Key Points
APC activates PAR3 in the presence of EPCR by noncanonical cleavage at Arg41.
APC-derived PAR3 tethered-ligand peptides induced APC-like vascular barrier protective effects in vitro and in vivo.
Abstract
The direct cytoprotective activities of activated protein C (APC) on cells convey therapeutic, relevant, beneficial effects in injury and disease models in vivo and require the endothelial protein C receptor (EPCR) and protease activated receptor 1 (PAR1). Thrombin also activates PAR1, but its effects on cells contrast APC’s cytoprotective effects. To gain insights into mechanisms for these contrasting cellular effects, protease activated receptor 3 (PAR3) activation by APC and thrombin was studied. APC cleaved PAR3 on transfected and endothelial cells in the presence of EPCR. Remarkably, APC cleaved a synthetic PAR3 N-terminal peptide at Arg41, whereas thrombin cleaved at Lys38. On cells, APC failed to cleave R41Q-PAR3, whereas K38Q-PAR3 was still cleaved by APC but not by thrombin. PAR3 tethered-ligand peptides beginning at amino acid 42, but not those beginning at amino acid 39, conveyed endothelial barrier-protective effects. In vivo, the APC-derived PAR3 tethered-ligand peptide, but not the thrombin-derived PAR3 peptide, blunted vascular endothelial growth factor (VEGF)-induced vascular permeability. These data indicate that PAR3 cleavage by APC at Arg41 can initiate distinctive APC-like cytoprotective effects. These novel insights help explain the differentiation of APC’s cytoprotective versus thrombin’s proinflammatory effects on cells and suggest a unique contributory role for PAR3 in the complex mechanisms underlying APC cytoprotective effects.
Introduction
Activated protein C (APC) is a dual-function blood coagulation enzyme with potent anticoagulant and cytoprotective activities.1,2 APC’s cytoprotective effects generally counteract cellular stress and promote cellular survival and, depending on cell type and cellular insult, may include antiapoptotic activity, antiinflammatory activity, alterations of gene expression profiles, and protection of endothelial barrier functions.3-8 The current paradigm for APC’s cytoprotective effects on cells involves binding of APC to endothelial protein C receptor (EPCR), permitting the APC-mediated proteolytic cleavage of the protease activated receptor 1 (PAR1) N-terminus and generation of a novel N-terminal tethered ligand to activate PAR1-meditated signaling pathways. Although incomplete, this model has been extremely helpful for the conceptual separation of APC’s anticoagulant and cytoprotective activities.9-12 The use of cytoprotective selective- and anticoagulant selective-APC mutants helped identify the relative contribution of APC’s dual activities to therapeutic, relevant, beneficial effects in injury and disease models in vivo. Collectively, these studies indicated a major role for APC’s cytoprotective effects for mortality reduction in various sepsis models and neuroprotective effects in ischemic stroke models.12,13 These cytoprotective effects of APC are not only important when exogenous APC is administered in vivo but also when the endogenous APC cytoprotective activities reduce susceptibility to lipopolysaccharide-induced endotoxemia.14 Thus, given the physiological importance of the APC cytoprotective pathway, it is imperative to get a better understanding of the mechanisms involved.
Recent evidence showed that cytoprotective signaling by APC occurs in PAR1- and EPCR-containing lipid rafts or caveolae and is dependent on caveolin-1.15,16 Furthermore, barrier-protective APC signaling via PAR1 requires β-arrestin recruitment and downstream signaling rather than the traditional G-protein–coupled signaling, thus providing a first clue that APC-mediated PAR1 signaling is clearly distinct from traditional PAR1 signaling.17 Despite these refinements of the molecular model for APC’s cytoprotective effects on cells, major questions remain to be answered. How can APC convey cytoprotective effects via PAR1, whereas thrombin uses the same receptor to mediate opposite proinflammatory effects?
PAR1 is a 7-transmembrane G protein-coupled receptor and part of a 4-member family (PAR1 through PAR4) that requires proteolytic cleavage of its N-terminus to reveal a tethered-ligand sequence for activation of intracellular signaling cascades.18 PAR1 is best known as the thrombin receptor on human platelets, whereas mouse platelets, which are devoid of PAR1, use protease activated receptor 3 (PAR3) and PAR4 to induce platelet activation in response to thrombin.19 PAR3 is the odd member of the PAR family, as it is considered a nonsignaling receptor and agonist peptides representing the tethered-ligand exposed after proteolytic cleavage by thrombin at Lys38 failed to induce PAR3-dependent intracellular signaling pathways but rather activated PAR1 and PAR2.20-22 Early studies suggested that PAR3, in addition to PAR1, contributed to APC’s neuroprotective effects in vivo, as blocking antibodies against murine PAR3 or genetic deletion of PAR3 partially abrogated the effects of APC.23 Recently, PAR3 was also implicated in nephroprotective effects of APC in vivo and in the antiapoptotic activity of APC on podocytes.24 Because both thrombin and APC seem to activate PAR3, we hypothesized that differences in thrombin-mediated vs APC-mediated PAR3 activation may provide novel insights into the opposing PAR1-dependent effects of APC vs thrombin signaling.
Methods
See supplemental Methods (on the Blood website) for details not found here.
PAR3 cleavage assays
Cleavage of the PAR3 synthetic peptide representing residues 21 to 65 of the PAR3 N-terminal tail by APC and thrombin was performed as described.9
On Cell Western assays for PAR3
PAR3 On Cell Western assays were done according to the manufacturer’s recommendations using anti-PAR3 antibodies M14, H103, or Mab19b and the In Cell Western module of Odyssey Imager (LI-COR) with Image Studio Software v2.0.
Endothelial barrier assay
Permeability of endothelial cell (EA.hy926) barrier function was determined as described using APC (20 nM) or PAR3 peptides (all 50 μM) and thrombin (10 nM) induced permeability.11 Alternatively, endothelial permeability was induced by histones (10 μg/mL; from calf thymus [Roche]).
In vivo vascular permeability assay
The study was approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute and complies with National Institutes of Health guidelines. SKH1-E hairless male (6-8 weeks old) were from Charles River Labs (Wilmington, MA). Vascular permeability was determined using a vascular endothelial growth factor (VEGF)-induced leakage model with some modifications.25,26 Peptides, phosphate-buffered saline control, and Evans blue were injected intravenously and VEGF was injected subcutaneously. Evans blue extravasation in the skin was quantified using the Odyssey Imager (LI-COR).
Statistical analysis
Statistical significance (P < .05) was determined using a Student t test or one-way ANOVA and Bonferroni multiple comparison post-tests as appropriate (Prism 5.0; Graphpad Software, La Jolla, CA).
