Yeasts and filamentous fungi can be true pathogens infecting healthy persons or they can be opportunistic pathogens that cause infections in humans with compromised immune systems due to cancer chemotherapy, organ transplantation treatment protocols, or HIV-1 infection, among other causes. Fungal infections can be characterized as systemic, indicating that the infection is deep, or localized cutaneous, meaning that the infection is superficial as it occurs on the skin, nails, hair or mucous membranes. Skin, nails, and hair infections by dermatophytes (Trichophyton sp., Microsporum sp., Epidermophyton sp.) and skin, nails, mucous membranes and systemic infections by Candida albicans and other Candida sp. may occur. Also, deep fungal infections caused by Histoplasma capsulatum (Histoplasmosis), Blastomyces dermatitidis (Blastomycosis), and Aspergillus fumigatus (Aspergillosis) can affect the lungs and other internal organ systems. Over the years, the number of opportunistic fungal infections has increased dramatically due to the increase in immune-compromised patients, the use of broad-spectrum antibiotics, and the availability of more invasive medical procedures resulting in an increase in prescriptions of antifungal drugs and the emergence of drug-resistant pathogenic strains. Fungal infections are, therefore, an important modern medical problem causing morbidity and mortality among hospitalized patients. The increasing frequency of infections caused by C. albicans, A. fumigatus and Cryptococcus neoformans has also led to an expansion in the number of studies on the use of antifungal drugs in clinical practice (1-2). In the quest for new antifungal drugs, the budding yeast Saccharomyces cerevisiae has served as a valuable model system because many aspects of its genetics, cell wall construction, and stress signaling are also seen among other pathogenic fungal species, in particular C. albicans, a major opportunistic pathogen (2). Despite remarkable similarities between many fungal and mammalian metabolic and signal transduction pathways, promising antifungal targets are currently under study focusing on cell wall biosynthesis, lipid composition of the plasma membrane, and synthesis of DNA and protein.
Because of the limited success of traditional drug screening strategies and the mainstreaming of high throughput molecular genetic technologies, the search for more potent, specific, and non-toxic antifungal therapies has moved towards a wider use of applications such as gene expression profiling, insertional mutagenesis and RNA mediated gene silencing for identification of novel target proteins (3). The availability of DNA-microarrays for genome wide analysis of gene expression has provided a very important tool for research on host-pathogen interactions and the analysis of expression profiles of pathogens exposed to different physiological conditions (4-5). Gene expression profiling studies are currently being applied to the major pathogens C. albicans, Cr. neoformans and A. fumigatus, which can cause severe infections among the immune-compromised populations. These studies have been conducted using four main experimental strategies: 1) treatment of cells with an antifungal drug before profiling to obtain a drug-specific gene expression signature, 2) expression profiling of fungal strains containing gene deletions in suspected target genes or transcription factors as a strategy to identify cellular targets playing a central role in the control of growth and metabolism; 3) comparison of global expression profiles of phenotypic variants of the same pathogenic strain as a strategy to identify expression patterns of genes associated with drug resistance and virulence, and 4) RNA isolation for direct profiling of the pathogen while infecting the host (3). These methods have been applied in studies of differential gene expression in oral Candidiasis (6), identification of differentially expressed genes of the ergosterol pathway after treatment with itraconazole, and the role of transcription factors associated with virulence in C. albicans (3, 7).
Insertion mutagenesis by genetic recombination has been frequently employed to generate important information through genetic analysis of null mutant strains. In C. albicans the essentiality of the RAM2 gene in the prenylation pathway was demonstrated using this technique (8) and transient loss of genes has enabled researchers to select essential gene targets required for cell viability (9). Using these many different variations of mutagenesis, more than 600 potentially essential genes have been identified for A. fumigatus. Though less widely used, gene silencing using antisense RNA or RNA interference followed by gene expression profiling, has also helped researchers identify critical genes required for growth in C. albicans (10) and the discovery of potential novel drug targets in Cr. neoformans and Aspergillus (11-12). Bioinformatic tools are of vital importance to these molecular genetic approaches for mining of fungal genome databases, analysis of gene expression, and sequence databases (3).
