In Kawasaki disease (KD), molecular-based detection of human adenovirus (HAdV) infection is not uncommon, may represent persistence of HAdV-C, and should be interpreted with caution. Together, quantitative polymerase chain reaction and HAdV typing may aid in distinguishing HAdV disease mimicking KD from KD with concomitant HAdV detection.
Keywords: adenovirus, Kawasaki disease, PCR
Abstract
Background. Human adenovirus (HAdV) infection mimics Kawasaki disease (KD) but can also be detected in KD patients. Evidence suggests that HAdV-C species can persist in pediatric adenoids and/or tonsils. We sought to determine (1) the frequency of HAdV detection by real-time polymerase chain reaction in KD patients, (2) the differences in HAdV semiquantitative nasopharyngeal viral loads between KD patients with detectable HAdV vs those with HAdV disease, and (3) whether nasopharyngeal HAdV-C shedding is occurring in KD.
Methods. From August 2009 through April 2011, HAdV-positive patients were identified in 1 of the following groups: group I, complete or incomplete KD as defined by the American Heart Association (AHA); group II, treated for incomplete KD but not fulfilling AHA criteria; and group III, otherwise healthy children with some KD-like features ultimately diagnosed with HAdV disease.
Results. Among 77 KD patients diagnosed, 8.8% (5/57) of group I and 25% (5/20) of group II KD patients had HAdV detected. Viral loads were significantly lower in group I (n = 5) vs group III (n = 26; P = .034). Of the 13 specimens available for HAdV typing, 7 of 7 group III and 1 of 3 group II specimens were determined to be HAdV-B using viral culture. The remaining 5 KD samples were unable to be cultured and molecular typing showed either HAdV-C (n = 3) or were nontypeable (n = 2).
Conclusions. In KD, molecular-based HAdV detection is not uncommon, may represent persistence of HAdV-C, and should be interpreted with caution. Together, quantitative polymerase chain reaction and HAdV typing may aid in distinguishing HAdV disease mimicking KD from KD with concomitant HAdV detection.
(See the Editorial Commentary by Rowley and Shulman, on pages 65–6.)
Kawasaki disease (KD) is a febrile vasculitis of unknown etiology and represents the most common cause of pediatric acquired heart disease in the developed world. In the United States the estimated annual incidence of KD is approximately 10–21 per 100 000 children aged <5 years [1–3]. Given the clinical similarities, human adenovirus (HAdV) infection is one of the most frequent conditions included in the differential diagnosis when considering KD [4]. Recent studies show that up to 8.8% of children treated for KD have respiratory viruses identified in the upper respiratory tract, including a patient with HAdV infection who eventually developed coronary artery aneurysm [5]. HAdV has also been isolated from a lymph node in a patient with fatal KD [6]. The introduction of highly sensitive molecular methods allows for enhanced detection of HAdV in the upper respiratory tract. However, few data are available regarding frequency, viral load, and types of HAdV in KD patients.
HAdVs belong to the Adenoviridae family and are divided into 7 subgroups designated “species” (A–G) according to their immunologic, biologic, and biochemical characteristics [7]. Primary infections with species C (HAdV-C) occur commonly in children younger than age 5; in addition to primary infection, HAdV-C (especially serotypes 1, 2, and 5) can also persist in pediatric tonsils for years with low-grade replication [8]. The present study was designed to determine whether there are differences in the nasopharyngeal (NP)/throat HAdV burden as indicated by semiquantitative cycle thresholds (Cts) between children determined to have HAdV disease (with some KD-like features) vs those determined to have KD with incidental detection of HAdV, and to determine if HAdV-C shedding may be occurring in patients with KD.
