Abstract
All Gram-negative bacteria studied to date have been shown to produce outer membrane vesicles (OMVs), which are budded, released spheres of outer membrane with periplasmic content. OMVs have been implicated in the delivery of virulence factors in pathogenesis. However, OMVs also benefit non-pathogenic species by delivering degradative enzymes to defend an ecological niche against competing bacterial species, and they can serve as an envelope stress response. Despite these important roles, there is very little known about the mechanism of production of OMVs. Here we review the advantage of vesiculation, particularly in a non-pathogenic context, as well as the hurdles that have to be overcome in Gram-negative envelope architecture before a vesicle can form and bud. Lastly, we address the question of whether OMV production is a stochastic or regulated process.
Introduction
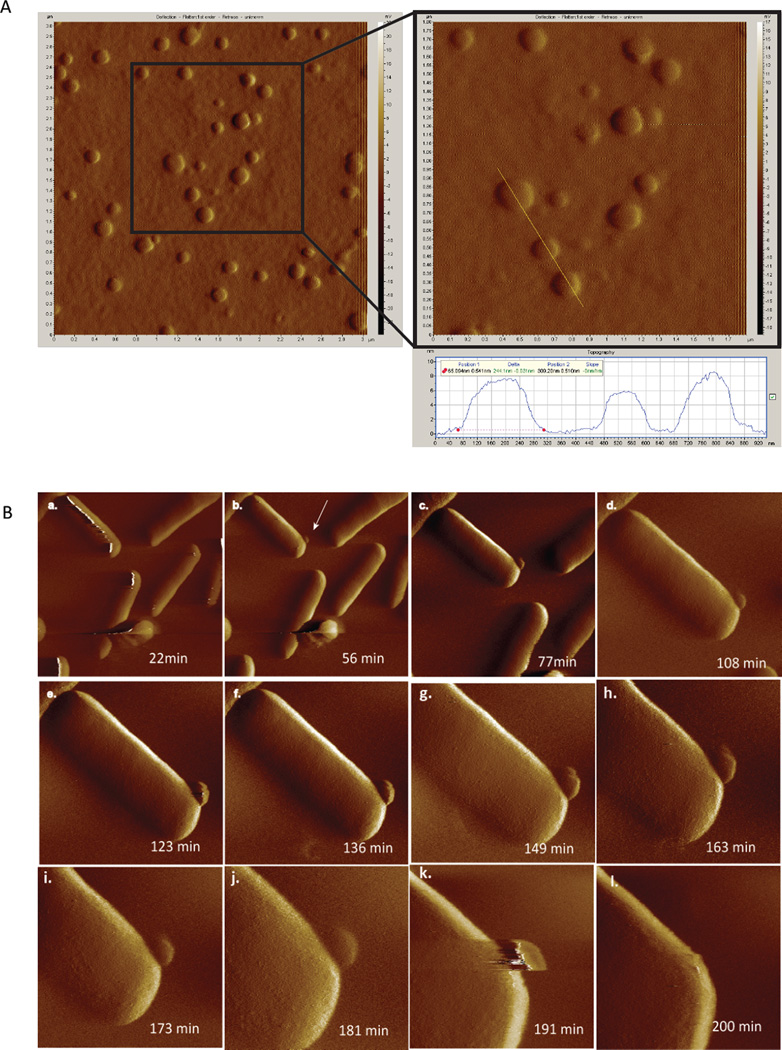
Outer membrane vesicles (OMVs) are secreted, ~ 20 – 250 nm diameter spherical structures of the Gram-negative bacterial outer membrane (OM) with periplasmic soluble components entrapped in their lumen (Fig. 1, Fig 2) [1–8]. Vesiculation appears to be a ubiquitous physiological process: In vitro planktonic [8] and agar-grown laboratory cultures , bacteria living in fresh water environments [7], biofilms [9, 10], and Gram-negative pathogens in animal hosts have been shown to produce OMVs [11, 12]. Biochemical assays and atomic force microscopy imaging of live E. coli shows that OMV production occurs without a disruption in the integrity of the cell [13] (Fig 1B).
Figure 1. Atomic force microscopy images of pure OMVs and OMV budding by E. coli.
A. Purified OMVs from an E. coli nlpI transposon mutant [95]. OMVs were purified from an overnight culture as described [95]. A 50 µl aliquot of the vesicle suspension was applied to gelatin-coated mica for 20 minutes before thoroughly rinsing with deionized water. The sample was dried under a stream of dry nitrogen and imaged in air using contact mode atomic force microscopy. Scanning speed ranged from 6–10 microns per second.
B. Time-course of an E. coli nlpI transposon mutant [95] producing an OMV. The bacteria were grown to log phase (shaking, Luria Broth, 37°C). Cells were immobilized on gelatin-coated mica as described previously [96]. Continuous MacMode™ atomic force microscopy was performed in buffer. The images shown were collected at room temperature at the indicated times using speeds ranging from 1–7 microns per second.
