Abstract
Purpose
To evaluate the impact of miR-155 on the outcome of adults with cytogenetically normal (CN) acute myeloid leukemia (AML) in the context of other clinical and molecular prognosticators and to gain insight into the leukemogenic role of this microRNA.
Patients and Methods
We evaluated 363 patients with primary CN-AML. miR-155 levels were measured in pretreatment marrow and blood by NanoString nCounter assays that quantified the expression of the encoding gene MIR155HG. All molecular prognosticators were assessed centrally. miR-155–associated gene and microRNA expression profiles were derived using microarrays.
Results
Considering all patients, high miR-155 expression was associated with a lower complete remission (CR) rate (P < .001) and shorter disease-free survival (P = .001) and overall survival (OS; P < .001) after adjusting for age. In multivariable analyses, high miR-155 expression remained an independent predictor for a lower CR rate (P = .007) and shorter OS (P < .001). High miR-155 expressers had approximately 50% reduction in the odds of achieving CR and 60% increase in the risk of death compared with low miR-155 expressers. Although high miR-155 expression was not associated with a distinct microRNA expression profile, it was associated with a gene expression profile enriched for genes involved in cellular mechanisms deregulated in AML (eg, apoptosis, nuclear factor-κB activation, and inflammation), thereby supporting a pivotal and unique role of this microRNA in myeloid leukemogenesis.
Conclusion
miR-155 expression levels are associated with clinical outcome independently of other strong clinical and molecular predictors. The availability of emerging compounds with antagonistic activity to microRNAs in the clinic provides the opportunity for future therapeutic targeting of miR-155 in AML.
INTRODUCTION
MicroRNAs are short noncoding RNAs that regulate the expression of proteins involved in pivotal cellular mechanisms. Initially encoded as premicroRNAs, they undergo several steps of maturation before being incorporated into RNA-induced silencing complexes and hybridizing to target mRNAs.1,2 This process results in either degradation or translation inhibition of the target mRNAs and, in turn, protein downregulation.
Aberrant expression levels of microRNAs and their target mRNAs occur in human cancer and likely contribute to malignant transformation. In acute myeloid leukemia (AML), microRNAs are involved in disruption of hematopoietic mechanisms of cell differentiation, proliferation, and survival; contribute to the molecular heterogeneity of the disease; and impact on treatment response and outcome.2,3
The MIR155HG gene located at chromosome band 21q21.3 encodes miR-155. In the normal host, this microRNA is induced in hematopoietic stem cells and myeloid progenitor cells during inflammatory responses under the transcriptional control of nuclear factor-κB (NF-κB) and activator protein 1 (AP-1).4 By preferentially targeting inhibitors of inflammation, miR-155 sustains normal mechanisms of the innate immune response.4–7 The potential oncogenic role of miR-155 was identified early by showing its enhanced expression levels in lymphoma, aggressive molecular subtypes of chronic lymphocytic leukemia, and solid tumors.4 Subsequently, high levels of miR-155 were demonstrated to cause aggressive pre–B-cell leukemia and myeloproliferative disorders in murine models.8,9
In AML, we and others have reported that higher expression of miR-155 is often associated with FLT3 internal tandem duplication (FLT3-ITD), a genetic marker predicting poor outcome.10–13 However, whether miR-155 upregulation impacts on clinical outcome in patients with AML independently from FLT3-ITD and other established prognosticators is unknown. This is likely to be relevant not only for patients' risk stratification, but also for establishing specific clinical approaches. Indeed, compounds with antagonistic activity to microRNAs are emerging for clinical use, and miR-155 could represent a novel therapeutic target in AML.14 Thus, we assessed the clinical impact of this microRNA in a cohort of adult patients with primary (de novo) cytogenetically normal AML (CN-AML) who were well characterized molecularly at diagnosis. Furthermore, to gain biologic insights, we performed genome-wide gene and microRNA expression analyses.
PATIENTS AND METHODS
Patients, Treatment, and Cytogenetic Analysis
Three hundred sixty-three adults, including 153 younger (age < 60 years) and 210 older (age ≥ 60 years) patients, with untreated, primary CN-AML who received intensive first-line therapy on Cancer and Leukemia Group B (CALGB) trials and had diagnostic bone marrow (BM) or blood specimens available for molecular analyses were included. The diagnosis of normal cytogenetics was based on the analysis of ≥ 20 metaphases in BM specimens subjected to short-term culture and confirmed centrally.15 All patients received cytarabine-daunorubicin–based induction chemotherapy. Younger patients were treated on CALGB trials 9621 or 19808.16–18 Older patients were treated with less intense regimens on CALGB protocols 8525, 8923, 9420, 9720, and 10201.19–24 Per protocol, no patient received allogeneic stem-cell transplantation during first complete remission (CR), defined according to published criteria.25 See Data Supplement for treatment details.