MATERIALS AND METHODS
Study Patient Definitions and Design
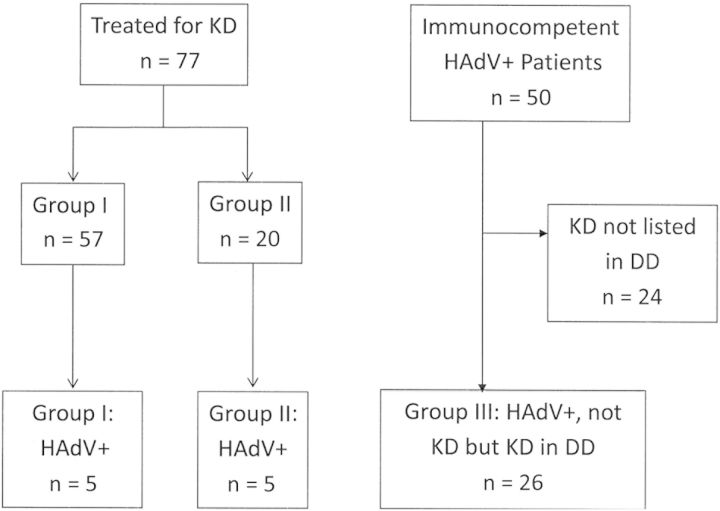
With institutional review board approval, we prospectively identified all KD patients (aged 1 month to 18 years) hospitalized from August 2009 to April 2011 whose throat or NP specimens were tested for HAdV infection by polymerase chain reaction (PCR). Using microbiology records, we retrospectively identified all non-KD immunocompetent, hospitalized, or emergency department patients who were diagnosed with an HAdV throat or NP infection by PCR during the same time period (Figure 1). HAdV Cts were compared between children with complete or incomplete KD in whom HAdV was detected vs those diagnosed with HAdV disease in which KD was considered in the differential diagnosis. HAdV typing was done on all available frozen specimens.
Figure 1.
Classification of patients. Group I included complete or incomplete Kawasaki disease (KD) that met American Heart Association (AHA) criteria. Patients in group II were treated for incomplete KD but did not meet full AHA criteria, and group III consisted of patients who were not treated for KD but who had documented consideration of KD in the differential diagnosis. Abbreviations: DD, differential diagnosis; HAdV, human adenovirus; KD, Kawasaki disease.
Kawasaki Disease Definition
All patients treated for Kawasaki disease during the study period were identified prospectively. Viral testing for HAdV in patients with KD was at the discretion of the treating physician and was performed in 92% of cases. We stratified the KD patients based on likelihood of correct diagnosis: group I (complete or incomplete KD as defined using American Heart Association [AHA] guidelines) and group II (incomplete KD with insufficient lab criteria to fulfill AHA guidelines) [4]. The treating clinician was also asked to describe his/her level of confidence in the diagnosis of KD at the time of ordering treatment for KD: “highly or very likely KD,” “probable KD,” or “possibly KD and benefits of treating for KD outweigh risks of not treating.”
HAdV Infection Patient Definition
All immunocompetent patients hospitalized or evaluated in the emergency department with detection of HAdV in the throat and/or NP specimens by PCR were identified using microbiology database records. Patients with HAdV infection admitted to the pediatric intensive care unit (ICU) and those with an underlying chronic or immunocompromised state (ie, patients taking immunosuppressive medications, transplant patients, or those with malignancy or a history of malignancy) were excluded. Other infections identified in those patients (either viral or bacterial) were recorded. Within this cohort of patients with HAdV infection, we selected those who most clinically resembled KD (group III) on the basis of documentation of KD as part of the differential diagnosis in their medical records. Clinical, echocardiographic, and laboratory data were collected using a standardized form for all patients.
HAdV PCR and Typing
The microbiology laboratory at Nationwide Children's Hospital has developed and validated a semiquantitative, real-time HAdV PCR that targets a conserved DNA region of the viral hexon gene [9]. Using laboratory-cultured virus, the assay has been shown to detect 20 of 20 serotypes representing the 6 major HAdV species (A–F). Total nucleic acid was extracted from swabs using NucliSENS easyMAG (bioMérieux). PCR was performed on 10 µL of extract in 40 µL reactions which were amplified to 45 cycles using the Applied Biosystems 7500. Each specimen was run in duplicate; samples were considered positive if both replicates had Ct values ≤45; average HAdV Cts per run(s) were recorded.
