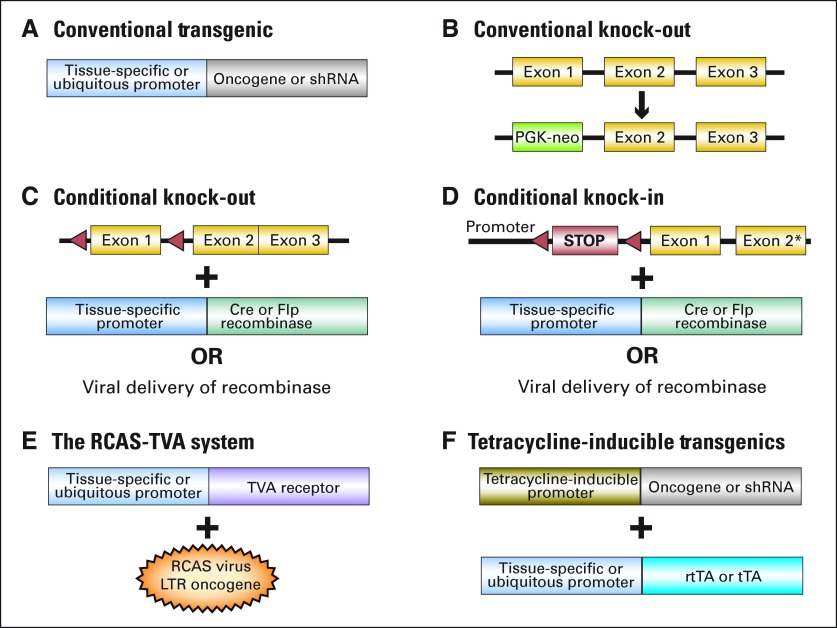
The first mouse models of cancer were created with the advent of technologies to generate transgenic mice by pronuclear injection of DNA.17 These transgenic constructs carried cDNAs encoding an oncogene downstream of a ubiquitous or tissue-specific promoter (Fig 1A), allowing for overexpression of a specific oncogene. Initially, these efforts were aimed at understanding which genes (eg, SV40 early region, MYC) could give rise to tumors and in which tissues.18,19 Subsequently, gene targeting in embryonic stem cells provided the means to knock out genes (eg, Trp53, Rb, Nf1) and test directly the effects of tumor suppressor gene loss in mice (Fig 1B).20–22 Early models knocked out the genes in all tissues. Today, GEMMs allow for inducible, tissue-specific expression of oncogenes as well as conditional, tissue-specific deletion of tumor suppressors. Strategies to generate various models are described in detail in Figure 1.
Fig 1.
Schematic representation of DNA constructs used to generate genetically engineered mouse models (GEMMs) of human cancer. (A) In conventional transgenic animals, transcription of a cDNA encoding an oncogene or a sequence encoding a short hairpin RNA (shRNA; to knockdown expression of a target gene) is driven by a tissue-specific or ubiquitous promoter. (B) In this example of conventional gene targeting, the gene is disrupted by replacing its first exon with an antibiotic selection gene (eg, a phosphoglycerate kinase-neomycin [PGK-neo] cassette). (C,D) To generate conditional knock-outs and knock-ins, recombinases (bacteriophage P1-derived Cre recombinase and Saccharomyces Cerevisiae–derived Flp recombinase) are used to eliminate or activate expression of a functional gene at a specific time. For example (C), an exon of a target gene is cloned between specific short direct repeats (loxP sites for Cre, and Frt sites for Flp). The target gene is functional until it is exposed to the recombinase, at which time the flanked DNA is excised by the recombinase. Expression of the recombinase can be directed to a defined tissue by generating transgenic mice that express the enzyme under the control of a tissue-specific promoter or by using viruses to deliver the enzyme to a tissue of interest. (D) The Cre/loxP and Flp/FRT systems can also be used to conditionally express a mutant in a tissue of interest from its endogenous promoter or an oncogene/shRNA from a housekeeping promoter.83,84 The main advantage of using recombinase-based systems is that they allow for tissue-specific expression of the sequence of interest. A potential disadvantage is that, once recombination has occurred, the excision is irreversible. The recombinase can also be delivered using a viral vector (eg, Adenovirus-cre), allowing the event to occur only in a small subset of cells within an otherwise normal tissue, which recapitulates the scenario that most frequently occurs in human tumors.82 (E) An avian retrovirus–based method to deliver oncogenes somatically to subsets of cells within a tissue of interest has been used in mice.85 Here, a transgenic mouse is generated that carries the receptor for the avian leukosis virus (ALV) under the control of a tissue-specific promoter. The mouse is then infected with an ALV-pseudotyped retrovirus (RCAS) carrying an oncogene or, as shown recently, a microRNA.86 Mouse cells do not normally express the viral receptor; thus, normal cells are not affected. In addition to ensuring that a subset of cells in the defined organ is infected, this method allows oncogenes to be conditionally expressed in different tissues. It is particularly suited to highly proliferative tissues, because the replication-competent RCAS retrovirus only infects cycling cells. (F) One of the most widely used methods to inducibly express oncogenes or shRNAs is to express them under the control of a tetracycline-inducible promoter. Activation of the tetracycline-inducible promoter in the Tet-ON system occurs when the animal is exposed to tetracycline (or the tetracycline analog doxycycline) and expresses the reverse tetracycline transactivator (rtTA) in the tissue(s) of interest. Withdrawal of tetracycline causes expression of the transgene to shut off. On the contrary, in the Tet-OFF system, the tetracycline transactivator (tTA) induces expression from the tetracycline regulated promoter in the absence of tetracycline. Expression is then repressed on addition of the drug. The Tet systems have been widely used in tumor maintenance studies to determine whether tumors have become dependent on continuous expression of an oncogene or loss of a tumor suppressor gene. An alternative strategy to inducibly regulate the function of a protein encoded by a transgene in vivo is to modify the protein to contain an estrogen-responsive moiety so that its activity can be induced by addition of the estrogen analog, tamoxifen.87 tTa, tetracycline transactivator.