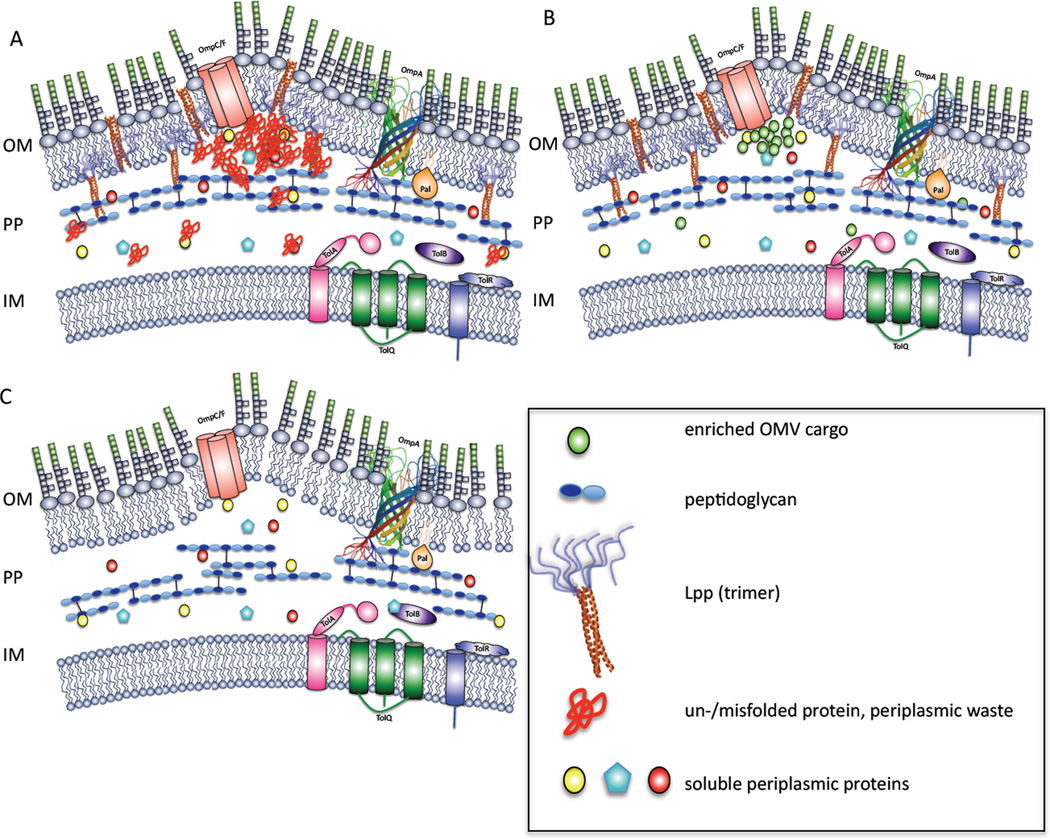
Figure 2. OMV production model.
Overview of Gram-negative envelope architecture in the context of OMV production.
Why might bacteria expend so much energy (in the form of synthesized macromolecules) by secreting entire portions of their envelope? Diverse functions have been ascribed to OMVs, many of which are means by which Gram-negative bacteria interact with their environment. One important function attributed to OMVs is that they serve as virulence mediators for pathogens by carrying virulence factors, such as toxins and proteases, as well as other proinflammatory molecules and antigens, such as flagellin and peptidoglycan (PG) [14–17]. However, OMV production occurs at a constitutive level for a wide variety of bacteria, suggesting this is a highly conserved process which is not only important for pathogens [7]. It has been shown that OMVs can facilitate the transfer of antibiotic resistance enzymes and exogenous DNA between strains and even between species [18, 19], an advantage for both pathogenic and non-pathogenic species. Furthermore, OMVs can act in a general predatory fashion that aids the parent bacterium by generating room in an ecological niche and to acquire nutrients. By the secretion, adherence, and in some cases, subsequent fusion of OMVs containing degradative factors, cell lysis of both Gram-negative and Gram-positive species has been observed [20, 21]. Finally, one of the emerging functional roles of OMVs is that they provide a general advantage in bacterial survival. Increased OMV production correlated with increased survival upon both antimicrobial peptide treatment and T4 bacteriophage infection [22]. Under these conditions, OMVs act as decoy cells absorbing the harmful agents. Survival is also improved by OMV hyperproduction in situations of OM and periplasmic stress [23]. We will discuss in more detail the OMV production response to envelope stress, since studies of this process have also generated mechanistic and regulatory insight into OMV production.
Numerous studies have been dedicated to determining OMV composition in various species and conditions [24–27], as well as the mechanism and regulation of their biogenesis [28]. OMV content is likely derived from the proteins and lipids present at the site of budding, therefore their composition could yield clues as to their mechanism of production. Kulp and Kuehn recently reviewed experimental techniques and isolation of OMVs [8], which are critical to evaluate the compositional analyses, so we will not address those topics here. Instead, we will focus on the architectural requirements for OMV generation and the data supporting several hypothetical models.
The Gram-negative envelope
In order to discuss the mechanics of OMV production, we must introduce the architecture of the Gram-negative bacterial envelope from which the vesicles originate (Fig. 2). The envelope consists of an OM, an inner membrane (IM) and a periplasm between the two membranes, which contains a thin layer of PG [29, 30]. Lipoproteins are membrane anchored via a covalently attached lipid moiety and are most frequently destined for the OM but are also found in the IM [30, 31]. Integral OM proteins (OMPs) almost exclusively fold as β-barrels whereas integral IM proteins are predominantly α-helical structures [32, 33]. Unlike the IM which has phospholipids in both leaflets, in the OM, lipopolysaccharide (LPS) predominates in the outer leaflet and phospholipids in the inner leaflet [34]. Facing the extra-cellular milieu, LPS can act as a barrier against the potentially harmful environment [35]. LPS is also termed endotoxin, based on its highly inflammatory effects on the mammalian immune system [36]. OMVs contain integral OM and OM-anchored lipoproteins, periplasmic protein, as well as the OM phospholipids and LPS [8]. As discussed in more detail later in this review, enrichment and exclusion of several envelope components in OMVs in comparison to their abundance in the cell [14–16, 37–39], support the concept that at least in some cases, OMV production is a regulated mechanism.
