Abstract
Recent observations have suggested that classic antibiotics kill bacteria by stimulating the formation of reactive oxygen species. If true, this notion might guide new strategies to improve antibiotic efficacy. In this study the model was directly tested. Contrary to the hypothesis, antibiotic treatment did not accelerate the formation of hydrogen peroxide in Escherichia coli and did not elevate intracellular free iron, an essential reactant for the production of lethal damage. Lethality persisted in the absence of oxygen, and DNA repair mutants were not hypersensitive, undermining the idea that toxicity arose from oxidative DNA lesions. We conclude that these antibiotic exposures did not produce reactive oxygen species and that lethality more likely resulted from the direct inhibition of cell-wall assembly, protein synthesis, and DNA replication.
Results
In recent decades the growing number of antibiotic-resistant pathogens has spurred efforts to further understand and improve the efficacy of the basic antibiotic classes. Most clinically used antibiotics target cell-wall assembly, protein synthesis, or DNA replication. However, recent reports have raised the possibility that although these antibiotics block growth by directly inhibiting the targets mentioned above, they may owe their lethal effects to the indirect creation of reactive oxygen species that then damage bacterial DNA (1–10).
The evidence supporting this proposal included the observation that cell-penetrating dyes were oxidized more quickly inside antibiotic-treated bacteria (3–8). Furthermore, iron chelators (3, 4, 7–9), which suppress hydroxyl-radical-generating Fenton chemistry, and thiourea (4, 6, 8–10), a potential scavenger of hydroxyl radicals, lessened toxicity. Mutations that diminish fluxes through the tricarboxylic acid cycle were protective (3–5), suggesting a key role for respiration, and DNA-repair mutants were somewhat sensitive (4, 8). Systems analysis of aminoglycoside-treated Escherichia coli suggested a model that fits these data (5). It was postulated that interference with ribosome progression would release incomplete polypeptides, some of which are translocated to the cell membranes where they might trigger envelope stress. The Arc regulatory system is perturbed, potentially accelerating respiration and thereby increasing the flux of superoxide and hydrogen peroxide into the cell interior. These two oxidants have known sequelae that ultimately lead to DNA damage. Specifically, superoxide and hydrogen peroxide damage the iron-sulfur clusters of dehydratases (11, 12), releasing iron atoms and elevating the pool of intracellular unincorporated iron (13, 14). This iron can then react with hydrogen peroxide in the Fenton reaction, generating hydroxyl radicals that either directly damage DNA (15) or indirectly oxidize the deoxynucleotide pool, which is subsequently incorporated into DNA (16). This scenario could explain the observed oxidation of intracellular dyes, protection by scavengers and chelators, a requirement for respiration, and the sensitivity of DNA-repair mutants.
This model is plausible, and so we devised experiments to directly test the molecular events that underpin it. The bacterial strain (Escherichia coli MG1655), growth medium (LB), and antibiotic doses were chosen to match those of previous studies (4). Kanamycin was used to target translation, ampicillin to block cell-wall synthesis, and norfloxacin to disrupt DNA replication.