Adenovirus Typing
Thirteen specimens were available from frozen stores for HAdV typing by molecular methods. Clinical specimens were inoculated onto A549 cell monolayers for viral isolation and typing by restriction enzyme analysis of viral genomic DNA and by amplification and sequencing of hexon and fiber genes as previously described [10–12]. For specimens for which an isolate could not be obtained, total DNA was extracted from 100 µL after the first passage and processed for PCR amplification and sequencing of the hexon hypervariable region 7 as previously described [13]. Amplicons were sequenced and molecular identity was inferred from phylogenetic analysis.
Statistical Analysis
Descriptive analysis was performed using frequency distributions. Means and medians were used to summarize patient demographic and baseline characteristics. Groups were compared using Student t test or Mann-Whitney U test for continuous variables as appropriate. A 2-tailed P value of ≤ .05 was considered significant. SigmaPlot 12.0 was used for the analysis.
RESULTS
KD Patients With HAdV Detection
During the 20 months of the study, 77 patients (47 [61%] male) <18 years of age were treated for KD (mean age, 3.3 ± 1.9 years): 57 in group I and 20 in group II; 71 (92%) were tested for HAdV infection by PCR in NP or throat samples. Five of 57 patients (8.8%) in group I KD, and 5 of 20 (25%) in group II KD had detectable/tested positive for HAdV (Figure 1).
All 5 HAdV-positive patients in group I (Table 1) had complete KD. The treating clinician confidence in the KD diagnosis was “highly or very likely” in 4 patients and “probable” in 1 patient. One patient with “probable” KD required intubation in the ED, had clinically significant myocarditis and shock on presentation, and remained febrile 36 hours after the completion of treatment with intravenous immunoglobulin (IVIG). Because of the detection of HAdV in the NP sample (Ct = 25.6), HAdV-associated myocarditis was also considered, but KD was eventually confirmed when coronary artery ectasia developed. HAdV PCR was negative in the blood. For the 5 HAdV-positive patients in group II (incomplete KD), clinician confidence in the KD diagnosis was “highly or very likely” in 1, “probable” in 2, and “possible and benefits of treatment for KD outweigh risks of not treating” in 2. One patient thought to have probable KD also had parainfluenza virus detected by direct immunofluorescence assay (DFA). Two of 5 patients required therapy with a second dose of IVIG. No coronary abnormalities were detected in this group, but 3 patients did not return for follow-up echocardiography at our institution. One child had documented periungual desquamation 2 weeks after the initial hospitalization.
Table 1.
Comparison of Human Adenovirus (HAdV)–Positive Group I and Group II Kawasaki Disease (KD) vs HAdV Disease With KD-Like Features (Group III)
| Group I: KD (n = 5) | Group II: KD (n = 5) | Group III: HAdV Disease (n = 26) | *P Value 1 | ^P Value 2 | |
|---|---|---|---|---|---|
| Age, y, median (range) | 2.4 (2.1–4.4) | 2.0 (1.7–4.1) | 3.0 (2.0–6.0) | NS | NS |
| Fever on treatment in hospital or ED, d, median (IQR) | 6 (5–9) | 8 (4–10) | 5 (4–7) | NS | NS |
| Bilateral conjunctival injection, No. (%) | 5 (100%) | 4 (80%) | 21 (81%) | NS | NS |
| With exudate or tearing, No. (%) | 0 | 3 (60%) | 7 (27%) | ||
| Mucosal changes of tongue/lips, No. (%) | 5 (100%) | 4 (80%) | 16 (62%) | NS | NS |