When patients with CN-AML in the current study were compared with patients enrolled onto the same CALGB protocols who were not studied because they had no material available for analysis (n = 499), there were no statistically significant differences in any of the outcome end points analyzed (for other clinical characteristics, see Data Supplement). All patients provided written informed consent, and study protocols were in accordance with the Declaration of Helsinki and approved by Institutional Review Boards at each center.
Molecular Analyses
We measured the expression of MIR155HG transcript (hereafter referred to as miR-155) using an enzyme-independent probe-based quantification system that allows digital counting of individual mRNA molecules (nCounter; NanoString Technologies, Seattle, WA), as previously reported.26 The method allows for direct measurement of actual levels of RNAs with no need for target amplification as used by other polymerase chain reaction (PCR) –based assays. The sensitivity, dynamic range, linearity, and reproducibility of the nCounter system have been validated previously,26 and mRNA expression levels measured with this approach show excellent correlation with data obtained by oligonucleotide microarrays or quantitative PCR. miR-155 levels were normalized using ABL as an internal control. Target/internal control ratios were log2 transformed for downstream analyses.
The presence or absence of additional molecular markers, that is, FLT3-ITD, FLT3 tyrosine kinase domain mutations (FLT3-TKD), and MLL partial tandem duplication; mutations in NPM1, CEBPA, WT1, TET2, ASXL1, DNMT3A, RUNX1, IDH1, and IDH2; and expression levels of BAALC, ERG, and miR-181a were assessed centrally, as previously reported.12,27–40
Microarray Profiling
Gene and microRNA expression profiling was performed using HG-U133 plus 2.0 oligonucleotide microarrays (Affymetrix, Santa Clara, CA) and Ohio State University custom microRNA microarrays, respectively, as previously reported.39–41 Expression signatures were identified by correlating the expression levels of MIR155HG with those of protein-coding genes and microRNAs using Spearman rank correlation.42 The false discovery rate (FDR) was used to assess the multiple testing errors. A permutation test was computed based on 1,000 random permutations. The CI of FDR assessment was 80%, and the maximum allowed proportion of false-positive genes was 1%. Gene Ontology analysis to assess enrichment of genes in the miR-155–associated signature in distinct biologic processes was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID).43
Definition of Clinical End Points and Statistical Analyses
For definitions of outcome end points, see the Data Supplement. The main objective of our study was to evaluate the association of miR-155 expression levels with clinical and molecular characteristics and the impact of aberrant miR-155 expression on outcome. We first divided patients into quartile groups based on the expression levels of miR-155 and assessed the outcome associations by the trend test for disease-free survival (DFS; P < .001) and overall survival (OS; P < .001). This justified the use of the median cut to dichotomize patients into high and low expressers of miR-155.
The associations of miR-155 expression status (low/high) with baseline clinical, demographic, and molecular features were compared using the Wilcoxon rank sum test and the Fisher's exact test for continuous and categorical variables, respectively.42,44 Univariable logistic regression models were constructed to evaluate miR-155 expression for achievement of CR, and univariable Cox proportional hazards models were used to evaluate the associations of miR-155 expression with DFS and OS.45,46 Multivariable logistic regression models were generated for attainment of CR, and multivariable proportional hazards models were constructed for DFS and OS, using a limited backward elimination procedure. Clinical variables that were considered for univariable analyses, in addition to miR-155 expression, were age, sex, race, hemoglobin, platelet count, WBC count, and the centrally assessed molecular variables. Variables significant at α = .20 from the univariable analyses were considered for multivariable analyses. For the time-to-event end points, the proportional hazards assumption was checked for each variable individually. All models considering both age groups were adjusted for an age group effect (≥ 60 v < 60 years).
RESULTS
Associations of miR-155 Expression With Clinical and Molecular Characteristics in Patients With CN-AML
At diagnosis, high miR-155 expressers had higher WBC counts (P < .001) and higher percentages of blood (P = .004) and BM blasts (P < .001) than low expressers (Table 1). High miR-155 expressers were also more frequently FLT3-ITD positive (P < .001), RUNX1 mutated (P < .001), WT1 mutated (P = .03), and high ERG (P = .02) and BAALC (P = .002) expressers, and less frequently CEBPA mutated (P = .003), IDH2 mutated (P = .004), and FLT3-TKD positive (P = .08; Table 1). However, when younger and older patients were considered separately, the frequencies of these molecular variables in high miR-155 expressers versus low expressers varied slightly according to the age group considered (Data Supplement).
Table 1.