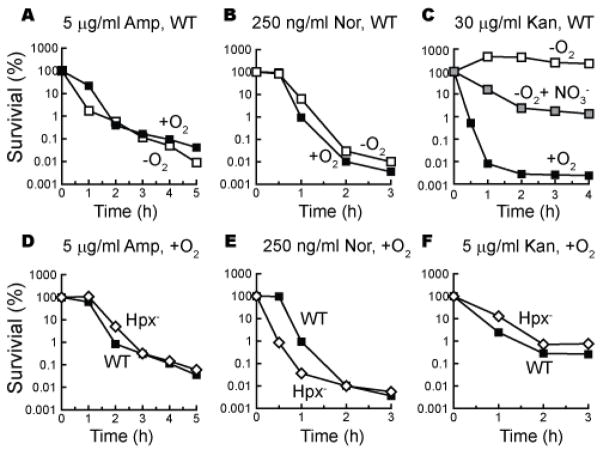
Superoxide and hydrogen peroxide are generated inside cells when flavoenzymes inadvertently transfer a fraction of their electron flux directly to molecular oxygen (15, 17). Thus neither of these reactive oxygen species can be formed under anoxic conditions. We found, however, that ampicillin and norfloxacin were as lethal to cells in an anaerobic chamber as to cells in air-saturated medium (Fig. 1A–C). Anoxia sharply diminished the toxicity of kanamycin, but lethal activity was partially restored at high kanamycin doses when nitrate was provided as an alternative respiratory substrate. This pattern mirrors the effects of oxygen and nitrate upon kanamycin import, which is governed by the magnitude of the proton motive force (18, 19). Thus reactive oxygen species were not required for the lethal actions of kanamycin, and they made no apparent contribution at all to the toxicity of ampicillin and norfloxacin.
Figure 1. Antibiotic efficacy does not require oxygen or H2O2.
(A–C) Wild type cells were treated with ampicillin (A), norfloxacin (B) or kanamycin (C) in the presence (solid squares) or absence (open squares) of oxygen. In panel C anoxic killing was also tested in the presence of 40 mM KNO3 (gray squares). (D–F) Wild-type cells (solid squares, MG1655) and congenic Hpx− cells (open diamonds, AL427) were treated with antibiotics. Results are representative of at least three biological replicates.
Hydroxyl radicals are formed inside cells when ferrous iron transfers electrons to H2O2, so hydroxyl-radical stress is greatest when either H2O2 or ferrous concentrations are high. An E. coli mutant that lacks catalase and peroxidase activities (denoted Hpx−) cannot scavenge H2O2 and more easily accumulates it to toxic levels; substantial damage to proteins and to DNA ensues (12, 20, 21). However, this mutant was not more sensitive to ampicillin or kanamycin than were wild-type cells (Fig. 1D, F). It was more sensitive to norfloxacin, an effect that might result from the pre-existing DNA damage in the scavenger mutant, coupled with the inhibitory effect of norfloxacin upon DNA metabolism (22) (Fig. 1E).
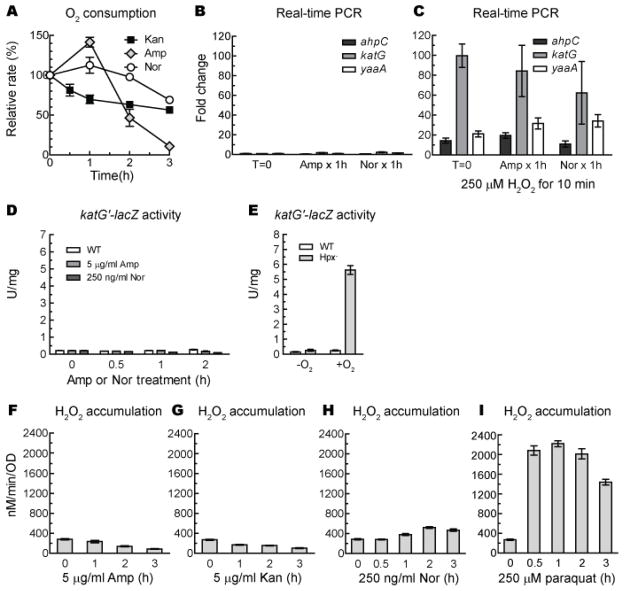
We directly tested the prediction that antibiotic treatment would augment respiration and thereby increase superoxide and H2O2 formation. The respiratory chain is normally a minor source (< 20%) of these oxidants (17), so a very large flux increase would be needed to substantially amplify total intracellular oxidant production. However, measurements of oxygen consumption with a Clark-type electrode showed that respiration slowly declined, rather than increased, when cells were treated with kanamycin (Fig. 2A). This trend persisted throughout the period during which the number of viable cells declined by > 99%. Norfloxacin had little effect on respiration. Ampicillin transiently increased respiration, perhaps by causing envelope damage that dissipated the backpressure of the proton motive force, before cell lysis ended metabolism. In no case did respiration accelerate enough to be a sufficient explanation for toxicity.
