Abstract
The number of membrane protein structures in the Protein Data Bank is becoming significant and growing. Here, the transmembrane domain structures of the helical membrane proteins are evaluated to assess the influences of the membrane mimetic environments. Toward this goal, many of the biophysical properties of membranes are discussed and contrasted with those of the membrane mimetics commonly used for structure determination. Although the mimetic environments can perturb the protein structures to an extent that potentially gives rise to misinterpretation of functional mechanisms, there are also many structures that have a native-like appearance. From this assessment, an initial set of guidelines is proposed for distinguishing native-like from nonnative-like membrane protein structures. With experimental techniques for validation and computational methods for refinement and quality assessment and enhancement, there are good prospects for achieving native-like structures for these very important proteins.
Keywords: structure determination, membrane biophysical properties, structural perturbations, bilayer and detergent environments, native structures, structural enhancement
SCOPE AND MOTIVATION
The solubilizing environments of membrane proteins can influence their structures. All membrane protein structures, except bacteriorhodopsin (33), have been characterized in nonnative, membrane mimetic environments. Here, we survey the bitopic oligomeric and polytopic α-helical membrane protein structures from the Protein Data Bank (PDB) in an attempt to assess and explain the influences of the membrane mimetic environments on protein structures and hence the perturbations from native-like structures. Nearly four decades ago Anfinsen (3) described the structure of a protein as resulting from the amino acid sequence in a given environment, dictated by interactions both within the structure and between the protein and its environment. We describe why this latter set of interactions can become so significant for membrane proteins. Of course, for very large membrane protein complexes the ratio of internal interactions to interactions with the environment becomes large, and consequently the influence of the environment is somewhat diminished. We consider both large and small membrane protein structures but exclude from this survey those proteins with multiple internal ligands, which can provide additional tertiary structure stabilization. Furthermore, this survey is limited to the transmembrane (TM) domains of these proteins and does not deal with the influence of the membrane mimetic environments on the water-soluble loops and domains of these proteins.
This effort was motivated by a comparison of structures from various constructs of the influenza A M2 protein determined by solid-state and solution NMR spectroscopy as well as X-ray crystallography. These structures, obtained from liquid crystalline lipid bilayers, detergent micelles, and detergent-based crystals, showed significant differences in helical packing (16, 18). The M2 protein as a tetramer forms an acid-activated and proton-selective channel and is a proven drug target (76). The TM domain alone, consisting of a single TM helix from each monomer, forms an ion channel that is blocked by the drugs amantadine and rimantadine. The conductance properties are identical to those of the full-length protein when an amphipathic helix just after the TM helix is included in the construct (known as the conductance domain) (58, 85). Some of the differences among the M2 structures, illustrated in Figure 1, have been proposed as representing different functional states. Yet closer scrutiny of the structures and the sample conditions suggests that the sample conditions, specifically the membrane mimetics, appear to be responsible for the structural differences. The observation that the membrane mimetic environment significantly perturbs the M2 structure and that such perturbations can be interpreted as representing functional states was the motivation for our examination of other membrane protein structures in the PDB.
Figure 1.
Comparison of M2 structures determined in different membrane mimetic environments. (a) Two detergent-based crystal structures of the transmembrane domain, 3BKD (pink) (91) and 3LBW (cyan; carrying the G34A mutation) (2), superimposed to 2L0J (green) (85), the solid-state NMR structure of the conductance domain in lipid bilayers. (b) Two micelle-based solution NMR structures of the conductance domain, 2RLF (yellow) (81) and 2KWX (blue; carrying the V27A mutation) (75), superimposed to 2L0J.
Our aim is to identify native-like versus nonnative-like helical membrane protein structures and to further our understanding of the environmental influences on these structures. This task is complicated by the fact that the membrane protein structure and the mimetic environment mutually influence each other, with the result that each protein structure is stable in its own mimetic environment. Moreover, many membrane proteins, ion channels (including the M2 protein) in particular, cycle through several functional states, so that there may be not one but several native structures for each protein. To proceed, we start with a description of the biophysical properties of membrane environments that are expected to be important for preserving native-like structures. On the basis of these properties we propose a set of qualitative guidelines for identifying native-like versus nonnative-like membrane protein structures. The goal is not so much to ascertain whether these structures represent functional states as to assess whether they are compatible with native membrane environments. We find that a significant number of structures appear to be perturbed by their membrane mimetic environments, but more importantly we can identify in many cases why the perturbations have taken place. We recognize that even nonnative-like structures can be valuable, but hope that the insight gained here helps avoid the overinterpretation of structures. We propose that validation of TM domain structures can be achieved with limited distance/orientation restraints from techniques that probe the proteins in a lipid bilayer environment and that computational modeling in a native-like environment can potentially correct some of the structural perturbations. We also emphasize that all the structure determination techniques have led to some excellent native-like structures.
MEMBRANE PROTEINS AND THEIR MEMBRANE ENVIRONMENTS
Biophysical Properties of the Membrane Environments
The amphipathic nature of lipids in an aqueous solution results in their assembly into bilayers. Native membranes have a very broad range of lipids (87). The inner and outer leaflets of the membrane often have substantially different lipid compositions. Recently, Sanders & Mittendorf (80) have argued convincingly that specific membrane proteins function in a wide variety of lipid environments during the lifetime of a cell and that significant variations in lipid compositions may not have major functional consequences. Here, we focus on the general biophysical properties of cell membranes as opposed to the lipid composition. Different membranes have somewhat different hydrophobic thicknesses. Mitochondrial inner membranes are somewhat thinner than plasma membranes. Raft-like domains that are high in cholesterol and sphingomyelin are substantially thicker and more hydrophobic with less aqueous permeation (87). Such substantial differences in membrane biophysical properties are known to influence membrane protein function. The influenza M2 protein functions as a proton channel in the viral envelope, which is high in sphingomyelin and cholesterol. Yet synthetic membranes with high concentrations of these lipids do not support drug binding or the unique chemistry of the His37 tetrad critical for proton conductance (7). Recently, it has been shown that M2 is localized at the cholesterol-poor periphery of raft-like domains during viral budding (79, 82); this localization presumably is preserved in mature virus particles and obviously allows for drug binding. In the synthetic cholesterol-rich membranes, the thicker hydrophobic dimension could induce a reduced tilt of the TM helices and hence a reduced size of the opening into the proton conductance pore, thereby preventing drug binding in the pore. Possibly a similar situation occurs in dihexanoyl phosphatidylcholine (DHPC) micelles (2RLF; Figure 1b), where the helical tilt was observed to be smaller and the drug did not bind in the pore (81), counter to the observations in liquid crystalline lipid bilayers (85).
As a result of the amphipathic nature of the membrane, there occurs a dramatic change in dielectric constant across the membrane, which first increases from ~80 (at 25°C) in the aqueous phase to a value two- to threefold higher in the charged lipid headgroup region, and then decreases sharply to ~4–5 in the lipid interfacial region and finally down to ~2 in the hydrophobic core of the lipid bilayer (69, 90). In a palmitoyloleoyl phosphatidylcholine bilayer the last very low dielectric region spans approximately 24 Å, whereas the last two low dielectric regions together extend to 34 Å. The two orders of magnitude variation in dielectric constant across the membrane has a profound effect on the magnitude and distance dependence of electrostatic interactions. In particular, near the bilayer center significant electrostatic interactions are difficult to break without a catalyst (101). This is the primary reason why helices are formed in the headgroup region and then inserted across the membrane (78); the polypeptide has little chance of recouping from any nonnative hydrogen bonds formed in the hydrophobic core of the membrane. In addition, across membranes the water concentration varies over many orders of magnitude, with an especially large gradient when significant concentrations of cholesterol, sphingomyelin, and methylated acyl chains are present (26, 62, 86). The combination of the lack of water in the hydrophobic core and the low dielectric constant suggests that both hydrogen bond exchange and amide hydrogen/deuterium (H/D) exchange are rare in such an environment.
The amphipathic nature of lipid molecules also means that, across the membrane, different types of forces are at play in stabilizing the bilayer structure. While the headgroups form multiple hydrogen bonds with water molecules, a barrier to prevent water penetration is formed in the lipid inter-facial region. In the hydrophobic core, the dry acyl chains tend to stay away from each other in order to maximize chain entropy. Consequently, a lateral pressure profile is created, with negative lateral pressure in the lipid interfacial region and positive lateral pressure in the hydrophobic core (10, 34).
A feature that distinguishes phospholipids from other amphiphiles is the former's extremely low critical concentration for self-assembly. This concentration is of the order of nanomolar, meaning that when lipids assemble into bilayers, monomeric lipids are present in the aqueous phase only in the nanomolar range (12). In contrast, the critical concentration for detergents to assemble into micelles is typically above 1 mM. The significantly higher monomer concentration could increase the chance of monomeric detergents being inserted into membrane protein structures.
Differences Between Membrane and Water-Soluble Proteins
As a result of strengthened electrostatic interactions in the membrane interstices, the intrahelical hydrogen bonds are shorter (4, 47) and there is much less competition for the backbone amide and carbonyl sites from water and hydrophilic side chains. In addition, the average torsion angles are shifted such that the peptide planes are closer to being parallel to the helical axis (8° tilt) compared with those that are typical of water soluble helices (12° tilt), so as to minimize exposure of the backbone polar groups (47, 72). Consequently, TM helices generally adhere more closely to ideal geometry and have significantly enhanced stability.