Comparison of Clinical and Molecular Characteristics With miR-155 Expression in Patients With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Characteristic | Low miR-155* (n = 182) | High miR-155* (n = 181) | P | ||
|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | ||
| Age, years | .60 | ||||
| Median | 63 | 62 | |||
| Range | 19-83 | 18-81 | |||
| Male sex | 94 | 52 | 90 | 50 | .75 |
| Race | .73 | ||||
| White | 160 | 89 | 164 | 91 | |
| Nonwhite | 19 | 11 | 17 | 9 | |
| WBC, × 109/L | < .001 | ||||
| Median | 20.7 | 37.9 | |||
| Range | 0.8-295.0 | 1.0-450.0 | |||
| Blood blasts, % | .004 | ||||
| Median | 50 | 64 | |||
| Range | 0-97 | 0-99 | |||
| Bone marrow blasts, % | < .001 | ||||
| Median | 63 | 74 | |||
| Range | 4-97 | 6-97 | |||
| Hemoglobin, g/dL | .75 | ||||
| Median | 9.3 | 9.4 | |||
| Range | 4.9-14.5 | 4.6-15.0 | |||
| Platelet count, × 109/L | .22 | ||||
| Median | 70 | 61 | |||
| Range | 4-481 | 8-850 | |||
| Extramedullary involvement | 43 | 24 | 51 | 29 | .34 |
| NPM1 | .32 | ||||
| Mutated | 109 | 61 | 117 | 66 | |
| Wild type | 70 | 39 | 60 | 34 | |
| FLT3-ITD | < .001 | ||||
| Present | 37 | 21 | 97 | 55 | |
| Absent | 143 | 79 | 80 | 45 | |
| CEBPA | .003 | ||||
| Mutated | 38 | 21 | 17 | 10 | |
| Single mutated | 14 | 12 | |||
| Double mutated | 24 | 5 | |||
| Wild type | 141 | 79 | 160 | 90 | |
| ELN genetic group† | < .001 | ||||
| Favorable | 119 | 66 | 55 | 31 | |
| Intermediate-I | 60 | 34 | 121 | 69 | |
| TET2 | .62 | ||||
| Mutated | 42 | 24 | 46 | 26 | |
| Wild type | 134 | 76 | 129 | 74 | |
| ASXL1 | .86 | ||||
| Mutated | 16 | 9 | 17 | 10 | |
| Wild type | 160 | 91 | 157 | 90 | |
| DNMT3A | .36 | ||||
| Mutated | 57 | 33 | 63 | 38 | |
| R882 | 36 | 44 | |||
| Non-R882 | 21 | 19 | |||
| Wild type | 118 | 67 | 104 | 62 | |
| RUNX1 | < .001 | ||||
| Mutated | 9 | 5 | 33 | 20 | |
| Wild type | 157 | 95 | 128 | 80 | |
| IDH1 | .17 | ||||
| Mutated | 15 | 8 | 23 | 13 | |
| Wild type | 163 | 92 | 151 | 87 | |
| IDH2 | .004 | ||||
| IDH2 mutated | 44 | 25 | 22 | 13 | |
| Codon R140 mutation | 36 | 20 | |||
| Codon R172 mutation | 8 | 2 | |||
| Wild type | 134 | 75 | 152 | 87 | |
| FLT3-TKD | .08 | ||||
| Present | 24 | 13 | 13 | 7 | |
| Absent | 155 | 87 | 164 | 93 | |
| WT1 | .03 | ||||
| Mutated | 11 | 6 | 23 | 13 | |
| Wild type | 168 | 94 | 154 | 87 | |
| MLL-PTD | .53 | ||||
| Present | 10 | 6 | 14 | 8 | |
| Absent | 170 | 94 | 167 | 92 | |
| ERG expression group* | .02 | ||||
| High | 79 | 43 | 102 | 56 | |
| Low | 103 | 57 | 79 | 44 | |
| BAALC expression group* | .002 | ||||
| High | 76 | 42 | 106 | 59 | |
| Low | 106 | 58 | 75 | 41 | |
Abbreviations: ELN, European LeukemiaNet; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene.
The median expression value was used as a cut point. Gene expression was measured using NanoString.
Within patients with cytogenetically normal acute myeloid leukemia (CN-AML), the ELN favorable genetic group comprises patients with mutated CEBPA and/or mutated NPM1 without FLT3-ITD. All remaining patients with CN-AML (ie, those with wild-type CEBPA and NPM1 mutation with FLT3-ITD or with wild-type NPM1 with or without FLT3-ITD) belong to the ELN intermediate-I genetic group.
Prognostic Value of miR-155 Expression
When all patients were considered and analyses were adjusted for age, high miR-155 expressers had lower odds of achieving CR than low expressers (P < .001; odds ratio, 0.41; 95% CI, 0.25 to 0.68). With a median follow-up time for patients alive of 7.9 years (range, 2.3 to 12.9 years), high miR-155 expressers had shorter DFS (P < .001; hazard ratio, 1.59; 95% CI, 1.20 to 2.09) and OS (P < .001; hazard ratio, 2.00; 95% CI, 1.58 to 2.53) than low expressers (Fig 1).