We will review areas of research that have shown promise for identification of new drug targets. For the purpose of this discussion we will organize these areas into four categories: i) Cell wall architecture, ii) Plasma membrane composition, iii) Cellular machinery, and iv) Signaling pathways. Where appropriate, we will describe our own recent efforts and contributions to this research being conducted in our laboratory.
i. Cell wall architecture
The cell wall is an essential structure for the fungal cell, which participates in multiple cellular processes such as nutrient transport, metabolism, and communication with the extracellular environment. It is the principal barrier to the surrounding environment because it controls cell turgor which maintains the normal osmolarity necessary for cellular biochemical reactions (2) and it sustains cell integrity that is essential for budding and cytokinesis (13-14). This structure is composed of glucans, chitin and mannoproteins. Since the cell wall is essential for fungal cell survival and because human cells do not have walls, this structure continues to be considered a principal target for the development of antifungal agents (2). Caspofungin, anidulafungin, and micafungin are echinocandins approved by the FDA in 2001 after several decades of intensive research (15-17). Echinocandins are natural cyclic lipopeptides that interfere with cell-wall synthesis by noncompetitive inhibition of β-1, 3-glucan synthase, an enzyme present in most pathogenic fungi (18). As a result, this inhibition leads to a depletion of cell wall glucan and its resultant osmotic instability. The echinocandin compounds LY 303366, MK 0991, and FK 463 are typically administered in combination with available standard antifungal agents because of their distinct mechanism of action (19). Another antifungal agent, nikkomycin Z (NZ), is a pyrimidine nucleoside linked to a peptide moiety that is a potent competitive inhibitor of chitin synthase III leading to inhibition of growth and osmotic fragility (19-20). Although the clinical importance of NZ is currently limited because high doses are required for effectiveness, we continue to explore its usefulness as a sensitizing agent for standard antifungal compounds. Our laboratory is currently analyzing the global expression profile of S. cerevisiae cells containing mutations that cause cell wall stress (21) in order to apply this knowledge in understanding the stress-response in clinically important fungi. Our approach involves genetic deletion of two components of the cytokinetic machinery, myosin II and chitin synthase II that cause cell wall stress and sensitize yeast cells to NZ (22). We have reported that sensitivity to NZ is also enhanced in a dose-dependent manner by co-treatment with the compound 2,3-Butanedione monoxime (BDM) (22), a disruptor of the cortical cytoskeleton in mammalian and fungal cells (23). As we will discuss later, the PKC1 signaling pathway is essential for cell survival under these conditions (21). Inhibitors of PKC1 or downstream effectors of this pathway may therefore have potential as sensitizing agents for standard antifungal agents.
ii. Plasma membrane composition
The plasma membrane is the permeability barrier of the cell composed mainly of sterols and phospholipids that determine its fluidity. Fungal plasma membranes are similar to mammalian plasma membranes in their structure except for having ergosterol rather than cholesterol as the principal sterol. Ergosterol is the site of interaction for polyene antibiotics while azoles interfere with ergosterol synthesis. The polyene antibiotics are produced by Streptomyces species and bind directly to ergosterol to form complexes that increase membrane permeability. As a result, there is a rapid leakage of cytoplasmic content including cellular potassium, other ions, and small molecules leading to the inhibition of glycolysis, respiration and cell death. Amphotericin B, nystatin, and pimaricin are polyene antibiotics widely used clinically because of their high affinity for ergosterol and relatively low affinity for its mammalian counterpart, thereby limiting their toxicity in mammalian cells (20). The azole antifungal agents are classified as imidazoles (ketoconazole and miconazole) or triazoles (itraconazole and fluconazole) as determined by the number of nitrogens (either two or three respectively) in the five-membered azole ring (19). Their mechanism of action consists in the inhibition of the ergosterol synthesis by interfering with the cytochrome P-450-dependent enzyme lanosterol demethylase (18-19). The resulting ergosterol depletion leads to plasma membrane permeability changes and inhibition of growth. A limitation of the azole antifungal drugs is their frequent interactions with co-administered drugs, which may result in adverse clinical consequences (24).
