Abstract
Purpose
To determine the association of RUNX1 mutations with therapeutic outcome in younger and older patients with primary cytogenetically normal acute myeloid leukemia (CN-AML) and with gene/microRNA expression signatures.
Patients and Methods
Younger (< 60 years; n = 175) and older (≥ 60 years; n = 225) patients with CN-AML treated with intensive cytarabine/anthracycline-based first-line therapy on Cancer and Leukemia Group B protocols were centrally analyzed for RUNX1 mutations by polymerase chain reaction and direct sequencing and for established prognostic gene mutations. Gene/microRNA expression profiles were derived using microarrays.
Results
RUNX1 mutations were found in 8% and 16% of younger and older patients, respectively (P = .02). They were associated with ASXL1 mutations (P < .001) and inversely associated with NPM1 (P < .001) and CEBPA (P = .06) mutations. RUNX1-mutated patients had lower complete remission rates (P = .005 in younger; P = .006 in older) and shorter disease-free survival (P = .058 in younger; P < .001 in older), overall survival (P = .003 in younger; P < .001 in older), and event-free survival (P < .001 for younger and older) than RUNX1 wild-type patients. Because RUNX1 mutations were more common in older patients and almost never coexisted with NPM1 mutations, RUNX1 mutation–associated expression signatures were derived in older, NPM1 wild-type patients and featured upregulation of genes normally expressed in primitive hematopoietic cells and B-cell progenitors, including DNTT, BAALC, BLNK, CD109, RBPMS, and FLT3, and downregulation of promoters of myelopoiesis, including CEBPA and miR-223.
Conclusion
RUNX1 mutations are twice as common in older than younger patients with CN-AML and negatively impact outcome in both age groups. RUNX1-mutated blasts have molecular features of primitive hematopoietic and lymphoid progenitors, potentially leading to novel therapeutic approaches.
INTRODUCTION
Acute myeloid leukemia (AML) is a cytogenetically and molecularly heterogeneous disease characterized by maturation arrest and uncontrolled proliferation of abnormal myeloid precursors. Cytogenetically normal AML (CN-AML) is the largest cytogenetic group among both younger (< 60 years) and older (≥ 60 years) patients with AML and the one that is best characterized molecularly.1–3 Several recurring mutations with prognostic significance in genes such as FLT3,4,5NPM1,6,7CEBPA,8,9 WT1,10,11MLL,12,13IDH1,14,15IDH2,14,15 and TET216 have been identified in CN-AML. These mutations are increasingly being used both as single markers and in combination to refine the outcome prediction of patients with CN-AML and to stratify them to risk-adapted treatments.17 In addition to refining prognostication, mutations in some of these genes are associated with distinct gene and microRNA expression signatures9,18 that may provide clues regarding mutation-specific leukemogenic mechanisms.
The runt-related transcription factor 1 (RUNX1) gene encodes the α-subunit of core binding factor, a heterodimeric transcription factor required for definitive hematopoiesis.19 Monoallelic germline mutations in this gene occur in rare cases of familial platelet disorder with predisposition to AML,20,21 and acquired mutations have been identified in myelodysplastic syndromes22,23 and AML.24–34 Acquired RUNX1 mutations have been associated with poor clinical outcome in younger patients with CN-AML31,32; however, these mutations were not found to impact outcome in older patients with CN-AML.31 Moreover, previous studies analyzed patients heterogeneous with regard to cytogenetics,31–33 AML type (primary or secondary),32 and treatment received (including allogeneic stem-cell transplantation [alloSCT] in first complete remission [CR1]),31–33 leaving the prognostic impact and molecular signatures associated with RUNX1 mutations in patients with primary CN-AML as open questions that remain to be fully explored.
The aims of this study were to investigate the prognostic impact of RUNX1 mutations in relatively large and well-characterized cohorts of younger and older patients with primary CN-AML treated similarly with intensive cytarabine/anthracycline-based first-line therapy and to assess whether RUNX1 mutations are associated with specific gene/microRNA expression signatures in CN-AML.
PATIENTS AND METHODS
Patients, Treatment, and Sample Collection
Pretreatment bone marrow or blood samples were obtained from 175 younger patients (age 18 to 59 years) and 225 older patients (age 60 to 83 years) with primary CN-AML, who received intensive cytarabine/anthracycline-based first-line therapy on Cancer and Leukemia Group B trials. Per protocol, no patient received an alloSCT in CR1. For details regarding treatment protocols and sample collection, see the Data Supplement. All patients provided written informed consent, and all study protocols were in accordance with the Declaration of Helsinki and approved by institutional review boards at each center.
Cytogenetic and Mutational Analyses
The diagnosis of CN-AML was based on the analysis of ≥ 20 metaphases in bone marrow specimens subjected to short-term cultures and confirmed by central karyotype review.35 Exons 3 to 8 of RUNX1 were amplified from genomic DNA by polymerase chain reaction and analyzed by direct sequencing. All mutations were validated by repeat polymerase chain reaction and sequencing. Patients were also characterized centrally for FLT3 internal tandem duplication (ITD),4FLT3 tyrosine kinase domain mutations,36MLL partial tandem duplication,13,37NPM1,6,18WT1,10CEBPA,9IDH114, IDH2,14TET2,16ASXL1,38 and DNMT3A39 mutations as previously reported. Methods regarding germline analysis and determination of phase in double-mutant patients are provided in the Data Supplement.
Microarray Experiments
Gene expression profiling was performed using Affymetrix (Santa Clara, CA) oligonucleotide microarrays, and microRNA expression profiling was performed using a custom microarray, as previously reported.18 Differentially expressed probe sets or probes were identified by comparing RUNX1-mutated and RUNX1 wild-type patients, using univariable significance levels of P < .001 for gene expression and P < .005 for microRNA expression profiles. For details regarding microarray and gene set analysis, see the Data Supplement.
Statistical Analyses
Baseline characteristics were compared between RUNX1-mutated and RUNX1 wild-type patients using Fisher's exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Definitions of clinical end points (ie, complete remission [CR], disease-free survival [DFS], overall survival [OS], and event-free survival [EFS]) are provided in the Data Supplement. For time-to-event analyses, survival estimates were calculated using the Kaplan-Meier method, and groups were compared with the log-rank test. We constructed multivariable logistic regression models to analyze factors for the achievement of CR and multivariable Cox proportional hazards models for factors associated with survival end points. For details regarding statistical analyses, see the Data Supplement. All analyses were performed by the Alliance for Clinical Trials in Oncology Statistics and Data Center.
RESULTS
RUNX1 Sequence Variations and Description of Mutations
Of 400 patients, 60 (15%) had at least one RUNX1 sequence variation. Patients with synonymous variations (n = 3) were considered equivalent to RUNX1 wild type. Patients with intron sequence variations of more than 10 base pairs from an exon junction (n = 3) were considered unclassifiable between RUNX1 mutated and RUNX1 wild type and, therefore, were excluded from the analysis. Patients with the missense variation p.L56S (n = 5) were also excluded because of controversy over whether this is a polymorphism32,34 or a true mutation.40 The presence of p.L56S in the germline was confirmed in three of four patients with material available.