Results
Cleavage of PAR3 by APC is dependent on EPCR
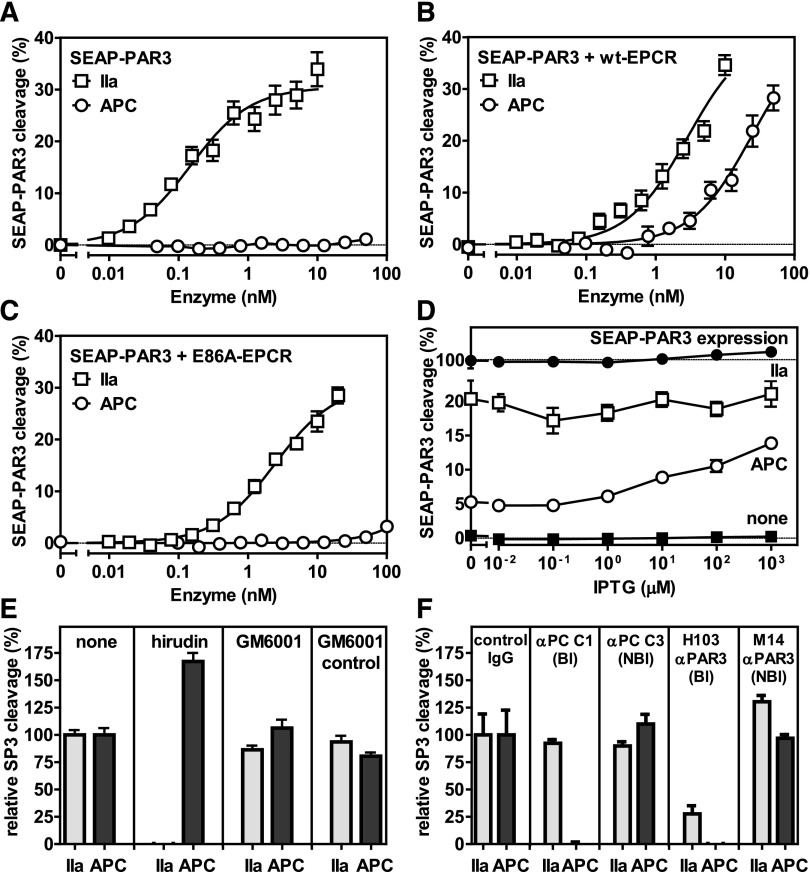
Using a secreted embryonic alkaline phosphatase (SEAP)-PAR3 fusion construct expressed in HEK-293 cells, no cleavage of PAR3 by APC was observed in the absence of EPCR (Figure 1A), whereas APC potently cleaved PAR3 in the presence of wild-type (wt)-EPCR (Figure 1B). Cleavage of PAR3 by APC was observed at APC concentrations between 5 and 50 nM, which are comparable with APC concentrations required for cytoprotective activities in vitro.10,11 In contrast, thrombin cleaved PAR3 efficiently both in the absence and presence of EPCR, as anticipated. Cleavage of PAR3 by APC required APC binding to EPCR, as APC failed to cleave PAR3 in the presence of E86A-EPCR that is defective in APC binding (Figure 1C).27 To determine the requirement of EPCR for APC-mediated PAR3 cleavage in more detail, wt-EPCR expression in SEAP-PAR3 cells was regulated using an IPTG-inducible vector (EPCR-pTUNE).28 Very little EPCR was expressed in the absence of IPTG as determined by On Cell Western or traditional western blot but at saturating concentrations of IPTG expression of EPCR increased twofold, yet was still fourfold lower than the level in wt-EPCR/SEAP-PAR3 cells (supplemental Figure 1A-B). APC-mediated PAR3 cleavage increased proportional to the expression of EPCR (Figure 1D), indicating that APC can cleave PAR3 at low expression levels of EPCR that are comparable with those found in endothelial cells (supplemental Figure 1B). Reflecting the divalent cation profile of APC binding to EPCR,29 PAR3 cleavage by APC required CaCl2 and was enhanced by low concentrations of ZnSO4 at saturating concentrations of CaCl2 and MgCl2 (supplemental Figure 2). Neither hirudin nor the matrix metalloproteases inhibitor, GM6001, inhibited APC-mediated PAR cleavage, indicating that cleavage of PAR3 was not due to traces of thrombin in the APC preparation or to APC-mediated induction of cell-derived matrix metalloproteases (Figure 1E). Inhibition of APC-mediated PAR3 cleavage by the APC activity-blocking antibody C1 indicated that the activity of APC was required. Inhibition of thrombin- and APC-mediated PAR3 cleavage by the PAR3 cleavage-blocking H103 antibody confirmed the specificity of the observed PAR3 cleavage, whereas the nonblocking M14 anti-PAR3 antibody against extracellular loop 3 was without effect (Figure 1F). Thus, cleavage of PAR3 by APC requires the presence of functional EPCR and is directly proportional to the EPCR expression level on the cell surface.
Figure 1.
EPCR-dependent PAR3 cleavage by APC. APC-mediated cleavage of PAR3 was analyzed by the proteolytic release of SEAP from a SEAP-PAR3 fusion protein expressed in HEK-293 cells in the absence or presence of stable EPCR coexpression. Dose response of PAR3 cleavage by thrombin (IIa) (□) or APC (○) in the (A) absence of EPCR and in the presence of (B) wt-EPCR or (C) E86A-EPCR defective in APC binding. (D) PAR3 cleavage by 2 nM thrombin (□) or 20 nM APC (○) in SEAP-PAR3 HEK-293 cells in the presence of isopropyl β-D-1-thiogalactopyranoside (IPTG) tunable wt-EPCR coexpression (EPCR-pTUNE). IPTG did not affect SEAP release from cells in the absence of protease (■) or the total SEAP-PAR3 expression on cells (●). PAR3 cleavage was expressed as a percentage of the total available SEAP-PAR3 on the cells. (E-F) Specificity controls of PAR3 cleavage by 10 nM thrombin or 20 nM APC in wt-EPCR/SEAP-PAR3 cells (E). Inhibitors used were directed against thrombin (20 U/mL hirudin) and matrix metalloproteinases (10 μM GM6001 or GM6001 inactive control). (F) Blocking (Bl) and nonblocking (NBl) antibodies (all 20 μg/mL) were directed against APC or PAR3. PAR3 cleavage was expressed relative to the cleavage in the absence of inhibitors (E) or in the presence of nonimmune IgG antibodies (F). Data points represent the mean ± SEM (n ≥ 3). Single amino acid abbreviations denote A, Ala; and E, Glu.
Identification of the APC cleavage site in PAR3
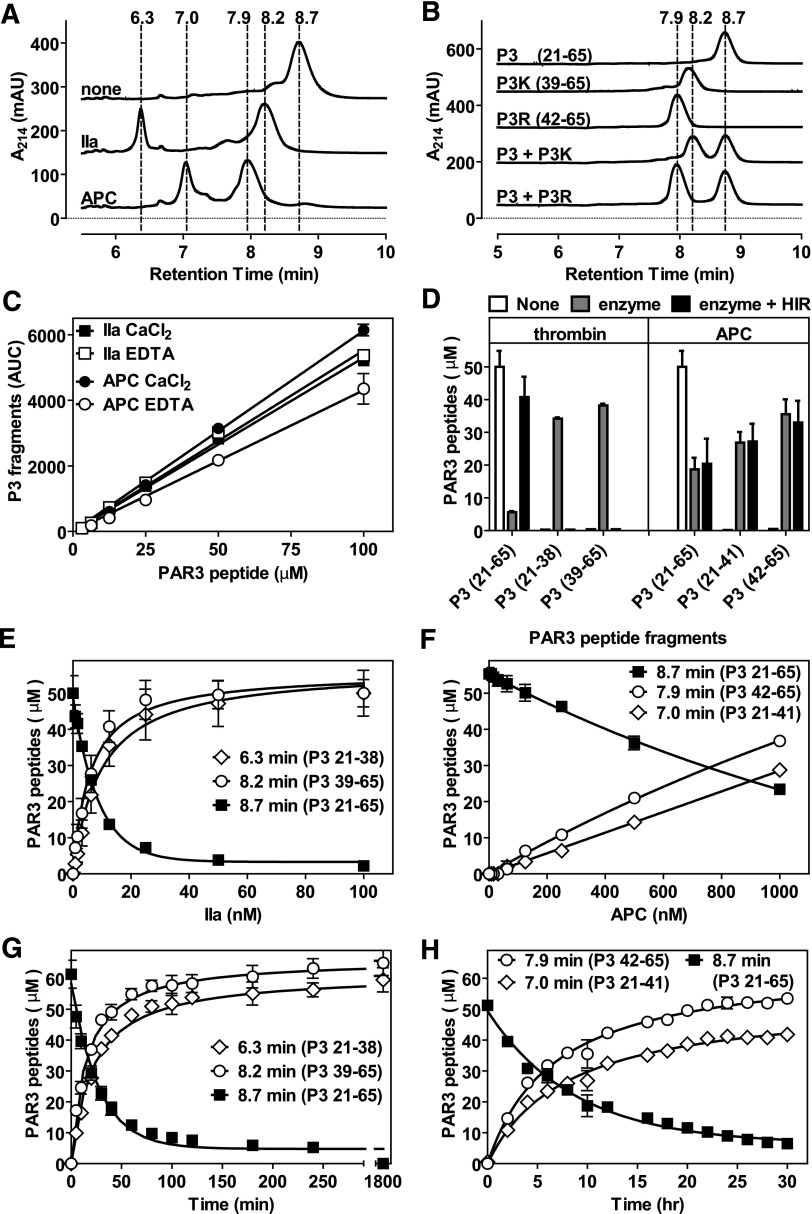