Besides targeting the ergosterol in the membrane, current efforts are also focusing on enzymes involved in the biosynthesis of phospholipids or sphingolipids that are exclusive of fungi (25). Fungal sphingolipids synthesized from phytoceramide are essential for growth and cell viability. Although the metabolism of sphingolipids is similar among fungi and animals, certain enzymes involved in sphingolipids synthesis are considered potential antifungal drug targets due to specific differences with their mammalian counterparts (1). Serine palmitoyltransferase is an enzyme that catalyzes the committed step in sphingolipid biosynthesis conserved from fungi to humans. Inhibition of this enzyme by the natural compounds myriosins, sphingofungins, lipoxamycins, viridiofungins, and the non-natural product cycloserine block the production of all sphingolipids resulting in fungal cell death (1). Inositol phosphorylceramide synthase (IPC-synthase) is the enzyme catalyzing the first fungus-specific step in sphingolipids synthesis and is therefore an ideal target of antifungal drugs (25). There are several natural compounds described that inhibit the activity of this enzyme, including aureobasidin A, khafrefungin, and rustmycin (1). Moreover, the AUR1 gene of S. cerevisiae encodes a protein that is necessary for IPC synthase activity and confers resistance to the antifungal drug aureobasidin-A when mutated (26). Aur1 protein may represent a new target for screening for IPC synthase inhibitor compounds.
a. Novel fatty acids
Though not yet proven to interact with the plasma membrane, novel fatty acids derived from marine sponges have been found to be fungitoxic, either by plasma membrane disruption that eventually disintegrates the cell (27) or by inhibiting fatty acids biosynthesis (28). Acetylenic fatty acids (8-16 carbons) are one of the most promising groups of fatty acids to exert antifungal effects (29), while α-methoxylated fatty acids are less toxic (30). These represent a group of compounds with antifungal drug-sensitizing properties that will be investigated in order to elucidate their mechanism of action.
iii. Cellular machinery
Components of the cellular machinery have been proposed as potential drug targets in fungi. Specific examples are topoisomerases that belong to the nuclear DNA replication and RNA transcription machinery, where they are key components in DNA-RNA synthesis. Topoisomerases are enzymes that act in several biochemical processes such as chromosome replication, transcription, recombination, and chromosome segregation by controlling the topological state of DNA (31). Camptothecin, a topoisomerase-specific inhibitor, stabilizes topoisomerase/DNA complexes, leading to DNA damage and cell death (31). Several studies have demonstrated that Topoisomerase 1 (TOP1) is essential for life and is a virulence factor for some fungi (32). Its deletion in C. albicans induces slow cellular growth and aberrant cell morphology (32). The fungal TOP1 gene has a considerable amount of coding sequence not present in human homologs (33) suggesting differences in their protein structure and presenting the possibility of exploring these topoisomerases as drug targets.
The elongation factors required for mRNA translation in eukaryotes are promising targets for antimycotic drugs. For example, the elongation factor III (EF3) is unique to fungi and is essential for protein synthesis (31). Sordarin and sordarin-like compounds act by blocking the protein elongation cycle at the initial steps of ribosomal translocation, prior to GTP hydrolysis. They have in vitro activity against a wide range of pathogenic fungi, including Aspergillus spp, Candida spp, Cr. neoformans, other filamentous fungi, (34-35) and Saccharomyces cerevisiae, making them promising antimycotic agents.
Pyrimidine analogs also have been used as antifungal agents. The fluoropyrimidine 5-fuorocytosine (5-FC) is an FDA-approved drug in this class although it has a limited spectrum of activity and significant potential for toxic effects (36-37). The mechanism of action of 5-FC involves intracellular deamination to 5-fuorouracil followed by its incorporation into RNA that causes miscoding and blocking of RNA and DNA biosynthesis (20).
The ubiquitin (Ub) system is involved in protein degradation in eukaryotic cells including fungi. It was reported that ubiquitins (Ubs) in the fungus Aspergillus nidulans were strongly expressed in the presence of antifungal drugs like amphotericin B and miconazole (38). Another study suggested that the expression of Ub genes in fungi might be enhanced by different kinds of antifungal agents, correlating with resistance to drugs (39). Investigators have suggested that Ub might be associated with different receptor-like components at the cell surface and these could be related to antifungal drug resistance (39-40). We have accumulated substantial experimental evidence to confirm that resistance to nikkomycin Z can be correlated to an elevated expression of the ubiquitin conjugating enzyme UBC4 and the ubiquitin gene UBI4 (41) while the actual mechanism for this resistance is not defined.