Figure 2. Antibiotics do not create H2O2 stress.
(A) O2 consumption rates were not substantially elevated by antibiotic treatment. Oxygen consumption was measured with a Clark-type electrode before and during exposure to 5 μg/ml kanamycin, 5 μg/ml ampicillin, or 250 ng/ml norfloxacin. Respiration rates were normalized to optical density. (B–C) The OxyR regulon was not induced by ampicillin or norfloxacin. (B) mRNA was collected from naïve and antibiotic-exposed cells, and the expression of OxyR-responsive genes was quantified by RT-PCR. Signals were normalized to that of the housekeeping gene gapA, and fold inductions were calculated. Where indicated, cultures were additionally treated with exogenous H2O2 at room temperature for 10 min prior to mRNA collection. (C, D) Expression of a katG′::lacZ fusion was measured in exponential-phase wild-type cells (AL441) before and after antibiotic incubations (C). As a positive control, expression was measured in wild-type and Hpx− cells (AL494) before (−O2) and after 1 hour aeration (+O2) (D). (E–H) Classic antibiotics did not promote H2O2 formation. Aerobic Hpx− cells (AL427) were treated with antibiotic, and at designated time points samples were collected and the ongoing rate of H2O2 production was measured. The redox-cycling compound paraquat (H) was included as a positive control; note that this dose was insufficient to cause cell death (viable counts increased for > 3 hours). Data in this figure are reported as the means and SEM from at least 3 independent experiments.
E. coli features a transcription factor, OxyR, that is activated by H2O2 whenever its levels approach the concentrations (0.5–1 μM) sufficient to damage enzymes or DNA (23). The OxyR regulon includes genes whose products assist in H2O2 scavenging (katG, ahpCF), free-iron control (dps, fur, yaaA), and enzyme protection (mntH, sufABCDE) (15, 24). When cells were treated with ampicillin and norfloxacin, they accumulated lethal damage within one hour; however, RT-PCR analysis indicated that OxyR-controlled genes were not induced (Fig 2B). These antibiotic-treated cells robustly activated katG, ahpC, and yaaA when exogenous H2O2 was added as a control. (After one hour, kanamycin-treated cells failed to respond even to authentic H2O2, perhaps because OxyR protein was depleted in these translationally blocked cells.) This result was confirmed by measurements of katG::lacZ expression (Fig. 2C, D). Thus, neither ampicillin nor norfloxacin create enough H2O2 stress to trigger the natural H2O2 sensor within the cell.
The rate of intracellular H2O2 formation can be directly measured using catalase/peroxidase mutants. Endogenous H2O2 rapidly diffuses from these cells out into the growth medium, and the rate of its accumulation can be quantified by direct assay (Fig. S1) (17). When cells were treated with lethal concentrations of kanamycin and ampicillin, H2O2 production did not increase (Fig. 2E–H). In norfloxacin-treated cells H2O2 formation increased slightly (< 2-fold) after one hour but not at all within the first 30 min when > 99% of cells accumulated lethal damage. In contrast, when cells were treated with a non-lethal dose of the redox-cycling drug paraquat, H2O2 production was stimulated ~ 7-fold. We conclude that these classic bactericidal antibiotics did not promote H2O2 formation.