| Polymorphous rash, No. (%) | 5 (100%) | 4 (80%) | 13 (50%) | NS | NS |
| Changes of extremities, No. (%) | 5 (100%) | 3 (60%) | 7 (27%) | .047 | NS |
| Unilateral neck swelling, No. (%) | 3 (60%) | 1 (20%) | 1 (3.9%) | .008 | NS |
| Cough, No. (%) | 0 (0%) | 4 (80%) | 13 (50%) | NS | NS |
| Rhinorrhea, No. (%) | 2 (40%) | 3 (60%) | 12 (46%) | NS | NS |
| IVIG resistance, No. (%) | 1 (20%) | 2 (40%) | NA | NA | NA |
| WBC count, median (IQR) | 12.5 (9.1–17.0) | 10.6 (10.1–16.0) | 9.7(6.2–13.0) | NS | NS |
| Platelets, median (IQR) | 290 (218–467) | 367 (265–411) | 265 (247–312) | NS | NS |
| ESR, median (IQR) | 41 (10–56) | 37 (20–76) | 44 (32–51) | NS | NS |
| Albumin, median (IQR) | 3.7 (3.1–3.9) | 3.7 (3.5–4.1) | 3.8 (3.5–4.2) | NS | NS |
| ALT, median (IQR) | 130 (54–165) | 17 (12–202) | 10 (3–17.3) | .001 | NS |
| C-reactive protein, median (IQR) | 4.6 (2.9–16.1) | 4.1 (0.9–4.6) | 3.0 (2.1–5.3) | NS | NS |
| WBCs in urine/HPF, median | 23 | 2 | 1 | .004 | NS |
| Virological data | |||||
| Specimens available, No. | 3 | 3 | 7 | NA | NA |
| AdV threshold count, mean ± SD | 34.4 ± 5.4 | 31.6 ± 9.3 | 27.0 ± 6.6 | .034 | NS |
| Growth in culture, No. (%) | 0/3 (0%) | 1/3 (33%) | 7/7 (100%) | NS | NS |
| HAdV type | C-2 (n = 2), nontypeable (n = 1) | C-1 or C-2 (n = 1), B-3 (n = 1), nontypeable (n = 1) | B-3 (n = 6), B-7 (n = 1) | NA | NA |
Abbreviations: AdV, adenovirus; ALT, alanine aminotransferase; ED, emergency department; ESR, erythrocyte sedimentation rate; HAdV, human adenovirus; HPR, high power field; IQR, interquartile range; IVIG, intravenous immunoglobulin; KD, Kawasaki disease; NA, not applicable; NS, not significant; SD, standard deviation; WBC, white blood cell.
* Group I KD vs AdV disease.
^ Group II KD vs AdV disease.
Characteristics of HAdV-Positive Patients
During the same time period, HAdV infection was identified by PCR in 50 otherwise healthy children (Figure 1). All 50 patients had fever. Thirty (60%) patients had upper respiratory tract illness, 11 (22%) had lower respiratory tract illness, and 29 (58%) had gastrointestinal symptoms including nausea/vomiting, diarrhea, or abdominal pain. Three patients had intussusceptions during or around the time of HAdV detection in the upper respiratory tract, and 1 was diagnosed with mesenteric adenitis by computed tomography.
Of these 50 patients, we then identified the 26 patients in whom KD was documented in the differential diagnosis (group III). Clinical, virologic, and laboratory characteristics were compared between group I and II KD patients and the 26 patients included in group III (Table 1). Among group III patients, 13 (50%) presented with upper respiratory complaints (cough or rhinorrhea), 15 (57%) had gastrointestinal complaints (nausea, vomiting, diarrhea, or abdominal pain), 21 (81%) had conjunctivitis, 13 (50%) had rash, and 14 (54%) had posterior pharyngeal/tonsillar inflammation documented.
Viral Load/Ct Comparison
KD patients in group I had significantly higher Ct values (ie, lower viral loads) compared with group III patients (mean, 34.4 ± 5.4 vs 27.0 ± 6.6; P = .034; Figure 2A). One group I KD patient who presented in shock and developed coronary artery ectasia had detection of HAdV in the NP sample with a Ct of 25.6. There was no significant difference in the Ct values between group II KD and group III (mean, 31.6 ± 9.3 vs 27.0 ± 6.6).
Figure 2.