Sumoylation is a post-translational modification pathway composed of SUMO (Small Ubiquitin-like Modifiers) enzymes that regulate diverse cellular processes such as intracellular trafficking, cell cycle, DNA repair and replication, RNA metabolism, and cell signaling through its reversible and covalent attachment to target proteins (42). Among the SUMO substrates are a large number of regulators of gene expression, particularly transcription factors, co-activators or repressors (43). An analysis of SUMO-mediated stress response in S. cerevisiae revealed that ethanol stress increases sumoylation while affecting cell growth (44). Further studies on the regulation of this pathway in pathogenic conditions can offer insights into sumoylation as a potential drug target.
Lindquist and colleagues have established that the molecular chaperone Hsp90 enables the emergence and maintenance of fungal drug resistance (45). For Candida albicans, Hsp90 mediates resistance to azoles while for Aspergillus terreus; Hsp90 is required for basal resistance to echinocandins. These investigators employed combination therapy with an Hsp90 inhibitor successfully as an experimental therapeutic strategy against these fungi in model systems. They have concluded from this work that the use of Hsp90 inhibitors “enhances the efficacy of existing antifungals, blocks the emergence of drug resistance, and exerts broad-spectrum activity against diverse fungal pathogens.”
iv. Signaling pathways
a. PKC1-dependent cell wall integrity pathway (CWIP)
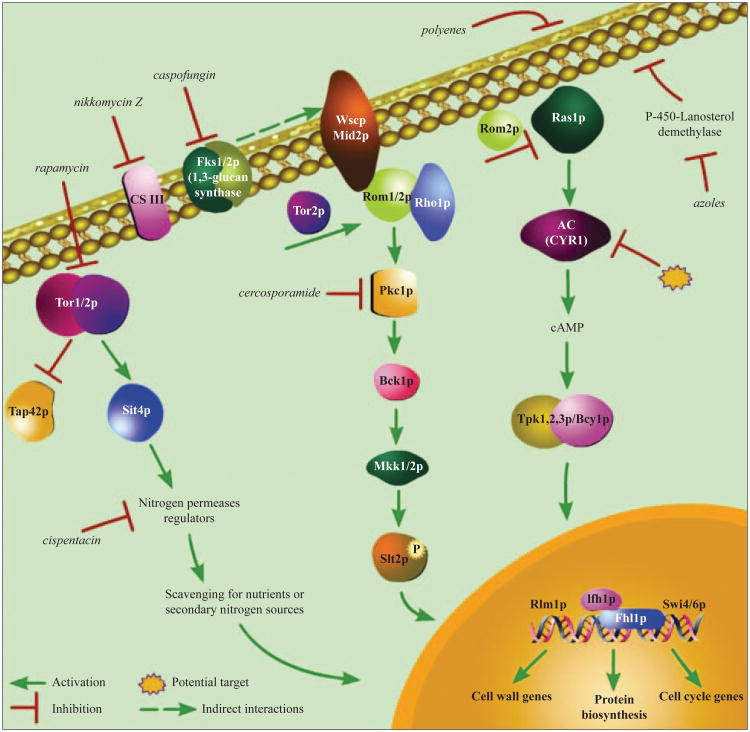
Integrity of the cell wall during fungal growth is essential for survival and adaptation to environmental stresses. Although several signaling pathways may contribute to the maintenance of cellular integrity, the CWIP is considered essential for sensing cell wall perturbations under many cell stress conditions. The signaling activity of this pathway is initiated at the cell surface through sensor-transducer proteins and is transmitted to downstream effectors through Rom2p-Rho1p complexes culminating with the activation of the MAPK/Slt2p. A family of sensors constituted by members of the Wsc-family, Mid2p, and its homologue Mtl1p, are localized upstream of the MAP kinase cascade (46). This signaling cascade is a linear pathway comprised of calcium-dependent protein kinase (PKC1), a MEKK (BCK1), a pair of redundant MEKs (MKK1/2) and a MAP kinase (MPK1/SLT2) (Figure 1) (2). The targets of Mpk1p include two transcription factors, Rlm1p and Swi4/6p, involved in the regulation of multiple cell wall components and cell cycle genes respectively.
Figure 1.
The stress-response signaling pathways in Saccharomyces cerevisiae contain many targets for current and future antifungal agents. Inhibitors for specific components of these signal transduction cascades are shown in italics.