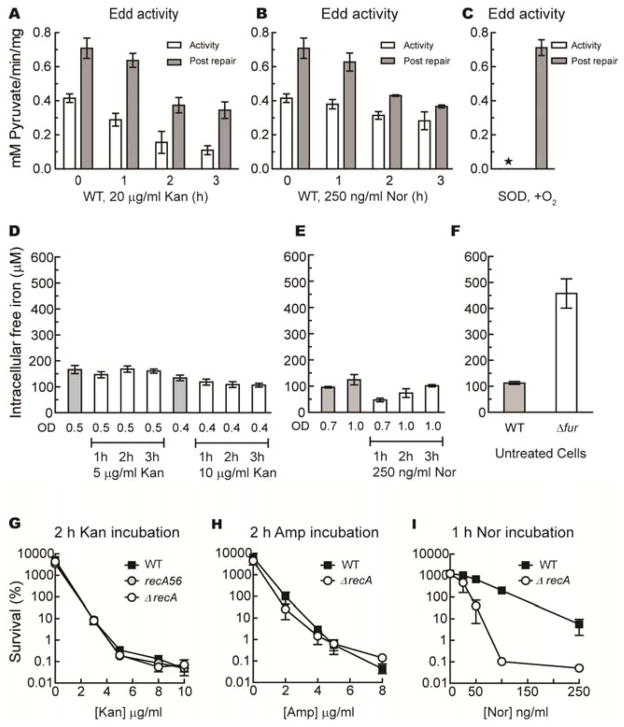
There is one other way that hydroxyl-radical formation might be accelerated: by increases in the amount of unincorporated iron. Iron can accumulate inside cells under conditions of superoxide stress, due to the destruction of labile enzymic iron-sulfur clusters (13, 14), and this mechanism was postulated to occur during antibiotic exposure (4). We monitored the status of 6-phosphogluconate dehydratase, an iron-sulfur enzyme that quickly loses activity in superoxide-stressed cells. We observed that the overall enzyme activity diminished during hours of antibiotic treatment, but the fraction of enzymes with incomplete clusters did not significantly rise (Fig. 3A–C). In contrast, in control mutants that lack superoxide dismutase, the entire enzyme population was inactive due to cluster damage.
Figure 3. Antibiotic lethality does not correlate with levels of unincorporated iron.
(A–C) The superoxide-sensitive enzyme Edd was not damaged by cell exposure to kanamycin or norfloxacin. Cells growing in aerobic LB with 0.2% gluconate were incubated with lethal doses of kanamycin (20 μg/ml, A) or norfloxacin (250 ng/ml, B), and Edd activity was measured before (open bar) and after (closed bar) in vitro cluster repair with ferrous iron/DTT. Panel C represents control data from an SOD-deficient strain (KI232). The asterisk denotes < 4% normal activity. (D–F) Norfloxacin and kanamycin did not increase intracellular iron levels. The levels of intracellular unincorporated iron were determined by whole-cell EPR analysis before and during antibiotic exposure. Gray bars represent iron levels in untreated cells at similar optical densities. (F) Levels of unincorporated iron were measured in untreated congenic wild type cells (MG1655) and Δfur mutants (Jem913). (G–I) DNA-repair mutants are not hypersensitive to antibiotic doses that kill wild-type cells. Wild-type cells (solid squares) and recA mutants (open circles) were treated with the indicated doses of kanamycin (G) or ampicillin (H) for 2 hours, or norfloxacin (I) for one hour. Fig. S3A demonstrates the hypersensitivity of recA mutants to authentic H2O2. Data in this figure are reported as the means and SEM from at least 3 independent experiments.
Levels of intracellular unincorporated iron were directly imaged by whole-cell electron paramagnetic resonance (EPR) spectroscopy (14). These levels did not rise in kanamycin- or norfloxacin-treated cells (Fig. 3D–E). (Levels could not be measured after ampicillin treatment, as lysing cells could not be concentrated for the measurement.) In contrast, free-iron levels were increased 4-fold in fur regulatory mutants that over-synthesize iron import systems (Fig. 3F). Notably, while these fur mutants were slightly more sensitive to ampicillin, they were actually more resistant to kanamycin and norfloxacin (Fig. S2). This result, also observed by others (3), indicates that intracellular free-iron levels do not correlate with antibiotic sensitivity.