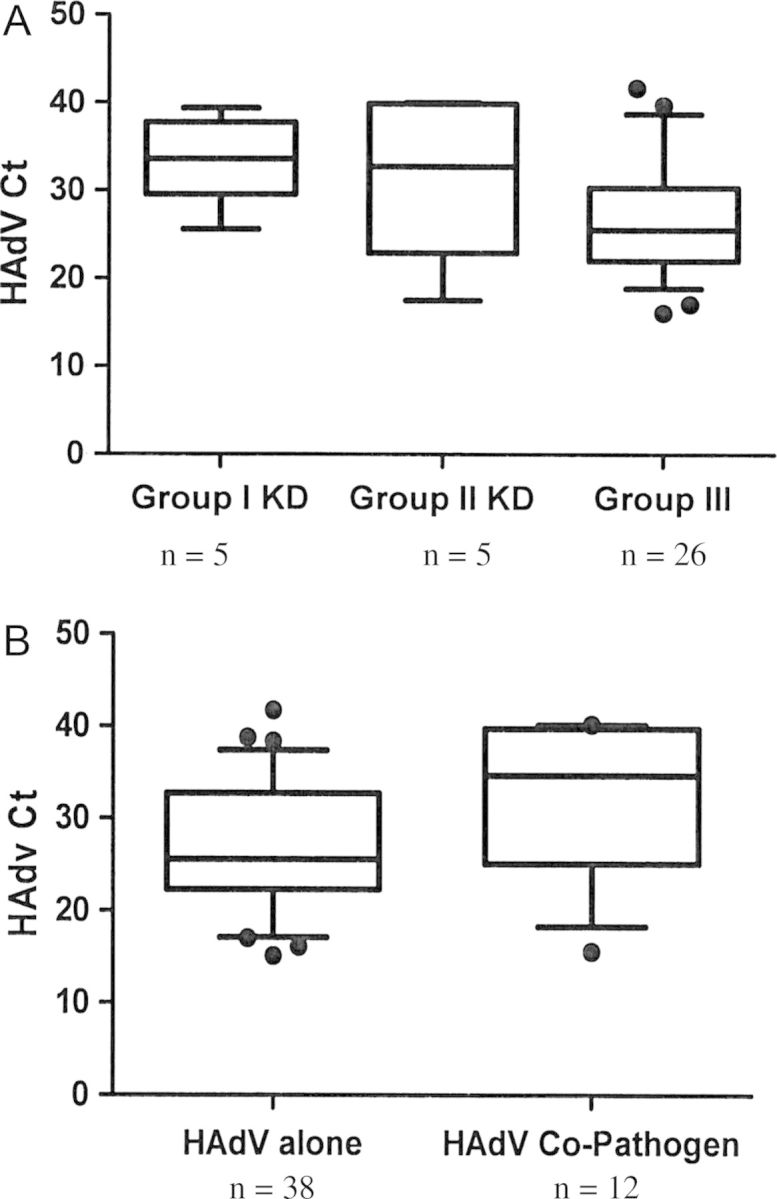
A, Comparison of human adenovirus (HAdV) cycle threshold (Ct) among 3 groups. Group I, HAdV-positive KD (all complete); group II, HAdV-positive KD (incomplete); and group III, HAdV disease. Group I Kawasaki disease (KD) patients had significantly higher Cts compared to those with Group III (mean, 34.4 ± 5.4 vs 27.0 ± 6.6; P = .034). There was no significant difference in the Cts between group II KD and group III (mean, 31.6 ± 9.3 vs 27.0 ± 6.6). B, Comparison of Ct values among patients with HAdV alone vs HAdV with another pathogen identified. All patients were immunocompetent, non-KD patients. Those with HAdV alone (n = 38) had significantly lower Cts compared to those with HAdV and another identifiable pathogen (n = 12; mean, 27.1 ± 1.1 vs 34.6 ± 6.9; P = .038). Abbreviations: Ct, cycle threshold; HAdV, human adenovirus; KD, Kawasaki disease.