These techniques have allowed nearly every cancer type to be modeled in the mouse, including common adult cancers like breast, prostate, and lung cancers and rarer tumors like pancreatic cancer. Moreover, childhood tumors like medulloblastomas and rhabdomyosarcomas have also been modeled in GEMMs. For detailed descriptions of several of these models, the reader is referred to recent comprehensive reviews.23–28
GEMMs Versus Xenografts
Historically, xenografts have been the most extensively used mouse models in preclinical drug testing. In xenografts, human cancer–derived cells or tumor fragments are implanted into immunodeficient mice subcutaneously or orthotopically (ie, into the organ of interest) to propagate tumors that are monitored for response to drug treatment. Xenografts are useful for these studies, because many different drugs and dosing schedules can be tested against the same set of cells (in different mice). One advantage of tumor xenografts is that the starting material is usually derived from advanced cancers or metastases; the cells presumably represent real human tumors replete with genetic complexity. However, tumor-derived cell lines may not completely recapitulate intratumor heterogeneity, because the cells that grow may represent only a subpopulation of tumor cells. Another disadvantage of xenografts is that tumor growth occurs in a host with an impaired immune system and usually (in subcutaneous models) in an artificial site where the original human tumor did not develop. GEMMs circumvent these issues, because tumors arise in situ where immune function, angiogenesis, and inflammatory processes can all interact normally with the developing tumor. Thus, GEMMs allow for the analysis of tumors as they develop through defined stages of tumorigenesis, facilitating studies of the biology of the tumors early and late in the process.
DEVELOPING NEW THERAPEUTIC STRATEGIES TO TREAT CANCERS AND OVERCOME DRUG RESISTANCE
Testing New Targeted Therapies
As more is learned about the molecular alterations present in individual tumors, the concept of oncogene addiction and the success of kinase inhibitors have prompted the development of a plethora of targeted agents. Consequently, the testing of agents in accurate preclinical models reflective of human tumors has gained importance. As an example, we describe how multiple new GEMMs of lung cancer are providing valuable insight into treatment of the disease.
Lung adenocarcinomas frequently harbor mutations in genes encoding ERBB family members and in components of their downstream signaling pathways, such as EGFR, ERBB2, KRAS, BRAF, PIK3CA, and LKB1.29 Mutations are often mutually exclusive (eg, EGFR and KRAS mutations are almost never found in the same tumor.) and arise in different subgroups of patients with lung cancer (eg, EGFR mutations are mostly found in lung tumors from patients who have a limited smoking history in contrast to KRAS mutations that are more frequently smoking-associated). Many groups have generated GEMMs on the basis of mutations in these different genes. In most cases, the mice develop lung adenocarcinomas, validating the importance of the initiating oncogene in the genesis of this type of tumor. Although the mice develop histopathologically similar tumors, preclinical studies have shown that treatment strategies need to be tailored to the specific mutation driving the tumor (Table 1).30–47 Mice engineered to express the tyrosine kinase inhibitor (TKI) –sensitive EGFR mutants (ie, an exon 19 deletion and an L858R mutant) develop lung adenocarcinomas and respond to reversible and irreversible EGFR TKIs.30,32,34 In contrast, as observed in humans, tumors carrying mutant Kras or the TKI-resistant EGFRT790M mutant do not respond to treatment with these drugs.30,31,35 These mouse models of primary and acquired resistance to TKIs are now being used to test new drugs and drug combinations, because effective therapies for these tumors remain elusive.
Table 1.
