Abstract
Induction of tumor-specific immune responses results in the inhibition of tumor development. However, tumors recur because of the tumor immunoediting process that facilitates development of escape mechanisms in tumors. It is not known whether tumor escape is an active process whereby anti-tumor immune responses induce loss or downregulation of the target antigen in the antigen-positive clones. To address this question, we used rat neu-overexpressing mouse mammary carcinoma (MMC) and its relapsed neu antigen-negative variant (ANV). ANV emerged from MMC under pressure from neu-specific T cell responses in vivo. We then cloned residual neu antigen-negative cells from MMC and residual neu antigen-positive cells from ANV. We found marked differences between these neu-negative clones and ANV, demonstrating that the residual neu-negative clones are probably not the origin of ANV. Since initial rejection of MMC was associated with the presence of IFN-γ-secreting T cells, we treated MMC with IFN-γ and showed that IFN-γ could induce downregulation of neu expression in MMC. This appears to be due to methylation of the neu promoter. Together, these data suggest that neu antigen loss is an active process that occurs in primary tumors due to the neu-targeted anti-tumor immune responses.
Keywords: Cytokines, Immune evasion, Tumor immunology
Introduction
Tumor escape and recurrences are major challenges in immunotherapy of cancers, including breast carcinomas. Therefore, understanding the mechanisms by which primary tumors escape from the host immune responses may offer critical insights into the improvement of cancer immunotherapy and lead to the development of new immunotherapeutic approaches. A variety of molecular alterations in tumors have been reported. These include, but are not limited to, the loss or downregulation of MHC class I antigens in tumors, defects in antigen presentation machinery such as TAP and/or β-2 microglobulin, expression of Fas ligand and/or loss of Fas in tumors, expression of HLA-E or Qa1 as killer inhibitory ligands in tumors, and loss of tumor antigens [1-4].
While immune responses can be induced against a variety of cancers, resulting in the inhibition of tumor development, molecular alterations in tumors can also occur under immune pressure, resulting in tumor escape. In other words, anti-tumor immune responses can function as a “double-edged” sword exerting both host-protective and tumor-evading effects on developing cancers; “cancer immunoediting” has been coined to accurately describe the latter phenomena. It was reported that chemically induced methylcholanthrene (Meth A) sarcomas derived from immunocompetent animals were more tumorogenic when inoculated into naive wild-type mice than tumors similarly derived from immunodeficient animals [5-7]. Based on these findings, it has been envisaged that cancer immunoediting is a result of three processes: elimination, equilibrium, and escape [8]. At the equilibrium phase between immune response and tumor growth, Darwinian selection has been suggested to be the mechanism for cancer immunoediting [9]. According to Darwinian selection, the tumor-specific immune responses eliminate highly immunogenic tumor cells, leaving behind tumor variants of reduced immunogenicity that have a better chance of surviving in the immunocompetent host. There are also reports suggesting that immune-mediated induction of epigenetic changes in primary tumors leads to tumor antigen loss [10, 11]. Using a neu-over-expressing primary tumor (mouse mammary carcinoma, MMC) and its relapsed neu antigen-negative variant (ANV), we describe how neu-specific immune responses may induce tumor escape. We show that IFN-γ is involved in downregulation of neu expression in primary tumors by inducing methylation of the mouse mammary tumor virus (MMTV) promoter.