DNA is substantially damaged when hydroxyl radicals are rapidly formed inside cells, and mutants that are deficient in the excision or recombinational repair of oxidative DNA lesions are particularly vulnerable (15). The extreme sensitivity of recA and polA mutants to exogenous H2O2 is illustrated in Fig. S3A. However, recA strains exhibited little or no hypersensitivity to killing doses of kanamycin or ampicillin, suggesting that these antibiotics have little effect upon the rate of DNA damage (Fig. 3G–H). The recA mutants were hypersensitive to norfloxacin (Fig. 3I), as expected, since this antibiotic directly introduces DNA strand breaks by interrupting the catalytic cycle of topoisomerases. The polA mutants were actually more resistant to kanamycin and ampicillin than were wild-type strains (Fig. S3B), a phenomenon that likely reflects the slower growth rate of this mutant. In sum, these data indicate that, unlike H2O2 stress, these antibiotics do not introduce lethal damage into the DNA of cells.
Other workers noted that lower doses (1–2 μg/ml) of ampicillin slowly kill recA mutants while being merely bacteriostatic to wild-type strains (4, 8). We reproduced that result (Fig. S4A). However, this behavior also occurred under anoxic conditions, when reactive oxygen species are non-existent (Fig. S4B). The slight sensitivity of recA mutants may reflect defects in septation from their innate replication problems. At higher doses of ampicillin, i.e., at doses sufficient to kill wild-type bacteria, recombination proficiency had little or no impact upon death rate.
Collectively, these data indicate that the known mechanisms of oxidative stress by reactive oxygen species were not involved in causing the death of antibiotic-treated cells. Oxygen was dispensable for toxicity, and neither OxyR activity nor direct measurements of H2O2 formation indicated that reactive oxygen species were formed at accelerated rates. Further, no customary markers of oxidative damage—enzyme damage, increases in free-iron levels, or hypersensitivity of DNA-repair mutants—were detected.
Fluorescein-based dyes such as HPF are more quickly oxidized within antibiotic-treated cells than in control cells, and this fact has been regarded as evidence that hydroxyl radicals are present (25). Indeed, thiourea, a presumptive scavenger of hydroxyl radicals, diminished the antibiotic-augmented fluorescence. However, we observed that although thiourea efficiently blocked dye oxidation in a simple in vitro Fenton system, ethanol—another hydroxyl-radical scavenger of similar efficiency (26)—failed to do so, even at far higher doses (Fig. S5A–B). This result implies that the dye was actually oxidized by the high-valence iron (FeO2+) initially formed by the Fenton reaction, rather than by the hydroxyl radical to which it decomposes; thiourea, a sulfur compound, may have re-reduced the metal before it oxidized the dye. Indeed, the dye was very rapidly oxidized by horseradish peroxidase (Fig. S5C), which has an analogous high-valence metal intermediate but does not release hydroxyl radicals (27). The dye was even oxidized by ferricyanide, a ferric-iron chelate of moderate oxidizing potential (+ 0.44 V) (Fig. S5E). Finally, the fluorescence of the oxidized dye product was itself quenched after the fact by addition of the same doses of thiourea that were used in antibiotic studies (Fig. S5D); this effect cannot be due to hydroxyl-radical scavenging activity. Thus it is plausible that fluorescein-based dyes are predominantly oxidized in vivo by oxidized enzymic metal centers, which may be more prominent in antibiotic-treated cells as metabolism fails. In any case, one must be cautious in interpreting the oxidation of these dyes inside cells.