The most dramatic difference in amino acid composition between TM helices and water-soluble helices is the reduction in charged and highly polar (His, Gln, and Asn) residues, typically by a factor of 3 or 4 (23). Furthermore, when present in TM helices they are most often at the termini of the helices where the side chains interact with the lipid headgroups (Figure 2a,b). The hydrophobic dimension of helical membrane proteins is typically 25–30 Å, although it appears that there can be situations where it is thinned to somewhat less than 25Å (89). Weakly polar Ser and Thr side chains are sometimes found on the hydrophobic surface of these membrane proteins, probably because they can hydrogen-bond back to the backbone of the helix, thereby concealing most of the hydrophilic group from the lipid acyl chains (Figure 2c). These residues could be used in alternative functional states to form interhelical hydrogen bonds. Their propensity in TM helices appears to be somewhat greater than that of their water-soluble counterparts. The overall result of the reduced prevalence of charged and polar residues is a reduction in tertiary and quaternary stability for the TM region. The predominance of hydrocarbon side chains suggests that nonspecific van der Waals and at-a-distance electrostatic interactions are largely responsible for the tertiary and quaternary structures of these proteins. Of course the occasional hydrogen bonds and close-range electrostatic interactions between charged and polar side chains can also be important, but they are much scarcer. Such interactions may be most frequently observed when they are preserved across functional states.
Figure 2.
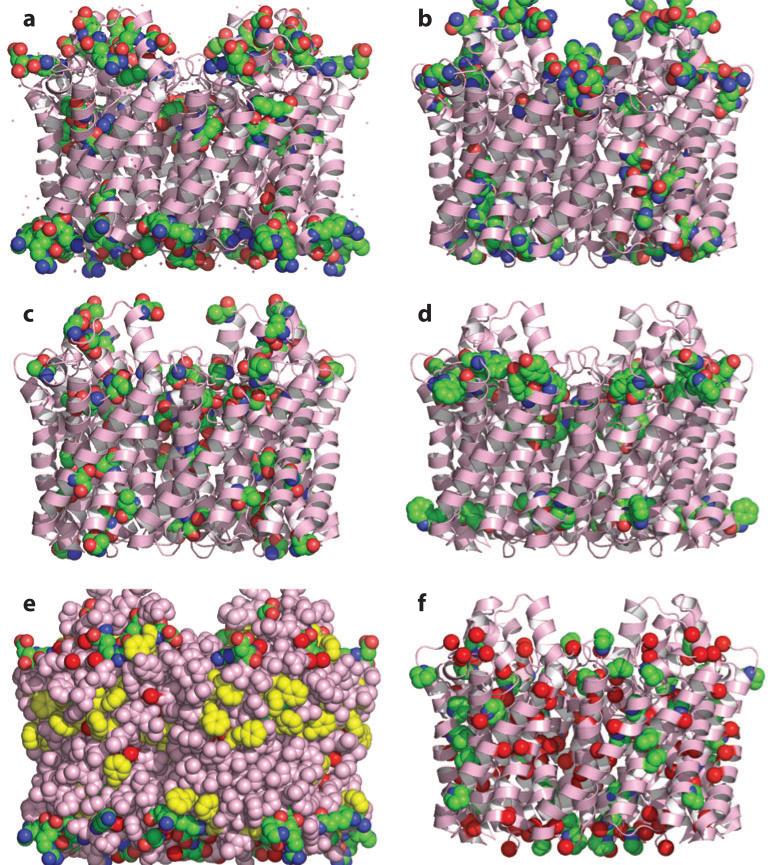
Amino acid distribution in the transmembrane domain of the glycerol facilitator (1FX8) (29) shown as pink ribbon except in panel e, where the surface is emphasized by showing an all-atom representation (pink except for highlighted residues in yellow). For the space-filling atoms: carbon, green spheres; nitrogen, blue spheres; and oxygen, red spheres, unless otherwise noted. (a) Charged residues Asp, Glu, Arg, and Lys. (b) Highly polar residues His, Asn, and Gln. (c) Polar residues Ser and Thr. (d ) Polar aromatic residues Trp and Tyr. (e) Charged residues (green carbons), Phe (yellow carbons), and Gly Cα (red carbons). (f ) Gly Cα (red carbons) and Pro ring atoms (green carbons).
The three residues that appear to be most enriched in TM helices relative to water-soluble helices are Phe, Gly, and Trp (23). In addition, Pro is much more common in the core of TM helices as opposed to being involved just in capping helices. The explanation for Trp is well known and well studied (65). Membrane proteins must be properly positioned and oriented with respect to the membrane, and Trp as well as Tyr seem to be involved in anchoring the proteins by forming a ring around them in the membrane interfacial region (Figure 2d). In addition, numerous charged residues also delimit the region positioned in the hydrophobic core of the membrane (Figure 2a). Phe is less well studied but appears to shield polar groups from the hydrophobic core of the lipid bilayer (Figure 2e). In the structure of a rhomboid family protease (2IC8) (98), an amphipathic loop is positioned below the membrane surface and serves as a lateral gate to the active site (Supplemental Figure 1a, follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org); three Phe side chains, one Tyr side chain, and one Trp side chain shield the backbone polar groups from exposure to the hydrophobic environment (Supplemental Figure 1b).
Gly and Pro are known as helix breakers in most biochemistry textbooks and yet these residues are present at very high levels throughout TM helices (Figure 2f ). It is well known (43, 84) that Gly residues facilitate interhelical interactions, especially in the form of Gly motifs (e.g., GxxxG and GxxxGxxxG). In Gly-mediated interhelical interactions, the backbone polar groups of the Gly residues can come into close contact (22). In addition, Gly residues can be important for facilitating conversion between functional states. The concept of prokinks has been introduced (73); i.e., Gly residues can induce kinks in helices. Potentially, a kink could be present in one functional state and not in another (45, 102). Gly and Pro residues destabilize TM helices by permitting kinks and bends that also lead to increased surface contact between helices and hence increased tertiary structural stability. In other words, the presence of the helix-breaking residues allows the excessive secondary structural stability generated by the low dielectric environment and lack of water in the membrane hydrophobic core to be sacrificed for increased tertiary structural stability (22). This increased dependence on interhelical side chain–side chain van der Waals interactions and backbone-backbone electrostatic interactions for tertiary stability, necessitated by the lack of polar residues and their stabilizing interactions, suggests that to achieve adequate tertiary structural stability, the helices should be well packed and devoid of substantial cavities unless they are needed for functional activities. In addition, in native-like membrane protein structures, Gly residues rarely face the hydrophobic environment, as this would expose the polar backbone atoms to the lipid environment, thereby destabilizing the membrane protein structure (22) (Figure 2e,f).
RECOGNIZING NATIVE-LIKE VERSUS NONNATIVE-LIKE HELICAL MEMBRANE PROTEIN STRUCTURES
The success of structural biology in providing atomic-resolution structural details has been an awesome scientific success story over the past 60 years, to the extent that X-ray crystal structure and, more recently, solution NMR structure have become synonymous with native structure. This reputation has been built upon tens of thousands of water-soluble protein structures crystallized in a homogeneous aqueous environment or observed directly in an isotropic aqueous sample. However, the heterogeneous and anisotropic environment of a membrane presents a new and challenging situation for structural biology. As a result, all the structure determination techniques have relied on membrane mimetics. Now that we have numerous membrane protein structures in the PDB, obtained not only from X-ray crystallography and solution NMR spectroscopy but also from electron crystallography and solid-state NMR spectroscopy, there is the opportunity to evaluate the native-likeness of these structures.
The preceding section suggests that helical membrane protein structures are potentially sensitive to the membrane mimetic environments. Our survey of the membrane protein structures in the PDB, to be presented in the next section, has indeed identified many possible structural perturbations by mimetics. The identification was guided by a set of initial considerations based in part on our understanding of the biophysical properties of native membranes and membrane mimetics. These were then refined during the structural survey. We list them here as a set of guidelines for recognizing native-like versus nonnative-like structures. None of these guidelines is absolute, but each violation can suggest cause for concern, and when several of them are combined they can indicate that a structure or a region of the structure is nonnative-like. Conversely, if a structure passes all these guidelines, then there is some confidence in the native-likeness of the structure. It is important to emphasize that the guidelines are for evaluating the TM domain of the protein and not for the connecting loops or any water-soluble domain.
The hydrophobic dimension of the TM domain needs to be approximately equal to that of the native membrane. The hydrophobic dimension can be assessed by the separation of charged residues, e.g., by measuring the separation of the Cα carbons coupled with a distance estimate for the snorkeling of the side chain to the membrane interfacial region. For most membrane protein structures the hydrophobicity of the membrane mimetic environment is much less than that in a native membrane environment, and so these side chains are often not in a native-like conformation, hence the need to estimate the snorkeling distance. Alternatively, the hydrophobic dimension can be estimated by the separation of the tryptophan and tyrosine aromatic belts in the would-be membrane interfacial region.
The acyl-facing surface of the TM domain needs to have little exposure of hydrophilic groups. Here, it is important to know the oligomeric state of the protein. Charged residues should not be exposed to the hydrophobic core of the membrane, and even polar residues are rarely exposed in native-like structures, although Ser and Thr hydroxyls can hydrogen-bond to the backbone and in that way conceal much of their hydrophilicity. Fenestrations from the hydrophobic environment into the hydrophilic interior of the protein are not typically observed in native-like structures. The hydrophilic backbone atoms are generally shielded from the fatty acyl environment by bulky hydrocarbon side chains (e.g., the glycerol facilitator in Figure 2e and rhomboid protease in Supplemental Figure 1). Conserved Gly residues are typically not observed on the lipid-facing surface (22) (Figure 2e).