Located just underneath the OM, the PG consists of a three-dimensional mesh-like network of glycan strands crosslinked by short peptides. The PG gives the bacteria their characteristic shape and also serves a protective function, preventing cell lysis due to changes in osmolarity and mechanical stress [40]. It has been known for some time that PG is associated with OMVs [3], and this has been confirmed more recently by the functional analysis of OMVs from the pathogens Helicobacter pylori, Pseudomonas aeruginosa and Neisseria gonorrhoeae [41]. These were found to stimulate PG-specific immune responses in vitro as well as in a mouse model of infection.
The layers of the Gram-negative envelope are “stitched” together and stabilized via protein crosslinks reaching from the IM through PG to the OM (Fig. 2). Lpp, an abundant lipoprotein in Escherichia coli, or its counterpart in other Gram-negative bacteria, covalently crosslinks the OM and PG, providing structural envelope integrity [42, 43]. Additionally, OmpA and the Tol-Pal system add non-covalent stability to the envelope [44, 45]. OmpA is an OM porin containing a PG interaction motif [46, 47]. The Tol-Pal system has been attributed as part of the cell division machinery necessary for OM invagination and is highly conserved among Gram-negative bacteria [48, 49]. The Tol-Pal system is essential in Caulobacter crescentus, which does not have an Lpp homologue to form the typical OM-PG covalent crosslinks [50, 51]. In other organisms, mutations and deletions in lpp or pal lead to a very fragile envelope accompanied by the leakage of periplasmic proteins, implying that membrane integrity as well as envelope stability is compromised [44, 52].
Like other bacterial compartments, the envelope is dynamic. Membrane biogenesis, cell division, and remodeling to adapt to new environments require that the bacterial envelope components rearrange while maintaining cellular integrity [32, 53, 54]. Here we consolidate the data on the dynamics of the envelope structure in light of the mechanism of OMV production.
OMVs and envelope crosslinks
Because of the crosslinked architecture of the envelope, it is very likely that an essential, initial step in generating a vesicle bud is the liberation of the OM from the covalent and non-covalent OM-PG-IM crosslinks without concomitant damage and loss of membrane integrity. Historically, it was found that mutations and deletions in Lpp, Tol-Pal and OmpA yield hypervesiculation phenotypes accompanied with cellular leakage as a consequence in membrane instability [44, 52, 55]. However, neither wild-type nor numerous hypervesiculation mutants are necessarily accompanied by membrane instability [13], leading us to believe that naturally occurring OMVs are produced by a more subtle process that depends on random or regulated disruptions in the density of the crosslinks (Fig 3).
Figure 3. Involvement of cargo and PG-OM crosslinks in OMV production.
A. OMVs as cellular garbage cans. Proteinaceous waste is shed via OMVs and the accumulation of misfolded protein may aid in vesicle formation.
B. OMV cargo enrichment. Cargo is secreted via OMVs and its localized accumulation may aid in OMV formation by occurring in regions lacking crosslinks and/or preventing crosslink formation.
C. Destabilized envelope due to the lack of Lpp. The lack of this major OM component results in a discontinuous, leaky OM, and the absence of its ability to crosslink the OM with the PG results in a looser OM.
Before further speculating on possible mechanisms by which Gram-negative species might modulate the positioning of covalent and non-covalent envelope crosslinks, the relevant envelope players will be introduced.
Lpp
Lpp is the most abundant E. coli protein with an estimated 750,000 copies per cell [56]. The lipid moiety of this lipoprotein anchors it to the OM, but it exists in what has been historically referred to as “free” and “bound” forms. The free form is solely OM-anchored, whereas the bound form refers to the Lpp that is covalently crosslinked to PG (Fig. 2). The covalent crosslink occurs between the last residue of Lpp (Lys 58 in E. coli) and diaminopimelic acid of PG. The ratio of the free to the bound form was estimated to be 2:1 [43, 57–60]. Lpp has been shown to form a stable trimeric helical structure in solution in vitro [61, 62]. Shu et al. suggest a model in which one of three helices is covalently attached to PG [62]. Free Lpp monomers most likely also associate into trimers [63]. Immunogold labeling has demonstrated that newly synthesized Lpp is homogeneously distributed across EDTA-permeabilized cells for the free form, and across purified PG sacculi for the bound form [64]. It has also been suggested that the conversion of free to bound form is reversible and that they are in a dynamic equilibrium [60, 65].
There have been studies examining if Lpp mutants behave like the full deletion strain. A lipid-anchor deficient variant of Lpp, as well as a mutation that decreased expression, have been shown to have similar defects as the full deletion strain [66, 67]. These results are not surprising since the absence of the lipid moiety would presumably generate a soluble protein, abolish OM targeting, and consequently any structural contribution native Lpp may give to the envelope, despite its remaining capacity to form covalent PG crosslinks.