Based on the canonical Lys38 cleavage site for thrombin, cleavage of PAR3 by APC at Lys38 is highly unusual, as proteolysis of the scissile bond by APC generally occurs after an Arg residue. Therefore, the APC proteolysis profile of a synthetic PAR3 peptide, representing residues 21 to 65 of the PAR3 extracellular domain (peptide P3; supplemental Figure 4) was determined and compared with thrombin. Upon proteolysis of the P3 peptide by APC, 2 distinct fragments became apparent that were different from the parental peptide (Figure 2A). Interestingly, the APC-generated fragments were different from the 2 fragments generated by thrombin, indicating that proteolysis occurred at different cleavage sites. Analysis of the fragments by mass spectrometry (supplemental Figure 3) revealed that proteolysis of the P3 peptide by APC generated fragments A (Rt = 6.3 minutes; molecular mass (Mr) = 1943.3 Da) and B (Rt = 8.2 minutes; Mr = 2930.7 Da) vs parental P3 peptide (Rt = 8.7 minutes; Mr = 4856 Da). In contrast, proteolysis of the P3 peptide by thrombin generated fragments D (Rt = 7.0 minutes; Mr = 2347.2 Da) and E (Rt = 7.9 minutes; Mr = 2526.0 Da). Matching of the fragments’ molecular weight to the peptide sequence revealed that APC cleaved P3 at Arg41, whereas thrombin cleaved P3 at Lys38 (Table 1). Chromatography of the synthetic peptides (supplemental Figure 4) representing the C-terminal fragment after cleavage by APC (P3RL residues 42-65) or thrombin (P3KL residues 39-65) resulted in identical peaks compared with the corresponding fragments generated by proteolysis, indicating the accuracy of the Mass-Spec determination and identification of the P3 cleavage sites (Figure 2B). APC-mediated cleavage of the P3 peptide was enhanced in the presence of CaCl2, whereas CaCl2 or EDTA did not influence cleavage by thrombin (Figure 2C). No evidence was found that the presence of CaCl2 or EDTA altered the cleavage site for APC or thrombin. Cleavage of the P3 peptide by thrombin at Lys38 was inhibited by hirudin, but cleavage by APC at Arg41 was not (Figure 2D). To determine whether proteolysis of the P3 peptide by APC or thrombin occurred at sites other than Lys38 and Arg41, the generation of concentration- and time-dependent cleavage fragments was determined. Half-maximal cleavage of the P3 peptide by thrombin in 1 hour required ∼6 nM thrombin (Figure 2E), whereas half-maximal cleavage by APC for 10 hours required closer to 1000 nM APC (Figure 2F). Similarly, half-maximal cleavage of the P3 peptide by 10 nM thrombin was achieved in ∼18 minutes (Figure 2G), whereas half-maximal cleavage by 500 nM APC required ∼7 hours (Figure 2H). Importantly, no additional cleavage fragments for either thrombin or APC could be detected, indicating that cleavage of the P3 peptide at Arg41 by APC is not preceded by cleavage at Lys38, as the cleavage of the PAR1 peptide at Arg46 by APC is preceded by cleavage at Arg41.30
Figure 2.
Proteolysis of a synthetic PAR3 peptide by APC and thrombin. (A) Chromatogram of P3 fragments generated after 48 hours in the absence (none) or presence of APC (500 nM) or thrombin (IIa) (10 nM). (B) Chromatogram of the P3 parental peptide and synthetic peptides representing the predicted C-terminal cleavage fragments generated by APC (P3R) and thrombin (P3K). (C) Generation of P3 N-terminal cleavage fragments by APC (500 nM, 9 hours; fragment [21-41], Rt 7.0 minutes) and thrombin (10 nM, 2 hours; fragment [21-38], Rt 6.3 minutes) in the absence of divalent metal ions (2 mM EDTA) or in the presence of 2 mM CaCl2 and 0.6 mM MgCl2. (D) Generation of P3 N- and C-terminal cleavage fragments by APC (500 nM, 10 hours) and thrombin (10 nM, 2 hours) in the absence and presence of 100 U/mL hirudin (HIR). (E) Concentration-dependent cleavage of the P3 peptide by thrombin (1 hour). (F) Concentration-dependent cleavage of the P3 peptide by APC (10 hours). (G) Time-dependent cleavage of the P3 peptide by thrombin (10 nM). (H) Time-dependent cleavage of the P3 peptide by APC (500 nM). (A-B) Representative chromatograms. (C-G) Data points represent the mean ± SD (n ≥ 3).
Table 1.
PAR3 peptide fragments generated by APC or thrombin proteolysis
| Fragment | Rt | Cleaved by | Experimental mass | Calculated mass | PAR3 peptide (21-65) | Cleavage site |
|---|---|---|---|---|---|---|
| A | 6.3 | IIa | 1943.2 | 1943.3 | SGMENDTNNLAKPTLPIK-38 | K38 |
| B | 8.2 | IIa | 2930.7 | 2931.3 | 39-TFRGAPPNSFEEFPFSALEGWTGATIT | K38 |
| C | 8.7 | — | 4856 | 4856.6 | SGMENDTNNLAKPTLPIKTFRGAPPNSFEEFPFSALEGWTGATIT | — |
| D | 7.0 | APC | 2347.3 | 2347.8 | SGMENDTNNLAKPTLPIKTFR-41 | R41 |
| E | 7.9 | APC | 2526.0 | 2526.9 | 42-GAPPNSFEEFPFSALEGWTGATIT | R41 |
| F | 8.7 | — | 4856 | 4856.6 | SGMENDTNNLAKPTLPIKTFRGAPPNSFEEFPFSALEGWTGATIT | — |
Rt, retention time.