Results
Rejection of MMC in FVB mice is mediated by host T cell responses
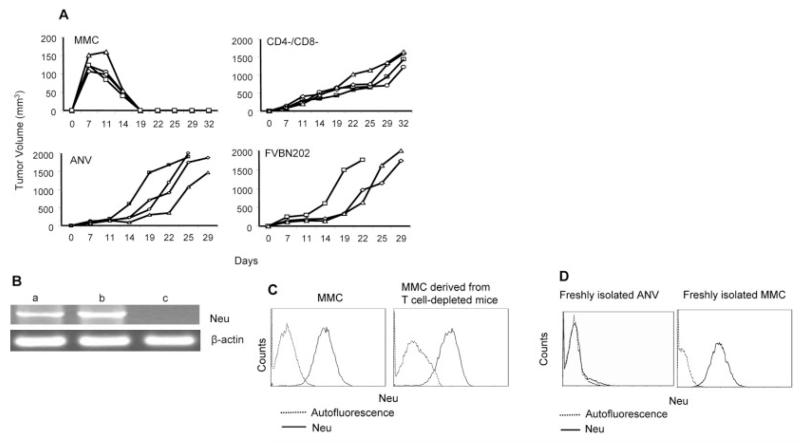
Wild-type FVB mice spontaneously reject MMC because the tumor cells overexpress rat neu antigen. In order to determine whether spontaneous rejection of MMC in FVB mice is mediated by T cells, animals were depleted of CD4+ and CD8+ T cells by the injection of GK1.5 and 2.43 Ab, respectively [12]. Animals were then inoculated with MMC (5 × 106 cells/mouse). Depletion of T cell subsets continued until the end of the trial. As shown in Fig. 1A, all wild-type mice rejected MMC within 3 wk, while animals depleted of CD4+ and CD8+ T cells (CD4−/CD8−) progressively developed tumors. In order to determine the neu specificity of the tumor rejection, wild-type mice were inoculated with ANV. All the mice failed to reject ANV. As an additional control for the neu specificity of MMC rejection, FVBN202-transgenic mice that tolerate neu protein were inoculated with MMC. All the FVBN202 mice progressively developed tumors. Semi-quantitative RT-PCR analysis of the tumors isolated from CD4−/CD8− FVB mice showed no neu antigen loss in the absence of effector Tcells (Fig. 1B). The MMC cell line and the relapsed ANV tumors were used as neu-positive (b) and neu-negative (c) controls, respectively. Expression of neu mRNA in the MMC cell line was higher (100%) than expression in freshly isolated mRNA from solid tumors (61%) because of the presence of other infiltrating cells in the tumor microenvironment. When these solid tumors were cultured in vitro to establish viable tumor clones, neu overexpression in the clones was consistently comparable with neu overexpression in the MMC line (MFI: 276 versus 265, respectively; Fig. 1C). Freshly isolated MMC or ANV were positive or negative for neu expression, respectively (Fig. 1D).
Figure 1.
Rejection of MMC in FVB mice and downregulation of neu antigen are mediated by T cell immune responses. (A) Wild-type immunocompetent (MMC) or CD4+ and CD8+ T cell-depleted (CD4−/CD8−) FVB mice (n=4) were inoculated with MMC. One group of FVB mice was inoculated with ANV (ANV). As a control, FVBN202-transgenic mice (n=3), which are tolerant to neu protein and fail to reject MMC, were inoculated with MMC (FVBN202). (B) RT-PCR analysis for detection of neu mRNA in tumors derived from MMC-challenged CD4−/CD8− FVB mice (a) show no neu antigen loss. Expression of neu mRNA in neu-overexpressing MMC (b) and neu-negative ANV (c) was determined as positive and negative controls. (C) Detection of neu expression by flow cytometry analysis of a viable MMC line and MMC cells isolated from CD4−/CD8− FVB mice. (D) Expression of the neu protein in freshly isolated ANV or MMC.