Antifungal drugs such as caspofungin, block cell wall synthesis by inhibiting β-1, 3-glucan synthases (47). Synergistic effects are observed in vivo and in vitro when this echinocandin is combined with other antifungal drugs such as fluconazole or amphotericin B (48). Research has shown that caspofungin activates the PKC1 pathway leading to Slt2p phosphorylation, and has identified Wsc1p cell surface protein as the sensor for the stress induced by caspofungin (46). Like the inactivation of β-1, 3-glucan synthases, a PKC1 deletion mutant causes loss of osmotic integrity (49). The antifungal natural product cercosporamide is a highly selective and potent PKC1 inhibitor (50) that in combination with caspofungin has a synergistic effect against the opportunistic pathogen C. albicans, as well as against budding yeast S. cerevisiae. In our own studies with S. cerevisiae, we have found that loss of myosin type ii function by genetic deletion (myo1Δ) activates the PKC1 pathway. Function of the PKC1 pathway was shown to be essential because a null mutation in SLT2, the final MAP kinase effector of this pathway, exhibited a synthetic lethality phenotype in a myo1Δ strain (21). In this light, one of our current working hypotheses is that inhibition of myosin II function can sensitize yeast cells to antifungal compounds that interfere with cell wall biogenesis like caspofungin and nikkomycin Z.
b. TOR signaling pathway
The targets of rapamycin (TOR) proteins are members of a ubiquitous family of signaling proteins. These proteins have essential conserved roles in transducing signals in response to exogenous or endogenous cellular stimuli (1). TOR proteins were discovered by mutations that control cell growth by conferring resistance to the growth inhibitory effect of the potent antifungal agent rapamycin. Since this drug interferes with TOR signaling in eukaryotic cells, it has been used as a chemical probe to delineate TOR-dependent responses in these cells (51). Rapamycin is toxic to some pathogenic yeast and fungi, including C. albicans, Cr. neoformans, and A. fumigatus (52). In S. cerevisiae, rapamycin inhibits the TOR pathway by disrupting the stable complex formed between the phosphatase regulator Tap42p and the protein phosphatase Sit4p which results in cell cycle arrest and antiproliferative effects (53), downregulation of translation initiation (54), and repression of ribosome biogenesis (Figure 1) (55). Our ongoing studies have shown that myo1Δ cells are hypersensitive to the inhibitory effects of rapamycin suggesting that combined defects in cell wall biogenesis and cytokinesis can sensitize cells to this drug. The amino acid analog cispentacin that inhibits homoserine dehydrogenase (56) has been considered for use as an inhibitor of amino acid biosynthesis in fungi. A novel use of this compound may provide a strategy to inactivate the TOR pathway in fungal cells by inducing nutrient starvation while avoiding the immunosuppressive action of rapamycin observed in mammalian model organisms (Figure 1).
c. PKA signaling pathway
The signaling cascade mediated by cyclic AMP (cAMP)-dependent protein kinase A (PKA) is involved in the regulation of virulence, morphogenesis, and development in various fungi (57). This pathway is composed of Ras proteins (encoded by RAS1 and RAS2 GTPases) that consecutively activate adenylate cyclase (encoded by CDC35 in C. albicans and CYR1 in S. cerevisiae), which generates cAMP resulting in PKA activation (58). The regulatory subunit of PKA is encoded by BCY1 while the PKA catalytic subunit is encoded by two functionally redundant genes TPK1 and TPK2 (58). The PKA pathway was also implicated in fungal resistance to certain antibiotics, including polymyxin B, dicarboximide, and azoles (59). CDC35 and CYR1 mutations demonstrated a hyper-susceptibility to azole and other sterol biosynthesis inhibitors (59). Down regulation of the adenylate cyclase regulator RAS1 was observed in myosin II-deficient (myo1Δ) cells (21). We are currently analyzing the correlation between hypersensitivity to nikkomycin Z and rapamycin, and the activation status of the RAS/PKA signaling pathway in this mutant.
Conclusions
Development of new antifungal strategies requires detailed knowledge of the fundamental controls governing fungal cell division, intracellular signaling, and growth. We have discussed diverse approaches used to develop new compounds into viable drugs yet the mechanisms of action of many compounds with development potential remain undetermined. The use of non-pathogenic model systems such as Saccharomyces cerevisiae for such studies can contribute a great deal to our knowledge of basic fungal biology which can then be extrapolated to clinically relevant fungi and accelerate the development of translational applications.