Noncanonical PAR3 cleavage at Arg41 by APC on cells
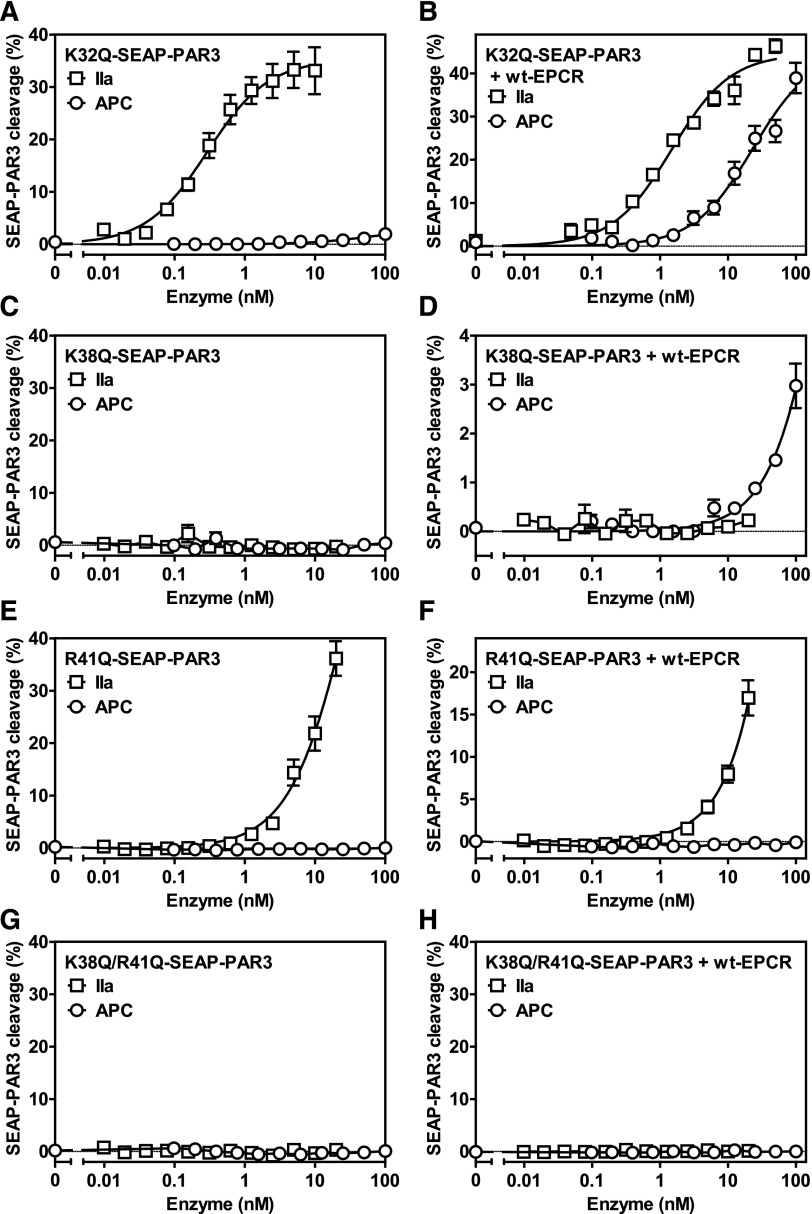
To confirm that APC also cleaved PAR3 at Arg41 and not at Lys38 on cells, the corresponding mutations were introduced in SEAP-PAR3. Mutation of Lys38 abolished thrombin-mediated PAR3 cleavage (Figure 3C), but in the presence of wt-EPCR, APC could still cleave PAR3 despite the Lys38 cleavage site mutation (Figure 3D). Conversely, mutation of the Arg41 allowed for efficient cleavage of PAR3 by thrombin (Figure 3E), whereas APC failed to cleave Arg41-mutated PAR3 in the presence of EPCR (Figure 3F). Both thrombin and APC did not induce PAR3 cleavage when both Lys38 and Arg41 cleavage sites were mutated regardless of the absence (Figure 3G) or presence (Figure 3H) of EPCR, indicating that Lys38 and Arg41 are the only cleavage sites available in PAR3 for thrombin and APC, respectively. Accordingly, mutation of Lys32 resulted in PAR3 cleavage by thrombin and APC in the absence (Figure 3A) and presence of EPCR (Figure 3B) that was indistinguishable from cleavage of the wt-SEAP-PAR3 cleavage by these proteases (Figure 1A-B). Analysis of PAR2 cleavage by APC provides additional evidence on the specificity of APC cleaving PAR3 at Arg41. APC showed appreciable PAR2 cleavage in the presence of wt-EPCR, albeit at APC concentrations that were 10-fold higher than required for PAR3 cleavage by APC (supplemental Figure 5). Similar to cleavage of PAR3 by APC, cleavage of PAR2 by APC was dependent on the presence of functional EPCR, as E86A-EPCR did not support APC-mediated PAR2 cleavage. No SEAP-PAR2 cleavage was observed in the presence of thrombin (data not shown). Despite the presence of several basic residues neighboring the PAR2 canonical cleavage site at Arg36, mutation of Arg36 abolished EPCR-dependent APC proteolysis of PAR2 (supplemental Figure 5). Thus, proteolysis of canonical and noncanonical cleavage sites in PAR2 and PAR3 by APC are PAR specific and sequence specific, although both cleavages require APC binding to EPCR.
Figure 3.
Proteolysis of PAR3 cleavage site mutants by APC and thrombin on cells. Cleavage of SEAP-PAR3 mutants by APC and thrombin (IIa) in the absence (A,C,E,G) and presence (B,D,F,H) of wt-EPCR. (A-B) Lys32Gln-SEAP-PAR3. (C-D) Lys38Gln-SEAP-PAR3. (E-F) Arg41Gln-SEAP-PAR3. (G-H) Lys38Gln/Arg41Gln-SEAP-PAR3. SEAP released in the media was determined 1 hour after the addition of APC or thrombin and corrected for background and spontaneous SEAP release in the absence of protease (<2%). The activity of released SEAP was expressed as a percentage of total available SEAP activity on the cells determined in the same volume. No SEAP activity was detected on HEK-293 or wt-EPCR HEK-293 cells in the absence of SEAP-PAR3. Data points represent the mean ± SEM (n ≥ 3). Single amino acid abbreviations denote K, Lys; R, Arg; and Q, Gln.
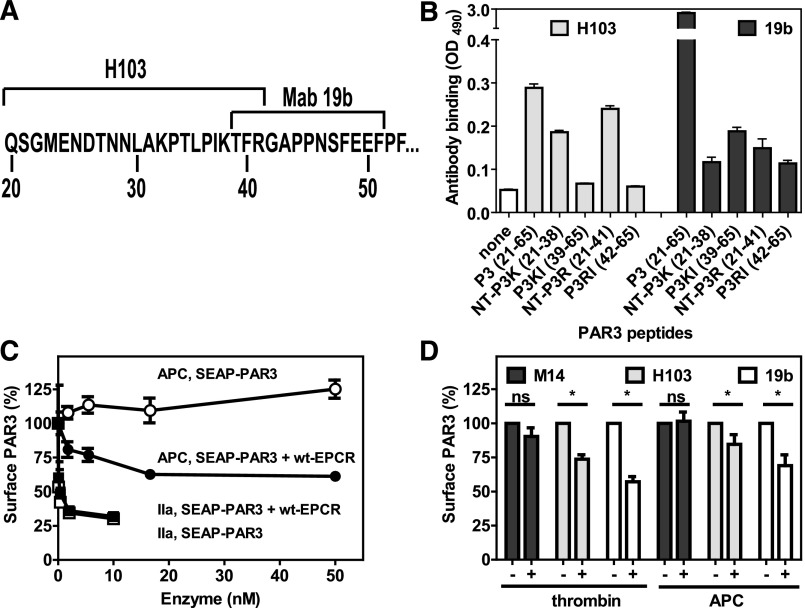
To determine whether APC can cleave PAR3 on endothelial cells expressing endogenous PAR3 and EPCR, available PAR3 antibodies were screened for recognition of cleavage-sensitive epitopes. Cleavage-blocking antibody H103 recognized an epitope N-terminal of Arg41 (Figure 4A), as it did not recognize the P3KL and P3RL peptides C-terminal of the thrombin and APC cleavage site (Figure 4B). However, the presence of an additional epitope at residues 66 to 103 could not be excluded, because H103 was raised against a PAR3 fragment comprising residues 1 to 103. H103 showed little preference for the N-terminal APC-derived fragment compared with the thrombin-derived fragment. Recognition of SEAP-PAR3 cells with H103 after incubation with thrombin confirmed the loss of an H103 epitope after PAR3 cleavage (Figure 4C). Furthermore, incubation of cells with APC resulted in a loss of the H103 epitope (indicating PAR3 cleavage) in the presence of EPCR but not in the absence of EPCR, thus confirming EPCR-dependent cleavage of PAR3 by APC (Figure 1A-B). On endothelial cells, both thrombin and APC induced a significant reduction of the H103 epitope, whereas reactivity with anti-PAR3 antibody M14 against the third extracellular loop of PAR3 remained unchanged after thrombin or APC incubation, indicating that the reduction of H103 reactivity was due to cleavage and not internalization of PAR3 (Figure 4D). Cleavage of endothelial PAR3 was confirmed using antibody Mab19b with its epitope previously mapped to PAR3 residues 39 to 51.31 As anticipated, Mab19b did not recognize the P3RL peptide C-terminal of the APC cleavage site, but recognition of the suggested epitope encompassing P3KL peptide was <5% of the parental P3 peptide, suggesting that recognition of PAR3 by Mab19b was cleavage sensitive and that additional residues N-terminal of Thr39 likely contribute to its epitope (Figure 4B). Confirming the H103 antibody results, reactivity of endothelial cells with Mab19b significantly diminished after thrombin or APC treatment (Figure 4D). Thus, APC cleaved PAR3 on endothelial cells expressing endogenous PAR3 and EPCR.
Figure 4.