Heterogeneity of MMC and ANV in the expression of neu
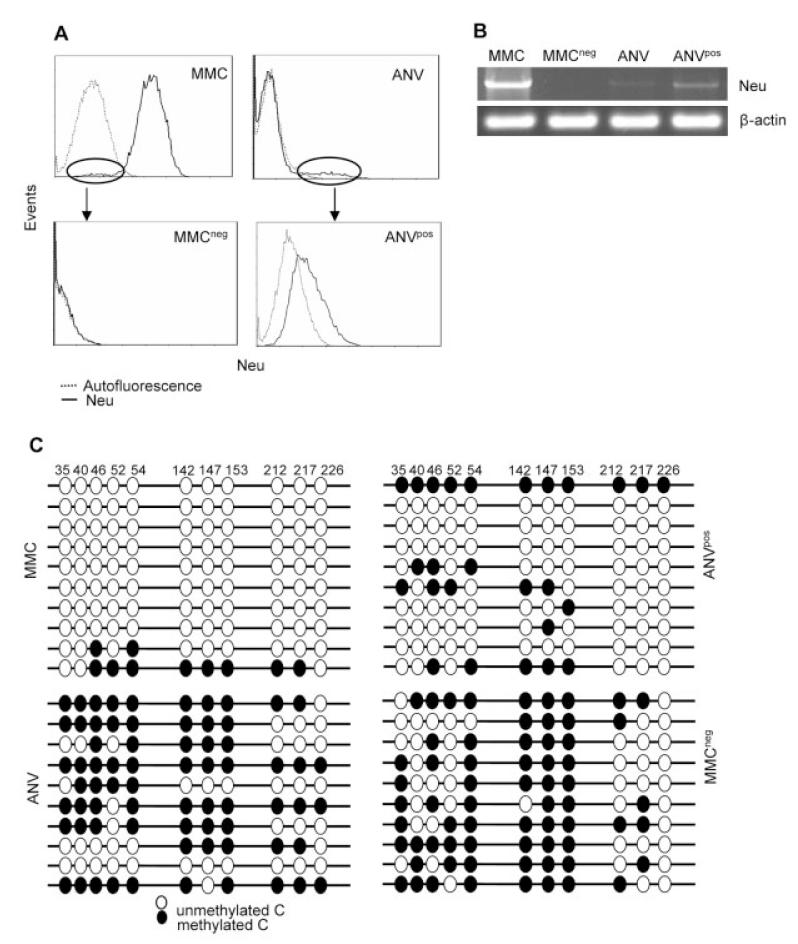
We have established several lines of primary MMC and relapsed ANV cells. Flow cytometry analyses of MMC and ANV lines showed residual neu-negative clones among MMC cells and neu-positive clones among ANV (Fig. 2A). We sought to determine whether ANV emerged from the residual neu-negative clones in MMC because of the elimination of neu-overexpressing clones by anti-neu immune responses. We sorted neu-negative clones of MMC (MMCneg) and neu-positive clones of ANV (ANVpos); MMCneg cells were 100% neu-negative, while ANVpos cells were 100% neu-positive, with intermediate expression of neu as compared to MMC (Fig. 2A). These cells were stable for the absence or presence of neu expression over 45 passages. Semi-quantitative RT-PCR analysis also showed that ANVpos and MMCneg cells were positive (23%) and negative (0%), respectively, for the expression of neu mRNA (Fig. 2B). MMC had the highest expression of neu mRNA (100%), while ANV had minimal neu expression (5%). Since neu expression in the FVBN202-transgenic mouse is regulated by hypomethylation of MMTV promoter [13], we used bisulfite methylation assays and detected hypomethylation and hypermethylation of the CpG-rich sites within region 1 (corresponding to nucleotides 56-316) of MMTV in MMC or ANVpos and ANV or MMCneg, respectively (Fig. 2C). There was no mutation in the neu gene or MMTV promoter (data not shown).
Figure 2.
Establishment of MMCneg and ANVpos tumor lines from MMC and ANV. (A) MMC or ANV were stained with anti-neu Ab and subjected to flow cytometry analyses. Residual neu-negative clones of MMC and neu-positive clones of ANV were sorted using the Beckman Coulter EPICS Elite sorter. Sorted cells were cultured and cloned in vitro and subjected to flow cytometry for further analyses of neu expression. (B) RT-PCR analysis of the indicated tumor lines for the expression of neu mRNA using β-actin as an internal control. (C) Bisulfite genomic sequencing of the indicated cell lines. A total of ten clones per sample was sequenced.