Deatherage et al. examined Δlpp complementation with a truncated Lpp version lacking the C-terminal lysine (Lys 58) in Salmonella and found that it behaved like a deletion strain in terms of OMV production and detergent sensitivity [55]. We conducted a complementary experiment in E. coli. Three L,D-transpeptidases YcfS, YbiS and ErfK have been shown to independently form the covalent crosslink between Lpp and PG in E. coli [68]. This triple mutant (ΔycfSΔybiSΔerfK) expresses wild-type Lpp but lacks the enzymes that allow it to catalyze the covalent crosslink. OMV production of this strain was elevated ~ 30-fold over the wild-type, but significantly lower than OMV production by Δlpp, which was ~150-fold over the wild-type (Fig. 4). These results suggest that the free form of OM-localized Lpp adds to membrane stability. The reason behind the different outcomes of these data with Deatherage et al. is not completely clear, but a couple of points are noteworthy. Most obviously, the work was done in highly-related, but nonetheless, different bacterial species. Further, Cowles et al. has shown that the C-terminal portion of the free form of Lpp is exposed to the extracellular surface, and therefore that at least some population of Lpp adopts a transmembrane conformation [63]. It is possible that the Lys 58 deletion altered the charge or conformation of the protein such that the amount of Lpp in the transmembrane state was reduced or prevented and any membrane stabilizing effect with it. Since free Lpp is a highly abundant protein, it is very plausible that its precise localization could substantially contribute to membrane stability.
Figure 4. Free Lpp contributes to membrane integrity.
OMVs were purified from the indicated bacterial mutants grown overnight in LB at 37°C. OMVs were quantified by densitometry of major Omps as per [95] and compared to OMV production by the wild-type (WT). n=4, *, p=0.01, **, p=0.001.
A regulatory link is already known to exist between Lpp expression and σE, a modulator of OMV production (A. J. McBroom, I. A. MacDonald, and M. J. Kuehn, unpublished results, and [69]). In E. coli, σE is a transcription factor that is activated as part of the heat shock response to the accumulation of misfolded OMPs in the envelope and is also essential during normal growth conditions [70, 71]. A small RNA, Reg26, downregulates cellular Lpp concentrations and is under positive control of σE [72]. Regulation of the network of Lpp crosslinks through Reg26 upon σE-activating envelope stress allows for positive regulation of the OMV stress response [73], which will be discussed in more detail later.
Pal
Pal is an OM lipoprotein that has been shown to associate non-covalently with PG via a conserved α-helical PG interaction motif [74–76]. It is part of the Tol-Pal system, which consists of IM proteins, TolA, TolQ and TolR, and the periplasmic protein TolB. TolA-Q-R interact in the membrane via transmembrane helices [77], whereas TolB forms a complex with Pal [76]. The two complexes interact with each other via TolA and Pal in a proton motive force-dependent manner [78]. Additionally, it has been shown that Pal forms independent complexes with Lpp, OmpA and TolB [79], however the Pal-Lpp interaction has not been defined in detail. It has been demonstrated that Pal preferentially localizes to the septum and the new daughter poles, which appears to be more stringent in C. crescentus than in E. coli [48, 49]. This specificity of Pal placement, and particularly the areas of the envelope which are deficient in Tol-Pal complexes, could help explain how Pal plays a role in OMV formation.
Besides its role in envelope stability, the Tol-Pal complex has been shown to aid in OM invagination during the constriction phase of cell division [48, 49]. There appears to be a general correspondence of mutations in proteins involved in cell division and large circular structures emanating from the septum [55, 80, 81]. Although these structures have been labeled as “OMVs,” their size and site of budding are not what we typically see for OMVs produced by wild-type Gram-negative bacteria. Consequently, defining how Pal plays potentially separate roles in cell division and OMV production provides a challenge in the field.
OmpA
Besides its interaction with Pal, OmpA plays additional roles in envelope stability. The carboyxl-terminal domain of Acinetobacter baumanii OmpA (the OmpA-like domain) has been crystallized bound to diaminopimelic acid, confirming the direct interaction with PG [47]. Two strictly conserved residues facilitate the binding of OmpA to PG, implying that this interaction is not species-specific, but is rather a general interaction between PG and OmpA-like proteins [47]. Additionally, multiple groups have shown that the lack of OmpA results in hypervesiculation in Salmonella, Vibrio cholerae and E. coli [55, 73, 82]. Similar to the regulation of Lpp by Reg26, a small RNA discovered in V. cholerae, VrrA, has been shown to downregulate the expression of OmpA, which in turn increases OMV production [73]. This mechanism is conserved across Gram-negative species [83]. MicA, the E. coli homologue as well as VrrA are regulated by σE [73, 84]. Thus, as in the case of Lpp mentioned above, this set of relationships may further explain how Gram-negative bacteria can increase OMV production under σE-activating conditions [8].
OMVs and envelope stress
Increased OMV production was determined to be beneficial to bacteria challenged with stressors that lead to the periplasmic accumulation of misfolded protein waste, such as protease impairment, denaturant, or overexpressed toxic proteins [37]. These results lead to the hypothesis that harmful proteinaceous waste may be shed via OMVs (Fig 3A), and that OMV production is an envelope stress response [37].