Because only a limited number of polar and charged side chains are in the TM region, it is necessary for tertiary structural stability that helices pack closely together. Large and/or numerous cavities within the TM domain coupled with poor utilization of Gly and Ala motifs are indications of a nonnative-like tertiary structure. Extraneous organics such as detergents embedded in the protein structure may also indicate nonnativeness.
Because the interaction energy associated with crystal contacts between proteins in a crystal lattice can be significant in comparison with the tertiary stability of the TM domain, it is important to evaluate these contacts, both those involving the TM domain alone and those involving the water-soluble domain, the latter especially for electrostatic interactions.
Oligomeric structures, in general, should possess oligomeric symmetry. Although some functional states may require deviations from oligomeric symmetry, functionally unexplained deviations may suggest asymmetric interactions with the environment, leading to potentially nonnative-like structures.
In X-ray crystallography the observation of water and/or hydrophilic organics in the vicinity of the hydrophobic surface of the protein indicates a weak hydrophobic environment, which can lead to a poorly defined hydrophobic dimension and potential structural perturbations.
In detergent micelle samples the H/D exchange of amide protons in the TM domain indicates a weak hydrophobic environment that again can lead to structural perturbations. Other signs include helical rotations that expose hydrophilic residues and an outward curvature of helices, suggesting their exposure to the micelle surface.
MEMBRANE PROTEIN STRUCTURES DETERMINED IN MIMETIC ENVIRONMENTS
It is a great challenge to solubilize membrane proteins in mimetic environments that enable structure determination by any of the techniques, and preserve the native structures. On the one hand, all the structure determination techniques have produced some excellent native-like structures. On the other hand, many structures appear to be influenced by the deficiencies of mimetics in modeling the biophysical properties of native membranes.
3D Crystalline Environment
The 3D crystal lattice for X-ray diffraction studies can take on the appearance of repeating bilayers with definable hydrophobic and hydrophilic layers. The hydrophobic layers can have an appropriate thickness and can be largely devoid of water and hydrophilic organics. The aquaporin structure (27) (2W1P; Figure 3a) is a good example of such a lattice. The observed water molecules and the glucoside detergent headgroups are located in a hydrophilic layer (not shown). Although protein loops and termini in two adjacent hydrophilic layers in principle could interleave, this does not occur in 2W1P. In addition, there is no hydrophobic-hydrophobic contact between monomers except for the formation of a parallel tetramer, which appears to be a native oligomeric state. Therefore, the crystal contacts generate minimal perturbations on the aquaporin structure. In contrast, although the crystal lattice for the influenza M2 tetrameric TM domain (91) (3BKD; Figure 3b) takes a bilayer-like form, there are substantial crystal contacts between tetramers, leading to more significant perturbations, as discussed below. Contrary to the parallel orientation of the protein molecules in the aquaporin structure (as in membrane bilayers), if the protein molecules pack in an antiparallel configuration as for the M2 tetramers in the 3BKD crystal, then apparently there is less likelihood of achieving a native-like structure.
Figure 3.
Bilayer-like lattices in crystals. (a) Aquaporin (2W1P) (27) lattice structure with a parallel orientation of tetramers and minimal contacts between tetramers. (b) Antiparallel packing of M2 transmembrane domain tetramers (3BKD) (91), highlighting contacts between charged residues (space filling).
Crystal lattices may also take on nonbilayer forms, often leading to weaker hydrophobic environments and the potential for more structural perturbations. The human leukotriene C4 synthase, belonging to the MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) family, has four TM helices per monomer and functions as a trimer (2UUH) (60). The trimers in this lattice occupy a tetrahedral array, meaning that the hydrophobic domains are at an angle of 109° with respect to each other (Figure 4a). The weak hydrophobic environment generated by such a lattice is suggested by the abundance of water molecules surrounding the TM domain (see below) and by the antiparallel packing between the C-terminal segments of two neighboring trimers, each reaching deeply into the would-be membrane hydrophobic core around the neighboring trimer, despite the presence of three Arg residues in the C-terminal segment. Another nonbilayer crystal lattice is presented by the structure of estrone sulfatase (1P49) (38), whose TM domain is composed of a pair of helices. Here, the helices from three monomers interact in a nearly orthogonal and hence obviously nonnative arrangement (Figure 4b). Although the length of the two helices is appropriate for spanning the native membrane, the helices expose multiple hydrophilic residues, suggesting that the helical rotation may not be native-like. There are also a few water molecules as well as a glucoside detergent headgroup in what would be the bilayer hydrophobic region. These nonbilayer crystal lattices apparently have not provided a hydrophobic environment that adequately mimics that of the native membrane. The structural consequences just noted suggest that bilayer-like lattices are more likely than nonbilayer ones to harbor native-like structures.
Figure 4.
Nonbilayer lattices. (a) Leukotriene C4 synthase (2UUH) (60), showing a tetrahedral arrangement of trimers. (b) Estrone sulfatase (1P49) (38), showing helix pairs from three monomers interacting in a nearly orthogonal arrangement.
The homotrimeric acid sensing ion channel 1 (ASIC1) (2QTS) (42) has a large threefold symmetric water-soluble domain, but the TM domain, with a pair of helices from each monomer, lacks such symmetry (Figure 5a). Electrostatic interactions between the TM helices of one trimer and the water-soluble domain of another seem to contribute to the TM domain asymmetry (Figure 5a,b). Apparently, the weak interactions that provide tertiary and quaternary structural stability for the TM domain cannot compete with the interactions that provide the structural stability for the water-soluble domain. Hence the water-soluble domain retains its symmetry, whereas the TM domain does not. Significant electrostatic interactions are also observed in the crystal lattice of Mhp1, a nucleobase cation symporter 1 (NCS1) family transporter (100) (2JLN; Figure 5c). Here, as in many other structures, it is not clear whether the TM structure has been significantly perturbed by these interactions. Perhaps perturbations can be suggested in 2Q7M (25), the structure of 5-lipoxygenase-activating protein (FLAP) (Figure 5d), a member of the MAPEG family. Here, similar to 2UUH (Figure 4a), there are substantial hydrophobic and electrostatic interactions between the C-terminal helices of two neighboring trimers. These interactions appear to result in the exposure of Lys116 and Arg117 in the hydrophobic core of what would be the membrane interior. In addition, the last helix starts in the middle of the would-be membrane and terminates in the middle of a would-be membrane around the neighboring trimer. It is as if the interactions with the neighboring trimer have pulled the last helix halfway through the would-be membrane. In the recently determined dimer structure of the saccharide transporter ChbC (3QNQ) (11), multiple electrostatic contacts in the crystal lattice also appear to pull two outlying helices (TM8 and TM10) away from what would be the planar hydrophobic region of the native membrane (Supplemental Figure 2a, b). These possibly displaced helices contribute to a highly concave surface facing the cytosolic side.
Figure 5.
Electrostatic crystal contacts. (a) Crystal contacts in 2QTS (42), between the transmembrane domain of one ASIC1 trimer (green) and the water-soluble domain of a neighboring trimer (tan). (b) Enlarged view of the boxed region in panel a, highlighting the electrostatic contacts. (c) Crystal contacts in 2JLN (100), involving multiple charged residues at the interface between different Mhp1 molecules. (d) Intertrimer interactions between charged residues (Arg117, D142, and E144) in the apparently displaced last helices by neighboring FLAP (5-lipoxygenase-activating protein) trimers (2Q7M) (25).
The crystal contacts in 3BKD (for the M2 TM domain) (Figure 3b) include an extensive hydrophobic interface between two helices from neighboring antiparallel tetramers as well as salt bridges between an Arg residue and two Glu residues, one from the same tetramer and the other from a neighboring tetramer. The result is an asymmetric tetrameric helix bundle. When the antiviral drug amantadine is bound to the G34A mutant of this M2 construct (3C9J) (91) in the pore between the helices, neither an intrahelical nor an interhelical salt bridge is formed and the tetrameric structure is nearly symmetric. However, there are still significant hydrophobic interactions between the antiparallel tetramers. And the helices remain severely splayed (Figure 6a). The resulting fenestrations to the hydrophilic pore from the would-be hydrophobic environment are untenable from a functional perspective and perhaps indicate perturbations by the hydrophobic interactions between neighboring tetramers and other environmental factors. A more recent crystal structure of the same construct without the bound drug (3LBW) (2) has the channel pore sealed from the hydrophobic environment, similar to the structure of the M2 conductance domain determined in lipid bilayers (2L0J) (85) (Figure 1a). The bilayer-like lattice for the rhomboid protease (2IC8) (98) generates trimers composed of parallel monomers, although it is unknown whether the physiologically relevant oligomeric state is a trimer; the trimers in turn are packed in an antiparallel orientation (Figure 6b). Significant intertrimer interactions are mediated through helix 3 and the functionally important amphipathic loop (Figure 6b,c). Within the trimer, helix 5 of one monomer forms extended contacts with helices 1 and 2 of an adjacent monomer, such that the interactions between monomers through helix 5 are greater than the interactions within the monomer by this helix. Therefore, helix 5 is poorly packed within the monomer. Overall, despite the extensive intratrimer and intertrimer interactions, the structural fold of the monomer appears to be intact, except for the packing of helix 5.