The remaining 392 patients were classified as either RUNX1 wild type (n = 343) or RUNX1 mutated (n = 49; Table 1; Data Supplement). Mutations were present in 8% of younger (< 60 years) patients (14 of 173 patients) and 16% of older (≥ 60 years) patients (35 of 219 patients; P = .02). RUNX1-mutated patients had either a single mutation (n = 39) or two distinct mutations (n = 10). Among the latter, mutations were biallelic in all patients with tissue available (n = 6). Five RUNX1-mutated patients with single mutations had a homo-/hemizygous pattern, consistent with loss of heterozygosity for the wild-type RUNX1 allele. Among RUNX1-mutated patients with germline material available (n = 35), mutations were rarely found in the germline (n = 3). Coding sequence mutations were missense (n = 24), nonsense (n = 4), frameshift (n = 22), and in-frame insertions/deletions (n = 2). A list of the specific mutations and germline status in each patient is provided in the Data Supplement.
Table 1.
Comparison of Demographics and Clinical and Molecular Characteristics by RUNX1 Mutation Status in All Patients (N = 392) With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Demographic or Characteristic | RUNX1-Mutated Patients (n = 49) | RUNX1 Wild-Type Patients (n = 343) | P | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Age, years | < .001 | |||||
| Median | 68 | 61 | ||||
| Range | 30-81 | 18-83 | ||||
| Age group, years | .02 | |||||
| < 60 | 14 | 29 | 159 | 46 | ||
| ≥ 60 | 35 | 71 | 184 | 54 | ||
| Male sex | 28 | 57 | 169 | 49 | .36 | |
| Race | .29 | |||||
| White | 46 | 96 | 308 | 90 | ||
| Nonwhite | 2 | 4 | 33 | 10 | ||
| Hemoglobin, g/dL | .01 | |||||
| Median | 8.8 | 9.5 | ||||
| Range | 4.6-11.6 | 4.8-15.0 | ||||
| Platelet count, × 109/L | .46 | |||||
| Median | 77 | 60 | ||||
| Range | 8-309 | 4-850 | ||||
| WBC count, × 109/L | .04 | |||||
| Median | 21 | 28.6 | ||||
| Range | 0.9-434.1 | 0.9-450.0 | ||||
| Blood blasts, % | .006 | |||||
| Median | 37 | 58 | ||||
| Range | 0-96 | 0-99 | ||||
| Bone marrow blasts, % | .54 | |||||
| Median | 65 | 68 | ||||
| Range | 7-97 | 4-97 | ||||
| Extramedullary involvement | 11 | 23 | 90 | 27 | .73 | |
| NPM1 | < .001 | |||||
| Mutated | 3 | 6 | 238 | 69 | ||
| Wild type | 46 | 94 | 105 | 31 | ||
| FLT3-ITD | .87 | |||||
| Present | 17 | 35 | 125 | 36 | ||
| Absent | 32 | 65 | 218 | 64 | ||
| CEBPA | .06 | |||||
| Mutated | 3 | 6 | 60 | 17 | ||
| Single mutated | 2 | 26 | ||||
| Double mutated | 1 | 34 | ||||
| Wild type | 46 | 94 | 283 | 83 | ||
| ELN Genetic Group* | < .001 | |||||
| Favorable | 4 | 8 | 186 | 54 | ||
| Intermediate-I | 45 | 92 | 157 | 46 | ||
| FLT3-TKD | .15 | |||||
| Present | 1 | 2 | 29 | 9 | ||
| Absent | 47 | 98 | 303 | 91 | ||
| WT1 | .61 | |||||
| Mutated | 5 | 12 | 34 | 10 | ||
| Wild type | 43 | 88 | 309 | 90 | ||