Cleavage of PAR3 by APC and thrombin on endothelial cells. (A) Schematic representation of the PAR3 cleavage site sequence with the epitopes for the cleavage sensitive PAR3 antibodies H103 and Mab19b as indicated. (B) Peptide mapping of the cleavage site-sensitive epitopes of anti-PAR3 antibodies H103 and Mab19b. Peptides represent the N-terminal (NT-P3K) or C-terminal fragment (P3K) of PAR3 after cleavage at Lys38 or the N-terminal (NT-P3R) or C-terminal fragment (P3R) of PAR3 after cleavage at Arg41 or the entire cleavage site region of the PAR3 N-terminal tail (P3). (C) On Cell Western of anti-PAR3 antibody H103 on SEAP-PAR3 (open symbols) and SEAP-PAR3/wt-EPCR (closed symbols) HEK293 cells after incubation with APC (●, ○) or thrombin (IIa) (■, □) for 2 hours. (D) EA.hy926 endothelial cells after incubation with control buffer, APC (100 nM), or thrombin (0.25 nM) for 3 hours. The minus and plus refer to the presence or absence of thrombin and APC, respectively. Cell bound anti-PAR3 antibodies were detected using the On Cell Western assays with PAR3 cleavage site sensitive H103 and Mab19b antibodies and the cleavage-insensitive antibody M14 directed against the PAR3 extracellular loop 3. (B) Representative experiment in triplicates. (C-D) Data points represent the mean ± SEM (n ≥ 3). ns, not significant.
APC-derived PAR3 tethered-ligand peptides are endothelial barrier protective and reduce vascular leakage in vivo
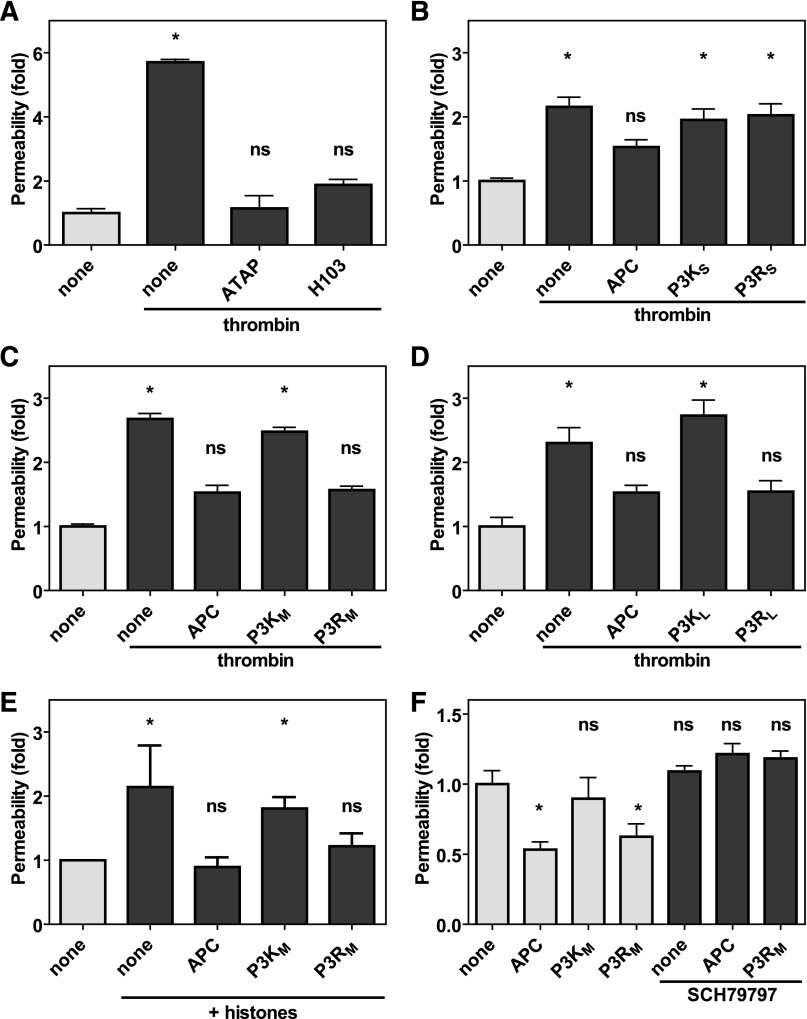
PARs are unique among G protein-coupled receptors in that they carry their own encrypted tethered ligand immediately N-terminal of the cleavage site only to be exposed after proteolytic activation of the PAR. Noncanonical cleavage of PAR3 by APC at Arg41 would therefore implicate either creation of a novel PAR3 tethered ligand or destruction of the PAR3 tethered ligand exposed by canonical cleavage of PAR3 at Lys38 by thrombin. Barrier-disruptive effects by thrombin are known to involve PAR1, but the contribution of PAR3 is not as well appreciated.32 However, the PAR3 cleavage blocking antibody H103 prevented thrombin-induced endothelial permeability similar to the PAR1 cleavage-blocking antibody ATAP2 (Figure 5A), indicating that thrombin’s endothelial barrier-disruptive effects required both PAR1 and PAR3. Thus, APC could conceivably cleave PAR3 to prevent thrombin-mediated PAR3 effects. To determine whether PAR3 cleavage at Arg41 created a novel tethered ligand, peptides representing the new N-terminus after APC cleavage at Arg41 were generated and compared with peptides representing the N-terminus after thrombin cleavage at Lys38 (supplemental Figure 4). Insights gleaned from TRAP peptides suggested that the first 6 residues are a minimal requirement to induce PAR1 activation,33 but 6-mer PAR3 peptides either starting at Thr39 (P3KS) or Gly42 (P3RS) did not prevent thrombin-induced endothelial barrier disruption (Figure 5B). In contrast, longer PAR3 peptides derived from APC cleavage starting at Gly42 and ending at Ser54 (P3RM) or Thr65 (P3RL) both prevented thrombin-induced endothelial barrier disruption similar to APC (Figure 5C-D). Remarkably, the same peptides derived from thrombin cleavage starting at Thr39 and ending at Ser54 (P3KM) or Thr65 (P3KL) did not reduce thrombin-induced barrier disruption. Thus, APC cleavage of PAR3 at Arg41 created a novel tethered ligand that conveyed APC-like barrier-protective effects on endothelial cells. Because PAR3 was involved in both barrier-protective effects by APC and barrier-disruptive effects by thrombin, histones were used to induce endothelial permeability independently of thrombin.34 Histones induced a rapid increase in endothelial permeability, which could be prevented by incubation of the cells with APC (Figure 5E). Similar to APC, the APC-derived P3RM peptide, but not the thrombin-derived P3KM peptide, prevented histone-induced endothelial barrier disruption. In another thrombin-independent endothelial barrier assay where the endothelial cells were near confluent and spontaneously leaky, both APC and the P3RM but not the P3KM peptide improved endothelial barrier function (Figure 5F). Collectively, these results demonstrated that PAR3 peptides (≥13-mer) derived from APC cleavage of PAR3 at Arg41 acted as APC mimetics and conveyed APC-like endothelial barrier-protective effects on cells. PAR3 cannot signal on its own but instead modulates PAR1-dependent signaling via the formation of PAR1 heterodimers.24,32 Accordingly, the barrier-protective effects of P3RM on spontaneously leaky near-confluent endothelial cells were abrogated by the specific PAR1 antagonist SCH79797, indicating that the barrier-protective effects of APC-derived PAR3 peptides required PAR1 similar to the barrier-protective effects mediated by APC (Figure 5F).
Figure 5.
The novel tethered ligand P3R peptide improves endothelial barrier functions. Endothelial barrier function in vitro was determined by the permeability of an endothelial cell layer to Evans blue-albumin complexes. (A) Contribution of PAR1 and PAR3 using the cleavage-blocking antibodies ATAP-2 (25 μg/mL) against PAR1 or H103 (25 μg/mL) against PAR3 to thrombin-induced endothelial permeability. (B-D) Modulation of thrombin-induced endothelial permeability by PAR3 peptides (50 μM) derived from cleavage at Lys38 (P3K) or Arg41 (P3R) of various lengths (supplemental Figure 4) compared with APC (20 nM). Shown are data for (B) short peptides (P3KS and P3RS) comprised of 6 amino acids, (C) medium-length peptides ending at Ser54 (P3KM and P3RM), and (D) long peptides ending at Thr65 (P3KL and P3RL). (E) Protection of endothelial barrier function by PAR3 peptides (50 μM) derived from cleavage at Lys38 (P3KM) or Arg41 (P3RM) against permeability induced by histones. (F) Modulation of endothelial barrier function of near confluent endothelial by PAR3 peptides (50 μM) derived from cleavage at Lys38 (P3KM) or Arg41 (P3RM) in the absence (gray bars) and presence (dark bars) of the PAR1 antagonist SCH79797. Data points represent the mean ± SEM (n ≥ 3 independent inserts). Asterisk denotes a statistically significant difference (P < .05).