Figure 6.
Hydrophobic crystal contacts. (a) Two antiparallel tetramers of the M2 transmembrane domain in 3C9J (91), showing splayed helices. (b) Parallel (intratrimer; all green) and antiparallel (intertrimer; green versus yellow) arrangements of rhomboid protease monomers in the 2IC8 bilayer-like lattice (98). (c) Space-filling view of two antiparallel 2IC8 monomers at the intertrimer interface.
The hydrophobic thickness of a membrane protein in the native membrane is determined by the mutual influence of the protein and its environment. In a typical crystal lattice of a membrane protein without a large water-soluble domain, the hydrophobic domain and its neighbors typically pack densely with hydrophobic contacts (see Figure 6) and there is little opportunity for the amphipathic organic molecules to help define the hydrophobic dimension of the protein. For a protein with a large water-soluble domain, there is often more volume surrounding the hydrophobic domain that can be occupied by detergents, and therefore the potential for a detergent aggregate to help define the hydrophobic dimension of the protein, similar to what happens in solution NMR. In this case, the total protein mass per unit volume of the unit cell is reduced (83). Occasionally membrane proteins have been crystallized with a considerable number of lipids present, such as the Gloeobacter violaceus pentameric ligand-gated ion channel GLIC (5) (3EAM; Figure 7a). In this structure nearly an annulus of lipids diffract, clearly demarcating the hydrophobic thickness and demonstrating a hydrophobic match to the protein. In some other protein structures the hydrophobic dimensions are rather short, and it is difficult to determine whether this is a requirement for the functionality of the protein or a perturbation by the membrane mimetic environment. For example, a second structure of ASIC1 (3HGC) (31) has much shorter and more highly tilted helices compared with the earlier structure (2QTS) (42), resulting in a hydrophobic thickness that is barely 20 Å. Contrary to the lack of threefold symmetry in the earlier structure, the TM domain in this more recent structure is nearly symmetric. The structure of the P2X4 receptor (35) (4DW1; Figure 7b) in the same superfamily displays TM helices that are highly tilted and kinked, resulting in a hydrophobic thickness that is less than 20 Å. In both of these cases Gly residues are exposed on the lipid-facing surface, and in the latter case inter-monomer fenestrations leave the channel pore open to the fatty acyl environment (Figure 7c), even more so than in the 3BKD and 3C9J structures of the M2 TM domain (Figure 6a).
Figure 7.
Hydrophobic dimensions of transmembrane domains. (a) Gloeobacter violaceus pentameric ligand-gated ion channel (GLIC) (3EAM) (5) displaying a substantial lipid annulus along with an appropriate hydrophobic thickness. (b) P2X4 receptor (4DW1) (35) with a thin hydrophobic dimension resulting from kinked and highly tilted helices. (c) Surface representation of 4DW1 highlighting the significant opening from the hydrophobic environment into the channel pore.
Weak hydrophobic environments can result from the presence of water, organics (including detergent headgroups), and hydrophilic portions of neighboring protein molecules in the crystal lattice. Because many crystal structures do not resolve the detergent and water molecules, it is hard to directly assess the hydrophobicity of the environments, but increasingly this data is available. For instance, in 2IC8 the rhomboid protease monomers are surrounded by the polar and nonpolar groups of detergent molecules as well as water molecules, indicating that this bilayer-like lattice does not seem to have clearly defined hydrophilic and hydrophobic layers (Figure 8a). Numerous water molecules are observed throughout the surface of the leukotriene C4 synthase structure (2UUH; Figure 8b). This distribution of water molecules indicates a weak hydrophobic environment, which perhaps facilitates the packing between the C-terminal segments of two neighboring trimers noted above (Figure 4a). For some other proteins, the structures hint at a weak hydrophobic environment. In the structure of PepTSo (2XUT) (68), the last two helices appear to be poorly packed with the rest of the TM domain, resulting in an interhelical loop in what would be the membrane hydrophobic core (Figure 8c). The tertiary interactions with the rest of the structure are weak, as evidenced by substantial cavities between these helices and the rest of the structure. A similar situation occurs in the structure of the maltose transporter complex (3RLF) (70), in which the packing of the first two helices of the MalF subunit with the rest of the TM domain is not well established, and there are multiple Gly residues exposed to the would-be hydrophobic environment (fSupplemental Figure 3a). In the structure of UraA (3QE7) (56), an NCS2 transporter, the last two helices appear to terminate prematurely, again leaving an intrahelical loop in the would-be membrane hydrophobic core along with exposed Gly residues (Supplemental Figure 3b). The structure of a site-2 protease (3B4R) (24) provides another example of possible misfolding. Here, the first helix is capped by a β-sheet that has backbone polar groups exposed to the would-be hydrophobic environment (Supplemental Figure 4). This configuration does not appear tenable; instead, if the helix were continued, the structure would present a continued hydrophobic surface.
Figure 8.
Weak hydrophobic environments in crystal lattices. (a) Rhomboid protease monomers (2IC8) (98) surrounded by detergent and water molecules (carbon, green spheres; oxygen, red spheres). (b) Leukotriene C4 synthase (2UUH) (60) displaying water molecules throughout the surface of the structure. (c) PepTSo (2XUT) (68), with cavities showing poor packing of the last two helices with the rest of the transmembrane domain.
One of the most significant differences between the detergents used for protein crystallization and the lipids in native membranes is the critical monomeric concentration for self-assembly. The critical lipid concentration for bilayer assembly is in the nanomolar range, whereas the critical detergent concentration for micelle formation is typically in the millimolar range (12). The 106-fold-higher monomeric concentration could increase the chance of monomeric detergents being inserted into membrane proteins. Detergent molecules can line the surface of a membrane protein much like lipid molecules, with the nonpolar tail and polar head next to the hydrophobic and hydrophilic surfaces of the protein, respectively. This is illustrated by the structure of a proton-translocating pyrophosphatase (4A01) (53), which shows proper positioning of detergents on the hydrophobic surface of the protein (Figure 9a). In contrast, monomeric detergents are seen inserted into some membrane protein crystal structures where lipids are unlikely to be located. In the 3BKD structure of the M2 TM domain, β-octyl glucoside and polyethylene glycol molecules are observed both in the channel pore and between the helices (Figure 9b); it is not clear whether these organics or the crystal contacts are responsible for the flaring of the helices. The structures of ASIC1 (2QTS), leukotriene C4 synthase (2UUH), and GLIC (3EAM) all have detergents in the channel pores. The GLIC structure is thought to represent the open state of the channel (5, 39), so perhaps the detergents are simply filling a vacancy. In other structures, single detergent molecules are wedged between helices (Figure 9c–e), as in the NCS2 transporter (3QE7) (56), a fucose transporter (3O7P) (19), and a sodium/calcium exchanger (3V5U) (52), among others. In the structure of the ChbC saccharide transporter (3QNQ) (11), two detergent molecules have their nonyl chains buried at the dimer interface (Supplemental Figure 2c). The structure of a putative sulfate permease (3TX3) (55) has four TM helices, which form two pairs that cross at approximately 60°. Each pair forms its own crystal contacts with neighboring protein molecules. In this apparently perturbed structure, detergent molecules display a broad range of orientations much like a detergent micelle (Figure 9f).
Figure 9.
Detergent molecules in and around transmembrane (TM) domains. (a) Proton-translocating pyrophosphatase (4A01) (53), with detergents appropriately lining the protein surface. (b) M2 TM domain (3BKD) (91), with detergents in the pore and between helices. Detergents wedged between helices in (c) the NCS2 transporter (3QE7) (56), (d) a fucose transporter (3O7P) (19), and (e) a sodium/calcium exchanger (3V5U) (52). (f) Putative sulfate permease (3TX3) (55), with detergents oriented randomly around TM helices.
Along with a weaker hydrophobic environment, a typical crystalline lattice may also have a weak lateral pressure. The latter could contribute to poor packing of TM helices, such as splaying. Possibly one such example is provided by the structure of the YiiP zinc transporter (3H90) (57). Here, two six-helix bundles are splayed so far apart that, in a bilayer environment, several lipids could diffuse into the gulf between the helix bundles (Figure 10a). The presence of significant internal cavities in both of the six-helix bundles further supports the possibility that the helix packing may not be native-like. Interestingly, in another ABC transporter, the MsbA lipid flippase (3B60) (99), a significant opening between the six-helix bundles may be necessary for its function (Figure 10b). In this case, presumably a lipid or at least its hydrophilic headgroup does diffuse into the opening between the six-helix bundles for transport across the membrane. Despite this functional requirement, the opening between the two-helix bundles is considerably smaller than in the zinc transporter, suggesting that the flippase structure is native-like. Weak lateral pressure in crystalline lattices may also have an impact on a wide variety of other structures, including those for which outlying helices are not packed well with the TM domain core, such as PepTSo (2XUT) (68), the maltose transporter (3RLF) (70), the NCS2 transporter (3QE7) (56), and the site-2 protease (3B4R) (24).
Figure 10.
Potential connection between helix splaying and weak lateral pressure. (a) Zinc transporter (3H90) (57), with two-helix bundles separated by a large gulf and cavities within the bundles. (b) Lipid flippase (3B60) (99), with a smaller opening between the helix bundles that may be required for protein function. In panels a and b, charged residues are displayed to delimit the hydrophobic thickness.