Summary of Preclinical Trials in GEMMs of Lung Adenocarcinoma
| Drug Action | Drug | Pathway |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EGFRL858R | EGFRDEL | EGFRL+T | EGFRD+T | HER2YVMA | KrasG12D | PIK3CAH1047R | BRAFV600E | EML4-ALK | ||
| TKI | Erlotinib30–33,36 | R | R | PD | PD | PD | PD | ND | ND | ND |
| EGFR antibody | Cetuximab32,34 | R | ND | SD/TR | ND | ND | ND | ND | ND | ND |
| Irreversible TKI | BIBW-299234–36 | R | ND | SD/TR | ND | SD/TR | ND | ND | ND | ND |
| Irreversible TKI (BIBW2992), mTOR inhibitor (rapamycin) | BIBW-2992 +rapamycin36 | ND | ND | ND | ND | R | ND | ND | ND | ND |
| Irreversible TKI (BIBW2992), EGFR antibody (cetuximab) | BIBW-2992 +cetuximab34 | ND | ND | R | ND | ND | ND | ND | ND | ND |
| Irreversible TKI | HKI-27232,33 | R | ND | SD/TR | ND | ND | ND | ND | ND | ND |
| Irreversible TKI (BIBW2992), mTOR inhibitor (rapamycin) | HKI-272 +rapamycin33 | ND | ND | R | ND | ND | ND | ND | ND | ND |
| EGFRT790M inhibitor | WZ-400237 | ND | ND | R | R | ND | ND | ND | ND | ND |
| PI3K/mTOR inhibitor | NVP-BEZ23538–40 | ND | ND | PD | ND | R: GDC-941 | PD | R | ND | ND |
| MEK1/2 inhibitor | AZD-6244*38,39,45,46 | ND | ND | PD | ND | ND | SD/TR | ND | R: CI-1040/PD0325901 | ND |
| PI3K/mTOR inhibitor (NVP-BEZ235)MEK1/2 inhibitor (AZD-6244) | NVP-BEZ235 +AZD-624438,39,47 | ND | ND | R | ND | ND | R | ND | ND | PD47 |
| Multiple RTK inhibitor | Sunitinib41 | ND | ND | ND | ND | ND | R | ND | ND | ND |
| HSP90 inhibitor | 17-AAG31,44,47 | SD/TR | ND | SD/TR | ND | ND | ND | ND | ND | TR: DMAG |
| ALK inhibitor | 2,4-pyrimidinediamine derivative43,47 | ND | ND | ND | ND | ND | ND | ND | ND | R: TAE684 |
| ERBB3 antibody (MM-121)EGFR antibody (cetuximab) | MM-121 + cetuximab42 | ND | ND | R | ND | ND | ND | ND | ND | ND |
Abbreviations: GEMMs, genetically engineered mouse models; TKI, tyrosine kinase inhibitor; R, partial or complete response; PD, progressive disease; ND, not determined; EGFR, epidermal growth factor receptor; SD/TR, stable disease or very transient response; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol-3 kinase; RTK, receptor tyrosine kinase; HSP90, heat-shock protein 90; ALK, anaplastic lymphoma receptor tyrosine kinase; ERBB, erythroblastic leukemia viral gene homolog.
Mice described in study by Dankort et al46 mostly developed lung adenomas, rarely adenocarcinomas.
In mice bearing tumors expressing the EGFRT790M mutant, at least four novel treatment strategies have already been identified. In one study, the authors systematically tested the standard chemotherapeutic agents paclitaxel and pemetrexed, the irreversible TKI BIBW-2992 and the anti-EGFR antibody cetuximab in EGFRL858R+T790M mice with lung tumors detectable using magnetic resonance imaging.34 None of these agents reproducibly elicited responses when administered alone. However, treatment of mice with BIBW-2992 and cetuximab in combination induced dramatic tumor regression. Analysis of tumor lysates from mice treated with BIBW-2992 plus cetuximab revealed depletion of both total and phosphorylated EGFR in contrast to lysates from tumors derived from mice treated with either agent alone, in which a modest decrease in either phosphorylated EGFR (with BIBW-2992 treatment) or total EGFR (with cetuximab treatment) were observed. The combination of cetuximab with MM-121, an antibody that prevents ligand-dependent activation of ERBB3 by its heterodimerization partners EGFR and ERBB2, also led to tumor regression in mice with T790M-harboring tumors.42 A third strategy involves combining drugs that target pathways downstream of mutant EGFR like the phosphatidylinositol-3 kinase (PI3K) pathway and the RAS pathway. To do this a combination of a PI3K/mammalian target of rapamycin (mTOR) inhibitor and a MEK (MEK is downstream of RAS) inhibitor were used and were shown to be effective, although neither drug alone worked.39 Finally, a novel irreversible kinase inhibitor identified by virtue of its ability to selectively inhibit the growth of cells expressing EGFRT790M mutants is effective in mice with EGFRL858R+T790M– or EGFRDEL+T790M–induced tumors.37 Multiple clinical trials are being planned on the basis of these studies.
Promising strategies to treat KRAS mutant tumors have also been identified from preclinical studies using GEMMs. For example, the combination of a PI3K/mTOR inhibitor and an MEK inhibitor was effective against KrasG12D-induced tumors (approximately six-fold tumor shrinkage), whereas the MEK inhibitor alone led to only a two-fold reduction in tumor volume and the PI3K/mTOR inhibitor did not work at all.38 These preclinical data would predict that the ongoing phase II clinical trials of AZD6244 (MEK1/2 inhibitor) or ridaforolimus (mTOR inhibitor) in patients with KRAS-mutant lung adenocarcinomas may not show a significant effect. Trials designed to test the safety and efficacy of combining MEK and PI3K inhibitors are being planned.
Although it is difficult to predict how successful these strategies will be for treating human disease, they are based on targeting pathways on which many tumor cells are exquisitely dependent for their survival. Stratification of patients to be included in trials on the basis of accurate assessment of biomarkers that predict response to the drugs will be essential for positive outcomes to be observed.