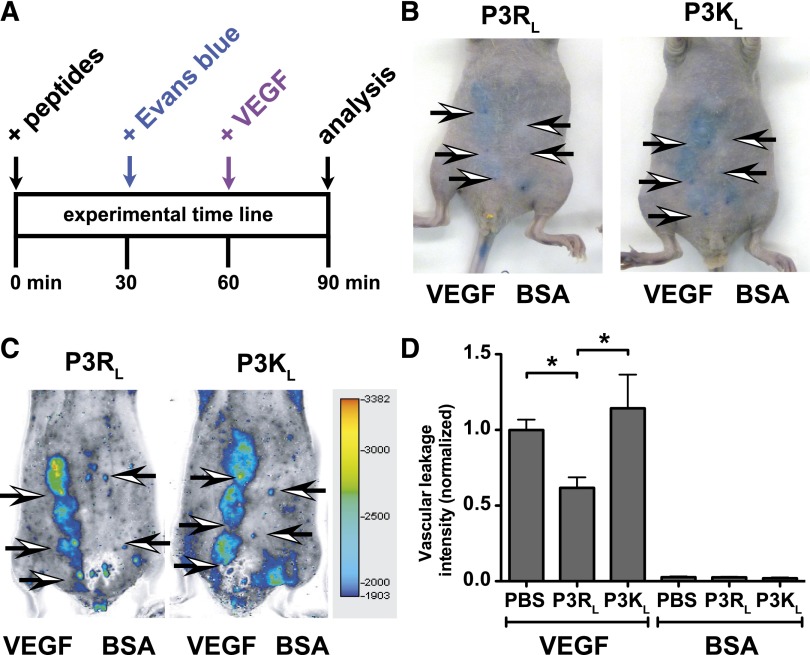
An acute VEGF-induced skin vascular leakage model was used to test if APC-derived PAR3 peptides modulated vascular permeability in vivo (Figure 6A). Mice were intravenously injected with Evans blue 30 minutes before subcutaneous injection of VEGF or bovine serum albumin control on the abdomen, and extravasation of Evans blue was determined by placing the mice on the Odyssey Infrared Fluorescence Imager and recording the fluorescence of Evans Blue in the skin at 700 nm. After injection of bovine serum albumin vehicle control, only the needlepoints marking the injection site could be observed, whereas VEGF induced clearly distinguishable areas of Evans blue extravasation (Figure 6). Areas of Evans blue extravasation were easily detectable in the 700-nm channel of the Odyssey Imager, which greatly facilitated reproducible quantification of Evans blue extravasation and eliminated the time-consuming need for Evans blue extraction from punch biopsy (Figure 6C). When the PAR3 peptides (P3RL or P3KL) or phosphate-buffered saline vehicle control were injected intravenously in the retro-orbital sinus 60 minutes before VEGF, P3RL but not P3KL decreased vascular leakage by 38% (P < .05, n = 8 mice) (Figure 6D). Neither P3RL nor P3KL affected vascular leakage in the absence of VEGF.
Figure 6.
TR47 decreases capillary vascular VEGF-induced permeability in the skin. VEGF-induced extravasation of Evans blue in the skin was determined in immune competent SKH1-E mice. (A) The experimental time line for the in vivo VEGF-induced vascular leakage model and administration of peptides. (B) Photographs for Evans blue extravasation in the skin of 2 representative mice that received the P3RL (left) or P3KL (right) peptide after subcutaneous injection with VEGF (3 left arrows) or with bovine serum albumin (BSA) control (2 right arrows). (C) Pseudo-colored heat map display of Evans blue extravasation quantified by the Odyssey near infrared imager at 700 nm. P3RL- (left) or P3KL- (right) treated mice were injected subcutaneously with VEGF (3 left arrows) or with BSA control (2 right arrows). (D) Quantification of in vivo VEGF-induced vascular leakage for mice treated with phosphate-buffered saline (PBS) control, P3RL (125 μg), or P3KL (125 μg) peptide. Data are also shown for BSA-treated control mice that received PBS, P3RL, or P3KL but no VEGF. (B-C) Representative experiments. (D) Data points represent the mean ± SEM (n ≥ 8). Asterisk denotes a statistically significant difference (P < .05).
In summary, the P3RL peptide representing the sequence of the novel N-terminus that is generated by cleavage of PAR3 at Arg41 exerts remarkable biologic activities in vitro and in vivo that reflects the general cytoprotective activity profile of APC but not that of thrombin. Based on these results, we propose a novel paradigm for the biochemical mechanisms of APC via PAR3 involving generation of a new N-terminal tethered ligand, which initiates APC-like cytoprotective endothelial barrier-protective effects in vitro and promotes vascular integrity in vivo.
Discussion
Thrombin signaling via PAR1 results in activation of RhoA and endothelial barrier-disruptive effects.35 In contrast, barrier-protective APC signaling via PAR1 occurs in cells expressing EPCR and requires β-arrestin 2 recruitment to PAR1 in lipid rafts or caveolae and downstream signaling that results in Akt and Rac1 activation.17,36 Thus, APC-mediated PAR1 activation is not only qualitatively different from thrombin-mediated PAR1 activation but also mechanistically different. However, the question remains how APC and thrombin can both use PAR1 to mediate these opposite cellular effects. Because PAR3 is implicated in contributing to APC’s cytoprotective effects and is a substrate for thrombin, we hypothesized that PAR3 might contribute to differentiation of APC’s vs thrombin’s effects on cells that are dependent on PAR1. Here, we show that APC cleaved PAR3 in the presence but not the absence of EPCR. Cleavage of PAR3 by APC uniquely occurred at the noncanonical Arg41 residue in contrast to thrombin-mediated cleavage of PAR3 occurring at the canonical Lys38.
Inherent to the biochemistry of PAR receptors, where receptor activation occurs via interactions of the receptor with the tethered ligand immediately following the protease cleavage site, a noncanonical cleavage site implies either the generation of a novel tethered ligand or the destruction of the canonical ligand. We confirmed that PAR3 contributed to thrombin-induced barrier disruption;32 thus, it is conceivable that noncanonical APC cleavage of PAR3 at Arg41 could contribute to destruction of the canonical PAR3 tethered ligand, thereby blunting the barrier-disruptive effects of thrombin. Furthermore, the evidence also strongly suggests that APC cleavage of PAR3 at Arg41 created a novel tethered ligand. The noncanonical APC-derived PAR3 tethered-ligand peptides (P3R) induced endothelial barrier-protective effects regardless of whether barrier disruption was induced by thrombin or histones or in semiconfluent cells. These barrier-protective effects were specific to the APC-generated PAR3 peptides, because the canonical, thrombin-derived, PAR3 tethered-ligand peptides (P3K) were without effect on barrier permeability. In vivo vascular protection of the APC-derived (P3RL) PAR3 peptide against VEGF-induced vascular breach confirmed the cellular agonism of PAR3 peptides starting at Gly42. Thus, APC cleavage of PAR3 at Arg41 creates a novel tethered ligand with APC-like barrier-protective effects. These striking observations raise many new questions that remain to be answered.