Detergent Micellar Environment
Micelles prepared from a wide variety of detergents have been characterized on the basis of their shape and size (54) and their suitability for preparing membrane protein samples for NMR spectroscopy (48). For a variety of reasons, including a weaker hydrophobic environment, it appears that the protein is far more responsible for defining its own hydrophobic thickness in a micelle environment than in a bilayer environment. In fact, the addition of a protein can alter the dimensions of a micelle more significantly than the dimensions of a bilayer. In contrast, a native membrane may force the protein to decrease its hydrophobic thickness by increasing the tilt of its helices, as occurs in the influenza M2 protein. The solution NMR structure of the M2 conductance domain (81) did not have the antiviral drug bound in the pore, apparently because the helical tilt was significantly less than that observed in liquid crystalline bilayer environments (6, 41, 85), and consequently the channel pore is too narrow to harbor the drug molecule (Figure 11). Here, the detergent was DHPC, which forms prolate-ellipsoid micelles with minor and major axes of approximately 20 and 40 Å, respectively, for the hydrophobic core (54). For the M2 conductance domain to maintain its fourfold symmetry, the protein would be oriented with its symmetry axis parallel to the major axis and hence there is as much as 40Å to accommodate the hydrophobic thickness of the protein. The incompatibility of this hydrophobic thickness in the detergent micelle environment with drug binding implies that the membrane protein is dependent on the native membrane environment to thin the hydrophobic dimension by inducing greater helical tilt.
Figure 11.
Different pore sizes and helix tilts in M2 structures. (a) 2RLF (81), conductance domain in DHPC (dihexanoyl phosphatidylcholine) micelles. (b) 2L0J (85), conductance domain in lipid bilayers. (c) 2KQT, transmembrane domain in lipid bilayers (6, 41). Two horizontal lines delimit the amantadine (AMT) binding site in the channel pore.
As with the M2 conductance domain in DHPC micelles, the hydrophobic core of detergent micelles in the absence of proteins is often larger than the hydrophobic thickness of the proteins. Another example is KdpD, a histidine kinase receptor (2KSF) (61) with two short helices in a four-helix bundle (Figure 12a). One short helix has only 14 residues, corresponding to a length of 21 Å; the second has 16 residues, or a length of 24 Å. The detergent was LMPG (lyso-myristoyl phosphatidylglycerol), which probably forms oblate-ellipsoid micelles similar to LPPG (lyso-palmitoyl phosphatidylglycerol). For LPPG the minor and major axes of the hydrophobic core are approximately 40 and 60 Å, respectively (54); for LMPG the minor axis may be reduced by as much as 5Å because of a shorter alkyl chain. The two short helices of KdpD may be due partly to a weak hydrophobic environment in the LMPG micelles. Because they are significantly shorter than the minor axis of the LMPG micelles, the helices could result in a decrease in the hydrophobic dimension of the micelles. It is also worth noting that these helices would be too short to span the hydrophobic core of a native membrane.
Figure 12.
KdpD histidine kinase receptor (2KSF) (61) with two short helices. (a) Two exposed Gly residues (red spheres). (b) Amide protons (yellow spheres) displaying significant hydrogen/deuterium exchange. (c) Exposed hydrophilic side chains.
For many years there was considerable debate in the literature about the penetration of water into micelles. Such studies appear to have been dependent on the techniques, including a variety of spectroscopic tools (17, 63, 77) and other biophysical approaches (21, 94). Although it appears as if a relatively small core is highly hydrophobic in detergent micelles, the interfacial region appears to be broad and hence there is neither a steep dielectric gradient nor a sharply varying lateral pressure profile as in bilayers. For KdpD, the resulting weak hydrophobic environment is demonstrated by significant amide backbone H/D exchange in three of the four helices (Figure 12b). Interestingly, the three helices that display H/D exchange are those that have the greatest number of hydrophilic residues (Figure 12c). By contrast, in a bilayer environment the number of hydrophilic residues would have little if any influence on the H/D exchange of the amide protons. Similar H/D exchange is observed in another histidine kinase receptor, ArcB (61) (2KSD; Figure 13a,b), where once again the helix with the greatest number of hydrophilic residues undergoes H/D exchange, whereas the helix that is more hydrophobic does not.
Figure 13.
ArcB histidine kinase receptor (2KSD) (61). (a) Curved helix with amide hydrogen/deuterium exchange sites highlighted by yellow spheres. (b) Exposed hydrophilic side chains. (c) Exposed Gly residue (red sphere).
The ellipsoidal shape of a detergent micelle means that one side of a helix positioned in the micelle can be in a considerably more hydrophilic environment than the other side, and hydrophilic residues in the middle of a TM helix can interact with the micelle interfacial region without dragging the charged residues at the terminus of a TM helix into the hydrophobic environment as they would in a lipid bilayer. Indeed, the TM helix of ArcB that undergoes H/D exchange is curved apparently to conform to the micelle ellipsoidal shape, such that many hydrophilic side chains along the length of the TM helix can interact with the micelle surface (Figure 13a,b). As a result, the interactions that could be expected to occur between the two helices of this structure are nonexistent. The interactions with the micelle surface explain why residues with hydrophilic side chains undergo H/D exchange. Although the TM helices of KdpD are not curved, most of the hydrophilic side chains throughout the TM domain are oriented toward the micelle surface. The result is a nearly hydrophobic side chain population for the core of this four-helix bundle instead of a hydrophobic surface facing the would-be membrane hydrophobic region. In addition, two of the three Gly residues in the middle of the TM helices are exposed (Figure 12a). The ArcB structure also exposes a Gly residue (Figure 13c). It appears as if the helix-helix interactions in these structures have been significantly perturbed by the detergent micelle environments.
While the ArcB and KdpD structures expose most of their hydrophilic residues on their surfaces, in the dimeric structure of BNip3 (2KA2) (92) two contiguous hydrophilic residues (Ser172 and His173) in the middle of the single TM helix of each monomer form interhelical hydrogen bonds to stabilize the dimeric structure (Figure 14a). In addition, the dimer takes advantage of a GxxxG motif for tight packing at the C termini of the TM helices. The CD3 ζζ dimer also relies on interhelical hydrogen bonds between two residues, Tyr12 and Thr17, in each monomer for quaternary stability (8) (2HAC; Figure 14b). The closely packed Asp2 side chains, mediated by either a water molecule or a shared proton, also provide further stabilization.
Figure 14.
Stabilization of helix-helix packing by interhelical hydrogen bonds between side chains. (a) BNip3 dimer (2KA2) (92) stabilized by two His173-Ser172 hydrogen bonds and close packing afforded by a GxxxG motif. (b) CD3 ζζ dimer (2HAC) (8) stabilized by two Tyr12-Thr17 hydrogen bonds and close packing of the Asp2 side chains.
The diacylglycerol kinase (DAGK) structure (2KDC) (95) is a trimer with three TM helices per monomer. It also has outward curved helices, but unlike the ArcB structure, the helices here do not have hydrophilic side chains exposed on the outer surface of the structure, except for the N-terminal 28 residues that are positioned in what would be the membrane hydrophobic region (Figure 15a). The curved helices expose backbone polar atoms to the would-be membrane hydrophobic region. In addition, the TM domain is poorly packed, with numerous cavities (Figure 15b). As with crystal structures, it is expected that the stability of TM helix bundles depends on numerous weak interactions derived from tight packing of the helices, except where necessitated for protein function. Here, there are no tightly packed helices, although two of the three helices have AxxxAxxxA motifs (Figure 15c).
Figure 15.
The diacylglycerol kinase (DAGK) trimer (2KDC) (95) with outward curved and poorly packed helices. (a) Charged residues, including those among the N-terminal 28 residues. (b) Large interhelical cavities. (c) Multiple Ala motifs not used for helix packing. In panels b and c, the N-terminal 28 residues are not displayed for clarity.
Amphipathic helices that interact with the bilayer surface have been known to be perturbed by the micellar environment (15). In the 2RLF structure of the influenza M2 conductance domain, the amphipathic helices from the tetramer formed a water-soluble bundle that was fully H/D exchangeable (81). Studies of the full-length M2 protein in a lipid bilayer environment showed that some of the amphipathic helix residues had the slowest amide H/D exchange in the entire protein, including the TM helix (93). A more recent solution NMR structure of the conductance domain, carrying the amantadine-resistant V27A mutation (2KWX) (75), showed the amphipathic helix interacting with the micelle surface in a position similar to that observed for the wild-type M2 conductance domain in lipid bilayers (85) (Figure 1b). The amphipathic helix (residues 1–25) of DAGK (2KDC) (95) is oriented such that the charged residues face what would be the membrane center (Figure 15a). The amphipathic helices in the solution NMR structure of the phospholamban pentamer (1ZLL) (71) are almost parallel to the TM helix bundle and oriented away from the would-be bilayer surface, while the structure (2KYV) (96) that integrated data from bilayer preparations showed the amphipathic helices in the bilayer interface (Figure 16).
Figure 16.
Different orientations of the amphipathic helices in two structures of the phospholamban pentamer. (a) 1ZLL (71). (b) 2KYV (96). For 2KYV, charged residues are shown in one amphipathic helix to indicate that it has an appropriate helical rotation.
As with the 3D crystalline environment, monomeric detergent concentrations in the millimolar range can be anticipated. Such concentrations may have deleterious effects on the water-soluble domains and even on the TM domains, as seen for the crystal structures into which detergent molecules may be inserted. However, in solution NMR such perturbations may not be detected unless the detergent was bound with high affinity, as the solution and high-temperature conditions favor dynamics.