Fig 1.
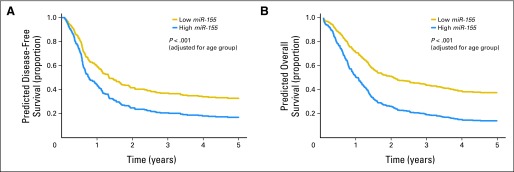
Age group–adjusted clinical outcome for patients with high and low miR-155 expression levels: (A) disease-free survival and (B) overall survival.
In multivariable analyses (Table 2), high miR-155 expressers were approximately 50% less likely to achieve CR (P = .007), after adjusting for NPM1 mutation status (P = .005), BAALC expresser status (P = .002), WBC (P < .001), and age group (P = .003). High miR-155 expressers also had a 60% increased risk of death (P < .001), after adjusting for BAALC expression status (P < .001), age group (P < .001), FLT3-ITD (P < .001), and race (P = .03). However, miR-155 expresser status did not remain in the multivariable model for DFS.
Table 2.
Multivariable Analyses in Patients With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Group | Complete Remission |
Disease-Free Survival |
Overall Survival |
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| All patients | |||||||||
| miR-155 expression, high v low | 0.46 | 0.27 to 0.81 | .007 | 1.20 | 0.89 to 1.61 | .23 | 1.62 | 1.25 to 2.09 | < .001* |
| NPM1, mutated v wild type | 2.42 | 1.31 to 4.48 | .005 | ||||||
| BAALC expression, high v low | 0.37 | 0.20 to 0.69 | .002 | 1.73 | 1.28 to 2.34 | < .001 | 2.16 | 1.68 to 2.76 | < .001 |
| WBC, each 50 units | 0.65 | 0.51 to 0.82 | < .001 | 1.17 | 1.02 to 1.34 | .02 | |||
| Age group, older v younger | 0.43 | 0.24 to 0.75 | .003 | 2.48 | 1.86 to 3.33 | < .001 | 2.38 | 1.84 to 3.08 | < .001 |
| FLT3-ITD, positive v negative | 1.96 | 1.45 to 2.65 | < .001 | 1.78 | 1.37 to 2.30 | < .001 | |||
| ERG expression, high v low | 1.38 | 1.01 to 1.86 | .04 | ||||||
| Race, white v nonwhite | 1.62 | 1.04 to 2.52 | .03 | ||||||
| Patients, age < 60 years | |||||||||
| miR-155 expression, high v low | 0.39 | 0.15 to 1.05 | .06 | 2.13 | 1.29 to 3.51 | .003 | 1.84 | 1.14 to 2.97 | .01 |
| RUNX1, mutated v wild type | 0.21 | 0.06 to 0.72 | .01 | ||||||
| Age, each 10-year increase | 0.45 | 0.26 to 0.77 | .004 | ||||||
| WBC, each 50 units | 1.49 | 1.22 to 1.81 | < .001 | ||||||
| FLT3-ITD, positive v negative | 2.82 | 1.70 to 4.66 | < .001 | 1.82 | 1.15 to 2.87 | .01 | |||
| FLT3-TKD, present v absent | 3.27 | 1.65 to 6.49 | < .001 | ||||||
| BAALC expression, high v low | 2.66 | 1.62 to 4.35 | < .001 | 2.33 | 1.46 to 3.72 | < .001 | |||
| Race, white v nonwhite | 2.81 | 1.19 to 6.66 | .02 | ||||||
| CEBPA, mutated v wild type | 0.47 | 0.26 to 0.86 | .02 | ||||||
| WT1, mutated v wild type | 2.25 | 1.28 to 3.95 | .005 | ||||||
| Patients, age ≥ 60 years | |||||||||
| miR-155 expression, high v low | 0.46 | 0.22 to 0.94 | .03 | 0.93 | 0.64 to 1.35 | .71 | 1.36 | 1.00 to 1.86 | .05* |
| NPM1, mutated v wild type | 2.45 | 1.10 to 5.47 | .03 | ||||||
| BAALC expression, high v low | 0.32 | 0.14 to 0.70 | .004 | 2.07 | 1.42 to 3.01 | .002 | 2.18 | 1.60 to 2.95 | < .001 |
| WBC, each 50 units | 0.65 | 0.48 to 0.88 | .005 | ||||||
| Age, each 10-year increase | 0.48 | 0.26 to 0.90 | .02 | ||||||
| FLT3-ITD, positive v negative | 1.83 | 1.25 to 2.69 | < .001 | 1.56 | 1.14 to 2.14 | .006 | |||
NOTE. ORs greater than or less than 1.0 mean higher or lower complete remission rates, respectively, for the higher values of the continuous variables and the first category listed for the categorical variables. HRs greater than or less than 1.0 indicate higher or lower risk, respectively, for relapse or death (DFS) or death (OS) for the higher values of the continuous variables and the first category listed for the categorical variables.