Why does APC cleave PAR3 at Arg41? Perhaps the pertinent question is: Why does thrombin cleave at Lys38? Both proteases clearly prefer a P1 Arg based on known substrate sequences. Cleavage at Lys38 by thrombin is likely dictated by the high-affinity binding of the PAR3 hirudin-like sequence to thrombin’s exosite I, which introduces spatial constraints that are incompatible with cleavage at Arg41. Because APC is not restricted by high-affinity binding to the PAR3 hirudin-like sequence, cleavage at Arg41 is not surprising considering that the PAR3 P5-P5′ (IKTFR/GAPPN) specificity subsite is very similar to that of protein C inhibitor (IFTFR/SARLN), with 80% identity in the P5-P1 residues and 40% in the P1′-P5′ residues.37 The PAR3 P5-P5′ region has been highly conserved (> 80%) between various species, except for rodents (supplemental Table 1). Remarkably, the equivalent of human Arg41 was Asn in both mouse and rat, whereas a basic cleavage site was completely absent in rat PAR3 that have Glu as the equivalent of human Lys38. The functional differentiation of PAR3 in rodents to restore platelet functionality to thrombin after PAR1 was lost from rodent platelets provides a possible explanation for this remarkable contrast between rodent PAR3 and that of other species.
Functional EPCR was required for cleavage of PAR3 by APC, similar to APC-mediated cleavage of PAR1.8,30 Previously, PAR3 cleavage by APC on podocytes was independent of EPCR.24 However, we found no evidence to support cleavage of human PAR3 by APC in the absence of EPCR. Other APC cofactors can possibly replace EPCR’s cofactor function for PAR3 cleavage by APC on podocytes.38-40 Alternatively, inflammatory mediators and various cytokines have known deleterious effects on EPCR expression and therefore, the effect on EPCR expression of continuous inclusion of interferon-γ in the culture media of conditionally immortalized podocytes to keep them proliferating and undifferentiated is difficult to predict.24,41,42
How do the APC-derived PAR3 tethered ligand peptides induce APC-like endothelial- and vascular-protective effects? Recently, we proposed a novel mechanism for APC-induced biased PAR1 signaling based on noncanonical PAR1 cleavage by APC at Arg46.30,43 A PAR1 peptide starting at Asn47 (TR47), representing the novel tethered ligand after APC cleavage at Arg46, conveyed endothelial barrier-protective effects in vitro and reduced VEGF-induced vascular permeability in the skin in vivo.30 Thus, functionally, the PAR1 and PAR3 noncanonical TR47 and P3R peptides seem to share similar endothelial- and vascular-protective effects; however, clear mechanistic differences were noted. For instance, the P3RL reduced vascular permeability in vivo when administered 1 hour, but not 5 minutes (data not shown), before VEGF, whereas TR47 was effective only when given 5 minutes, but not 1 hour, before VEGF,30 suggesting that P3RL and TR47 differ in their timing of cellular agonism resulting in vascular-protective effects. Abrogation of the barrier-protective effects of the P3R PAR3 peptide in the presence of the PAR1 antagonist SCH79797 indicated that PAR1 was involved. Although our data does not exclude the involvement of other PARs or even other non-PAR receptors, one interpretation would be that the APC-derived PAR3 peptide induced APC-like PAR1 activation. However, in contrast to TR47 that caused APC-like activation of Akt by phosphorylation of Ser473, neither P3K nor P3R peptides induced Akt or ERK1/2 phosphorylation in EA.hy926 endothelial cells (data not shown). Another interpretation would be that the APC-derived PAR3 peptide modulates PAR1-dependent signaling rather than inducing PAR1 signaling. Such a mechanism would be in agreement with the published literature. Early on, thrombin was found to induce PAR3-dependent signaling that required PAR1.21 PAR1 and PAR3 constitutively form PAR1-PAR3 heterodimers that after activation result in altered patterns of G-protein coupling compared with PAR1 homodimers.32 Furthermore, association of PAR3 with caveolin-1 suggests that PAR3, at least in part, resides in PAR1- and EPCR-containing caveolin-1–enriched micro domains or caveolae that are important for induction of APC cytoprotective signaling.24 Partial abrogation of APC’s endothelial barrier-protective effects by siRNA PAR3 knockdown is also consistent with modulation of PAR1 signaling by PAR3.44 Thus, despite functional similarities between PAR1 and PAR3 noncanonical TR47 and P3R peptides, the underlying mechanisms for cellular agonism seem to be different.
In summary, noncanonical cleavage of PAR3 by APC resulted in a tethered ligand sequence that is different and functionally distinct from that generated by thrombin. Although additional studies are needed to clarify the mechanisms for functional selectivity of canonical and noncanonical PAR3 agonist peptides, our data provide new insights into why and how thrombin- and APC-mediated PAR signaling are different and suggest a unique contributory role for PAR3 in the complex mechanisms underlying APC cytoprotective effects.
Supplementary Material
Acknowledgments
We are very grateful to Dr L. Brass (University of Pennsylvania, Philadelphia, PA) for the kind gift of the PAR1 and PAR3 antibodies, Dr C.J.S. Edgell (University of North Carolina, Chapel Hill, NC) for the EA.hy926 endothelial cells, and M. Mathias and D. Rozenshteyn for excellent technical assistance.
This work was supported by postdoctoral fellowships from the Swiss National Science Foundation, the Fondation Suisse pour les Bourses en Médecine et Biologie and Novartis PBGEP3-134242 and PASMP3_140065 (L.B.) and by National Heart Lung and Blood Institute, National Institutes of Health grants R00HL087618 and R01HL104165 (L.O.M.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.B. and L.O.M. performed experiments, designed the research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurent O. Mosnier, Department of Molecular and Experimental Medicine (MEM-180), The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: lmosnier@scripps.edu.
References
- 1.Esmon CT. The protein C pathway. Chest. 2003;124(3 Suppl):26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 2.Dahlbäck B, Villoutreix BO. The anticoagulant protein C pathway. FEBS Lett. 2005;579(15):3310–3316. doi: 10.1016/j.febslet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Mosnier LO, Zlokovic BV, Griffin JH. The cytoprotective protein C pathway. Blood. 2007;109(8):3161–3172. doi: 10.1182/blood-2006-09-003004. [DOI] [PubMed] [Google Scholar]
- 4.Rezaie AR. The occupancy of endothelial protein C receptor by its ligand modulates the par-1 dependent signaling specificity of coagulation proteases. IUBMB Life. 2011;63(6):390–396. doi: 10.1002/iub.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esmon CT. The endothelial protein C receptor. Curr Opin Hematol. 2006;13(5):382–385. doi: 10.1097/01.moh.0000239712.93662.35. [DOI] [PubMed] [Google Scholar]
- 6.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64(6):1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 7.Mosnier LO, Griffin JH. Inhibition of staurosporine-induced apoptosis of endothelial cells by activated protein C requires protease-activated receptor-1 and endothelial cell protein C receptor. Biochem J. 2003;373(Pt 1):65–70. doi: 10.1042/BJ20030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riewald M, Petrovan RJ, Donner A, Mueller BM, Ruf W. Activation of endothelial cell protease activated receptor 1 by the protein C pathway. Science. 2002;296(5574):1880–1882. doi: 10.1126/science.1071699. [DOI] [PubMed] [Google Scholar]
- 9.Mosnier LO, Gale AJ, Yegneswaran S, Griffin JH. Activated protein C variants with normal cytoprotective but reduced anticoagulant activity. Blood. 2004;104(6):1740–1744. doi: 10.1182/blood-2004-01-0110. [DOI] [PubMed] [Google Scholar]
- 10.Bae JS, Yang L, Manithody C, Rezaie AR. Engineering a disulfide bond to stabilize the calcium-binding loop of activated protein C eliminates its anticoagulant but not its protective signaling properties. J Biol Chem. 2007;282(12):9251–9259. doi: 10.1074/jbc.M610547200. [DOI] [PubMed] [Google Scholar]