The accumulation of proteinaceous waste in the periplasm is generally thought to be prevented by DegP, which is a periplasmic chaperone at low temperatures and a protease at high temperatures [85]. The loss of this dual functional protein was found to generate hypervesiculation phenotypes in E. coli, Salmonella enterica ssp. Typhimurium and P. aeruginosa [37, 69]. The hypervesiculation phenotype of the E. coli degP deletion strain was temperature dependent: at 30°C, vesiculation levels were comparable to the wild-type strain, but hypervesiculation was observed at 37°C [37]. Increasing temperatures lead to increasing amounts of misfolded proteins, which are expected to be the cause of the increase in OMV production (Fig. 3A). The addition of a hypovesiculating mutation to the degP mutant causes a more severe defect that is exacerbated at high temperature, supporting the hypothesis that the high levels of OMV production by the degP mutant relieves toxic proteinaceous stress (C. Schwechheimer and M. J. Kuehn, unpublished results). Furthermore, the degP null mutation is conditionally lethal but can be suppressed by an additional mutation in lpp, presumably by creating a leaky cell and relieving the periplasm from the toxicity of accumulated misfolded material [86]. Together, these data support the idea that the accumulated misfolded periplasmic material can be toxic and that OMV production can improve the survival of bacteria that have accumulated these toxic stressors.
Two studies apparently contradict this hypothesis. McMahon et al. recently showed that the amounts of OMVs produced by the opportunistic pathogen Serratia marcescens actually increased with decreasing temperature [87]. However, the inverse relationship between temperature and OMV production remained unexplained. This phenomenon may be specific to the envelope of S. marcescens as this has not been reported for other species examined thus far. Shibata and Visick examined a V. fischeri strain that was deleted for a degP homologue and reported a consequent decrease in OMV production [88]. The apparently opposite phenotype could be explained by several differences in the two experimental systems. We observed for E. coli that conditions in which DegP protease activity is required correlate with a growth defect as well as protein accumulation in the periplasm and hypervesiculation (C. Schwechheimer and M. J. Kuehn, unpublished results). By contrast, the degP homologue deletion cultures of V. fischeri were grown at 23°C, and the strain had no growth defect [88], which suggests that this protein is not critical under the conditions tested and that there is presumably a relatively low concentration of periplasmic waste. In addition, the degP homologue in V. fischeri plays a role in biofilm formation [88], and this or another function could contribute to its hypovesiculation phenotype.
Possible vesiculation mechanisms
Several mechanistic scenarios have been proposed that could lead to OMV formation, taking into account the crosslinked architecture of the Gram-negative envelope. These models are not mutually exclusive as specific lipid or protein contributions are likely occurring simultaneously with cell wall remodeling to cause bulging out of the OM. Further, different mechanisms may be used during different periods of the cell cycle.
OMV production could be dictated, or at least encouraged, by localization of curvature-inducing proteins such as those containing Inverted-BAR Domains found in eukaryotic cells [89], lipids such as cardiolipin [90], or small molecules such as pseudomonas quinolone signal (PQS) [91]. PQS produced and secreted by Pseudomonas species aids in curvature formation in LPS-liposomes [92] and PQS-producing strains have increased OMV production [69], however, PQS homologues have not been found in all Gram-negative bacteria that have been shown to vesiculate. In addition, polysaccharide-containing components of the OM have been shown to affect OMV production. For S. marscesens, inactivation of the synthesis of the enterobacterial common antigen, a surface exposed repeating sugar molecule, modulated vesiculation [87]. For wild-type P. aeruginosa, a minor species of LPS with highly charged O-antigen is enriched in OMVs [20, 21]. Despite the appeal of such single molecule-driven mechanisms, the connections between the envelope components cannot be ignored, and it is unlikely that stimulating membrane curvature would be sufficient to generate and release OMVs without simultaneously taking advantage of inconsistent intervals between envelope crosslinks (Fig. 2).
Several mechanisms are proposed by which OMV production could occur upon modulation of the covalent interactions between PG and the OM. First, non-uniform spatial distribution of the bonds could promote particular sites to become OMV bud sites. In this scenario, there would be localized breaking of bonds or localized downregulation of crosslinking proteins, and OM budding would occur more rapidly than diffusion of the bonds. Second, at particular sites in the envelope, OM biogenesis might occur more rapidly with respect to the underlying PG, generating a flexible OM and preventing the formation of covalent connections until the region matured. In this case, covalent bonds might be enriched in the more mature areas of the envelope. Third, regions of PG could be excised to create a localized excess of OM that could bud outwards and result in an OMV.
A general prerequisite for creating a OMV seems to be the generation of space between envelope crosslinks either to initiate a budding event, or to stimulate progression of a proto-bud. Space between PG and the OM may be achieved via preferential localization of Pal [48, 49], downregulation of OmpA and Lpp [73], and/or the absence of Lpp-PG crosslinking at a particular location. Pal not only forms non-covalent bonds with PG, but also with several other proteins, so an interaction with another protein could detach a portion of the OM to form a vesicle.
Once a bud has started to emerge, crosslinks could be broken during the development of the OMV, which would drive bud formation rather than resorption. As they form non-covalent bridges, it is not difficult to imagine that Pal and OmpA could dissociate from PG when the periplasmic dimension increases (discussed in more detail, below). Evidence for the lability of Lpp-PG crosslinks came from pulse chase experiments in which 40% of the pulse-labeled free form of Lpp was found in the bound form after one doubling time, and the radioactivity of the bound form, as well as the relative amount of the free form, did not change after a longer chase [60, 65]. These data showed that the Lpp-PG crosslinks must be transient: If conversion to the bound form were irreversible, the ratio of labeled free to labeled bound form would diminish over time, eventually leaving no pulse-labeled free form. To date however, no enzyme has been reported to cleave the covalent Lpp-PG crosslink, and further studies are needed to determine if such an enzyme exists.