Abbreviations: DFS, disease-free survival; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene; HR, hazard ratio; OR, odds ratio; OS, overall survival.
miR-155 expression did not meet the proportional hazards assumption. The model using a time-dependent covariate yielded similar results to the one using only the main effect and that is reported here.
Among younger patients, high miR-155 expressers, compared with low expressers, had a lower CR rate (P = .03; 76% v 90%, respectively) and shorter DFS (P < .001; 5-year DFS, 27% v 55%, respectively) and OS (P < .001; 5-year OS, 28% v 57%, respectively; Figs 2A and 2B; Data Supplement). In multivariable analyses (Table 2), high miR-155 expresser status was associated with a trend for a worse CR rate (P = .06) and remained significantly associated with shorter DFS (P = .003), after adjustment for FLT3-ITD (P < .001), FLT3-TKD (P < .001), BAALC expression status (P < .001), and race (P = .02); high miR-155 expresser status was also associated with shorter OS (P = .01), after adjustment for WBC (P < .001), FLT3-ITD (P = .01), BAALC expression status (P < .001), and CEBPA (P = .02) and WT1 (P = .005) mutation status. The risk of relapse or death of the younger high miR-155 expressers was approximately twice that of the low expressers.
Fig 2.
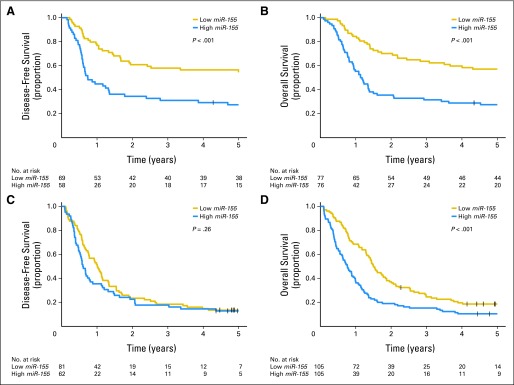
Survival curves according to age group and miR-155 expression (high v low): (A) disease-free survival and (B) overall survival of younger patients and (C) disease-free survival and (D) overall survival of older patients.
Among older patients (Data Supplement), high miR-155 expressers, compared with low expressers, had a lower CR rate (P = .008; 59% v 77%, respectively) and shorter OS (P < .001; 3-year OS, 15% v 25%, respectively; Fig 2D). DFS was not significantly different between the two expression groups (Fig 2C). In multivariable analyses (Table 2), high miR-155 expressers were 50% less likely to achieve CR (P = .03), after adjusting for NPM1 mutation status (P = .03), BAALC expression status (P = .004), WBC (P = .005), and age (P = .02), and had a 40% increased risk of death (P = .05), once adjusted for BAALC expression status (P < .001) and FLT3-ITD (P = .006).
We previously reported that low expression levels of miR-181a had a negative prognostic impact on CN-AML.40 Unfortunately miR-181a expression was available in only a subset of patients included in this study. However, in multivariable models limited to these patients (n = 298; Data Supplement), high levels of miR-155 remained independently associated with shorter OS (P < .001) after adjusting for miR-181a (P < .001), FLT3-ITD (P = .02), WT1 (P = .004), IDH1 (P = .03), ERG (P = .01), BAALC (P < .001), WBC (P = .02), and age (P < .001).
Impact on the European LeukemiaNet Genetic Groups
A modified classification of CN-AML has been recommended by an international expert panel on behalf of the European LeukemiaNet (ELN), in which the ELN favorable genetic group comprises patients with mutated CEBPA and/or mutated NPM1 without FLT3-ITD, whereas the ELN intermediate-I genetic group consists of patients without CEBPA mutations who are either NPM1 mutated with FLT3-ITD or have wild-type NPM1 with or without FLT3-ITD.47 When we analyzed the prognostic significance of miR-155 in patients classified in the ELN favorable genetic group, younger high miR-155 expressers, compared with low expressers, had a significantly lower CR rate (P = .03; 80% v 96%, respectively) and shorter DFS (P = .04; 5-year DFS, 45% v 66%, respectively) and OS (P = .02; 5-year OS, 48% v 71%, respectively; Data Supplement). In contrast, in the ELN intermediate-I genetic group, miR-155 expression did not impact on the probability of CR attainment (P = .99), DFS (P = .41), or OS (P = .20) duration. Among older patients, higher miR-155 expressers, compared with lower expressers, had a shorter OS both in the favorable (P = .06; 3-year OS, 23% v 34%, respectively) and intermediate-I genetic groups (P = .05; 3-year OS, 11% v 11%, respectively; median survival, 0.6 v 1.1 years, respectively).