- 11.Mosnier LO, Yang XV, Griffin JH. Activated protein C mutant with minimal anticoagulant activity, normal cytoprotective activity, and preservation of thrombin activable fibrinolysis inhibitor-dependent cytoprotective functions. J Biol Chem. 2007;282(45):33022–33033. doi: 10.1074/jbc.M705824200. [DOI] [PubMed] [Google Scholar]
- 12.Mosnier LO, Zampolli A, Kerschen EJ, et al. Hyperantithrombotic, noncytoprotective Glu149Ala-activated protein C mutant. Blood. 2009;113(23):5970–5978. doi: 10.1182/blood-2008-10-183327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerschen EJ, Fernandez JA, Cooley BC, et al. Endotoxemia and sepsis mortality reduction by non-anticoagulant activated protein C. J Exp Med. 2007;204(10):2439–2448. doi: 10.1084/jem.20070404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Ji Y, Zhang X, Drake M, Esmon CT. Endogenous activated protein C signaling is critical to protection of mice from lipopolysaccaride-induced septic shock. J Thromb Haemost. 2009;7(5):851–856. doi: 10.1111/j.1538-7836.2009.03333.x. [DOI] [PubMed] [Google Scholar]
- 15.Russo A, Soh UJ, Paing MM, Arora P, Trejo J. Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc Natl Acad Sci USA. 2009;106(15):6393–6397. doi: 10.1073/pnas.0810687106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae JS, Yang L, Rezaie AR. Receptors of the protein C activation and activated protein C signaling pathways are colocalized in lipid rafts of endothelial cells. Proc Natl Acad Sci USA. 2007;104(8):2867–2872. doi: 10.1073/pnas.0611493104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soh UJ, Trejo J. Activated protein C promotes protease-activated receptor-1 cytoprotective signaling through β-arrestin and dishevelled-2 scaffolds. Proc Natl Acad Sci USA. 2011;108(50):E1372–E1380. doi: 10.1073/pnas.1112482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407(6801):258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi-Matsui M, Zheng YW, Sulciner DJ, Weiss EJ, Ludeman MJ, Coughlin SR. PAR3 is a cofactor for PAR4 activation by thrombin. Nature. 2000;404(6778):609–613. doi: 10.1038/35007085. [DOI] [PubMed] [Google Scholar]
- 20.Hansen KK, Saifeddine M, Hollenberg MD. Tethered ligand-derived peptides of proteinase-activated receptor 3 (PAR3) activate PAR1 and PAR2 in Jurkat T cells. Immunology. 2004;112(2):183–190. doi: 10.1111/j.1365-2567.2004.01870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishihara H, Connolly AJ, Zeng D, et al. Protease-activated receptor 3 is a second thrombin receptor in humans. Nature. 1997;386(6624):502–506. doi: 10.1038/386502a0. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann R, Schulze B, Krause G, Mayr LM, Settmacher U, Henklein P. Proteinase-activated receptors (PARs): the PAR3 Neo-N-terminal peptide TFRGAP interacts with PAR1. Regul Pept. 2005;125(1-3):61–66. doi: 10.1016/j.regpep.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Liu D, Gelbard H, et al. Activated protein C prevents neuronal apoptosis via protease activated receptors 1 and 3. Neuron. 2004;41(4):563–572. doi: 10.1016/s0896-6273(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 24.Madhusudhan T, Wang H, Straub BK, et al. Cytoprotective signaling by activated protein C requires protease-activated receptor-3 in podocytes. Blood. 2012;119(3):874–883. doi: 10.1182/blood-2011-07-365973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanna MG, Wang SK, Gonzalez-Cabrera PJ, et al. Enhancement of capillary leakage and restoration of lymphocyte egress by a chiral S1P1 antagonist in vivo. Nat Chem Biol. 2006;2(8):434–441. doi: 10.1038/nchembio804. [DOI] [PubMed] [Google Scholar]
- 26.Miles AA, Miles EM. Vascular reactions to histamine, histamine-liberator and leukotaxine in the skin of guinea-pigs. J Physiol. 1952;118(2):228–257. doi: 10.1113/jphysiol.1952.sp004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liaw PCY, Mather T, Oganesyan N, Ferrell GL, Esmon CT. Identification of the protein C/activated protein C binding sites on the endothelial cell protein C receptor. Implications for a novel mode of ligand recognition by a major histocompatibility complex class 1-type receptor. J Biol Chem. 2001;276(11):8364–8370. doi: 10.1074/jbc.M010572200. [DOI] [PubMed] [Google Scholar]
- 28.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 29.Sen P, Sahoo S, Pendurthi UR, Rao LV. Zinc modulates the interaction of protein C and activated protein C with endothelial cell protein C receptor. J Biol Chem. 2010;285(26):20410–20420. doi: 10.1074/jbc.M110.111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosnier LO, Sinha RK, Burnier L, Bouwens EA, Griffin JH. Biased agonism of protease-activated receptor 1 by activated protein C caused by noncanonical cleavage at Arg46. Blood. 2012;120(26):5237–5246. doi: 10.1182/blood-2012-08-452169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cumashi A, Ansuini H, Celli N, et al. Neutrophil proteases can inactivate human PAR3 and abolish the co-receptor function of PAR3 on murine platelets. Thromb Haemost. 2001;85(3):533–538. [PubMed] [Google Scholar]
- 32.McLaughlin JN, Patterson MM, Malik AB. Protease-activated receptor-3 (PAR3) regulates PAR1 signaling by receptor dimerization. Proc Natl Acad Sci USA. 2007;104(13):5662–5667. doi: 10.1073/pnas.0700763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarborough RM, Naughton MA, Teng W, et al. Tethered ligand agonist peptides. Structural requirements for thrombin receptor activation reveal mechanism of proteolytic unmasking of agonist function. J Biol Chem. 1992;267(19):13146–13149. [PubMed] [Google Scholar]
- 34.Xu J, Zhang X, Pelayo R, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15(11):1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckers CM, van Hinsbergh VW, van Nieuw Amerongen GP. Driving Rho GTPase activity in endothelial cells regulates barrier integrity. Thromb Haemost. 2010;103(1):40–55. doi: 10.1160/TH09-06-0403. [DOI] [PubMed] [Google Scholar]
- 36.Bae JS, Yang L, Rezaie AR. Lipid raft localization regulates the cleavage specificity of protease activated receptor 1 in endothelial cells. J Thromb Haemost. 2008;6(6):954–961. doi: 10.1111/j.1538-7836.2008.02924.x. [DOI] [PubMed] [Google Scholar]
- 37.Berg DT, Gerlitz B, Shang J, et al. Engineering the proteolytic specificity of activated protein C improves its pharmacological properties. Proc Natl Acad Sci USA. 2003;100(8):4423–4428. doi: 10.1073/pnas.0736918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao C, Gao Y, Li Y, Antalis TM, Castellino FJ, Zhang L. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J Clin Invest. 2010;120(6):1971–1980. doi: 10.1172/JCI40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang XV, Banerjee Y, Fernández JA, et al. Activated protein C ligation of ApoER2 (LRP8) causes Dab1-dependent signaling in U937 cells. Proc Natl Acad Sci USA. 2009;106(1):274–279. doi: 10.1073/pnas.0807594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TC, Berny MA, Tucker EI, et al. Protein C supports platelet binding and activation under flow: role of glycoprotein Ib and apolipoprotein E receptor 2. J Thromb Haemost. 2008;6(6):995–1002. doi: 10.1111/j.1538-7836.2008.02979.x. [DOI] [PubMed] [Google Scholar]
- 41.Menschikowski M, Hagelgans A, Eisenhofer G, Siegert G. Regulation of endothelial protein C receptor shedding by cytokines is mediated through differential activation of MAP kinase signaling pathways. Exp Cell Res. 2009;315(15):2673–2682. doi: 10.1016/j.yexcr.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Sesin CA, Yin X, Esmon CT, Buyon JP, Clancy RM. Shedding of endothelial protein C receptor contributes to vasculopathy and renal injury in lupus: in vivo and in vitro evidence. Kidney Int. 2005;68(1):110–120. doi: 10.1111/j.1523-1755.2005.00385.x. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9(5):373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bae JS, Rezaie AR. Protease activated receptor 1 (PAR-1) activation by thrombin is protective in human pulmonary artery endothelial cells if endothelial protein C receptor is occupied by its natural ligand. Thromb Haemost. 2008;100(1):101–109. doi: 10.1160/TH08-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.