Distinct proliferation rates of ANV and MMCneg clones
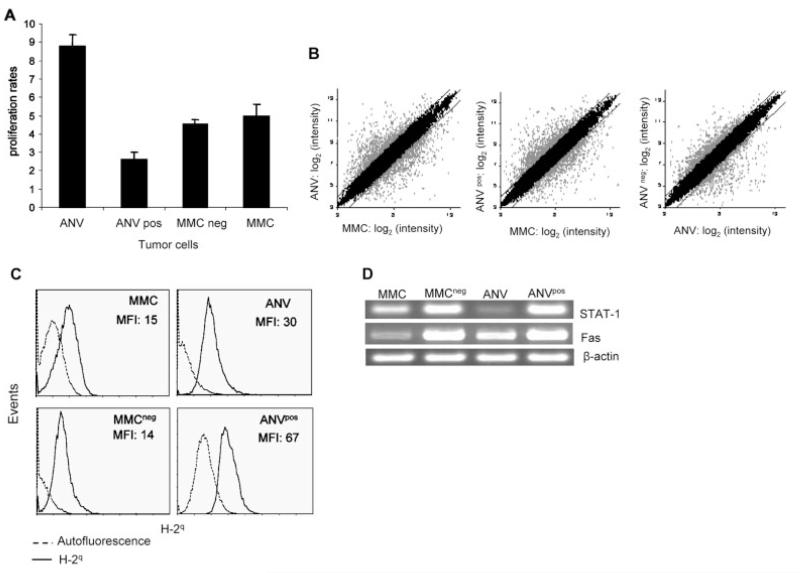
In order to determine proliferation rates in vitro, all the tumor lines (2.5 × 105 cells) were cultured for 3 days, and viable cells were counted in triplicates using trypan blue exclusion. The viability of cells was above 98%. Proliferation rates were calculated as follows: total cell numbers after 3-day culture divided by cell numbers on day 0. As shown in Fig. 3A, ANV had the highest rates of proliferation (8.8), while ANVpos had the lowest rates (2.6). Proliferation rates of MMC (5) and MMCneg (4.5) were comparable. Student’s t-test analysis showed significant differences in proliferation rates between ANV and MMCneg (p=0.01) or between MMC and ANVpos (p=0.03).
Figure 3.
MMC and ANV appear to have distinct morphology, proliferation rates, and gene expression profiles. (A) All the tumor lines were cultured at 2.5 × 105 cells/well in triplicates using tissue culture dishes. After 3 days in culture, adherent cells were detached using 0.25% Trypsin-EDTA. Cells were then counted using trypan blue exclusion. Proliferation rates were calculated as follows: total cell numbers after 3 days in culture divided by cell numbers on day 0. (B) Microarray analysis was performed on the indicated samples. Background correction, normalization, and expression summaries were calculated. Scatter plot of log2-transformed expression summaries of the 22 690 probe sets in the Mouse430A 2.0 array are plotted for the samples indicated in the axes of the graphs. Gray dots show genes that were at least 2-fold different for MMC versus ANV, MMC versus ANVpos, or ANV versus MMCneg. Similar results were obtained in independent experiments using two different microarray analyses on biological replicates of cells. (C) The indicated tumor lines were subjected to flow cytometry-based analyses using mouse anti-H-2q and FITC-conjugated anti-mouse Ig Ab. Isotype control Ab showed MFI similar to the autofluorescence (data not shown). Representative histograms are presented, and the MFI of quadruplicate experiments are shown after subtraction of the autofluorescence. (D) RT-PCR analysis of STAT-1 and Fas mRNA isolated from MMC, MMCneg, ANV, and ANVpos. Expression of β-actin was determined as an internal control.
Differential expression of H-2q, STAT-1 and Fas in ANV and MMCneg clones
There were substantial differences between MMC and ANV, ANVpos and MMC, or MMCneg and ANV in downregulation and upregulation of certain genes (shown in gray dots) (Fig. 3B). Since initial rejection of MMC in FVB mice was mediated by T cells, we evaluated expression of functional genes such as H-2q, the IFN-γ downstream signaling molecule STAT-1, and Fas in these tumor lines using three biological replicates. As shown in Fig. 3C, ANVpos had higher levels of H-2q expression (MFI: 67) compared to MMC (MFI: 15) (p=0.007). MMCneg appeared to have lower levels of H-2q (MFI: 14) compared to ANV (MFI: 30) (p=0.03). Staining of the tumor cells was performed at the same time, and there was no variation in autofluorescence. Therefore, MFI are presented after the subtraction of the autofluorescence.