| TET2 | 1.00 | |||||
| Mutated | 11 | 23 | 79 | 23 | ||
| Wild type | 37 | 77 | 260 | 77 | ||
| ASXL1 | < .001 | |||||
| Mutated | 17 | 35 | 21 | 6 | ||
| Wild type | 31 | 65 | 321 | 94 | ||
| DNMT3A | .15 | |||||
| Mutated | 12 | 24 | 121 | 36 | ||
| R882 | 8 | 16 | 78 | 23 | ||
| Non-R882 | 4 | 8 | 43 | 13 | ||
| Wild type | 37 | 76 | 214 | 64 | ||
| MLL-PTD | 1.00 | |||||
| Present | 3 | 7 | 19 | 6 | ||
| Absent | 42 | 93 | 286 | 94 | ||
| IDH1 | .24 | |||||
| R132 | 3 | 6 | 42 | 12 | ||
| Wild type | 46 | 94 | 299 | 88 | ||
| IDH2 | .84 | |||||
| IDH2 | 8 | 16 | 62 | 18 | ||
| R140 | 6 | 51 | ||||
| R172 | 2 | 11 | ||||
| Wild type | 41 | 84 | 279 | 82 | ||
Abbreviations: ELN, European LeukemiaNet; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene.
Favorable is defined as CEBPA mutated or FLT3-ITD negative and NPM1 mutated. Intermediate-I is defined as CEBPA wild type and FLT3-ITD positive and NPM1 mutated, FLT3-ITD negative and NPM1 wild type, or FLT3-ITD positive and NPM1 wild type.17
Association of RUNX1 Mutations With Other Clinical and Molecular Characteristics
In the combined cohort of younger and older patients, RUNX1 mutations were associated with a higher median age (P < .001) and lower hemoglobin (P = .01), WBC count (P = .04), and blood blasts (P = .006; Table 1). RUNX1-mutated patients harbored ASXL1 mutations more frequently (P < .001) and NPM1 (P < .001) and CEBPA mutations (P = .06) less frequently than RUNX1 wild-type patients.
Impact of RUNX1 Mutation Status on Treatment Outcome
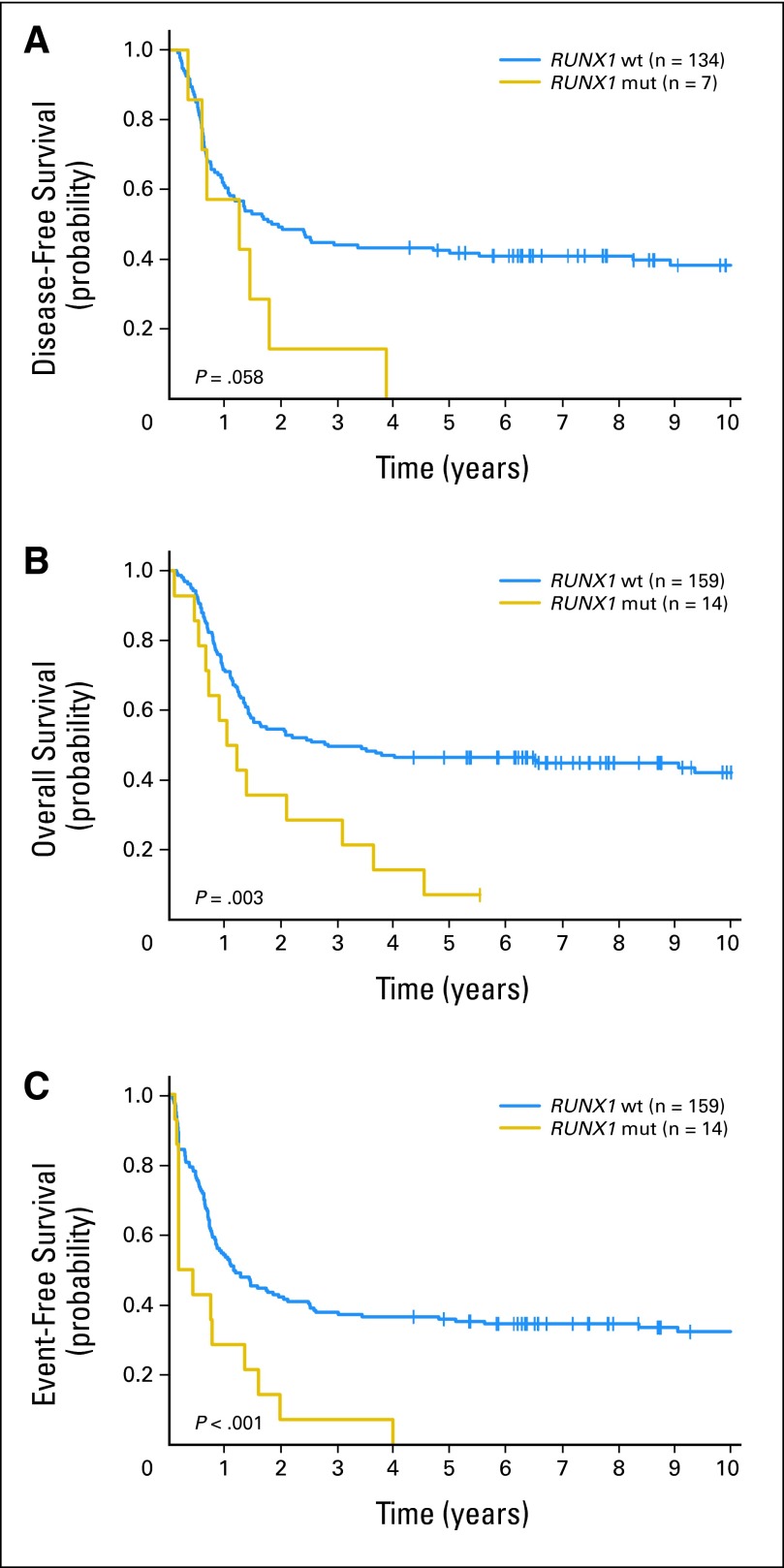
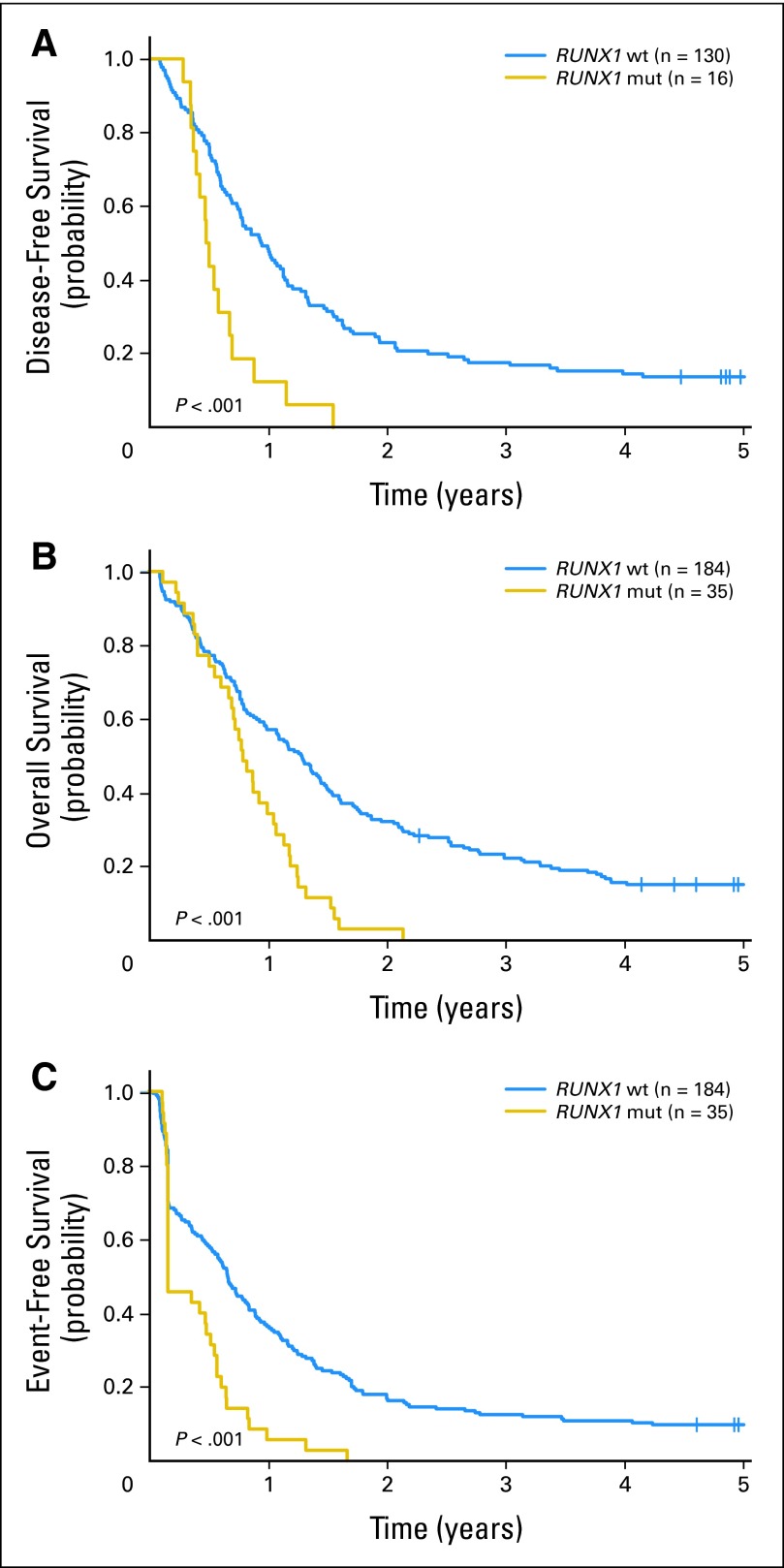
Among all patients, the median follow-up for patients alive was 7.8 years (range, 2.3 to 13.1 years). RUNX1-mutated patients, compared with RUNX1 wild-type patients, had an inferior CR rate (47% v 77%, respectively; P < .001) and shorter DFS (P < .001; 5-year DFS, 0% v 28%, respectively), OS (P < .001; 5-year OS, 2% v 30%, respectively), and EFS (P < .001; 5-year EFS, 0% v 22%, respectively; Table 2). Because consolidation treatment differed in intensity for younger and older patients, their outcomes were also analyzed separately. In younger patients, RUNX1 mutations, compared with RUNX1 wild-type status, were associated with an inferior CR rate (50% v 84%, respectively; P = .005) and worse DFS (P = .06; 5-year DFS, 0% v 42%, respectively), OS (P = .003; 5-year OS, 7% v 47%, respectively), and EFS (P < .001; 5-year EFS, 0% v 36%, respectively; Fig 1; Table 2). In older patients, RUNX1 mutations, compared with RUNX1 wild-type status, were also associated with an inferior CR rate (46% v 71%, respectively; P = .006) and worse DFS (P < .001; 3-year DFS, 0% v 18%, respectively), OS (P < .001; 3-year OS, 0% v 22%, respectively), and EFS (P < .001; 3-year EFS, 0% v 13%, respectively; Fig 2; Table 2).
Table 2.