Lastly, either by increasing OM synthesis, by excising PG, or by modulating the rate of PG turnover with respect to OM synthesis at a location between OM-PG crosslinks, a vesicle bud of the OM could emerge. This scenario suggests that the OM synthesis, the PG excision, and/or remodeling machinery is discretely localized and corresponds to sites of OMV production. Evidence has not yet been presented to either support or discredit this model.
OMV production: stochastic or regulated?
The short answer to this fundamental question is that it has not yet been established whether OMV production is a stochastic or regulated process. In fact, there is evidence for both, and it may depend on the conditions. Below, we summarize and interpret the evidence supporting the two routes.
Arguing against a stochastic process is the enrichment of some proteins in OMVs in comparison to their abundance in the bacterial envelope (Fig. 3B). It has been demonstrated that a chimeric protein mimicking a misfolded OMP, which is recognized by the cell as an σE envelope stress signal [93], is at least 10-fold enriched in OMVs with respect to its concentration in the periplasm [37]. Although this cargo was an artificial chimera, such enrichment is also expected for naturally-expressed unfolded proteins. In enterobacterial pathogens, Wai et al. demonstrated that the oligomeric pore-forming cytotoxin ClyA is enriched in the secreted OMVs [15]. Heat-labile enterotoxin [14], one of the toxins of enterotoxigenic E. coli, as well as leukotoxin from Actinobacillus actinomcetemcomitans [16] have been shown to be secreted and enriched in OMVs. Preferential sorting into OMVs has also been demonstrated for gingipains, proteases that constitute a major virulence factor of the human pathogen Porphyromonas gingivalis [38]. In this study, not only was preferential OMV inclusion observed, but also OMV exclusion of abundant OMPs. Importantly, in this study Haurat et al. also showed evidence that this sorting process is LPS O-antigen dependent. Cargo enrichment in OMVs from Myxococcus xanthus has also been reported, as alkaline phosphatase activity was almost exclusively found in OMVs [39]. These data support the concept that at least in some cases, OMVs formed under native conditions are the result of a regulated process involving a cargo sorting mechanism.
Non-native conditions highlight the possibility that OMVs can also be stochastically generated due to a significant decrease in envelope ties. As detailed earlier, cells lacking envelope crosslinking components exhibit unstable membranes and consequently hypervesiculate (Fig. 3C). Membrane-PG dissociation was indeed observed in high-resolution cryo electron tomography images of C. crescentus harboring deletions in the Tol-Pal system [81]. A mutation or deletion in degP also causes hypervesiculation without activating σE [37].Accumulated misfolded envelope material could downregulate crosslinking proteins in a σE-independent manner, but it is also possible that for these non σE-inducing conditions, the periplasmic dimension increases due to the accumulation of misfolded protein waste. This may disturb the distribution of non-covalent envelope crosslinks, simply because of their finite length or because periplasmic expansion distorts regions of the envelope that naturally lack crosslinks (Fig. 3A). Separation could become amplified further by the inability of crosslinked components to be generated or diffuse into that region because of the increased physical distance between PG and the OM. In either case, the result would be increased OMV production that is regulated by the amount of bulk protein in the periplasm. Indeed, overexpression of non-σE-activating periplasmic proteins cause OMV hyperproduction [23]. Notably, these overexpressed periplasmic proteins were not enriched in OMVs, further supporting the existence of a stochastic process.
Would periplasmic distension or changes in PG-OM crosslinking location or density ever occur naturally, without mutation or ectopic gene overexpression? Envelope stress that triggers OMP unfolding can cause σE-dependent downregulation of OmpA and Lpp by MicA and Reg26, respectively. Environmental stressors also could cause non σE-activating protein misfolding, which could overwhelm the other stress-response pathways, and lead to conditions of periplasmic bloating. Accumulated material could cause the periplasmic space to expand beyond the physical distance of the PG-OM crosslinks, as discussed above (Fig. 3A). Thus, similar mechanisms may come into play under native conditions and result in regulated vesicle production.
In sum, changes in the envelope composition can induce regulated OMV production or accentuate a stochastic process. OMV production can be modulated by σE-activation and consequent downregulation of OmpA and Lpp by MicA and Reg26, respectively, or by affecting the number or location of the ties between the PG and the OM through a physical increase in periplasmic expansion.
OMVs and PG turnover
Besides being involved in the envelope crosslinks, PG itself may play a role in OMV production. PG is a highly dynamic polymer that is constantly undergoing synthesis and degradation events through a highly regulated and often redundant repertoire of enzymes [40]. Hayashi et al. showed that a mutation in a PG amidase, an enzyme that cleaves PG amide bonds resulted in hypervesiculation in P. gingivalis [94]. This result coincides with our data generated from E. coli. Strains harboring mutations in PG hydrolases, such as the endopeptidases (which cleave peptide crosslinks) or the lytic transglycosylases (which cleave glycosidic bonds), also cause hypervesiculation (C. Schwechheimer and M. J. Kuehn, unpublished results). Whether these phenoytpes are also due to effects on PG-OM crosslinking remains to be determined, although it is clear that not all of the mutations that cause hypervesiculation cause a decrease in crosslinking and vice-versa (C. Schwechheimer and M. J. Kuehn, manuscript in preparation). These data not only reveal that altering the underlying PG can modulate vesicle formation of the OM, but also support the model that decreasing PG degradation may lead to an excess of OM material which could form OMVs.