MOUSE MODELS OF DRUG RESISTANCE AND TUMOR RELAPSE
A major clinical problem is the development of acquired drug resistance. Elucidation of mechanisms of acquired resistance could lead to new treatment strategies. Ideally, mechanisms of drug resistance could be studied on human tumor samples that have acquired resistance to a particular drug. Although this approach has been successful for understanding acquired resistance to EGFR TKIs in the case of lung cancer48–50 and to imatinib in CML51 and gastrointestinal stromal tumors,52 obtaining adequate numbers of samples and sufficient material for thorough molecular studies is challenging. Tumor cell lines offer an attractive alternative approach to study drug resistance, because unlimited material is available. However, the findings in cell lines do not always parallel the in vivo situation; hence, the clinical relevance of results obtained using cell lines is not always clear.53
For these reasons, GEMMs are emerging as important vehicles to study tumor relapse and drug resistance. Mouse tumors with defined genetic mutations are monitored serially for tumor regression and recurrence by using small animal imaging techniques (magnetic resonance imaging, computed tomography, positron emission tomography [PET], ultrasound). For example, cyclical long-term treatment of mice with lung adenocarcinomas harboring drug-sensitive EGFR mutants eventually gives rise to erlotinib-resistant tumors with a spectrum of secondary alterations almost identical to that observed in human tumors (ie, EGFR T790M and Met amplification), indicating that this model can be used in discovery efforts to identify novel mechanisms of resistance.54 Similarly, mice with KrasG12D-induced lung adenocarcinomas treated with cisplatin develop drug resistance.55 Compared with chemotherapy naive tumors, resistant tumors display an unstable genome, confirming that mechanisms hypothesized to account for cisplatin-resistance in human cancer actually occur in vivo.
Mouse models of acquired resistance to targeted therapies have also uncovered novel mechanisms of drug resistance.56 For example, mice with acute myelogenous leukemia (AML) arising as a result of a deficiency in the tumor suppressor gene Nf1 (Neurofibromin 1, a negative regulator of Ras signaling), and cooperating mutations introduced by retroviral insertional mutagenesis derive survival benefit from treatment with the MEK inhibitor, CI-1040. Eventually, however, all of the mice ultimately succumb to the disease because of outgrowth of AML clones with insertions in genes that modulate Ras pathway activation (Rasgrp1, Rasgrp4, and Mapk14). Whether these mechanisms occur in human tumors treated with MEK inhibitors remains to be determined, but they provide rational clues for investigation.
Inducible mouse models can also be used to study what leads to tumor relapse after an initiating oncogene is turned off. This situation could mimic resistance to targeted therapy, even before targeted therapeutics have been developed. For example, when tumors are induced in the mammary gland by expression of MYC, oncogene de-induction leads to tumor regression in 50% of cases. Tumors that do not regress harbor Kras mutations and high levels of Ras pathway activation.57,58 In contrast, mammary tumors induced by Wnt 1 (an oncogene upstream of MYC) frequently harbor Hras mutations that are not associated with high levels of Ras pathway activation, and the tumors regress even in the presence of the secondary mutations.58 These experiments demonstrate how some but not all activating mutations can render a tumor unresponsive to a specific, targeted therapy. Additional studies have confirmed the preferential cooperation between MYC and oncogenic Kras in the mammary gland by inducing expression of both oncogenes simultaneously. Accelerated tumor development was observed, and de-induction of both oncogenes led to more sustained regression than either oncogene alone, suggesting that combination therapy is beneficial in the appropriate context.59 Nevertheless, disease relapsed even when both oncogenes were turned off. These data highlight how resistance to therapy can still occur even when both members of a cooperating pair of oncogenes are targeted.
IMPROVING THE EFFECTIVENESS OF STANDARD CHEMOTHERAPY
For most advanced cancers, standard-of-care treatment regimens rely heavily on cytotoxic chemotherapy. Although these drugs can be effective at slowing down progression of the disease, they rarely lead to cures. Recently, GEMMs have been used to develop potentially more effective methods of administering chemotherapies to target tumor cells and the tumor microenviroment. For example, in a mouse model of neuroendocrine cancer (insulinoma) that reliably progresses from early- (hyperplastic islets) to late-stage (islet cell carcinomas) disease, the addition of a platelet-derived growth factor receptor (PDGFR) inhibitor [imatinib] known to disrupt cells supportive of the tumor microenvironment (ie, pericytes and endothelial cells) enhanced the effectiveness of cyclophosphamide (CTX) treatment. Additional tumor regression and an increase in survival was observed when the maximum-tolerated dose (MTD) regimen was switched to maintenance therapy with a metronomic regimen (continuous low-dose CTX) plus imatinib or the vascular endothelial growth factor receptor (VEGFR) and PDGFR inhibitor sunitinib (to disrupt endothelial cells).60 On the basis of these preclinical studies in the mouse, a phase II study in renal cell carcinoma has been initiated that uses gemcitabine given at its MTD followed by metronomic capecitabine plus the kinase inhibitor sorafenib (that inhibits both VEGFR and PDGFR), and this regimen has shown promising results.61 Similar experiments performed in the same mouse model have shown that inhibition of cathepsin proteases (proteolytic enzymes that are often found overexpressed at the invasive edges of tumors) in combination with the chemotherapy-switch CTX regimen led to a reduced tumor burden and increased survival.62
Another GEMM was recently used to show that the efficacy of gemcitabine in human pancreatic adenocarcinoma is probably limited by its inability to readily perfuse in situ tumors as a result of poor vascularization and dense stromal matrix.63 Interestingly, tumor stroma could be disrupted by inhibition of the Hedgehog pathway, which leads to an increase in angiogenesis and to higher levels of gemcitabine in the tumor. Phase II clinical trials are in progress to test the Hedgehog inhibitor GDC-0449 in combination with chemotherapy in pancreatic cancer. However, resistance is likely to occur, because mice treated with both drugs only showed a transient response to therapy.