Acknowledgments
José R. Rodríguez-Medina is supported by a PHS grant from NIGMS/NIAID (SC1AI081658-01) with partial support from NCRR-RCMI (G12RR03051). Marielis E. Rivera-Ruiz and Glorivee Pagán-Mercado receive support from MBRS-RISE (R25GM061838). Frances Segarra-Román is supported by institutional funds from the UPR Medical Sciences Campus.
Footnotes
The authors have no conflict of interest to disclose.
References
- 1.Cardenas ME, Cruz MC, Del Poeta M, Chung N, et al. Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin Microbiol Rev. 1999;12:583–611. doi: 10.1128/cmr.12.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Backer MD, Van Dijck P. Progress in functional genomics approaches to antifungal drug target discovery. Trends Microbiol. 2003;11:470–478. doi: 10.1016/j.tim.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Murad AM, d'Enfert C, Gaillardin C, Tournu H, et al. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors caTup1, caMig1 and caNrg1. Mol Microbiol. 2001;42:981–993. doi: 10.1046/j.1365-2958.2001.02713.x. [DOI] [PubMed] [Google Scholar]
- 5.Kato-Maeda M, Gao Q, Smal PM. Microarray analysis of pathogens and their interaction with hosts. Cell Microbiol. 2001;3:713–719. doi: 10.1046/j.1462-5822.2001.00152.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhao XJ, Newsome TJ, Cihlar R. Up-regulation of two candida albicans genes in the rat model of oral candidiasis detected by differential display. Microb Pathog. 1998;25:121–129. doi: 10.1006/mpat.1998.0218. [DOI] [PubMed] [Google Scholar]
- 7.De Backer MD, Ilyina T, Jun-Xiao M, Vandoninck S, et al. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob Agents Chemother. 2001;45:1660–1670. doi: 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song JL, White TC. RAM2: an essential gene in the prenylation pathway of candida albicans. Microbiology. 2003;149:249–259. doi: 10.1099/mic.0.25887-0. [DOI] [PubMed] [Google Scholar]
- 9.Michel S, Ushinsky S, Klebl B, et al. Generation of conditional lethal candida albicans mutants by inducible deletion of essential genes. Mol Microbiol. 2002;46:269–280. doi: 10.1046/j.1365-2958.2002.03167.x. [DOI] [PubMed] [Google Scholar]
- 10.De Backer MD, Nelissen B, Logghe M, Viane J, et al. An antisense-based functional genomics approach for identification of genes critical for growth of candida albicans. Nat Biotechnol. 2001;19:235–241. doi: 10.1038/85677. [DOI] [PubMed] [Google Scholar]
- 11.Gorlach JM, Mcdade HC, Perfect J, Cox GM. Antisense repression in Cryptococcus neoformans as a laboratory tool and potential antifungal strategy. Microbiology. 2002;148:213–219. doi: 10.1099/00221287-148-1-213. [DOI] [PubMed] [Google Scholar]
- 12.Bautista LF, Aleksenko A, Hentzer M, Santerre-Henriksen A, Nielsen J. Antisense silencing of the creA gene in Aspergillus nidulans. Appl environ Microbiol. 2000;66:4579–4581. doi: 10.1128/aem.66.10.4579-4581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cid VJ, Duran A, Rey F, Snyder MP, et al. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 15.Arikan S, Rex JH. New agents for the treatment of systemic fungal infections--current status. Expert Opin Emerg Drugs. 2002;7:3–32. doi: 10.1517/14728214.7.1.3. [DOI] [PubMed] [Google Scholar]
- 16.Hossain MA, Ghannoum MA. New investigational antifungal agents for treating invasive fungal infections. Expert Opin Investig Drugs. 2000;9:1797–1813. doi: 10.1517/13543784.9.8.1797. [DOI] [PubMed] [Google Scholar]
- 17.Zaas AK, Steinbach WJ. Micafungin: the US perspective. Expert Rev Anti Infect Ther. 2005;3:183–190. doi: 10.1586/14787210.3.2.183. [DOI] [PubMed] [Google Scholar]
- 18.Denning DW. Echinocandins and pneumocandins- a new antifungal class with a novel mode of action. J Antimicrob chemoter. 1997;40:611–614. doi: 10.1093/jac/40.5.611. [DOI] [PubMed] [Google Scholar]