Outcomes According to RUNX1 Mutation Status in Primary Cytogenetically Normal Acute Myeloid Leukemia
| End Point | RUNX1 Mutated | RUNX1 Wild Type | P* | OR† | 95% CI | HR‡ | 95% CI |
|---|---|---|---|---|---|---|---|
| All patients (N = 392) | |||||||
| No. of patients | 49 | 343 | |||||
| CR | < .001 | 0.27 | 0.14 to 0.49 | ||||
| No. of Patients | 23 | 264 | |||||
| % | 47 | 77 | |||||
| Disease-free survival | < .001 | 2.43 | 1.57 to 3.76 | ||||
| Median, years | 0.6 | 1.1 | |||||
| Disease free at 5 years, % | 0 | 28 | |||||
| 95% CI | — | 23 to 33 | |||||
| Overall survival | < .001 | 2.32 | 1.69 to 3.18 | ||||
| Median, years | 0.9 | 1.4 | |||||
| Alive at 5 years, % | 2 | 30 | |||||
| 95% CI | 0 to 9 | 25 to 35 | |||||
| Event-free survival | < .001 | 2.41 | 1.76 to 3.28 | ||||
| Median, years | 0.2 | 0.8 | |||||
| Event free at 5 years, % | 0 | 22 | |||||
| 95% CI | — | 18 to 26 | |||||
| Younger patients (n = 173) | |||||||
| No. of patients | 14 | 159 | |||||
| CR | .005 | 0.19 | 0.06 to 0.58 | ||||
| No. of Patients | 7 | 134 | |||||
| % | 50 | 84 | |||||
| Disease-free survival | .06 | 2.09 | 0.96 to 4.54 | ||||
| Median, years | 1.2 | 1.9 | |||||
| Disease free at 5 years, % | 0 | 42 | |||||
| 95% CI | — | 33 to 50 | |||||
| Overall survival | .003 | 2.34 | 1.30 to 4.20 | ||||
| Median, years | 1.1 | 2.8 | |||||
| Alive at 5 years, % | 7 | 47 | |||||
| 95% CI | 0 to 28 | 39 to 54 | |||||
| Event-free survival | < .001 | 2.56 | 1.46 to 4.49 | ||||
| Median, years | 0.3 | 1.1 | |||||
| Event free at 5 years, % | 0 | 36 | |||||
| 95% CI | — | 28 to 43 | |||||
| Older patients (n = 219) | |||||||
| No. of patients | 35 | 184 | |||||
| CR | .006 | 0.35 | 0.17 to 0.73 | ||||
| No. of Patients | 16 | 130 | |||||
| % | 46 | 71 | |||||
| Disease-free survival | < .001 | 2.61 | 1.52 to 4.50 | ||||
| Median, years | 0.5 | 0.9 | |||||
| Disease free at 3 years, % | 0 | 18 | |||||
| 95% CI | — | 12 to 25 | |||||
| Overall survival | < .001 | 2.25 | 1.53 to 3.30 | ||||
| Median, years | 0.8 | 1.3 | |||||
| Alive at 3 years, % | 0 | 22 | |||||
| 95% CI | — | 16 to 28 | |||||
| Event-free survival | < .001 | 2.17 | 1.49 to 3.18 | ||||
| Median, years | 0.2 | 0.7 | |||||
| Event free at 3 years, % | 0 | 13 | |||||
| 95% CI | — | 8 to 18 |
Abbreviations: CR, complete remission; HR, hazard ratio; OR, odds ratio.
P value is from Fisher's exact test for CR rate and from the log-rank test for time-to-event end points.
An OR of < 1 or > 1 means a lower or higher CR rate, respectively, for RUNX1-mutated patients compared with RUNX1 wild-type patients.
An HR of > 1 or < 1 corresponds to a higher or lower risk of an event, respectively, for RUNX1-mutated patients compared with RUNX1 wild-type patients.
Fig 1.
(A) Disease-free survival, (B) overall survival, and (C) event-free survival of patients younger than age 60 years with cytogenetically normal acute myeloid leukemia according to RUNX1 mutation status. RUNX1 mut, RUNX1 mutated; RUNX1 wt, RUNX1 wild type.
Fig 2.
(A) Disease-free survival, (B) overall survival, and (C) event-free survival of patients age 60 years and older with cytogenetically normal acute myeloid leukemia according to RUNX1 mutation status. RUNX1 mut, RUNX1 mutated; RUNX1 wt, RUNX1 wild type.
Because RUNX1 and NPM1 mutations are nearly mutually exclusive and prognostically favorable NPM1 mutations are prevalent in CN-AML (61% in our cohort), the prognostic impact of RUNX1 mutations was assessed in NPM1 wild-type patients separately. In younger NPM1 wild-type patients, RUNX1 mutations, compared with RUNX1 wild-type status, were associated with a lower CR rate (50% v 84%, respectively; P = .02), borderline worse DFS (P = .10; 5-year DFS, 0% v 38%, respectively) and worse OS (P = .04; 5-year OS, 8% v 40%, respectively) and EFS (P = .004; 5-year EFS, 0% v 34%, respectively; Data Supplement). In older NPM1 wild-type patients, RUNX1 mutations, compared with RUNX1 wild-type status, were associated with a shorter DFS (P = .005; 3-year DFS, 0% v 14%, respectively), OS (P = .002; 3-year OS, 0% v 13%, respectively), and EFS (P = .04; 3-year EFS, 0% v 7%, respectively; Data Supplement). RUNX1 mutations did not impact CR rate in older NPM1 wild-type patients.
We next examined the prognostic impact of RUNX1 mutations within the context of other molecular markers increasingly being used in the clinic. Recently, an international expert panel on behalf of the European LeukemiaNet (ELN) recommended a classification scheme for CN-AML based on NPM1, CEBPA, and FLT3-ITD mutations.17 According to this classification, patients with CN-AML are assigned to either the Favorable Genetic Group (NPM1 mutated without FLT3-ITD and/or CEBPA mutated) or the Intermediate-I (all remaining patients) Genetic Group. Because RUNX1 mutations rarely coexist with either CEBPA or NPM1 mutations, RUNX1-mutated patients fall almost exclusively into the ELN Intermediate-I Group (Table 1). Even within this already high-risk molecular group, RUNX1 mutations, compared with RUNX1 wild-type status, were associated with an inferior CR rate in younger patients (50% v 77%, respectively; P = .05) and worse EFS in older patients (P = .03; 3-year EFS, 0% v 6%, respectively; Data Supplement).
Because there is an association between RUNX1 and ASXL1 mutations, and ASXL1 mutations have a negative impact on outcome in older patients,38 we assessed their relative prognostic impact in this age group. Among ASXL1 wild-type patients, RUNX1-mutated patients, compared with RUNX1 wild-type patients, had an inferior CR rate (40% v 74%, respectively; P = .003), DFS (P = .007), OS (P < .001), and EFS (P < .001; Data Supplement). Among RUNX1 wild-type patients, ASXL1-mutated patients, compared with ASXL1 wild-type patients, had an inferior CR rate (42% v 74%, respectively; P = .01), borderline inferior DFS (P = .12), and inferior OS (P = .01) and EFS (P < .001; Data Supplement). Older patients harboring both mutations had a similar prognosis to those harboring either mutation alone (Data Supplement).
Multivariable Analyses
To assess whether RUNX1 mutations provide additional prognostic value in the context of other clinical and molecular prognosticators, we constructed multivariable models including all patients (N = 392; Table 3). RUNX1-mutated patients were four times less likely to achieve a CR (P < .001), after adjustment for IDH2 mutation status (P = .04), WBC (P = .004), and age group (P = .02). For DFS, patients with RUNX1 mutations were more than twice as likely to experience relapse or die (P < .001) after adjustment for FLT3-ITD status (P < .001), WBC (P = .02), and age group (P < .001). For OS, RUNX1-mutated patients had a 65% higher risk of death (P < .001) after adjustment for FLT3-ITD status (P < .001), WT1 mutation status (P < .001), WBC (P = .02), and age group (P < .001). For EFS, RUNX1-mutated patients were more than twice as likely to experience an event (P < .001) after adjustment for FLT3-ITD status (P < .001), WT1 mutation status (P = .04), WBC (P = .006), and age group (P < .001).