USING GEMMs TO IDENTIFY BIOMARKERS AND IMAGING STRATEGIES FOR EARLY DETECTION AND RESPONSE PREDICTION
For most cancers, outcomes are significantly better when tumors are diagnosed at early stages. However, many epithelial cancers are more frequently diagnosed at advanced stages of the disease because of a lack of useful biomarkers for the early detection of these tumors. New approaches to find blood biomarkers for cancer or to develop imaging techniques to screen high-risk individuals and the general population for these cancers are necessary. One such approach has been to use GEMMs, which allow for collection of blood from mice with genetically defined tumors at specific stages of tumorigenesis to identify plasma biomarkers of disease.
In a mouse model of pancreatic cancer driven by mutant Kras and loss of Ink4A/Arf (both of which are found in most human pancreatic adenocarcinomas), analysis of the plasma proteome revealed an increase in 165 proteins (> 1.5-fold increase; P < .05) in the plasma of tumor-bearing mice relative to controls.64 This list of proteins was additionally narrowed to exclude those that are likely to be unrelated to cancer (ie, complement, coagulation, and acute phase reactant proteins) and to focus on those that had a human ortholog and showed increased levels of gene expression in both murine and/or human pancreatic cancer. After validation in mouse plasma and tissue samples, a panel of proteins, including ALCAM, ICAM1, LCN2, TIMP1, REG1A, REG3 and IGFBP4, could distinguish sera among individuals with pancreatic cancer, individuals with chronic pancreatitis, and healthy controls. Moreover, a smaller panel consisting of LCN2, TIMP1, REG1A, REG3 and IGFBP4 was able to predict from precancer sera samples which individuals from a clinical study (ie, the Beta-Carotene and Retinol Efficacy Trial [CARET] trial) would develop pancreatic cancer. Similar studies in mouse models of ovarian and colon cancer have led to the identification of promising markers for those diseases.65,66
The need for accurate and reliable tests to quickly identify patients who respond and who experience relapse to specific treatments is paramount. Studies in GEMMs for which defined genetic mutations are engineered to cause cancer are likely to be useful for the identification of plasma protein changes that occur on treatment with specific drugs. If these proteins are shown to reliably change in abundance on oncogene induction and then again on treatment, they can be considered likely candidates that will distinguish patients who are responding to treatment with the drug from those who are not. Presumably, on continuous drug treatment, if drug-resistant tumors were to emerge, this same panel of proteins would begin to show changes, returning to the levels observed when a tumor was present. Monitoring of plasma protein levels in patients who are undergoing treatment would be an easy and relatively inexpensive way to assess the effectiveness of therapy.
GEMMs also have the potential to be important assets for the development of molecular imaging techniques to monitor disease progression, response to therapy, and eventual disease relapse. Such techniques could also pinpoint the biologic characteristics of individual tumors (eg, proliferation rate, signaling pathway changes). To date, the use of GEMMs for the development of molecular imaging has been limited, perhaps because of the need for both specialized imaging equipment and mice carrying appropriate mutations. In recent years, however, the rapid growth in the number of clinically relevant GEMMs and the acquisition by many institutions of small animal imaging modalities make this an auspicious moment for the preclinical development of imaging modalities. As an example, in a GEMM of high-grade glioma, the thymidine analog 3′-deoxy-3′-18F-fluorothymidine (18F-FLT) was used for PET imaging to quantify tumor cell proliferation.67 This imaging technique can now be used to assess the effectiveness of different therapies in this particular model and may be developed clinically to monitor how brain tumors respond to specific drugs.
METASTATIC DISEASE IN GEMMS
One of the most frequent criticisms of the use of GEMMs in cancer research is that they rarely recapitulate the metastatic phenotype seen within human tumors. Several explanations may account for this deficiency. First, human cancers are genetically more complex than tumors arising in GEMMs, which generally carry alterations only in one or two genes. Second, GEMMs are often engineered to develop tumors quickly (for experimental reasons). Tumor burdens, therefore, may increase so rapidly that animals have to be euthanized before metastatic tumors even have the possibility to emerge. Third, intrinsic differences between mice and humans may also contribute to the lack of metastases observed in GEMMs. Recognition of these shortcomings has sparked efforts to more effectively model tumor progression in GEMMs, especially by combining specific mutations observed in human advanced cancers. In particular, combination of mutations in oncogenes and tumor suppressor genes—a situation often observed in human tumors—has proven to be an effective strategy to observe metastases in GEMMs.