Semi-quantitative RT-PCR analyses showed that expression of STAT-1 was higher in ANVpos (95.4%) and MMCneg (100%) cells than in MMC (45.9%) and ANV (18.6%) cells. While MMC showed higher expression of STAT-1 than ANV, expression of Fas was higher in ANV (64.2%) than in MMC (38.5%) cells. ANVpos (96.3%) and MMCneg (100%) had higher levels of Fas than MMC or ANV (Fig. 3D).
IFN-γ-induced methylation of the MMTV promoter downregulates neu expression in MMC
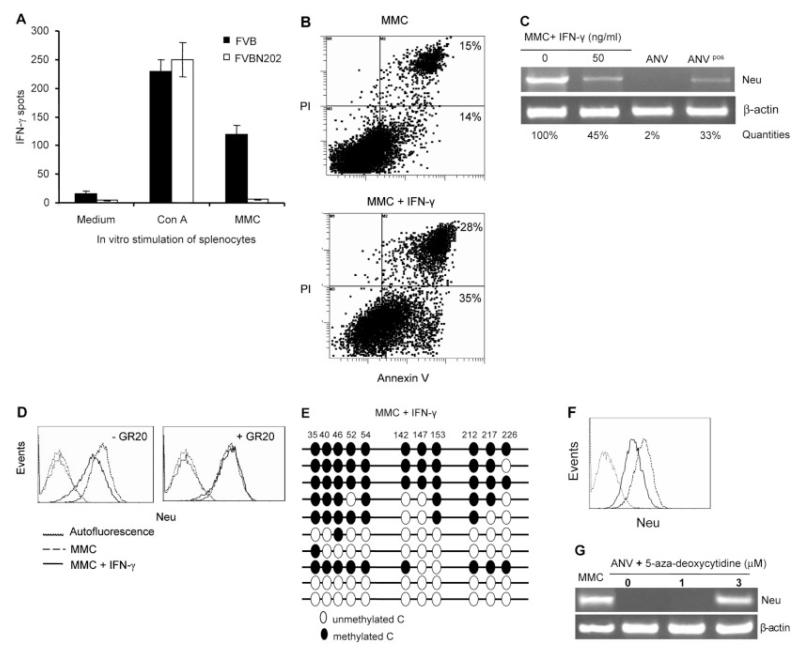
Initial rejection of MMC in FVB mice occurred in the presence of MMC-specific IFN-γ-secreting T cells, while splenocytes of FVBN202-transgenic mice showed no MMC-specific IFN-γ secretion (Fig. 4A). MMC and ANV appeared to have similar levels of expression of IFN-γ receptor [14]. Therefore, we cultured MMC in the presence or absence of IFN-γ in vitro to determine whether IFN-γ may induce neu antigen loss. At the end of day 3, the viability of adherent MMC was above 99%, as determined by trypan blue exclusion, and the total number of cells reached 21.7 × 106 in the absence of IFN-γ. IFN-γ induced apoptosis in the majority of MMC, so that total cell numbers reached 9.38 × 106, with only 28% viability (2.6 × 106 viable MMC). Flow cytometry analyses of the viable cells showed that IFN-γ increased early and late apoptotic cells from 14% and 15% to 35% and 28%, respectively (Fig. 4B). IFN-γ-treated viable MMC showed downregulation of neu expression both at the mRNA (Fig. 4C) and the protein (MFI: 329 versus 154; Fig. 4D) level after a 3-day culture. Such effects were abolished when the IFN-γ receptor was blocked using GR20 Ab (Fig. 4D). Bisulfite genomic sequencing showed that downregulation of neu expression by IFN-γ was due to IFN-γ-induced methylation of the MMTV promoter in MMC (Fig. 4E). In order to evaluate the status of neu expression ex vivo at the time of MMC rejection, tumors were removed 10–14 days after the challenge. Flow cytometry analyses of MMC showed downregulation of neu expression. However, 3- to 4-wk cultures of these MMC in the absence of IFN-γ resulted in upregulation of neu expression ex vivo (Fig. 4F). In order to determine whether hypomethylation of the MMTV promoter in ANV may reverse neu expression, we treated ANV with 1 and 3 μM 5-aza-deoxycytidine for 3 days. As shown in Fig. 4G, 3 μM 5-aza-deoxycytidine reversed neu expression in ANV as determined by RT-PCR. A higher concentration of 10 μM 5-aza-deoxycytidine was toxic and failed to reverse neu expression in ANV (data not shown).