Lipid Bilayer Environment
Synthetic lipid bilayers can satisfy many of the native membrane biophysical properties that appear to have implications for membrane protein structures. Once again, studies of the M2 proton channel have facilitated our understanding of how well the native environment needs to be modeled to obtain a native-like structure for a protein (18) (Figure 1), which as shown above is very sensitive to perturbations by its membrane mimetic environment. Many techniques can be used to study membrane proteins in a lipid environment, but only two have been used to determine 3D structures of membrane proteins in such an environment: electron crystallography (EC) and solid-state NMR (ssNMR) spectroscopy.
The first membrane protein EC structure, at a resolution of 7 Å, was determined for bacteriorhodopsin in 1975 (37), followed by a 3.5Å structure in 1990 (36). Since then, structures of aquaporin, H+ ATPase, H+/K+ ATPase, and EmrE, as well as two structures from the MAPEG superfamily, have been determined (1, 28, 40, 44, 49, 66). Importantly, the 1996 structure of bacteriorhodopsin (33) was obtained in its native purple membrane environment, showing a native trimeric state with lipids on the trimeric axis as well as on the external surface, leaving only the hydrophobic portion of the monomer-monomer interface bare (2BRD; Figure 17a,b). This EC structure overlays very well with the X-ray crystallography structure 2NTU (50), even to the rotameric states of many side chains (Figure 17c).
Figure 17.
Bacteriorhodopsin structures. (a) Electron crystallography (EC) structure of bacteriorhodopsin (2BRD) (33) in the native purple membrane environment, viewed down the bilayer normal to highlight lipids in the matrix surrounding the trimeric protein (one trimer and other monomers shown). (b) View into the bilayer plane, showing lipids surrounding a trimer. (c) Comparison of the EC structure (yellow) to an X-ray crystal structure (2NTU, orange) (50).
The EC structures of microsomal glutathione transferase 1 (3.2Å resolution; 2H8A) (40) and microsomal prostaglandin E synthase 1 (3.5Å resolution; 3DWW) (44) display the typical trimeric state of the MAPEG superfamily of proteins with four TM helices per monomer (Figure 18a), as observed by X-ray crystallography (Figures 4a and 5d). However, there are two significant enhancements in the structures determined by EC in a lipid environment. First, no charged side chains are exposed to the hydrophobic environment; although the two EC structures do not have the same charged residues, they do have multiple charged residues that are oriented toward the protein interior or in (or within reach of) the lipid interfacial region. Second, TM4 spans the entire hydrophobic dimension of the bilayer. So, the EC structures do not display the perturbations noted above for the X-ray structures of the MAPEG proteins (Figure 18b).
Figure 18.
Trimeric structures from the MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) superfamily, with charged residues highlighted in a monomer. (a) Electron crystallography structure of glutathione transferase 1 (2H8A) (40). (b) Comparison of 2H8A (green monomer) to the X-ray structure of FLAP (5-lipoxygenase-activating protein) (yellow and gray monomers, displaying charged residues) (2Q7M) (25).
The first ssNMR structure was deposited into the PDB in 1997 and was that of dimeric gramicidin A (gA) solubilized in liquid crystalline lipid bilayers (46) (1MAG; Figure 19a). Since then, more than 50 ssNMR structures have been deposited (though not all are membrane proteins); like gA, most of them are classified as peptides. However, structures of bona fide protein domains, including the M2 conductance domain (85) (2L0J; Figure 1), phospholamban (96) (2KYV; Figure 16b), the TM domain of the mercury transporter MerF (20) (2LJ2; Figure 19b), and the viral coat protein of Pf1 (74) (2KSJ; Figure 19c), have been recently determined in lipid environments. As illustrated in Figure 19, helices in the ssNMR structures are typically of appropriate lengths, with charged and Trp residues positioned in the lipid interfacial region. These are relatively small structures, albeit some of them are oligomeric. Most of the structures are backbone-only as a result of 15N-based spectroscopy of aligned samples. The gA structure is the only structure for which all side chains were characterized (46). The approach, through the combination of aligned sample and magic angle spinning spectroscopy, is rapidly developing today and is resulting in many more side chain restraints and larger membrane protein structures (9, 20, 32, 67). In addition, structural restraints can be obtained from proteins as they are expressed and inserted by the cellular machinery into host cell membranes (30, 64). This latter approach represents yet another validation tool for structural biology, as the proteins characterized in this fashion are never exposed to detergents and do not need to be purified.
Figure 19.
Three structures determined by solid-state NMR (ssNMR) spectroscopy in lipid environments, with residues in the lipid interfacial region highlighted. (a) Gramicidin A (1MAG) (46). (b) MerF Hg transporter (2LJ2) (20). (c) Pf1 coat protein (2KSJ) (74).
NATIVE-LIKE VERSUS NONNATIVE-LIKE STRUCTURES
All the structural techniques described here have determined some excellent structures that satisfy the guidelines for native-like structures listed above. These structures include the glycerol facilitator (1FX8) (29), the rhomboid protease (2IC8) (98), aquaporin (2W1P) (27), the proton-translocating pyrophosphatase (4A01) (53), the BNip3 dimer (2KA2) (92), the CD3 ζζ dimer (2HAC) (8), the M2 conductance domain (2L0J) (85), and bacteriorhodopsin (2BRD) (33), among others. However, some structures appear to violate these guidelines multiple times, for instance, the leukotriene C4 synthase (2UUH) (60), the ChbC saccharide transporter (3QNQ) (11), and the KdpD histidine kinase receptor (2KSF) (61). We recognize that even a nonnative-like structure can provide valuable insight. For example, we noted the possibly displaced outlying helices due to electrostatic crystal contacts and the insertion of detergent molecules into the dimer interface in the structure of the ChbC saccharide transporter. This structure nevertheless clearly defines the substrate-binding site and provides the basis for proposing a mechanism of transport (11).
An important use of the guidelines for distinguishing native-like from nonnative-like structures is to reduce the chance of overinterpreting structures. With membrane proteins, it can be difficult to ascertain whether structural changes represent different functional states or are a result of the perturbations by the membrane mimetic environment. For example, the 3B4R structure presents the site-2 protease as a dimer composed of antiparallel, asymmetric monomers (Supplemental Figure 4). One monomer (molecule B) is relatively compact, but the other monomer (molecule A) has a wide opening between helix 1 and helix 6. The two monomers were interpreted as representing the closed and open states, respectively (24). However, in this bilayer-like crystal lattice, sets of three dimers are arranged in a parallel orientation to form a trimer; in the trimer, three copies of helix 6, one from molecule A of each dimer, form a bundle stabilized by hydrophobic contacts, and helix 1 from molecule A also forms contacts with helices 1 and 6 of molecule B in a neighboring dimer (Supplemental Figure 4). These crystal contacts might be responsible for pulling apart helices 1 and 6 in molecule A. In any event, with such a wide opening between helices 1 and 6 over their entire TM length, what would prevent lipids in a native membrane from being lodged inside? As noted above, the capping of helix 1 by a β-sheet already indicates incompatibility with the native membrane environment. To their credit, the authors (24) recognized that the crystalline environment might have induced the observed perturbations. Clearly, structural validation in a lipid bilayer environment would be very useful.
STRUCTURE VALIDATION, REFINEMENT, AND QUALITY ASSESSMENT AND ENHANCEMENT
Structural restraints for membrane proteins in bilayer environments can be obtained from electron spin resonance and ssNMR. For instance, a few long distances from electron spin resonance on the Zn transporter (3H90, Figure 10a) would confirm or refute whether the helix bundles are as splayed as in the crystal structure. Helix tilt angles by ssNMR (59, 97) for a protein, such as the P2X4 receptor (4DW1, Figure 7b), with a structure that appears to have a thin hydrophobic dimension could either validate or refute such a structure. Indeed, the orientation of membrane proteins in their membrane environment is always an important question and often somewhat undefined by structures that are not in a bilayer-like crystal lattice or even those that are, such as the structure of the site-2 protease (3B4R; Supplemental Figure 4) where the threefold symmetry axis of the trimer of dimers is not perpendicular to the interfacial plane defined by the charged residues. Once again, ssNMR could be used to define the helix tilt angles with respect to a lipid bilayer normal. H/D exchange might also provide insights into defining the hydrophobic-hydrophilic interface of the native structure.
When distance and/or orientation restraints are obtained by ssNMR for most of the residues in the TM domain, this information can be used to calculate the structure, much like structure determination by solution NMR spectroscopy. Recently, it has become possible to use ssNMR data to restrain molecular dynamics simulations in lipid bilayers for structure refinement (14, 85; see sidebar, Structure Refinement by Restrained Molecular Dynamics Simulations). The explicit representation of lipid bilayers in these simulations ensures, at least in principle, that the membrane biophysical properties noted above are present to maintain the structural integrity of the membrane proteins. The accumulation of native-like membrane protein structures in the PDB will help improve the simulation protocols, including parameters in the molecular mechanics energy function and the relative weighting of the energy function versus the experimental restraints.