Table 3.
Multivariable Analysis of Outcome According to the RUNX1 Mutation Status in All Patients (N = 392) With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Variable | Complete Remission |
Disease-Free Survival |
Overall Survival |
Event-Free Survival |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| RUNX1: mutated v wild type | 0.25 | 0.13 to 0.47 | < .001 | 2.34 | 1.49 to 3.66 | < .001 | 1.65* | 1.13 to 2.42 | < .001 | 2.27 | 1.65 to 3.12 | < .001 |
| IDH2: mutated v wild type | 0.55 | 0.30 to 0.98 | .04 | — | — | — | — | — | — | — | — | — |
| FLT3-ITD: present v absent | — | — | — | 2.50* | 1.83 to 3.42 | < .001 | 1.56 | 1.22 to 1.99 | < .001 | 1.57 | 1.27 to 1.95 | < .001 |
| WT1: mutated v wild type | — | — | — | — | — | — | 1.99 | 1.39 to 2.84 | < .001 | 1.44 | 1.02 to 2.01 | .04 |
| WBC: continuous, 50-unit increase | 0.74 | 0.60 to 0.91 | .004 | 1.17 | 1.03 to 1.32 | .02 | 1.10 | 1.01 to 1.19 | .02 | 1.13 | 1.04 to 1.23 | .006 |
| Age group: ≥ 60 v < 60 years | 0.55 | 0.33 to 0.91 | .02 | 2.19 | 1.67 to 2.88 | < .001 | 2.46 | 1.93 to 3.15 | < .001 | 1.80 | 1.46 to 2.22 | < .001 |
NOTE. An OR of > 1 or < 1 corresponds to a higher or lower odds, respectively, of achieving a complete remission for higher values of continuous variables and the first level listed of a dichotomous variable. For time-to-event end points, an HR of > 1 or < 1 corresponds to a higher or lower risk, respectively, for higher values of continuous variables and the first level listed of a dichotomous variable. Variables were considered for inclusion in the multivariable models if they had a univariable P < .20. See Data Supplement for a full list of variables evaluated in univariable analysis. Variables with insufficient overlap with RUNX1 mutations could not be evaluated by multivariable models investigating the impact of RUNX1 mutations. These were NPM1, CEBPA, and ASXL1 mutations, MLL partial tandem duplication, and the European LeukemiaNet Genetic Groups. On the basis of univariable analyses, variables considered in the model for complete remission were RUNX1 mutations, WT1 mutations, IDH2 mutations, hemoglobin, platelet count, WBC, and age group (≥ 60 v < 60 years). Variables considered in the model for disease-free and overall survival were RUNX1 mutations, presence of FLT3-ITD, WT1 mutations, WBC, and age group (≥ 60 v < 60 years).
Abbreviations: HR, hazard ratio; ITD, internal tandem duplication; OR, odds ratio.
This variable did not meet the proportional hazards assumption. The P value corresponds to the Wald statistic of a 2-df test evaluating whether the coefficients for the variable and an artificial time-dependent covariate were equal to 0, to account for nonproportionality. The HR estimate is provided at 6 months.
Gene and MicroRNA Expression Signatures Associated With RUNX1 Mutations in CN-AML
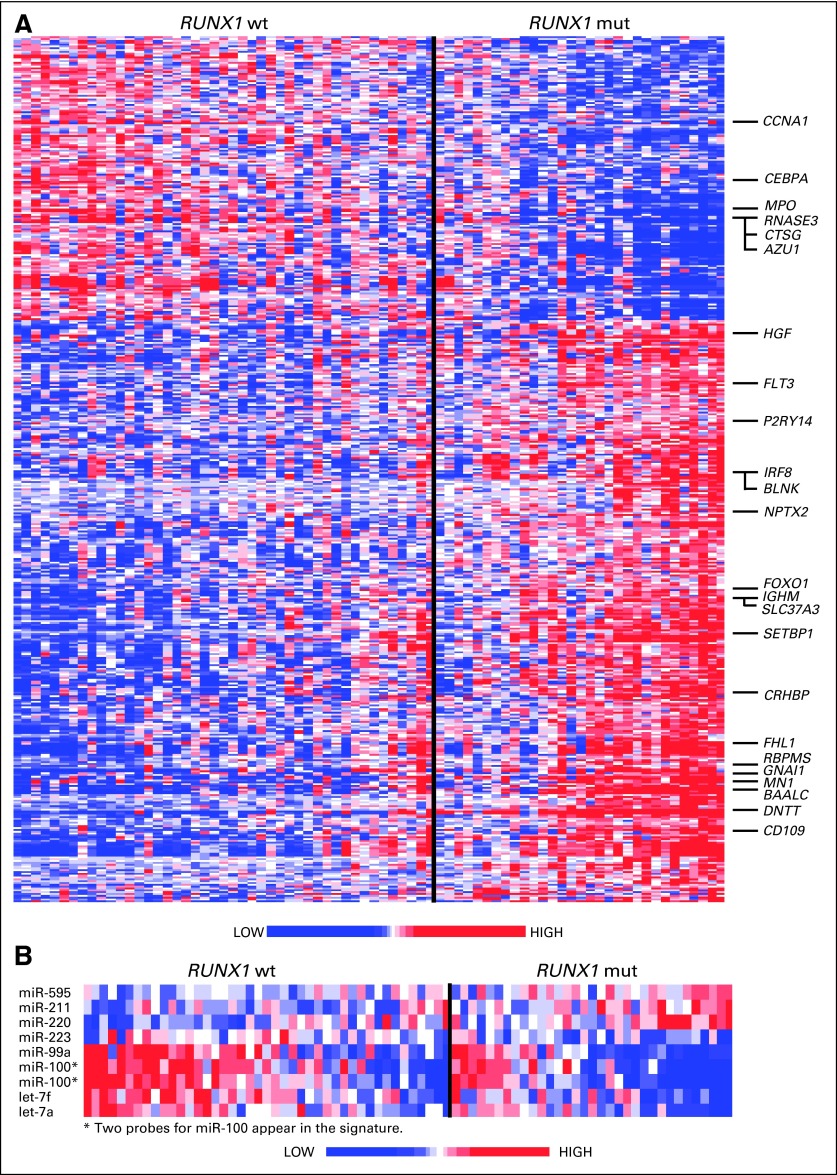
To gain molecular insight into RUNX1-mutated CN-AML, gene/microRNA expression signatures were derived. We focused only on older NPM1 wild-type patients because RUNX1 mutations are more common in this age group and almost never coexist with NPM1 mutations. This enabled the derivation of gene/microRNA expression signatures without interference from NPM1 mutations, which are themselves associated with strong gene/microRNA expression signatures.18 This yielded a signature composed of 484 probe sets representing 245 named genes differentially expressed between RUNX1-mutated (n = 31) and RUNX1 wild-type (n = 45) patients (Fig 3A; Data Supplement).
Fig 3.
(A) Heat map of the gene expression signature associated with RUNX1 mutations (mut) in older patients with NPM1 wild-type (wt) status. Upregulated and downregulated genes mentioned in the text are indicated. (B) Heat map of the microRNA expression signature associated with RUNX1 mutations in older patients with NPM1 wild-type status.