For example, in mouse lung adenocarcinomas arising as a result of KrasG12D expression, layering on an additional mutation, such as deletion of Trp53, leads to the development of lymph node metastases in 50% of the mice and to rare distant metastases.68 When point mutations at the TRP53 mutation hotspots R273 (R270 in the mouse) and R175 (R172 in the mouse) are introduced on one of the Trp53 alleles, an increase in metastatic disease is not observed. By using a slightly different strategy, however, another group used mice that spontaneously activate KrasG12D in the lung epithelium and crossed them with mice that carry the Trp53 R172H allele to generate compound KrasLA1/+; p53R172H/+ heterozygote mice. Strikingly, 36.5% of these mice with lung adenocarcinomas also develop metastatic disease, mainly to intrathoracic sites. Metastases were also observed in extrathoracic sites, such as the liver, adrenal gland, kidney, and body wall. Neither brain nor bone was affected by metastases, possibly indicating that tumors with this combination of genetic events are not effective at colonizing these tissues. In another example, deletion of the tumor suppressor Lkb1 (a serine/threonine kinase that regulates adenine monophosphate-activated protein kinase [AMPK]) in mice expressing oncogenic Kras in the lung accelerated tumor onset, and metastases were observed in two thirds of the mice examined.69 Finally, metastatic disease was observed in a mouse model of melanoma only when expression of the BrafV600E mutant was combined with deletion of the tumor suppressor Pten.70 Invasion of metastatic cells into the subcutis and lesions in the lymph nodes and lungs were found in almost all of the mice.
Significant advances in our understanding of the mechanisms that underlie metastatic spread have occurred thanks to the use of GEMMs. Namely, the notion that metastasis is a late step in tumor progression was challenged by two elegant studies that used breast cancer mouse models.71,72 In one study, the authors showed that dissemination of cancer cells occurs early in the tumorigenic process by transplanting premalignant mammary glands from MMTV-NeuT (ie, rat activated Her2/Neu under the control of the mouse mammary tumor virus promoter) transgenic mice into wild-type littermates. Immunostaining of bone marrow for human epidermal growth factor 2 (HER2) or cytokeratin revealed evidence of host-derived cells as early as the formation of atypical ductal hyperplasia in the mammary gland and before the development of invasive cancer, with no increase in the numbers of disseminated tumor cells (DTCs) with tumor progression. Evidence for early dissemination of tumor cells into both the lung and bone marrow was also found in MMTV-PyMT (ie, polyomavirus middle T antigen) transgenic mice. Importantly, with regard to human cancer, DTCs were observed in the bone marrow of patients who had breast cancer with ductal carcinoma in situ (and the DTCs were not present in controls). The numbers of DTCs did not change across different stages, which supports the murine studies. In the second study, untransformed mammary cells either from wild-type mice or from mice carrying inducible transgenes were able to colonize an ectopic site (in this case, the lung) and give rise to tumors at the site upon oncogene activation. Together, these studies demonstrate that two important steps in metastatic spread, cell dissemination and colonization, do not occur exclusively in late stages of tumorigenesis and that the functional requirements for these processes may be present in premalignant cells.
The select examples highlighted here underscore the feasibility of developing GEMMs of metastatic disease that are useful for understanding the molecular and cell biology basis of metastasis formation. Future efforts to expand the spectrum of GEMMs that develop metastases are necessary by the mouse modeling community to tackle this complex problem.
THE NEXT FIVE YEARS
Integrating Data From Human Oncogenomic Studies and Experiments in GEMMs
Many large integrative genomic projects have been undertaken with the goal of identifying mutations, expression changes, and copy number alterations present in human cancers.29,73-79 Understanding the relevance of the individual gene variations observed in these valuable data sets requires additional validation in model systems to distinguish driver (responsible for the growth of the tumor) and passenger (bystander) mutations and to determine whether specific genetic changes found together define molecular subsets of the disease that determine prognosis and treatment response. GEMMs will be valuable to test some of the hypotheses formulated through analysis of the genomic data. Information gleaned from such in vivo studies can be taken back directly to the clinic to impact cancer treatment. The additional data from large integrative studies will provide more information about human cancer and will enable the development of even more sophisticated mouse models of cancer.
Expanding Preclinical Studies in GEMMs
As described earlier in this review, xenograft models have historically been used for drug testing, but results from GEMMs have become increasingly insightful. Consequently, centralized core resources at academic and government research institutions are emerging to facilitate the use of mouse models for preclinical studies (eg, http://ccr.nci.nih.gov/research/Capr.aspx), which are labor intensive, are expensive, and require a variety of expertise from mouse imaging to the ability to do pharmacokinetic and pharmacodynamic studies. Such cores are likely to play an increasingly important role as the use of GEMMs in preclinical studies of lead compounds becomes more widely adopted.