Biologic Insights
To gain insight into the leukemogenic role of miR-155 expression in CN-AML, we first derived an Affymetrix gene expression signature associated with miR-155 expression in all patients. Using a conservative cutoff for significance (FDR < 0.001), we identified 196 mRNAs significantly correlated with miR-155 expression. Of these mRNAs, 154 were correlated positively (Data Supplement), and 42 were correlated negatively (Data Supplement). Among the positively correlated mRNAs, the most significant was MIR155HG, the primary RNA for miR-155, thereby validating the results of the nCounter assay. The Gene Ontology analysis revealed that genes involved in biologic processes related to antiapoptotic, proliferative, and inflammatory activities were enriched in high miR-155 expressers (FDR < 0.05; Table 3).
Table 3.
Gene Ontology Terms Positively Correlated With miR-155 Expression
| Biologic Process | FDR | Fold Enrichment | No. of Genes |
|---|---|---|---|
| GO:0006915–apoptosis | 7.18E-08 | 5.154073577 | 25 |
| GO:0012501–programmed cell death | 9.76E-08 | 5.078154327 | 25 |
| GO:0042981–regulation of apoptosis | 1.75E-07 | 4.322242001 | 28 |
| GO:0043067–regulation of programmed cell death | 2.18E-07 | 4.279658336 | 28 |
| GO:0010941–regulation of cell death | 2.37E-07 | 4.263904992 | 28 |
| GO:0008219–cell death | 2.66E-06 | 4.315371757 | 25 |
| GO:0016265–death | 3.05E-06 | 4.285569466 | 25 |
| GO:0006916–antiapoptosis | 1.52E-05 | 8.434666429 | 14 |
| GO:0006954–inflammatory response | 4.77E-04 | 5.72815808 | 15 |
| GO:0043066–negative regulation of apoptosis | 0.001336061 | 5.258902193 | 15 |
| GO:0009611–response to wounding | 0.001527465 | 4.21505972 | 18 |
| GO:0043069–negative regulation of programmed cell death | 0.001579 | 5.185658429 | 15 |
| GO:0060548–negative regulation of cell death | 0.001632093 | 5.171253823 | 15 |
| GO:0006952–defense response | 0.011381758 | 3.63249049 | 18 |
| GO:0006955–immune response | 0.013164608 | 3.417524265 | 19 |
| GO:0043122–regulation of I-κB kinase/NF-κB cascade | 0.037285223 | 9.279259196 | 8 |
Abbreviations: FDR, false discovery rate; GO, Gene Ontology; NF-κB, nuclear factor-κB.
For microRNA expression profiling, younger and older patients were separately analyzed to avoid confounding batch effects. Testing for expressed microRNAs correlated with miR-155 expression revealed no associated microRNAs other than miR-155 itself. The mature miR-155, quantified by microRNA microarrays, was the only microRNA significantly correlated with miR-155, as measured by the nCounter assay (P < .001). This finding again validated the results obtained with the nCounter assay.
DISCUSSION
MicroRNAs have been shown to play a role in leukemogenesis and to impact on clinical outcome.1–3 Furthermore, emerging proof-of-principle clinical studies support the potential therapeutic targeting of these small noncoding RNAs in human diseases including cancer.14 In this study, we focused on the clinical significance of miR-155, which has a pivotal role in leukemogenesis.6,9
We show that miR-155 expression levels constitute an independent prognostic factor in patients with primary CN-AML. Higher levels of miR-155 expression were associated with lower odds of achieving CR and higher risk for disease relapse or death. Although higher miR-155 expression impacted negatively on outcome of both younger and older patients, the association with outcome end points differed somewhat for the two age groups. For younger patients, higher miR-155 expresser status was associated with a lower CR rate and shorter DFS and OS. For older patients, higher miR-155 expresser status was associated only with a lower CR rate and shorter OS. This discrepancy between the age groups may be related not only to biologic differences but also to differences in the intensity of consolidation therapy administered to younger and older patients. Nevertheless, miR-155 emerged from our study as a single noncoding RNA with a strong and independent prognostic impact in CN-AML, even when considered in the context of other validated molecular and clinical prognosticators. Thus, our results validate in the clinic previous data from preclinical models supporting a crucial role of miR-155 in leukemia.6,9
We also tested the prognostic significance of miR-155 expression in the ELN genetic groups,47 separately in younger and older patients.48 In both age groups, miR-155 expression had a prognostic impact in the ELN favorable genetic group. Most of these patients do not harbor FLT3-ITD (except for the relatively rare patients with CEBPA mutations and FLT3-ITD), suggesting that the clinical impact of miR-155 is independent from the presence of FLT3-ITD. Indeed, high miR-155 expressers had worse OS than low expressers among FLT3-ITD–negative younger (P = .01; 3-year OS, 35% v 61%, respectively) and older (P = .02; 2-year OS, 24% v 41%, respectively) patients (data not shown). In the ELN favorable genetic group, miR-155 expression remained associated with CR rate (P = .02) and OS duration (P = .003) independently of TET2 and ASXL1 mutations, which we previously reported to impact negatively in this genetic group.33,34 In contrast, the lack of the impact of miR-155 overexpression on the ELN intermediate-I genetic group for younger patients could be related to the colinearity of high miR-155 expression and FLT3-ITD. This made it difficult to establish whether the prognostic impact of these two variables was independent from each other. In contrast, the negative impact of miR-155 overexpression on OS of older, ELN intermediate-I genetic group patients suggests a potential prioritized prognostic relevance of miR-155 over FLT3-ITD in this age group.