- 19.Chiou CC, Groll AH, Walsh T. New drugs and novel targets for treatment of invasive fungal infections in patients with cancer. Oncologist. 2000;5:120–135. doi: 10.1634/theoncologist.5-2-120. [DOI] [PubMed] [Google Scholar]
- 20.Georgopapadakou NH, Walsh TJ. Human mycoses: Drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Quiñones JF, Irizarry R, Díaz-Blanco N, Rivera-Molina FE, et al. Global mRNA expression analysis in myosin II deficient strains of Saccharomyces cerevisiae reveals an impairment of cell integrity function. BMc Genomics. 2008;9:34. doi: 10.1186/1471-2164-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivera-Molina FE, Gónzalez S, Maldonado Y, Ortiz J, et al. 2,3-Butanedione monoxime increase sensitivity to Nikkomycin Z in the budding yeast S.cerevisiae. World J Microbio Biotechnol. 2006;22:255–260. doi: 10.1007/s11274-005-9028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chon K, Hwang HS, Lee JH, Song K. The myosin ATPase inhibitor 2,3-butanedione-2 monoxime disorganizes microtubules as well F-actin in S.cerevisiae. Cell Biol Toxicol. 2001;17:383–393. doi: 10.1023/a:1013748500662. [DOI] [PubMed] [Google Scholar]
- 24.Albengres E, Louet H, Tillement JP. Drug interactions of systemic antifungal agents. Drug Saf. 1998;18:83–97. doi: 10.2165/00002018-199818020-00001. [DOI] [PubMed] [Google Scholar]
- 25.Mercer EI. Inhibitors of sterol biosynthesis and their applications. Prog Lipid Res. 1993;32:357–416. doi: 10.1016/0163-7827(93)90016-p. [DOI] [PubMed] [Google Scholar]
- 26.Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, et al. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AuR1 gene. J Biol chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 27.Avis TJ, Bélanger RR. Specificity and mode of action of the antifungal fatty acid cis-9-heptadecenoic acid produced by Pseudozyma focculosa. Appl Environ Microbiol. 2001;67:956–960. doi: 10.1128/AEM.67.2.956-960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood R, Lee T. Metabolism of 2-hexadecynoate and inhibition of fatty acid elongation. J Biol chem. 1981;256:12379–12386. [PubMed] [Google Scholar]
- 29.Gershon H, Shanks L. Antifungal properties of 2-alkynoic acids and their methyl esters. Can J Microbiol. 1978;24:593–597. doi: 10.1139/m78-096. [DOI] [PubMed] [Google Scholar]
- 30.Carballeira NM. New advances in fatty acids as antimalarial, antimycobacterial and antifungal agents. Prog lipid Res. 2008;47:50–61. doi: 10.1016/j.plipres.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein RJ. Topoisomerases in human disease. Lancet. 1988;1:521–524. doi: 10.1016/s0140-6736(88)91308-6. [DOI] [PubMed] [Google Scholar]
- 32.Fostel JM, Montgomery DA, Shen LL. Characterization of DNA topoisomerase I from candida albicans as a target for drug discovery. Antimicrob Agents chemother. 1992;36:2131–2138. doi: 10.1128/aac.36.10.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart L, Ireton GC, Champoux JJ. The domain arrangement of human topoisomerase I. J Biol chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 34.Andriole VT. Current and future antifungal therapy: new targets for antifungal therapy. Int J Antimicrob Agents. 2000;16:317–321. doi: 10.1016/s0924-8579(00)00258-2. [DOI] [PubMed] [Google Scholar]
- 35.Herreros E, Martínez CM, Almela MJ, Marriott MS, et al. Sordarins: In Vitro Activities of New Antifungal Derivatives against Pathogenic Yeasts, Pneumocystis carinii, and Filamentous Fungi. Antimicrob Agents Chemother. 1998;42:2863–2869. doi: 10.1128/aac.42.11.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennet JE. Flucytosine. Ann Intern Med. 1977;86:319–321. doi: 10.7326/0003-4819-86-3-319. [DOI] [PubMed] [Google Scholar]
- 37.Francis P, Walsh TJ. Evolving role of fucytosine in immunocompromised patients: new insights into safety, pharmacokinetics, and antifungal therapy. Clin Infect Dis. 1992;15:1003–1018. doi: 10.1093/clind/15.6.1003. [DOI] [PubMed] [Google Scholar]