Genes overexpressed in early hematopoietic stem/progenitor cells (HSPCs) relative to more mature progenitors, including BAALC, CD109, P2RY14, CRHBP, NPTX2, GNAI1, HGF, and FHL1, were upregulated in RUNX1-mutated patients.39–44 Genes upregulated (SETBP1, RBPMS, and SLC37A3) and downregulated (CCNA1 and RNASE3) in AML stem cells relative to AML progenitors were similarly deregulated in the RUNX1-mutated signature.45,46 Several genes normally expressed in early lymphoid precursors,47 including DNTT, BLNK, IGHM, IRF8, FOXO1, FLT3, and genes encoding multiple class II major histocompatibility complex molecules, were also upregulated in RUNX1-mutated patients, whereas CEBPA, a key promoter of granulopoiesis, and AZU1, MPO, and CTSG, components of neutrophil granules, were downregulated. Overexpression of genes known to negatively impact prognosis in CN-AML,48–50 including MN1 and the aforementioned BAALC, was also part of the RUNX1-mutated signature. Gene set analysis was performed to identify sets of genes representing canonical biologic pathways deregulated in patients with mutated RUNX1. Eighteen gene sets were significantly deregulated (Data Supplement), 16 of which were upregulated in RUNX1-mutated patients compared with RUNX1 wild-type patients. RUNX1-mutated blasts were more likely to overexpress genes involved in platelet activation, vascular endothelial growth factor signaling, G protein–coupled receptor signaling, and intestinal immune function relative to RUNX1 wild-type blasts.
Eight microRNAs were differentially expressed between RUNX1-mutated and RUNX1 wild-type patients (Fig 3B; Data Supplement). Two members of the let-7 tumor suppressor family, which represses self-renewal and promotes differentiation of stem cells,51 were downregulated, as was miR-223, a positive regulator of granulopoiesis.52MiR-99a and miR-100, microRNAs upregulated in AML with inv(16)(p13q22),53 were also downregulated, and miR-211, miR-220, and miR-595, with unknown functions in leukemogenesis, were upregulated in RUNX1-mutated blasts.
DISCUSSION
The identification of novel, prognostically relevant molecular markers is of great importance in CN-AML, because this is the largest cytogenetic subset in both younger and older patients2 and current molecular classification schemes do not fully capture the heterogeneity in outcome of these patients. Once a new marker is identified, its prognostic impact should be validated in both younger and older patients with CN-AML separately, because disease biology, treatment options, and outcomes differ between these age groups. Although prior studies have demonstrated a negative prognostic impact of RUNX1 mutations on EFS in younger patients with CN-AML,31,32 these mutations had no impact on outcome in older patients with CN-AML.31 Consequently, our study was designed to more fully explore how RUNX1 mutations impact on the prognosis of both younger and older patients with CN-AML and on global gene/microRNA expression.
We demonstrate that RUNX1 mutations occur not infrequently in older patients with CN-AML. To our knowledge, our study is the first to report a prevalence of RUNX1 mutations (16%) in a cohort of older patients with CN-AML. We confirm previous reports that RUNX1 mutations occur less frequently in younger patients with CN-AML32 and rarely coexist with either NPM1 or CEBPA mutations31–33 in either age group. This distinguishes RUNX1-mutated CN-AML primarily as a subset of NPM1 wild-type/CEBPA wild-type disease; among patients with wild-type NPM1 and CEBPA, RUNX1 mutations are relatively frequent, occurring in 38% and 44% of younger and older patients, respectively.
An important question is which RUNX1 sequence variations represent true, disease-associated RUNX1 mutations. We suspect that the germline RUNX1 mutations in our study are disease associated because they involve functional domains and have not been described previously as polymorphisms. In the case of p.L56S, there is evidence both in our study and in a study by Gaidzik et al32 that its presence is not always consistent between leukemic blasts and germline cells; thus, this variation can be disease associated. However, given the relatively few p.L56S cases in our study, we feel that the issue of whether this variant is truly a somatic disease allele or a polymorphism is still unresolved. Although the patients with p.L56S were excluded from our formal outcome analyses, inclusion of these patients in the RUNX1-mutated group did not change the overall results or conclusions (data not shown).
Our study shows that RUNX1 mutations portend a worse prognosis in both younger and older patients with CN-AML. To our knowledge, we demonstrate for the first time that in older patients with CN-AML, RUNX1 mutations are associated with a lower CR rate and shorter DFS, OS, and EFS relative to RUNX1 wild-type patients. Coupled with the high risk of treatment-related complications in older patients receiving intensive chemotherapy, these patients should be strongly considered for novel therapeutic approaches. The negative impact of RUNX1 mutations on EFS in the younger patients of our study confirms findings of others,32,31 who also found worse EFS associated with RUNX1 mutations in this age group. Unique to our study is the negative prognostic impact of RUNX1 mutations on other outcome end points in younger patients (CR rate, DFS, and OS). The shorter DFS and OS in our study may be related to the lack of alloSCT in CR1, because there is evidence that RUNX1-mutated patients achieving CR have a better outcome with postremission alloSCT than chemotherapy.32,33
As the number of molecular markers with prognostic impact increases in CN-AML, it becomes difficult to discern whether there is additional prognostic value of a new marker. Because the patients in this study have been extensively characterized for the presence of multiple prognostic markers, the relative impact of RUNX1 mutations could be determined. RUNX1 mutations remained prognostic in multivariable models; however, these models were limited by the exclusion of NPM1 and CEBPA mutational status, two well-characterized favorable prognostic markers,18,54 because of insufficient sample size and overlap with RUNX1 mutations. To address this limitation, analyses were conducted in NPM1 wild-type and ELN Intermediate-I Group patient subsets. The fact that RUNX1 mutations continued to associate with worse outcomes in both of these subsets suggests that they may add prognostic information to molecular markers currently being used in the clinic.
To gain insight into molecular features of RUNX1-mutated CN-AML, RUNX1 mutation–associated gene/microRNA expression signatures were derived in CN-AML for the first time. Several of the most strongly upregulated genes in the signature are also expressed in HSPCs and/or B-cell progenitors, whereas genes normally expressed in myeloid-committed cells are among the most downregulated. These findings are consistent with prior studies demonstrating that RUNX1 mutations occur more frequently in minimally differentiated (M0) AML24,55 and are associated with upregulation of B-cell lineage genes in this French-American-British subtype relative to RUNX1 wild-type AML M0 patients.56 In fact, a substantial number of the genes in the RUNX1 mutation–associated signature of our study were also associated with RUNX1 mutations in AML M0.56 In contrast, only five genes (2% of our signature) were common between the RUNX1 mutation–associated gene expression signature reported by Gaidzik et al32 and our signature. This is possibly because of age-related differences or because of the diverse cytogenetics of the patients in their study, including several patients with t(8;21)(q22;q22), inv(16)(p13q22), or t(15;17)(q24;q21), among others. The RUNX1 mutation–associated microRNA expression signature also lacks myeloid features, with downregulation of microRNAs normally expressed in either definitive myeloid progenitors (miR-223) or distinctly myeloid AML blasts (miR-99a and miR-100).52,53
In summary, our data demonstrate that RUNX1 mutations occur in a substantial proportion of patients with primary CN-AML with wild-type NPM1 and CEBPA. These mutations are associated with a poor outcome in both younger and older patients with CN-AML treated with intensive induction chemotherapy and not receiving alloSCT in CR1. Thus, patients harboring RUNX1 mutations warrant strong consideration of up-front novel therapies and/or early alloSCT. RUNX1 mutation–associated expression signatures are characteristic of HSPCs and lymphoid cells and provide candidate molecules to guide development of novel therapeutic approaches.
Supplementary Material
Acknowledgment
The Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participating in this study are provided in the Appendix. We thank Donna Bucci and the Cancer and Leukemia Group B Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center (Columbus, OH) for sample processing and storage services and Lisa J. Sterling and Colin G. Edwards, PhD, for data management.