Concluding Remarks
A vast amount of understanding has been gained over the years as to the role OMVs play in pathogenesis as well as their more general functional properties. The regulation and mechanism of their production is still very cryptic and the knowledge we have is fragmented, but what seems to become clear is that there may be multiple means by which OMVs are produced. Here, we touch on envelope protein accumulation, envelope crosslinks, and the biogenesis of PG and the OM. Nevertheless, major questions in the field remain. How is membrane curvature achieved? How do OMVs ultimately pinch off? How is OMV production regulated? How is cargo selectively enriched? What are the temporal characteristics of vesicle formation with respect to cell cycle? Further work will be necessary to answer these questions.
Acknowledgements
The authors acknowledge research support from NIH grants R01-GM099471, R01-AI79068, and R01-AI64464 and the Commonwealth Health Research Board of Virginia.
Abbreviations
- IM
inner membrane
- LPS
lipopolysaccharide
- OMV
outer membrane vesicle
- OM
outer membrane
- OMP
outer membrane protein
- PG
peptidoglycan
- PQS
pseudomonas quinolone signal
References
- 1.Gankema H, Wensink J, Guinee PA, Jansen WH, Witholt B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect Immun. 1980;29(2):704–713. doi: 10.1128/iai.29.2.704-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mug-Opstelten D, Witholt B. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli. Biochim Biophys Acta. 1978;508(2):287–295. doi: 10.1016/0005-2736(78)90331-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett. 1998;163(2):223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta. 1976;455(3):889–899. doi: 10.1016/0005-2736(76)90058-4. [DOI] [PubMed] [Google Scholar]
- 5.Mayrand D, Grenier D. Biological activities of outer membrane vesicles. Can J Microbiol. 1989;35(6):607–613. doi: 10.1139/m89-097. [DOI] [PubMed] [Google Scholar]
- 6.Kadurugamuwa JL, Beveridge TJ. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40(5):615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 7.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. JBacteriol. 1999;181(16):4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20(3–4):291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 10.Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188(16):5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandtzaeg P, Bryn K, Kierulf P, Ovstebo R, Namork E, Aase B, Jantzen E. Meningococcal endotoxin in lethal septic shock plasma studied by gas chromatography, mass-spectrometry, ultracentrifugation, and electron microscopy. J Clin Invest. 1992;89(3):816–823. doi: 10.1172/JCI115660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellman J, Loiselle PM, Zanzot EM, Allaire JE, Tehan MM, Boyle LA, Kurnick JT, Warren HS. Release of gram-negative outer-membrane proteins into human serum and septic rat blood and their interactions with immunoglobulin in antiserum to Escherichia coli J5. J Infect Dis. 2000;181(3):1034–1043. doi: 10.1086/315302. [DOI] [PubMed] [Google Scholar]
- 13.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188(15):5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275(17):12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115(1):25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 16.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32(1):1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 17.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74(1):81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other entericbacteria . Appl Environ Microbiol. 2000;66(10):4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171(5):2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens conceptually new antibiotics. J Bacteriol. 1996;178(10):2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Clarke AJ, Beveridge TJ. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J Bacteriol. 1996;178(9):2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63(2):545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson T, Matrakas D, Taylor YU, Manyam G, Morozov VN, Zhou W, van Hoek ML. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J Proteome Res. 10(3):954–967. doi: 10.1021/pr1009756. [DOI] [PubMed] [Google Scholar]
- 25.Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7(17):3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 26.Mullaney E, Brown PA, Smith SM, Botting CH, Yamaoka YY, Terres AM, Kelleher DP, Windle HJ. Proteomic and functional characterization of the outer membrane vesicles from the gastric pathogen Helicobacter pylori. Proteomics Clin Appl. 2009;3(7):785–796. doi: 10.1002/prca.200800192. [DOI] [PubMed] [Google Scholar]
- 27.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11(16):3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 28.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 80(6):1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raetz CR, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265(3):1235–1238. [PubMed] [Google Scholar]
- 30.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2(5) doi: 10.1101/cshperspect.a000414. a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim BiophysActa. 2004;1693(1):5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5) doi: 10.1101/cshperspect.a000414. a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luirink J, von Heijne G, Houben E, de Gier JW. Biogenesis of inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- 34.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15(12):2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 35.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63(2):545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286(2):1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of Myxococcus xanthus outer membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012 doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 40.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778(9):1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12(3):372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 42.Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta. 1975;415(3):335–377. doi: 10.1016/0304-4157(75)90013-1. [DOI] [PubMed] [Google Scholar]
- 43.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. EurJ Biochem. 1969;10(3):426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 44.Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol. 2002;184(3):754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y. The function of OmpA in Escherichia coli. Biochem Biophys Res Commun. 2002;292(2):396–401. doi: 10.1006/bbrc.2002.6657. [DOI] [PubMed] [Google Scholar]
- 46.Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. 2007;273(1):1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- 47.Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim SI, et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 26(1):219–228. doi: 10.1096/fj.11-188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2010;192(19):4847–4858. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63(4):1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sturgis JN. Organisation and evolution of the tol-pal gene cluster. J Mol Microbiol Biotechnol. 2001;3(1):113–122. [PubMed] [Google Scholar]