As native-like membrane protein structures accumulate in the PDB, it will also be possible to formalize the guidelines of the preceding section, in the form of a computational tool such as PROCHECK (51), for quality assessment of membrane protein structures. As a proof of concept, the calculation of Gly exposure on protein surfaces was recently found useful in discriminating between native-like and nonnative-like structures (22). A benchmark set of 26 putatively native-like structures essentially avoids Gly exposure on their lipid-facing surfaces. These structures include 1FX8, 2L0J, 2KYV, 3DWW, and 3EAM. In contrast, ASIC1 (Figure 5a,b), FLAP (Figure 5d), and KdpD (Figure 12), discussed above for other possible structural perturbations, as well as EmrD (2GFP) (103), all have conserved Gly residues exposed on the lipid-facing surfaces of the proteins. Here, we have also noted Gly exposure in the P2X4 receptor (4DW1), ArcB histidine kinase receptor (2KSD; Figure 13c), and maltose and NCS2 transporters (Supplemental Figure 3).
It may be possible to use molecular dynamics simulations in lipid bilayers to correct some of the structural perturbations. As mentioned above, it is common for charged side chains in the lipid interfacial region to have inappropriate orientations; it seems that this problem can be easily corrected by simulations in a lipid bilayer. It will be more interesting to explore the extent to which the more substantial perturbations discussed above can be corrected by molecular dynamics simulations. For example, a displaced helix (Figure 5d) in a lipid bilayer will experience a significant energetic penalty, which is expected to force the helix to at least move toward a native-like structure. Similarly, splayed helices (Figure 6a) or helix bundles (Figure 10a) are also expected to improve packing in molecular dynamics simulations in lipid bilayers.
Supplementary Material
PDB: Protein Data Bank
TM: transmembrane
Detergent micelle: assembly of tens to hundreds of amphipathic molecules, with hydrophobic tails buried at the core and headgroups in contact with water
DHPC: dihexanoyl phosphatidylcholine
Hydrogen/deuterium (H/D) exchange: reaction in which hydrogen, usually on the backbone amide, is replaced by deuterium, when H2O solvent is changed to D2O
Critical concentration: concentration at which an amphiphile starts to separate into a monomeric phase and an assembled phase (e.g., bilayer or micelle)
Crystal lattice: the arrangement of protein molecules into 3D periodic arrays, by repeating a unit cell, when forming a crystal
MAPEG: membrane-associated proteins in eicosanoid and glutathione metabolism
FLAP: 5-lipoxygenase-activating protein
LMPG: lyso-myristoyl phosphatidylglycerol
Electron crystallography (EC): a structure determination technique using electron diffraction of 2D crystals (and other non-3D crystal samples)
Solid-state nuclear magnetic resonance (ssNMR): NMR spectroscopic technique for characterizing samples that do not undergo isotropic motions, such as in liquid crystalline lipid bilayers
Restrained molecular dynamics simulations: simulations that include experimental restraints as pseudo energy terms to drive the Newtonian motion of protein, lipid, and water molecules
STRUCTURE REFINEMENT BY RESTRAINED MOLECULAR DYNAMICS SIMULATIONS.
Given the greater difficulty in obtaining structural information for membrane proteins prepared in lipid bilayers than in a detergent-based crystal or a detergent micelle sample, there is a greater need to maximize the use of the relatively sparse experimental restraints in structure calculation. Restrained molecular dynamics simulations are well suited for this purpose. The experimental restraints are represented as empirical energy terms and added to the regular molecular mechanics energy function. The membrane protein and the solubilizing environment (lipid and water molecules) can be represented at either a coarse-grained level (13, 88) or an all-atom level (14, 85). The former representation allows for more efficient conformational sampling, whereas the latter representation allows for more realistic modeling of the protein-environment interactions. In addition to structure refinement based on data from lipid bilayers, restrained molecular dynamics simulations can be used to improve X-ray and solution NMR structures of membrane proteins for better compatibility with the membrane environment.
SUMMARY POINTS.
The biophysical properties of many membrane mimetic environments used for membrane protein structure determination, such as hydrophobicity, hydrophobic dimensions, monomeric concentrations of amphiphiles, dielectric and water concentration gradient, and lateral pressure profile, can differ substantially from those of native membranes.
As a result of the low hydrophilic amino acid composition of the TM domains of helical membrane proteins, tertiary structural stability is compromised whereas secondary structural stability of helices is enhanced owing to the low dielectric membrane environment.
High Gly compositions are present in part to facilitate helix packing, and thus tertiary structural stability, while sacrificing excessive secondary structural stability. Hence, conserved Gly residues are not exposed to the hydrophobic lipid environment.
Crystal contacts can overwhelm tertiary structural stability, resulting in structural perturbations in the high protein density matrix.
The curved single hydrophilic surface of detergent micelles attracts hydrophilic residues in the middle of TM helices, resulting in an outward helical curvature, a perturbed orientation of TM helices, and extensive H/D exchange of the amide protons in TM helices.
Although some structures have perturbations that are easily recognized and many others appear to be native-like, a more challenging task is identifying those structures that are more modestly perturbed, but still perturbed to an extent that influences our understanding of the functional mechanisms of the proteins.
Membrane proteins are a particularly great challenge for structural biologists, but too often our focus has been on achieving a sample that crystallizes, tumbles isotropically, or is aligned, and not on the potential influences of the membrane mimetic environment on the native membrane protein structure.
A critical assessment with experimental validation and computational refinement of each structure should become the standard before claiming unwarranted functional conclusions that may mislead the biological community.
ACKNOWLEDGMENTS
This research has been supported in part by the National Institutes of Health through grants AI-023007, AI-074805, AI-073891, and GM-088187, and the National Science Foundation through Cooperative Agreement 0654118 between the Division of Materials Research and the State of Florida.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abe K, Tani K, Nishizawa T, Fujiyoshi Y. Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle. EMBO J. 2009;28:1637–43. doi: 10.1038/emboj.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acharya R, Carnevale V, Fiorin G, Levine BG, Polishchuk AL, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. USA. 2010;107:15075–80. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 4.Belrhali H, Nollert P, Royant A, Menzel C, Rosenbusch JP, et al. Protein, lipid water organization in bacteriorhodopsin: a molecular view of the purple membrane at 1.9Å resolution. Structure. 1999;7:909–17. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 5.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–14. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 6.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–92. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J. Am. Chem. Soc. 2011;133:4274–84. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–68. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Can TV, Sharma M, Hung I, Gor'kov PL, Brey WW, Cross TA. Magic angle spinning and oriented sample solid-state NMR structural restraints combine for influenza A M2 protein functional insights. J. Am. Chem. Soc. 2012;134:9022–25. doi: 10.1021/ja3004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–39. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Jin X, Levin EJ, Huang H, Zong Y, et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011;473:50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cevc G, Marsh D. Phospholipid Bilayers: Physical Principles and Models. Wiley; New York: 1987. [Google Scholar]
- 13.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc. Natl. Acad. Sci. USA. 2010;107:5435–40. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng X, Im W. NMR observable-based structure refinement of DAP12-NKG2c activating immunoreceptor complex in explicit membranes. Biophys. J. 2012;102:L27–L29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacryl-amide gel. J. Am. Chem. Soc. 2002;124:2450–51. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 16.Cross TA, Dong H, Sharma M, Busath DD, Zhou HX. M2 protein from influenza A: from multiple structures to biophysical and functional insights. Curr. Opin. Virol. 2012;2:128–33. doi: 10.1016/j.coviro.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross TA, Opella SJ. Structural properties of fd coat protein in sodium dodecyl sulfate micelles. Biochem. Biophys. Res. Commun. 1980;92:478–84. doi: 10.1016/0006-291x(80)90358-7. [DOI] [PubMed] [Google Scholar]
- 18.Cross TA, Sharma M, Yi M, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem. Sci. 2011;36:117–25. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang S, Sun L, Huang Y, Lu F, Liu Y, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–38. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 20.Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, et al. Structure determination of a membrane protein in proteoliposomes. J. Am. Chem. Soc. 2012;134:2047–56. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dill KA, Koppel DE, Cantor RS, Dill JD, Bendedouch D, Chen S- H. Molecular conformations in surfactant micelles. Nature. 1984;309:42–45. [Google Scholar]
- 22.Dong H, Sharma M, Zhou HX, Cross TA. Glycines: role in α-helical membrane protein structures and a potential indicator of native conformation. Biochemistry. 2012;51:4779–89. doi: 10.1021/bi300090x. [Conserved glycine residues are rarely exposed on the hydrophobic surface of membrane proteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eilers M, Patel AB, Liu W, Smith SO. Comparison of helix interactions in membrane and soluble α-bundle proteins. Biophys. J. 2002;82:2720–36. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Yan H, Wu Z, Yan N, Wang Z, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–12. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson AD, McKeever BM, Xu S, Wisniewski D, Miller DK, et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–12. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 26.Finkelstein A. Water Movement Through Lipid Bilayers, Pores, and Plasma Membranes. Wiley; New York: 1987. [DOI] [PubMed] [Google Scholar]
- 27.Fischer G, Kosinska-Eriksson U, Aponte-Santamaria C, Palmgren M, Geijer C, et al. Crystal structure of a yeast aquaporin at 1.15Å reveals a novel gating mechanism. PLoS Biol. 2009;7:e1000130. doi: 10.1371/journal.pbio.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 29.Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–86. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 30.Fu R, Wang X, Li C, Santiago-Miranda AN, Pielak GJ, Tian F. In situ structural characterization of a recombinant protein in native Escherichia coli membranes with solid-state magic-angle-spinning NMR. J. Am. Chem. Soc. 2011;133:12370–73. doi: 10.1021/ja204062v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopinath T, Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angew. Chem. Int. Ed. Engl. 2012;51:2731–35. doi: 10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron crystallo-graphic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [The only membrane protein structure in a native environment, for bacteriorhodopsin, determined by electron crystallography.] [DOI] [PubMed] [Google Scholar]