Technical Advances to Improve the Cost Effectiveness of GEMMs
Increasing knowledge of the multitude of genetic changes present in human tumors means that, to effectively understand the impact of these changes on tumorigenesis and on the response to specific drugs, we need to be able to mimic in the mouse, rapidly and cost effectively, the compendium of mutations deemed to be important. This represents a challenge, because the generation of knock-out, knock-in, and transgenic alleles is time consuming, as is breeding the different alleles. Several laboratories have devised strategies to overcome some of these problems. For example, multiple transgenic constructs can be introduced sequentially into embryonic stem (ES) cells (potentially already null for a specific tumor suppressor gene). The ES cells are then used to generate chimeric mice, which can be directly used as experimental animals.80 A major advantage of this system is that modified ES cells carrying multiple combinations of genes can be frozen and stored until needed, greatly reducing the costs that would be associated with maintaining these complex strains. Somatic delivery of viral vectors bearing one or more cDNAs or shRNAs of interest directly to organs is also likely to be an effective and widely used strategy to test the role of multiple genes in tumorigenesis in vivo.81,82
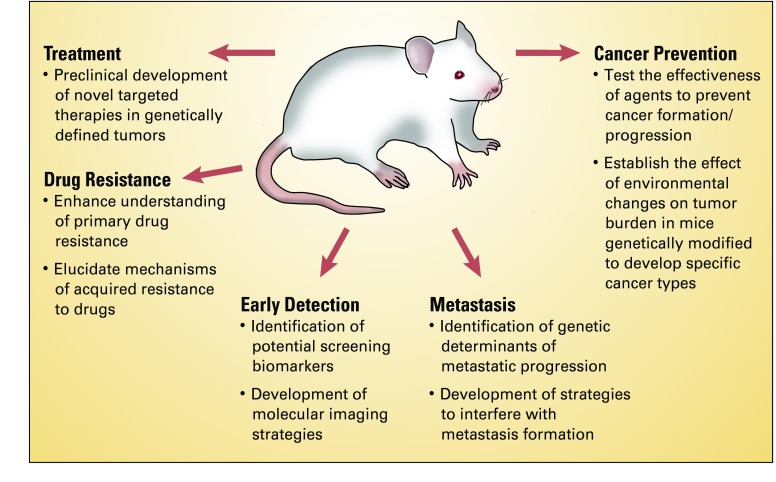
In conclusion, GEMMs of human cancer have contributed significantly to our understanding of basic cancer biology. The rapid development of new techniques to engineer mutations in the mouse genome with tissue and temporal specificity has now provided us with the tools to generate GEMMs that accurately mimic genetic changes present in human cancers and that can be used to test specific targeted therapies. As GEMMs continue to develop in sophistication, they will not only allow us to unravel complex cancer biologic problems but will also serve as platforms from which to develop strategies for early detection and treatment (Fig 2).
Fig 2.
Multiple uses of genetically engineered mouse models (GEMMs) of human cancer. Most clinically relevant problems in cancer biology studied by using GEMMs include treatment, drug resistance, metastasis, early detection, and cancer prevention (the last is not discussed in this review).
Glossary Terms
- Genetically engineered mouse model:
Mouse model in which the genetic make-up of the mouse has been modified by transgenic or gene-targeting technologies to affect expression of a gene of interest or to express a mutant.
- Oncogene dependence:
Also called “oncogene addiction”, these words are used to describe how some tumor cells require expression of a specific oncogene for their survival despite the fact that normal cells from which the tumor originated were viable in the absence of the oncogene.
- Oncogenomic:
Oncogenomics is the study of the cancer genome using high-throughput technologies.
- Targeted therapeutics:
Therapeutic agents that are specifically modeled to inhibit very specific molecules in signal transduction pathways implicated in the disease.
- Xenograft:
Host graft from a species that is not related to the recipient.
Footnotes
Supported by grants No. R00 CA131488 (K.P.), R01 CA120247 (K.P.), and R01 CA121210 (W.P.).
The rights to EGFR T790M mutation testing were licensed by both W.P. and K.P. to Molecular MD.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: William Pao, MolecularMD (C), AstraZeneca (C), Bristol-Myers Squibb (C) Stock Ownership: None Honoraria: Katerina Politi, OSI Pharmaceuticals Research Funding: None Expert Testimony: None Other Remuneration: Katerina Politi, MolecularMD, William Pao, MolecularMD
AUTHOR CONTRIBUTIONS
Conception and design: Katerina Politi, William Pao
Financial support: Katerina Politi, William Pao
Administrative support: Katerina Politi, William Pao
Collection and assembly of data: Katerina Politi, William Pao
Data analysis and interpretation: Katerina Politi, William Pao
Manuscript writing: Katerina Politi, William Pao
Final approval of manuscript: Katerina Politi, William Pao
REFERENCES
- 1.Chin L, Tam A, Pomerantz J, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 2.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 3.Pelengaris S, Littlewood T, Khan M, et al. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein IB. Cancer. Addiction to oncogenes: The Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 6.Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunther EJ, Moody SE, Belka GK, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debies MT, Gestl SA, Mathers JL, et al. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss. J Clin Invest. 2008;118:51–63. doi: 10.1172/JCI33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 13.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 16.Dickins RA, McJunkin K, Hernando E, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D, Wagner EF, Palmiter RD. The origins of oncomice: A history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 2007;21:2258–2270. doi: 10.1101/gad.1583307. [DOI] [PubMed] [Google Scholar]
- 18.Brinster RL, Chen HY, Messing A, et al. Transgenic mice harboring SV40 T-antigen genes develop characteristic brain tumors. Cell. 1984;37:367–379. doi: 10.1016/0092-8674(84)90367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T. Tumor suppressor gene mutations in mice. Annu Rev Genet. 1996;30:603–636. doi: 10.1146/annurev.genet.30.1.603. [DOI] [PubMed] [Google Scholar]
- 21.Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 22.Harvey M, McArthur MJ, Montgomery CA, Jr, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 23.Momota H, Holland EC. Mouse models of CNS embryonal tumors. Brain Tumor Pathol. 2009;26:43–50. doi: 10.1007/s10014-009-0253-0. [DOI] [PubMed] [Google Scholar]