- 51.Anwari K, Poggio S, Perry A, Gatsos X, Ramarathinam SH, Williamson NA, Noinaj N, Buchanan S, Gabriel K, Purcell AW, et al. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus. PLoS One. 2010;5(1):e8619. doi: 10.1371/journal.pone.0008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol Gen Genet. 1978;(1):167. 1–9. doi: 10.1007/BF00270315. [DOI] [PubMed] [Google Scholar]
- 53.Ernst RK, Guina T, Miller SI. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3(14–15):1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 54.Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2012;1277(1):8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 55.Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. Biogenesis of bacterial membrane vesicles. Mol Microbiol. 2009;72(6):1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikaido H. Outer Membrane. In: Neidhardt FC, Curtis RI, Ingraham JL, Lin ECC, Low KB, Magasnik B, et al., editors. E. coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 29–47. [Google Scholar]
- 57.Braun V, Bosch V. Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur J Biochem. 1972;28(1):51–69. doi: 10.1111/j.1432-1033.1972.tb01883.x. [DOI] [PubMed] [Google Scholar]
- 58.Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on themurein . Eur J Biochem. 1970;13(2):336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 59.Braun V, Wolff H. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem. 1970;14(2):387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 60.Inouye M, Shaw J, Shen C. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem. 1972;247(24):8154–8159. [PubMed] [Google Scholar]
- 61.Choi DS, Yamada H, Mizuno T, Mizushima S. Trimeric structure and localization of the major lipoprotein in the cell surface of Escherichia coli. J Biol Chem. 1986;261(19):8953–8957. [PubMed] [Google Scholar]
- 62.Shu W, Liu J, Ji H, Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J Mol Biol. 2000;299(4):1101–1112. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- 63.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol. 2011;79(5):1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hiemstra H, Nanninga N, Woldringh CL, Inouye M, Witholt B. Distribution of newly synthesized lipoprotein over the outer membrane and the peptidoglycan sacculus of an Escherichia coli lac-lpp strain. J Bacteriol. 1987;169(12):5434–5444. doi: 10.1128/jb.169.12.5434-5444.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inouye M, Hirashima A, Lee N. Discussion paper: biosynthesis and assembly of a structural lipoprotein in the envelope of Escherichia coli. Ann N YAcadSci. 1974;235(0):83–90. doi: 10.1111/j.1749-6632.1974.tb43258.x. [DOI] [PubMed] [Google Scholar]
- 66.Yem DW, Wu HC. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J Bacteriol. 1978;133(3):1419–1426. doi: 10.1128/jb.133.3.1419-1426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torti SV, Park JT. Lipoprotein of gram-negative bacteria is essential for growth and division. Nature. 1976;263(5575):323–326. doi: 10.1038/263323a0. [DOI] [PubMed] [Google Scholar]
- 68.Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, Marie A, Mengin-Lecreulx D, Arthur M, Gutmann L. Identification of the L,D- transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol. 2007;189(10):3927–3931. doi: 10.1128/JB.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tashiro Y, Sakai R, Toyofuku M, Sawada I, Nakajima-Kambe T, Uchiyama H, Nomura N. Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J Bacteriol. 2009;191(24):7509–7519. doi: 10.1128/JB.00722-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Las Penas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997;179(21):6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52(3):613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 72.Gogol E. Role of sRNAs in the sigmaE Dependent Cell Envelope Stress Response in Escherichia coli. San Francisco: University of California, San Francisco; 2011. [Google Scholar]
- 73.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70(1):100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mizuno T. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J Biochem. 1979;86(4):991–1000. doi: 10.1093/oxfordjournals.jbchem.a132631. [DOI] [PubMed] [Google Scholar]
- 75.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12(2):333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 76.Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni JC. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29(1):359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 77.Derouiche R, Benedetti H, Lazzaroni JC, Lazdunski C, Lloubes R. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270(19):11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 78.Cascales E, Gavioli M, Sturgis JN, Lloubes R. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol Microbiol. 2000;38(4):904–915. doi: 10.1046/j.1365-2958.2000.02190.x. [DOI] [PubMed] [Google Scholar]
- 79.Cascales E, Lloubes R. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol. 2004;51(3):873–885. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- 80.Chakraborti AS, Ishidate K, Cook WR, Zrike J, Rothfield LI. Accumulation of a murein-membrane attachment site fraction when cell division is blocked in lkyD and cha mutants of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1986;168(3):1422–1429. doi: 10.1128/jb.168.3.1422-1429.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol Microbiol. 2010;77(1):56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136(1):280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11(12):941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 84.Udekwu KI, Wagner EG. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007;35(4):1279–1288. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97(3):339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 86.Strauch KL, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171(5):2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McMahon KJ, Castelli ME, Vescovi EG, Feldman MF. Biogenesis of Outer Membrane Vesicles in Serratia marcescens Is Thermoregulated and Can Be Induced by Activation of the Rcs Phosphorelay System. J Bacteriol. 2012;194(12):3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shibata S, Visick KL. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol. 194(1):185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 21(4):340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 90.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2(11):e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 92.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. MolMicrobiol. 2008;69(2):491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113(1):61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 94.Hayashi J, Hamada N, Kuramitsu HK. The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol Lett. 2002;216(2):217–222. doi: 10.1111/j.1574-6968.2002.tb11438.x. [DOI] [PubMed] [Google Scholar]
- 95.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188(15):5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doktycz MJ, Sullivan CJ, Hoyt PR, Pelletier DA, Wu S, Allison DP. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy. 2003;97(1–4):209–216. doi: 10.1016/S0304-3991(03)00045-7. [DOI] [PubMed] [Google Scholar]