Figure 4.
IFN-γ downregulates expression of neu antigen by inducing methylation of the MMTV promoter. (A) Detection of MMC-specific IFN-γ secretion by splenocytes of wild-type FVB or FVBN202-transgenic mice (n=4) 2 wk after challenge with MMC using ELISPOT assay. Splenocytes were stimulated with irradiated MMC in vitro at 6:1 ratios. Con A stimulation was used as a positive control. (B) Flow cytometry analyses of early apoptotic (Annexin V-positive) or late apoptotic (PI- and Annexin V-positive) MMC in the absence or presence of IFN-γ for 3 days. (C) Semi-quantitative RT-PCR analysis of neu expression in MMC in the absence or presence of IFN-γ. ANV and ANVpos cell lines were used as controls for loss or intermediate expression of neu mRNA. Neu expression in the PCR products was quantitated in agarose gel using Quantity One 1-D analysis Software. (D) Flow cytometry analysis of the neu expression in MMC in the absence (dashed line) or presence (solid line) of IFN-γ in vitro. Left and right panels show in vitro culture conditions in the absence or presence of IFN-γ-blocking GR20 Ab (20 lg/mL), respectively. Viable cells were gated in all the FACS analyses. Autofluorescence is shown as dotted lines. Isotype control-induced fluorescence was similar to autofluorescence. (E) Bisulfite genomic sequencing of MMC cells after 3-day treatment with IFN-γ. A total of ten clones per sample was sequenced. (F) MMC were removed from FVB mice at the time of tumor rejection (day 10–14 post-challenge). Expression of the neu protein was detected by flow cytometry either immediately after the MMC removal (solid line) or after 3–4 wk ex vivo culture (dashed line). The dotted line indicates autofluorescence. (G) RT-PCR analysis of RNA isolated from untreated MMC or ANV as well as ANV treated with different concentrations of 5-aza-deoxycytidine for 3 days. β-actin was amplified as an internal control.
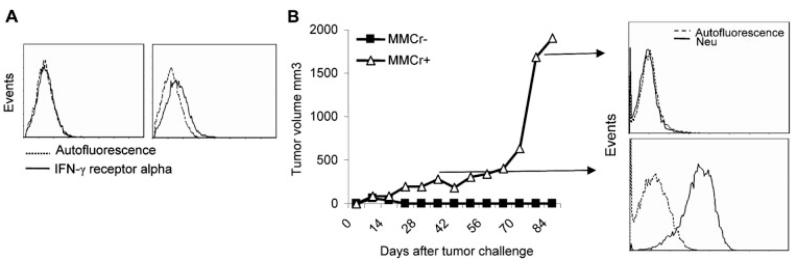
In order to determine the role of IFN-γ in the downregulation of neu expression in MMC in vivo, we used two clones of MMC: IFN-γ receptor-positive MMC (MMCr+) and IFN-γ receptor-negative MMC (MMCr−). Heterogeneity of MMC in the expression of IFN-γ receptor alpha allowed us to prepare MMCr+ and MMCr− clones through cell passages in vitro. Freshly isolated MMC from spontaneous mammary tumors were MMCr+, but they lost expression of IFN-γ receptor after a number of passages in vitro and became MMCr−. However, expression of neu antigen remained intact in MMCr−. In order to eliminate the role of Ab responses in the rejection of MMC and focus on CD8+ T cells as a major source of the neu-specific IFN-γ production, we depleted CD4+ T cells in vivo at the priming phase of the immune response. Animals were then inoculated with MMCr+ or MMCr− (Fig. 5A). While helpless CD8+ Tcells rejected MMCr− aggressively, MMCr+ remained at a plateau until 2–3 months after the challenge and then grew aggressively (Fig. 5B). Aggressive growth of MMCr+ was associated with the loss of neu expression and progression of MMC into ANV, while neu expression remained unchanged during the plateau phase (Fig. 5B). MMCr− did not relapse during this follow-up period. Rejection of MMCr−, but not MMCr+, in CD4-depleted mice is consistent with the IFN-γ blocking studies in vivo showing that IFN-γ is not the only cytokine involved in the rejection of MMC.