HAdV Typing
Thirteen HAdV clinical specimens were available from frozen stores for typing: 7 from group III and 6 from KD patients (group I, n = 3 and group II, n = 3). Virus isolation was readily achieved for all 7 specimens from the non-KD, HAdV group III. All 7 isolates were identified as HAdV-B (5 HAdV-3a-like, 1 novel genomic variant of HAdV-3, and 1 HAdV-7d2). An HAdV-3a-like virus was isolated from 1 group II KD patient. For the remaining 5 cases, viral isolation was not successful after 5 serial blind passages, so typing was carried out by PCR amplification of the hexon hypervariable region 7 (HVR-7) followed by sequencing and BLAST (Basic Local Alignment Search Tool) analysis (http://blast.ncbi.nlm.nih.gov). HAdV-C (HVR-7 type 2) DNA was detected in 3 samples: HAdV-C type 2 (n = 2) and HAdV-C (type 1 or 2; n = 1). For the remaining 2 samples, no amplicons were obtained from the analysis (Table 1).
To further validate the possibility of HAdV shedding in the context of other conditions, the effect of coinfection was then explored by comparing the HAdV Cts from all non-KD patients with HAdV infection (n = 50). Cts were compared between specimens detected as a coinfection vs those with HAdV infection alone. Twelve patients had another detectable infection, including herpes simplex virus stomatitis and Escherichia coli urinary tract infection (in the same patient), Epstein-Barr virus (VCA immunoglobulin M detection, n = 2), respiratory syncytial virus (n = 1), human metapneumovirus (n = 3), rhinovirus (n = 2), Clostridium difficile toxin detection in the stool, parvovirus viremia detected by PCR, and throat swab mycoplasma detection by PCR. Ct values were significantly higher (ie, lower viral loads) in patients who had a documented copathogen detected compared with those with HAdV alone (mean, 34.6 ± 6.9 vs 27.1 ± 1.1; P = .038; Figure 2B).
DISCUSSION
In an earlier retrospective study, we reported that 2.4% of children presenting with KD had HAdV in the upper respiratory tract that was detected using DFA or viral culture [5]. Four of the 6 HAdV-positive patients in that study were thought to have incomplete KD. In the present study, we clarified the rate of HAdV detection among those with complete or AHA-defined incomplete KD. We found that by using PCR, 8.8% of the patients of group I KD had detectable HAdV in NP samples (all with complete disease). Others have used detection of HAdV by DFA as a diagnostic test to exclude KD [14]. Our observations indicate that detection of HAdV in a patient with suspected KD should be interpreted with caution, as detection of HAdV is not uncommon and does not exclude the diagnosis of KD. This is particularly relevant when using PCR, which is highly sensitive and can detect low-level virus. The other clinical or laboratory data that were helpful in distinguishing KD from HAdV disease in our cohort included neck swelling, distal extremity changes, and presence of pyuria and/or hepatitis (Table 1), which is similar to previous reports [15].
Among the HAdVs capable of infecting the respiratory tract, HAdV-C (types 1, 2, 5, and 6) species are also capable of establishing low-level persistence or shedding. Garnett et al have shown that in 76% (186/243) of adenoid and/or tonsillar tissue specimens from pediatric patients aged 1–19 years (removed for tonsillar hypertrophy, recurrent bacterial infections, or recurrent otitis media), HAdV-C DNA was detected by PCR in 1 or both specimens, suggesting that HAdV is capable of establishing latency in these tissues [8]. The presence of HAdV-C DNA in adenoid or tonsillar tissues peaked at 4 years of age and declined thereafter, which coincides with the usual timing of KD diagnosis. Other DNA viruses, such as herpes simplex virus [16], cytomegalovirus, and Epstein-Barr virus, are commonly associated with intermittent shedding [17].
While in this cohort there were a limited number of frozen specimens available for HAdV typing, all the HAdV-C specimens were obtained from patients treated for KD. These specimens did not yield a significant amount of replicating virus by culture, and typing was only accomplished when molecular methods were used. The 2 nontypeable samples also did not grow in culture and likely did not contain enough DNA to support amplification. Garnett et al reported that infectious virus was detected in <15% of the tonsil/adenoidal samples collected, even among specimens with the highest level of HAdV DNA [8]. Culture-negative but PCR-positive HAdV detection has been reported previously in complete KD [18], and our observations support the findings of low amounts of infectious virus present in the respiratory specimens of 5 of the 6 KD patients with molecular detection of HAdV. In contrast, the non-KD samples yielded HAdV-B and grew easily in culture, indicating active infection.