We previously reported that miR-181a has prognostic impact only in molecular high-risk patients (ie, those with FLT3-ITD and/or wild-type NPM1), the majority of whom are classified in the ELN intermediate-I genetic group. Interestingly, in a multivariable model that included both miR-155 and miR-181a, in addition to other clinical and molecular prognosticators, higher levels of miR-155 and lower levels of miR-181a remained independently associated with shorter survival. Thus, our data indicate that expression levels of two distinct microRNAs, miR-155 and miR-181a, provide additional and complementary prognostic information for molecular subsets of patients with CN-AML and may potentially be used to refine molecular risk classifications and provide treatment guidance.
To our knowledge, this is the first clinical study where miR-155 was evaluated using nCounter, an amplification enzyme-independent quantification probe-based system that allows digital counting of RNA molecules. Using the nCounter assay as a primary quantification method has the potential to eliminate batch-related pitfalls intrinsic to quantitative PCR-based assays. This would make an accurate analysis of individual patients possible and allow the prospective use of miR-155 expression in the clinic. The accuracy of miR-155 measurements by the nCounter method was validated by their strong correlation with data obtained using two different microarray platforms (ie, Affymetrix oligonucleotide microarrays for the MIR155HG transcript and Ohio State University miR microarray for the mature miR-155).
To further understand how changes in miR-155 expression affect the aggressiveness of the disease, response to treatment, and outcome of patients with CN-AML, we used the combination of genome-wide gene and microRNA expression and Gene Ontology analyses. We show that expression of genes involved in antiapoptotic, proinflammatory, and NF-κB activation processes positively correlated with miR-155 expression. These results support the notion that miR-155 expression contributes to different degrees of disease aggressiveness in leukemia and other types of cancer by increasing cell proliferation rate and survival.7 Because miR-155 is relatively easy to measure at diagnosis, it is possible to envision it as a marker for risk stratification and guidance for targeting treatments such as emerging compounds with antagonistic activity to microRNAs.14 Furthermore, because miR-155 is reported to be a target of NF-κB,4,5 possible treatment approaches targeting this pathway may be a reasonable approach for patients with elevated miR-155 expression.
In summary, we report that the expression of miR-155 is independently associated with clinical outcome in CN-AML and may allow better characterization of molecular risk, especially in patients lacking FLT3-ITD, such as those classified in the ELN favorable genetic group. Although one of the limitations of the study was the retrospective nature of the analyses, it should be underscored that we were able to show independent prognostic significance of miR-155 expression in two distinct age groups (younger and older) that were similarly treated with cytarabine-anthracycline–based induction chemotherapy but received consolidation treatments of different intensity. Nevertheless, the study of additional independent cohorts of patients is required before the miR-155 quantification can be included in molecular panels adopted for risk stratification and treatment guidance for patients with CN-AML. Finally, the ongoing clinical development of compounds capable of downregulating microRNA expression in vivo offers hope for designing novel therapies for patients whose outcome is adversely impacted by high miR-155 expression levels.14
Supplementary Material
Footnotes
See accompanying editorial on page 2065 and article on page 2219; listen to the podcast by Dr Estey at www.jco.org/podcasts
Written on behalf of the Alliance for Clinical Trials in Oncology.