Glossary Terms
- Biallelic:
The condition in which both alleles of a gene are mutated.
- Cytogenetically normal acute myeloid leukemia (AML):
AML with a normal karyotype at diagnosis based on the microscopic analysis of ≥ 20 metaphase cells in bone marrow specimens subjected to short-term cultures; approximately 45% of patients with AML are cytogenetically normal.
- Gene-expression profiling:
Identifying the expression of a set of genes in a biologic sample (eg, blood, tissue) using microarray technology.
- Germline mutation:
An inherited variation in the lineage of germ cells. Germline mutations can be passed on to offspring.
- MicroRNAs:
Endogenous noncoding RNAs approximately 22 nucleotides long that regulate gene silencing by post-transcriptional mechanisms such as cleavage or translational repression.
- Missense:
A change (mutation) in one nucleotide that results in the coding of a different amino acid.
- Nonsense:
A mutation that changes a codon that codes for an amino acid into a stop codon, therefore terminating translation.
- Polymorphism:
Genetic polymorphisms are natural variations in the genomic DNA sequence present in greater than 1% of the population, with SNP representing DNA variations in a single nucleotide. SNPs are being widely used to better understand disease processes, thereby paving the way for genetic-based diagnostics and therapeutics.
- RUNX1:
This gene encodes a subunit of core binding factor, a heterodimeric transcription factor involved in normal hematopoiesis.
Appendix
The following Cancer and Leukemia Group B (CALGB) institutions, principal investigators, and cytogeneticists participated in this study: Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, P. Nagesh Rao, Wendy L. Flejter, and Mark J. Pettenati (Grant No. CA03927); The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, Diane Minka, and Nyla A. Heerema (Grant No. CA77658); North Shore–Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R.K. Koduru (Grant No. CA35279); University of Iowa Hospitals, Iowa City, IA: Daniel A. Vaena and Shivanand R. Patil (Grant No. CA47642); Duke University Medical Center, Durham, NC: Jeffrey Crawford, Sandra H. Bigner, Mazin B. Qumsiyeh, John Eyre, and Barbara K. Goodman (Grant No. CA47577); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (Grant No. CA02599); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett, Michael S. Watson, Eric C. Crawford, Peining Li, and Jaime Garcia-Heras (Grant No. CA77440); University of Chicago Medical Center, Chicago, IL: Hedy L. Kindler, Diane Roulston, Katrin M. Carlson, Yanming Zhang, and Michelle M. Le Beau (Grant No. CA41287); Dana-Farber Cancer Institute, Boston, MA: Harold J. Burstein, Ramana Tantravahi, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA32291); University of North Carolina, Chapel Hill, NC: Thomas C. Shea and Kathleen W. Rao (Grant No. CA47559); University of Massachusetts Medical Center, Worcester, MA: William V. Walsh, Vikram Jaswaney, Michael J. Mitchell, and Patricia Miron (Grant No. CA37135); Vermont Cancer Center, Burlington, VT: Steven M. Grunberg, Elizabeth F. Allen, and Mary Tang (Grant No. CA77406); Dartmouth Medical School, Lebanon, NH: Konstantin Dragnev, Doris H. Wurster-Hill, and Thuluvancheri K. Mohandas (Grant No. CA04326); Ft Wayne Medical Oncology/Hematology, Ft Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Weill Medical College of Cornell University, New York, NY: John Leonard, Ram S. Verma, Prasad R.K. Koduru, Andrew J. Carroll, and Susan Mathew (Grant No. CA07968); Minneapolis VA Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (Grant No. CA47555); Eastern Maine Medical Center, Bangor, ME: Harvey M. Segal and Laurent J. Beauregard (Grant No. CA35406); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (Grant No. CA04457); Rhode Island Hospital, Providence, RI: William Sikov, Teresita Padre-Mendoza, Hon Fong L. Mark, Shelly L. Kerman, and Aurelia Meloni-Ehrig (Grant No. CA08025); State University of New York (SUNY) Upstate Medical University, Syracuse, NY: Stephen L. Graziano, Larry Gordon, and Constance K. Stein (Grant No. CA21060); University of California at San Diego, Sand Diego, CA: Barbara A. Parker, Renée Bernstein, and Marie L. Dell'Aquila (Grant No. CA11789); Christiana Care Health Services, Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, and Jeanne M. Meck (Grant No. CA45418); Long Island Jewish Medical Center Community Clinical Oncology Program (CCOP), Lake Success, NY: Kanti R. Rai and Prasad R. K. Koduru (Grant No. CA11028); University of Puerto Rico School of Medicine, San Juan, Puerto Rico: Eileen I. Pacheco, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton; University of Maryland Cancer Center, Baltimore, MD: Martin J. Edelman, Joseph R. Testa, Maimon M. Cohen, Judith Stamberg, and Yi Ning (Grant No. CA31983); Western Pennsylvania Hospital, Pittsburgh, PA: John Lister and Gerard R. Diggans; Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA 12449); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim H. Huang (Grant No. CA12046); University of Nebraska Medical Center, Omaha, NE: Anne Kessinger and Warren G. Sanger (Grant No. CA77298); University of Minnesota, Minneapolis, MN: Bruce A. Peterson, Diane C. Arthur, and Betsy A. Hirsch (Grant No. CA16450); University of Illinois at Chicago: David J. Peace, Maureen M. McCorquodale, and Kathleen E. Richkind (Grant No. CA74811); Walter Reed Army Medical Center, Washington, DC: David C. Van Echo, Rawatmal B. Surana, and Digamber S. Borgaonkar (Grant No. CA26806); Georgetown University Medical Center, Washington, DC: Minnetta C. Liu and Jeanne M. Meck (Grant No. CA77597); McGill Department of Oncology, Montreal, Quebec, Canada: J. L. Hutchison and Jacqueline Emond (Grant No. CA31809); University of Cincinnati Medical Center, Cincinnati, OH: Orlando J. Martelo and Ashok K. Srivastava (Grant No. CA47515); Columbia-Presbyterian Medical Center, New York, NY: Rose R. Ellison and Dorothy Warburton (Grant No. CA12011); Virginia Commonwealth University MB CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (Grant No. CA52784); SUNY Maimonides Medical Center, Brooklyn, NY: Sameer Rafla and Ram S. Verma (Grant No. CA25119); Medical University of South Carolina, Charleston, SC: Mark R. Green and G. Shashidhar Pai (Grant No. CA03927); Southern Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John Ellerton and Marie L. Dell'Aquila (Grant No. CA35421); and University of California at San Francisco, San Francisco, CA: Charles J. Ryan and Kathleen E. Richkind (Grant No. CA60138).