Our study does suggest that there are differences in the HAdV NP/throat viral burden as measured semiquantitatively by the differences in Ct values between patients with HAdV disease and those with complete KD and HAdV detection. Ct values may be another helpful tool to distinguish KD with concurrent HAdV detection (lower viral burden) from HAdV-associated disease (higher viral burden) but future prospective studies that include standardized collection procedures are needed to confirm these initial observations. Coinfection, even among the non-KD patients with another pathogen (either bacterial or viral), was associated with a lower HAdV viral burden and raises the possibility that an acute illness may trigger low-level HAdV-C shedding. Reports indicate a low rate of HAdV detection by PCR in NP samples of asymptomatic children <6 years with a frequency of only 0.6% (1/157), but this rate increases among children with respiratory symptoms to 8.5% [18]. HAdV were also frequently detected as coinfections with another respiratory virus in NP swabs from children <6 years old with respiratory symptoms, again suggesting that HAdV-C shedding may be occurring during illness, though HAdV was not further typed [19].
Group II KD, HAdV-positive patients are particularly challenging to clinicians as the true diagnosis is oftentimes unclear but the consequences of missing KD may be grave. There were no significant differences in the detectable Ct values between group II and group III patients; however, 1 patient in group II KD had an easily cultured, identifiable HAdV-B (Ct = 32.8). Also documented was an otitis media. The clinician was not highly confident in the KD diagnosis and the DFA was also positive. Taken together, these findings suggest that HAdV may have caused the patient's acute disease. Group II likely includes patients who truly have KD with incidental HAdV detection, as well as some patients with HAdV disease. Further prospective study of group II, HAdV-positive patients with full molecular viral characterization along with complete follow-up, including documentation of periungual desquamation and coronary artery outcomes, will be helpful to differentiate between the 2 conditions.
The group I KD patient who presented in shock and eventually developed coronary artery ectasia had a fairly strong positive HAdV PCR with a Ct of 25, which might suggest active HAdV disease, but the detected HAdV could not be isolated in cell culture and PCR-based molecular typing was needed to confirm the presence of HAdV-C DNA. The blood HAdV PCR was negative, suggesting that there was not systemic dissemination of HAdV. This was the only HAdV-positive patient admitted to ICU in our cohort and currently it is unclear whether ICU patients have a different degree of HAdV shedding than non-ICU patients. This case also suggests that viral burden, culture, and typing may all be needed to clarify the etiologic role that HAdV plays in disease.
Our study has several limitations, including the retrospective clinical data analysis of the HAdV disease group. Several of our group II KD patients did not return for follow-up, so the true coronary artery outcomes are not known. Quality and site of sample collection (NP vs throat) may have affected the Ct counts. Although the PCR targeted a fairly conserved region of the hexon gene, it was not truly quantitative and may detect different types with varying efficiency.
In conclusion, our results indicate that (1) HAdV detection using PCR is not uncommon even in children with complete KD; (2) using semiquantitative real-time PCR, HAdV Ct values were higher (ie, lower viral burden) among patients with complete KD vs those thought to have true HAdV disease; and (3) even though HAdV-C causes acute disease in children <5 years of age, HAdV-C DNA shedding may occur in patients with KD. Further prospective studies with systematic measurement of viral load, molecular typing, and close clinical observation are needed to clarify the diagnostic challenge of HAdV disease vs KD with incidental HAdV detection.
Notes
Acknowledgments. We thank Doug Salamon, Kathy Mack, and Susan Core for technical assistance; the Nationwide Children's Hospital clinical research coordinators; and Monica Ardura for her critical review of the manuscript.