Finally, cell protection by either thiourea or iron chelators cannot be regarded as diagnostic of lethal Fenton chemistry. Both agents suppressed antibiotic sensitivity even under anaerobic conditions (Fig. S6), when death occurs through pathways that do not involve reactive oxygen species. We conjecture that they did so by reducing growth and/or metabolic rates. In general, it is unlikely that exogenous agents like thiourea could substantially diminish the lifetimes of hydroxyl radicals in vivo, since these radicals already react at nearly diffusion-limited rates with intracellular biomolecules that are collectively at molar concentrations (28). Iron chelators have pleiotropic effects, including the repression of TCA-cycle and respiratory enzymes (29), and may thereby diminish the membrane potential that drives aminoglycoside uptake. Synthesis of these enzymes is also repressed when cells are fed glucose, and this treatment analogously diminished kanamycin sensitivity (Fig. S7). Thus it is advisable to follow up experiments that rely upon these agents with ones that employ more-direct markers of oxidative stress.
We conclude that under our experimental conditions these major classes of antibiotics did not exert their lethal actions through the known mechanisms of oxidative stress. Of course, this does not preclude the possibility that antibiotics trigger novel mechanisms of stress that do not involve the reactive oxygen species that were tested here.
Supplementary Material
Acknowledgments
We thank Dan Hassett (University of Cincinnati) for helpful conversations about this topic, and Jim Collins, Graham Walker, and members of their laboratories for generously sharing ideas, unpublished data, and strains. We also thank Mark Nilges of the Illinois EPR Research Center for assistance. This work was supported by GM49060 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Materials and Methods
References (30–36)
References
- 1.Albesa I, Becerra MC, Battan PC, Páez PL. Biochem Biophys Res Commun. 2004;317:605. doi: 10.1016/j.bbrc.2004.03.085. [DOI] [PubMed] [Google Scholar]
- 2.Goswami M, Mangoli SH, Jawali N. Antimicrob Agents Chemother. 2006;50:949. doi: 10.1128/AAC.50.3.949-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. Cell. 2007;130:797. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Cell. 2008;135:679. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies BW, et al. Mol Cell. 2009;36:845. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeom J, Imlay JA, Park W. J Biol Chem. 2010;285:22689. doi: 10.1074/jbc.M110.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Science. 2012;336:315. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhao X, Malik M, Drlica K. J Antimicrob Chemother. 2010;65:520. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Proc Natl Acad Sci USA. 2012;109:12147. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo CF, Mashino T, Fridovich I. J Biol Chem. 1987;262:4724. [PubMed] [Google Scholar]
- 12.Jang S, Imlay JA. J Biol Chem. 2007;282:929. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liochev SI, Fridovich I. Free Radic Biol Med. 1994;16:29. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 14.Keyer K, Imlay JA. Proc Natl Acad Sci USA. 1996;93:13635. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay JA. Annu Rev Biochem. 2008;77:755. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada M, et al. Mol Microbiol. 2012;86:1364. doi: 10.1111/mmi.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seaver LC, Imlay JA. J Biol Chem. 2004;279:48742. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- 18.Vakulenko SB, Mobashery S. Clin Microbiol Rev. 2003;16:430. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taber HW, Mueller JP, Miller PF, Arrow AS. Microbiol Rev. 1987;51:439. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobota JM, Imlay JA. Proc Natl Acad Sci USA. 2011;108:5402. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, You X, Imlay JA. Proc Natl Acad Sci USA. 2005;102:9317. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drlica K. Curr Opin Microbiol. 1999;2:504. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 23.Aslund F, Zheng M, Beckwith J, Storz G. Proc Natl Acad Sci USA. 1999;96:6161. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng M, et al. J Bacteriol. 2001;183:4562. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. J Biol Chem. 2003;278:3170. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- 26.Anbar M, Neta P. Int J Appl Radiation and Isotopes. 1967;18:493. doi: 10.1016/0020-708x(63)90105-4. [DOI] [PubMed] [Google Scholar]
- 27.Ikemura K, et al. J Am Chem Soc. 2008;130:14384. doi: 10.1021/ja805735g. [DOI] [PubMed] [Google Scholar]
- 28.Davies MJ. Biochim Biophys Acta. 2005;1703:93. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Massé E, Vanderpool CK, Gottesman S. J Bacteriol. 2005;187:6962. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.