Footnotes
Supported in part by the National Cancer Institute (Grants No. CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657), the Coleman Leukemia Research Foundation, the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.), and the Conquer Cancer Foundation (J.H.M.).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Jason H. Mendler, Guido Marcucci, Clara D. Bloomfield
Financial support: Michael A. Caligiuri, Guido Marcucci, Clara D. Bloomfield
Administrative support: Richard A. Larson, Michael A. Caligiuri, Clara D. Bloomfield
Provision of study materials or patients: Bayard L. Powell, Thomas H. Carter, Meir Wetzler, Joseph O. Moore, Jonathan E. Kolitz, Maria R. Baer, Andrew J. Carroll, Richard A. Larson, Michael A. Caligiuri, Guido Marcucci, Clara D. Bloomfield
Collection and assembly of data: Jason H. Mendler, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Heiko Becker, Klaus H. Metzeler, Sebastian Schwind, Susan P. Whitman, Jihane Khalife, Jessica Kohlschmidt, Deedra Nicolet, Bayard L. Powell, Thomas H. Carter, Meir Wetzler, Joseph O. Moore, Jonathan E. Kolitz, Maria R. Baer, Andrew J. Carroll, Richard A. Larson, Michael A. Caligiuri, Guido Marcucci
Data analysis and interpretation: Jason H. Mendler, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Heiko Becker, Klaus H. Metzeler, Sebastian Schwind, Susan P. Whitman, Jessica Kohlschmidt, Deedra Nicolet, Guido Marcucci, Clara D. Bloomfield
Manuscript writing: All authors
Final approval of manuscript: All authors
Affiliations
Jason H. Mendler, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Heiko Becker, Klaus H. Metzeler, Sebastian Schwind, Susan P. Whitman, Jihane Khalife, Jessica Kohlschmidt, Deedra Nicolet, Michael A. Caligiuri, Guido Marcucci, and Clara D. Bloomfield, The Ohio State University Comprehensive Cancer Center, Columbus, OH; Kati Maharry, Michael D. Radmacher, Jessica Kohlschmidt, and Deedra Nicolet, Alliance for Clinical Trials in Oncology Statistics and Data Center, Mayo Clinic, Rochester, MN; Bayard L. Powell, Comprehensive Cancer Center of Wake Forest University, Winston-Salem; Joseph O. Moore, Duke University Medical Center, Durham, NC; Thomas H. Carter, University of Iowa, Iowa City, IA; Meir Wetzler, Roswell Park Cancer Institute, Buffalo; Jonathan E. Kolitz, Monter Cancer Center, Hofstra North Shore Long Island Jewish School of Medicine, Lake Success, NY; Maria R. Baer, Greenebaum Cancer Center, University of Maryland, Baltimore, MD; Andrew J. Carroll, University of Alabama at Birmingham, Birmingham, AL; and Richard A. Larson, The University of Chicago Medical Center, Chicago, IL.
REFERENCES
- 1.Gaidzik V, Döhner K. Prognostic implications of gene mutations in acute myeloid leukemia with normal cytogenetics. Semin Oncol. 2008;35:346–355. doi: 10.1053/j.seminoncol.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 3.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 5.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 6.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 7.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 8.Fröhling S, Schlenk RF, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paschka P, Marcucci G, Ruppert AS, et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virappane P, Gale R, Hills R, et al. Mutation of the Wilms' tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: The United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008;26:5429–5435. doi: 10.1200/JCO.2008.16.0333. [DOI] [PubMed] [Google Scholar]
- 12.Caligiuri MA, Strout MP, Lawrence D, et al. Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res. 1998;58:55–59. [PubMed] [Google Scholar]
- 13.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 16.Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 18.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda T, van Deursen J, Hiebert SW, et al. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 20.Michaud J, Wu F, Osato M, et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: Implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 21.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 22.Harada H, Harada Y, Niimi H, et al. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 23.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome: The mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol. 2007;139:405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 24.Preudhomme C, Warot-Loze D, Roumier C, et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2αB gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood. 2000;96:2862–2869. [PubMed] [Google Scholar]
- 25.Roumier C, Eclache V, Imbert M, et al. M0 AML, clinical and biologic features of the disease, including AML1 gene mutations: A report of 59 cases by the Groupe Français d'Hématologie Cellulaire (GFHC) and the Groupe Français de Cytogénétique Hématologique (GFCH) Blood. 2003;101:1277–1283. doi: 10.1182/blood-2002-05-1474. [DOI] [PubMed] [Google Scholar]
- 26.Matsuno N, Osato M, Yamashita N, et al. Dual mutations in the AML1 and FLT3 genes are associated with leukemogenesis in acute myeloblastic leukemia of the M0 subtype. Leukemia. 2003;17:2492–2499. doi: 10.1038/sj.leu.2403160. [DOI] [PubMed] [Google Scholar]
- 27.Silva FP, Morolli B, Storlazzi CT, et al. Identification of RUNX1/AML1 as a classical tumor suppressor gene. Oncogene. 2003;22:538–547. doi: 10.1038/sj.onc.1206141. [DOI] [PubMed] [Google Scholar]
- 28.Silva FP, Lind A, Brouwer-Mandema G, et al. Trisomy 13 correlates with RUNX1 mutation and increased FLT3 expression in AML-M0 patients. Haematologica. 2007;92:1123–1126. doi: 10.3324/haematol.11296. [DOI] [PubMed] [Google Scholar]
- 29.Dicker F, Haferlach C, Kern W, et al. Trisomy 13 is strongly associated with AML1/RUNX1 mutations and increased FLT3 expression in acute myeloid leukemia. Blood. 2007;110:1308–1316. doi: 10.1182/blood-2007-02-072595. [DOI] [PubMed] [Google Scholar]
- 30.Osato M, Asou N, Abdalla E, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 31.Schnittger S, Dicker F, Kern W, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 32.Gaidzik VI, Bullinger L, Schlenk RF, et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML Study Group. J Clin Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 33.Tang JL, Hou HA, Chen CY, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 34.Langabeer SE, Gale RE, Rollinson SJ, et al. Mutations of the AML1 gene in acute myeloid leukemia of FAB types M0 and M7. Genes Chromosomes Cancer. 2002;34:24–32. doi: 10.1002/gcc.10031. [DOI] [PubMed] [Google Scholar]
- 35.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 36.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 38.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia JS, Madzo J, Cooper D, et al. Pre-donor evaluation of an HLA matched sibling identifies a novel inherited RUNX1 mutation encoding a missense mutation found outside of the RUNT domain in familial platelet disorder. Blood. 2010;116:1116–1117. (abstr 2709) [Google Scholar]
- 41.Georgantas RW, 3rd, Tanadve V, Malehorn M, et al. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 42.He X, Gonzalez V, Tsang A, et al. Differential gene expression profiling of CD34+ CD133+ umbilical cord blood hematopoietic stem progenitor cells. Stem Cells Dev. 2005;14:188–198. doi: 10.1089/scd.2005.14.188. [DOI] [PubMed] [Google Scholar]
- 43.Murray LJ, Bruno E, Uchida N, et al. CD109 is expressed on a subpopulation of CD34+ cells enriched in hematopoietic stem and progenitor cells. Exp Hematol. 1999;27:1282–1294. doi: 10.1016/s0301-472x(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 44.Toren A, Bielorai B, Jacob-Hirsch J, et al. CD133-positive hematopoietic stem cell “stemness” genes contain many genes mutated or abnormally expressed in leukemia. Stem Cells. 2005;23:1142–1153. doi: 10.1634/stemcells.2004-0317. [DOI] [PubMed] [Google Scholar]
- 45.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 46.Gentles AJ, Plevritis SK, Majeti R, et al. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos PM, Borghesi L. Molecular resolution of the B cell landscape. Curr Opin Immunol. 2011;23:163–170. doi: 10.1016/j.coi.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldus CD, Tanner SM, Ruppert AS, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: A Cancer and Leukemia Group B study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 49.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langer C, Marcucci G, Holland KB, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2009;27:3198–3204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Büssing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 53.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 55.Osato M. Point mutations in the RUNX1/AML1 gene: Another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 56.Silva FP, Swagemakers SM, Erpelinck-Verschueren C, et al. Gene expression profiling of minimally differentiated acute myeloid leukemia: M0 is a distinct entity subdivided by RUNX1 mutation status. Blood. 2009;114:3001–3007. doi: 10.1182/blood-2009-03-211334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.