Financial support. This work was supported in part by the American Heart Association (grant number 11CRP5460003).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Belay ED, Holman RC, Clarke MJ, et al. The incidence of Kawasaki syndrome in West Coast health maintenance organizations. Pediatr Infect Dis J. 2000;19:828–32. doi: 10.1097/00006454-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Holman RC, Curns AT, Belay ED, Steiner CA, Schonberger LB. Kawasaki syndrome hospitalizations in the United States, 1997 and 2000. Pediatrics. 2003;112:495–501. doi: 10.1542/peds.112.3.495. [DOI] [PubMed] [Google Scholar]
- 3.Holman RC, Belay ED, Christensen KY, Folkema AM, Steiner CA, Schonberger LB. Hospitalizations for Kawasaki syndrome among children in the United States, 1997–2007. Pediatr Infect Dis J. 2010;29:483–8. doi: 10.1097/INF.0b013e3181cf8705. [DOI] [PubMed] [Google Scholar]
- 4.Newburger JW, Takahashi M, Gerber MA, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Pediatrics. 2004;114:1708–33. doi: 10.1542/peds.2004-2182. [DOI] [PubMed] [Google Scholar]
- 5.Jordan-Villegas A, Chang ML, Ramilo O, Mejias A. Concomitant respiratory viral infections in children with Kawasaki disease. Pediatr Infect Dis J. 2010;29:770–2. doi: 10.1097/INF.0b013e3181dba70b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embil JA, McFarlane ES, Murphy DM, Krause VW, Stewart HB. Adenovirus type 2 isolated from a patient with fatal Kawasaki disease. Can Med Assoc J. 1985;132:1400. [PMC free article] [PubMed] [Google Scholar]
- 7.Echavarria M. Adenoviruses in immunocompromised hosts. Clin Microbiol Rev. 2008;21:704–15. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnett CT, Erdman D, Xu W, Gooding LR. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76:10608–16. doi: 10.1128/JVI.76.21.10608-10616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol. 2003;70:228–39. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- 10.Kajon AE, Dickson LM, Fisher BT, Hodinka RL. Fatal disseminated adenovirus infection in a young adult with systemic lupus erythematosus. J Clin Virol. 2011;50:80–3. doi: 10.1016/j.jcv.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 11.Kajon AE, Dickson LM, Murtagh P, Viale D, Carballal G. Echavarria M. Molecular characterization of an adenovirus 3-16 intertypic recombinant isolated in Argentina from an infant hospitalized with acute respiratory infection. J Clin Microbiol. 2010;48:1494–6. doi: 10.1128/JCM.02289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selvaraju SB, Kovac M, Dickson LM, Kajon AE, Selvarangan R. Molecular epidemiology and clinical presentation of human adenovirus infections in Kansas City children. J Clin Virol. 2011;51:126–31. doi: 10.1016/j.jcv.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol. 2004;42:3963–9. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocholl C, Gerber K, Daly J, Pavia AT, Byington CL. Adenoviral infections in children: the impact of rapid diagnosis. Pediatrics. 2004;113:e51–6. doi: 10.1542/peds.113.1.e51. [DOI] [PubMed] [Google Scholar]
- 15.Barone SR, Pontrelli LR, Krilov LR. The differentiation of classic Kawasaki disease, atypical Kawasaki disease, and acute adenoviral infection: use of clinical features and a rapid direct fluorescent antigen test. Arch Pediatr Adolesc Med. 2000;154:453–6. doi: 10.1001/archpedi.154.5.453. [DOI] [PubMed] [Google Scholar]
- 16.Tronstein E, Johnston C, Huang ML, et al. Genital shedding of herpes simplex virus among symptomatic and asymptomatic persons with HSV-2 infection. JAMA. 2011;305:1441–9. doi: 10.1001/jama.2011.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hug M, Dorner M, Frohlich FZ, et al. Pediatric Epstein-Barr virus carriers with or without tonsillar enlargement may substantially contribute to spreading of the virus. J Infect Dis. 2010;202:1192–9. doi: 10.1086/656335. [DOI] [PubMed] [Google Scholar]
- 18.Shike H, Shimizu C, Kanegaye JT, et al. Adenovirus, adeno-associated virus and Kawasaki disease. Pediatr Infect Dis J. 2005;24:1011–4. doi: 10.1097/01.inf.0000183769.31951.1e. [DOI] [PubMed] [Google Scholar]
- 19.Jansen RR, Wieringa J, Koekkoek SM, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49:2631–6. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]