- 38.Noventa-Jordao AM, Nascimento AM, Goldman MHS, Terenzi HF, et al. Molecular characterization of ubiquitin genes from Aspergillus nidulans: mRNA expression on different stress and growth conditions. Biochim Biophys Acta. 2000;1490:237–244. doi: 10.1016/s0167-4781(99)00242-0. [DOI] [PubMed] [Google Scholar]
- 39.Kano R, Okabayashi K, Nakamura Y, Watanabe S, et al. Expression of Ubiquitin Gene in Microsporum canis and Trichophyton mentagrophytes Cultured with Fluconazole. Antimicrob Agents Chemother. 2001;45:2559–2562. doi: 10.1128/AAC.45.9.2559-2562.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sepulveda P, Lopez-Ribot JL, Gozalbo D, Cervera JP, et al. Ubiquitin-like epitopes associated with Candida albicans cell surface receptors. Infect Immun. 1996;64:4406–4408. doi: 10.1128/iai.64.10.4406-4408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Díaz-Blanco N, Rodríguez-Medina JR. Dosage rescue by UBC4 restores cell wall integrity in Saccharomyces cerevisiae lacking the myosin type II gene MYO1. Yeast. 2007;24:343–355. doi: 10.1002/yea.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:1–8. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ro K, Chattopadhyay A, Wohlschlegel J. Analysis of SUMO-mediated Stress Response in Saccharomyces cerevisiae. ASCB. 2008 Abstract #461 (CD-ROM) [Google Scholar]
- 45.Cowen LE, Singh SD, Köhler JR, Collins C, Zaas AK, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutics strategy for fungal infectious disease. Proc Natl Acad Sci U S A. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinoso-Martín C, Schüler C, Schuetzer-Muehlbauer M, Kuchler K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungi through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot Cell. 2003;2:1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz MB, Douglas CM. Lipopeptide inhibitors of fungal glucan synthase. J Med Vet Mycol. 1997;35:79–86. doi: 10.1080/02681219780000961. [DOI] [PubMed] [Google Scholar]
- 48.Stone EA, Fung HB, Abe M, Saka A, Watanabe D, et al. Caspofungi: an echinocandin antifungal agent. Clin Ther. 2002;24:351–377. doi: 10.1016/s0149-2918(02)85039-1. [DOI] [PubMed] [Google Scholar]
- 49.Levin DE, Bartlett-Heubusch E. Mutants in the S. Cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sussman A, Huss K, Chio LC, Heidler S, Shaw M, et al. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through High-Throughput screening. Eukaryot Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edinger AL, Linardic CM, Chiang GG, Thompson CB, et al. Differential effects of rapamycin on mammalian targets of Rapamycin signaling functions in mammalian cells. Cancer Res. 2003;63:8451–8460. [PubMed] [Google Scholar]
- 52.Cruz MC, Cavallo LM, Görlach JM, et al. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutton A, Immanuel D, Arndt KT. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulates the association of Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cervisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capobianco JO, Zakula D, Coen ML, Goldman RC. Anti Candida activity of cispentacin: The active transport by aminoacid permease and possible mechanisms of action. Biochem Biophys Res Commun. 1993;190:1037–1044. doi: 10.1006/bbrc.1993.1153. [DOI] [PubMed] [Google Scholar]
- 57.Bahn SY, Sundstrom P. CAP1, an Adenylate Cyclase-Associated Protein Gene, Regulates Bud-Hypha Transitions, Filamentous Growth, and Cyclic AMP levels and Is Required for Virulence of Candida albicans. J Bacteriol. 2001;10:3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/Cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain P, Akula I, Edlind T. Cyclic AMP signaling pathway modulates susceptibility of candida species and Saccharomyces cerevisiae to antifungal azoles and other sterol biosynthesis inhibitors. Antimicrob Agents chemother. 2003;47:3195–3201. doi: 10.1128/AAC.47.10.3195-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]