- 34.Gullingsrud J, Schulten K. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 2004;86:3496–509. doi: 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485:207–12. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 37.Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez-Guzman FG, Higashiyama T, Pangborn W, Osawa Y, Ghosh D. Structure of human estrone sulfatase suggests functional roles of membrane association. J. Biol. Chem. 2003;278:22989–97. doi: 10.1074/jbc.M211497200. [DOI] [PubMed] [Google Scholar]
- 39.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–18. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 40.Holm PJ, Bhakat P, Jegerschold C, Gyobu N, Mitsuoka K, et al. Structural basis for detoxification and oxidative stress protection in membranes. J. Mol. Biol. 2006;360:934–45. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 41.Hu J, Asbury T, Achuthan S, Li C, Bertram R, et al. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from influenza A virus. Biophys. J. 2007;92:4335–43. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9Å resolution and low pH. Nature. 2007;449:316–23. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 43.Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 1999;77:1609–18. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jegerschold C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, et al. Structural basis for induced formation of the inflammatory mediator prostaglandin E2. Proc. Natl. Acad. Sci. USA. 2008;105:11110–15. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Lee A, Chen J, Chadene M, Chait B, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;4176:523–52. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 46.Ketchem RR, Lee KC, Huo S, Cross TA. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. J. Biomol. NMR. 1996;8:1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- 47.Kim S, Cross TA. Uniformity, ideality, and hydrogen bonds in transmembrane α-helices. Biophys. J. 2002;83:2084–95. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, et al. An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomol. NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 49.Kuhlbrandt W, Zeelen J, Dietrich J. Structure, mechanism, and regulation of the Neurospora plasma membrane H+-ATPase. Science. 2002;297:1692–96. doi: 10.1126/science.1072574. [DOI] [PubMed] [Google Scholar]
- 50.Lanyi JK, Schobert B. Structural changes in the L photointermediate of bacteriorhodopsin. J. Mol. Biol. 2007;365:1379–92. doi: 10.1016/j.jmb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–91. [Google Scholar]
- 52.Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335:686–90. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 53.Lin SM, Tsai JY, Hsiao CD, Huang YT, Chiu CL, et al. Crystal structure of a membrane-embedded H+-translocating pyrophosphatase. Nature. 2012;484:399–403. doi: 10.1038/nature10963. [DOI] [PubMed] [Google Scholar]
- 54.Lipfert J, Columbus L, Chu VB, Lesley SA, Doniach S. Size and shape of detergent micelles determined by small-angle X-ray scattering. J. Phys. Chem. B. 2007;111:12427–38. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 55.Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–37. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu F, Li S, Jiang Y, Jiang J, Fan H, et al. Structure and mechanism of the uracil transporter UraA. Nature. 2011;472:243–46. doi: 10.1038/nature09885. [DOI] [PubMed] [Google Scholar]
- 57.Lu M, Chai J, Fu D. Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 2009;16:1063–67. doi: 10.1038/nsmb.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. USA. 2009;106:12283–88. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 2000;144:150–55. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez Molina D, Wetterholm A, Kohl A, McCarthy AA, Niegowski D, et al. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–16. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 61.Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, et al. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc. Natl. Acad. Sci. USA. 2010;107:10902–7. doi: 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathai JC, Sprott GD, Zeidel ML. Molecular mechanisms of water and solute transport across archaebacterial lipid membranes. J. Biol. Chem. 2001;276:27266–71. doi: 10.1074/jbc.M103265200. [DOI] [PubMed] [Google Scholar]
- 63.Menger FM, Boyer BJ. Water penetration into micelles as determined by optical rotatory dispersion. J. Am. Chem. Soc. 1980;102:5938–39. [Google Scholar]
- 64.Miao Y, Qin H, Fu R, Sharma M, Can TV, et al. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew. Chem. Int. Ed. Engl. 2012;51:8383–86. doi: 10.1002/anie.201204666. [Validation of the M2 transmembrane structure with observations of the full-length protein in E. coli membranes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michel H, Deisenhofer J. The photosynthetic reaction center from the purple bacterium Rhodopseudomonas viridis: aspects of membrane protein structure. Curr. Top. Membr. Transp. 1990;36:53–184. [Google Scholar]
- 66.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 67.Nevzorov AA. High-resolution separated local field spectroscopy with internuclear correlations. J. Magn. Reson. 2009;201:111–14. doi: 10.1016/j.jmr.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Newstead S, Drew D, Cameron AD, Postis VL, Xia X, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30:417–26. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nymeyer H, Zhou HX. A method to determine dielectric constants in nonhomogeneous systems: application to biological membranes. Biophys. J. 2008;94:1185–93. doi: 10.1529/biophysj.107.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oldham ML, Chen J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. USA. 2011;108:15152–56. doi: 10.1073/pnas.1108858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oxenoid K, Chou JJ. The structure of phospholambin pentamer reveals a channel-like architecture in membranes. Proc. Natl. Acad. Sci. USA. 2005;102:10870–75. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–97. doi: 10.1016/j.str.2008.02.018. [Transmembrane helices are remarkably uniform with strong hydrogen bonds and reduced peptide plane tilts.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Page RC, Li C, Hu J, Gao FP, Cross TA. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn. Reson. Chem. 2007;45:S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 74.Park SH, Marassi FM, Black D, Opella SJ. Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly. Biophys. J. 2010;99:1465–74. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pielak RM, Chou JJ. Solution NMR structure of the V27A drug resistant mutant of influenza A M2 channel. Biochem. Biophys. Res. Commun. 2010;401:58–63. doi: 10.1016/j.bbrc.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pinto LH, Lamb RA. Influenza virus proton channels. Photochem. Photobiol. Sci. 2006;5:629–32. doi: 10.1039/b517734k. [DOI] [PubMed] [Google Scholar]
- 77.Podo F, Ray A, Nemethy G. Structure and hydration of nonionic detergent micelles. High resolution nuclear magnetic resonance study. J. Am. Chem. Soc. 1973;95:6164–71. [Google Scholar]
- 78.Popot JL, Engelman DM. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4037–41. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 79.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–36. doi: 10.1016/j.virol.2010.12.003. [M2 is in a liquid crystalline environment in the viral coat dominated by raft-like lipids.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders CR, Mittendorf KF. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50:7858–67. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–95. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schroeder C, Lin T- I. Viral Membrane Proteins: Structure, Function, and Drug Design, ed. W Fischer. Kluwer/Plenum; New York: 2005. Influenza A virus M2 protein: proton selectivity of the ion channel, cytotoxicity, and a hypothesis on peripheral raft association and virus budding. pp. 114–30. [Google Scholar]
- 83.Schulz GE. A new classification of membrane protein crystals. J. Mol. Biol. 2011;407:640–46. doi: 10.1016/j.jmb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 84.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 2004;14:465–79. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Sharma M, Yi M, Dong H, Qin H, Peterson E, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–12. doi: 10.1126/science.1191750. [ssNMR characterization and computational refinement of the M2 conductance domain structure in liquid crystalline lipid bilayers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simon SA, McIntosh TJ, Latorre R. Influence of cholesterol on water penetration into bilayers. Science. 1982;216:65–67. doi: 10.1126/science.7063872. [DOI] [PubMed] [Google Scholar]
- 87.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011;3:1–17. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sompornpisut P, Roux B, Perozo E. Structural refinement of membrane proteins by restrained molecular dynamics and solvent accessibility data. Biophys. J. 2008;95:5349–61. doi: 10.1529/biophysj.108.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sonntag Y, Musgaard M, Olesen C, Schiott B, Moller JV, et al. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat. Commun. 2011;2:304. doi: 10.1038/ncomms1307. [A membrane protein and the membrane can adapt to each other by altering helix tilt and membrane thickness.] [DOI] [PubMed] [Google Scholar]
- 90.Stern HA, Feller SA. Calculation of the dielectric permittivity profile for a nonuniform system: application to a lipid bilayer simulation. J. Chem. Phys. 2003;118:3401–12. [Google Scholar]
- 91.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–99. doi: 10.1038/nature06528. [Crystal structures of the M2 transmembrane domain with and without bound drug.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sulistijo ES, Mackenzie KR. Structural basis for dimerization of the BNIP3 transmembrane domain. Biochemistry. 2009;48:5106–20. doi: 10.1021/bi802245u. [DOI] [PubMed] [Google Scholar]
- 93.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Turro NJ, Okubo T. Micellar microviscosity of ionic surfactants under high pressure. J. Am. Chem. Soc. 1981;103:7224–28. [Google Scholar]
- 95.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, et al. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–29. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. USA. 2011;108:9101–6. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J, Denny J, Tian C, Kim S, Mo Y, et al. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–67. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–80. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 99.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. USA. 2007;104:19005–10. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–13. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu F, Cross TA. Water: foldase activity in catalyzing polypeptide conformational rearrangements. Proc. Natl. Acad. Sci. USA. 1999;96:9057–61. doi: 10.1073/pnas.96.16.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yi M, Cross TA, Zhou HX. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc. Natl. Acad. Sci. USA. 2009;106:13311–16. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–44. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.