Supported in part by National Cancer Institute (Bethesda, MD) Grants No. CA101140, CA114725, CA31946, CA33601, CA16058, CA77658, CA129657, and CA140158; the Coleman Leukemia Research Foundation; the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.); the Pelotonia Fellowship Program (A.-K.E.); and the Conquer Cancer Foundation (J.H.M.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Guido Marcucci, Kati S. Maharry, Clara D. Bloomfield
Financial support: Guido Marcucci, Clara D. Bloomfield
Administrative support: Clara D. Bloomfield
Provision of study materials or patients: Guido Marcucci, Bayard L. Powell, Jonathan E. Kolitz, Michael A. Caligiuri, Richard M. Stone
Collection and assembly of data: Kati S. Maharry, Klaus H. Metzeler, Yue-Zhong Wu, Susan P. Whitman, Jason H. Mendler, Sebastian Schwind, Heiko Becker, Ann-Kathrin Eisfeld, Andrew J. Carroll, Michael A. Caligiuri
Data analysis and interpretation: Guido Marcucci, Kati S. Maharry, Klaus H. Metzeler, Stefano Volinia, Krzysztof Mrózek, Deedra Nicolet, Jessica Kohlschmidt, Bayard L. Powell, Jonathan E. Kolitz, Ramiro Garzon, Richard M. Stone, Clara D. Bloomfield
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcucci G, Mrózek K, Radmacher MD, et al. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 4.Tili E, Michaille JJ, Wernicke D, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connell RM, Chaudhuri AA, Rao DS, et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tili E, Croce CM, Michaille JJ. miR-155: On the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 9.Costinean S, Sandhu SK, Pedersen IM, et al. Src homology 2 domain–containing inositol-5-phosphatase and CCAAT enhancer-binding protein β are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R, Volinia S, Liu C-G, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitman SP, Maharry K, Radmacher MD, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 13.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 14.Obad S, dos Santos CO, Petri A, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 16.Kolitz JE, George SL, Marcucci G, et al. P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients under age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–1421. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolitz JE, George SL, Dodge RK, et al. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B Study 9621. J Clin Oncol. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 18.Kolitz JE, George SL, Barrier R, et al. A novel post-remission consolidation regimen for patients with acute myeloid leukemia (AML) < 60 years old with normal or unfavorable cytogenetics: Results from CALGB 9621. Blood. 2003;102:175a. (abstr 609) [Google Scholar]
- 19.Mayer RJ, Davis RB, Schiffer CA, et al. Intensive postremission chemotherapy in adults with acute myeloid leukemia. N Engl J Med. 1994;331:896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 20.Stone RM, Berg DT, George SL, et al. Postremission therapy in older patients with de novo acute myeloid leukemia: A randomized trial comparing mitoxantrone and intermediate-dose cytarabine with standard-dose cytarabine. Blood. 2001;98:548–553. doi: 10.1182/blood.v98.3.548. [DOI] [PubMed] [Google Scholar]
- 21.Lee EJ, George SL, Caligiuri M, et al. Parallel phase I studies of daunorubicin given with cytarabine and etoposide with or without the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age or older with acute myeloid leukemia: Results of Cancer and Leukemia Group B study 9420. J Clin Oncol. 1999;17:2831–2839. doi: 10.1200/JCO.1999.17.9.2831. [DOI] [PubMed] [Google Scholar]
- 22.Baer MR, George SL, Dodge RK, et al. Phase 3 study of the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age and older with acute myeloid leukemia: Cancer and Leukemia Group B study 9720. Blood. 2002;100:1224–1232. [PubMed] [Google Scholar]
- 23.Baer MR, George SL, Caligiuri MA, et al. Low-dose interleukin-2 immunotherapy does not improve outcome of patients age 60 years and older with acute myeloid leukemia in first complete remission: Cancer and Leukemia Group B study 9720. J Clin Oncol. 2008;26:4934–4939. doi: 10.1200/JCO.2008.17.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcucci G, Moser B, Blum W, et al. A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a proapoptotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients >60 years old. J Clin Oncol. 2007;25:360s. (suppl; abstr 7012) [Google Scholar]
- 25.Cheson BD, Cassileth PA, Head DR, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–819. doi: 10.1200/JCO.1990.8.5.813. [DOI] [PubMed] [Google Scholar]
- 26.Payton JE, Grieselhuber NR, Chang LW, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. 2009;119:1714–1726. doi: 10.1172/JCI38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 29.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker H, Marcucci G, Maharry K, et al. Mutations of the Wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:788–792. doi: 10.1182/blood-2010-01-262543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaidzik VI, Bullinger L, Schlenk RF, et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML Study Group. J Clin Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 37.Mendler JH, Maharry K, Radmacher MD, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwind S, Marcucci G, Maharry K, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwind S, Maharry K, Radmacher MD, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langer C, Marcucci G, Holland KB, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2009;27:3198–3204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollander M, Wolfe DA. New York, NY: John Wiley & Sons; 1999. Nonparametric Statistical Methods. [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Agresti A. New York, NY: John Wiley & Sons; 1996. An Introduction to Categorical Data Analysis. [Google Scholar]
- 45.Hosmer D, Lemeshow S. New York, NY: John Wiley & Sons; 2000. Applied Logistic Regression. [Google Scholar]
- 46.Hosmer D, Lemeshow S, May S. New York, NY: John Wiley & Sons; 2008. Applied Survival Analysis. [Google Scholar]
- 47.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 48.Mrózek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–4523. doi: 10.1200/JCO.2012.43.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