- 24.Huse JT, Holland EC. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009;19:132–143. doi: 10.1111/j.1750-3639.2008.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 26.Vargo-Gogola T, Rosen JM. Modelling breast cancer: One size does not fit all. Nat Rev Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 27.Jeet V, Russell PJ, Khatri A. Modeling prostate cancer: A perspective on transgenic mouse models. Cancer Metastasis Rev. 2010;29:123–142. doi: 10.1007/s10555-010-9212-9. [DOI] [PubMed] [Google Scholar]
- 28.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 29.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Politi K, Zakowski MF, Fan PD, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regales L, Balak MN, Gong Y, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS ONE. 2007;2:e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Shimamura T, Ji H, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A. 2009;106:474–479. doi: 10.1073/pnas.0808930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber AC, Li D, Song Y, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sos ML, Fischer S, Ullrich R, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandhi L, McNamara KL, Li D, et al. Sunitinib prolongs survival in genetically engineered mouse models of multistep lung carcinogenesis. Cancer Prev Res (Phila Pa) 2009;2:330–337. doi: 10.1158/1940-6207.CAPR-08-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoeberl B, Faber AC, Li D, et al. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer Res. 2010;70:2485–2494. doi: 10.1158/0008-5472.CAN-09-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimamura T, Li D, Ji H, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 46.Dankort D, Filenova E, Collado M, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Sasaki T, Tan X, et al. Inhibition of ALK, PI3K/MEK and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. doi: 10.1158/0008-5472.CAN-10-1671. [epub ahead of print on November 23, 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 51.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 52.Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 53.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 54.Politi K, Fan PD, Shen R, et al. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis Model Mech. 2010;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliver TG, Mercer KL, Sayles LC, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lauchle JO, Kim D, Le DT, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D'Cruz CM, Gunther EJ, Boxer RB, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 58.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006;26:8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Podsypanina K, Politi K, Beverly LJ, et al. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose “chemo-switch” regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol. 2005;23:939–952. doi: 10.1200/JCO.2005.07.093. [DOI] [PubMed] [Google Scholar]
- 61.Bellmunt J, Trigo JM, Calvo E, et al. Activity of a multitargeted chemo-switch regimen (sorafenib, gemcitabine, and metronomic capecitabine) in metastatic renal-cell carcinoma: A phase 2 study (SOGUG-02-06) Lancet Oncol. 2010;11:350–357. doi: 10.1016/S1470-2045(09)70383-3. [DOI] [PubMed] [Google Scholar]
- 62.Bell-McGuinn KM, Garfall AL, Bogyo M, et al. Inhibition of cysteine cathepsin protease activity enhances chemotherapy regimens by decreasing tumor growth and invasiveness in a mouse model of multistage cancer. Cancer Res. 2007;67:7378–7385. doi: 10.1158/0008-5472.CAN-07-0602. [DOI] [PubMed] [Google Scholar]
- 63.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faca VM, Song KS, Wang H, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pitteri SJ, JeBailey L, Faca VM, et al. Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery. PLoS ONE. 2009;4:e7916. doi: 10.1371/journal.pone.0007916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung KE, Faca V, Song K, et al. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila Pa) 2009;2:224–233. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradbury MS, Hambardzumyan D, Zanzonico PB, et al. Dynamic small-animal PET imaging of tumor proliferation with 3′-deoxy-3′-18F-fluorothymidine in a genetically engineered mouse model of high-grade gliomas. J Nucl Med. 2008;49:422–429. doi: 10.2967/jnumed.107.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackson EL, Olive KP, Tuveson DA, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 69.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 70.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Podsypanina K, Du YC, Jechlinger M, et al. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hüsemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 73.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leary RJ, Lin JC, Cummins J, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Y, Rideout WM, 3rd, Zi T, et al. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat Biotechnol. 2010;28:71–78. doi: 10.1038/nbt.1595. [DOI] [PubMed] [Google Scholar]
- 81.Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Politi K, Kljuic A, Szabolcs M, et al. 'Designer' tumors in mice. Oncogene. 2004;23:1558–1565. doi: 10.1038/sj.onc.1207275. [DOI] [PubMed] [Google Scholar]
- 84.Klinakis A, Szabolcs M, Chen G, et al. Igf1r as a therapeutic target in a mouse model of basal-like breast cancer. Proc Natl Acad Sci U S A. 2009;106:2359–2364. doi: 10.1073/pnas.0810221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orsulic S. An RCAS-TVA-based approach to designer mouse models. Mamm Genome. 2002;13:543–547. doi: 10.1007/s00335-002-4003-4. [DOI] [PubMed] [Google Scholar]
- 86.Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]