Figure 5.
IFN-γ-mediated neu antigen loss and tumor relapse in vivo. (A) Establishment of MMCr+ and MMCr− clones with the presence or lack of IFN-γ receptor alpha chain obtained from freshly isolated spontaneous mammary tumors or after a number of passages of MMC in vitro, respectively. (B) FVB mice (n=3) were depleted of CD4+ T cells and inoculated with MMCr+ or MMCr−. Tumor growth was monitored until 3 months after the challenge. RT-PCR analysis of neu mRNA expression in the tumors obtained from MMCr+-bearing FVB mice either at the plateau phase of tumor growth (day 40 post-challenge) or at the exponential growth phase (day 85 post-challenge).
Discussion
It was suggested that immunoediting of tumors is facilitated by the genetic instability of tumors [15], particularly in the genes encoding tumor antigens. In order to understand tumor antigen loss during tumor immunoediting, we used the wild-type FVB mouse and the FVBN202-transgenic mouse model of HER-2/neu-positive mammary carcinomas. Unlike wild-type FVB, FVBN202-transgenic mice express rat neu protein under the control of the MMTV promoter, rendering their immune system tolerant to the rat neu oncogene product. Wild-type FVB mice can reject primary MMC in T cell-dependent, neu-specific fashion as shown here with the in vivo CD4/CD8 T cell-depletion studies, induction of MMC-specific IFN-γ production, and detection of MMC-specific T cell responses [16]. Since CD4 depletion in vivo may also deplete CD4+ NK1.1 cells, we depleted NK1.1 cells in vivo to determine whether these cells may participate in MMC rejection. However, the presence of T cells in the absence of NK1.1 cells induced spontaneous rejection of MMC in FVB mice (data not shown). Despite T cell-mediated rejection of MMC, tumor relapse occurred due to neu antigen loss and emergence of ANV. Such neu antigen loss was induced by anti-tumor T cell responses, because expression of neu antigen remained intact in MMC cells derived from T cell-depleted mice. No marked differences were observed in the growth of MMC and ANV when these tumors were inoculated into FVBN202-transgenic mice [12].
It has been suggested that tumor immunoediting occurs due to Darwinian selection, where tumor-specific immune responses eliminate antigen-positive clones, and antigen-negative variants escape and grow [8, 9]. On the other hand, there are reports supporting the hypothesis that anti-tumor immune responses themselves induce changes in antigen-positive clones, converting them into antigen-negative clones [10, 11]. Using a neu-overexpressing primary tumor, MMC, and its relapsed neu-negative variants, ANV, our data in the present studies support the latter.
First of all, we show that neu antigen loss was due to the induction of epigenetic changes in neu-positive MMC in the presence of anti-tumor T cell responses. Initial rejection of MMC in wild-type FVB mice occurred in the presence of MMC-specific IFN-γ-producing Tcells. in vitro studies showed that IFN-γ was involved in downregulation of neu antigen in MMC. Three-day treatment of neu-overexpressing MMC with IFN-γ downregulated neu expression at both the mRNA level and the protein level in viable MMC. Sensitivity of MMC to IFN-γ-mediated methylation of the MMTV promoter depended on the status of the IFN-γ receptor in MMC. IFN-γ downregulated neu expression in the IFN-γ receptor-positive MMC but not in IFN-γ receptor-negative MMC clones. However, removal of IFN-γ from the culture resulted in normal proliferation of MMC and overexpression of neu after 3 to 4 wk in culture. Interestingly, expression of the neu antigen was downregulated at the time of MMC rejection and recovered after a 3- to 4-wk culture ex vivo. Considering that tumor relapse and emergence of ANV occur 1–3 months after the initial tumor rejection, IFN-γ treatment for 3 days might not be long enough to induce complete and/or stable neu loss. Alternatively, other components of the immune response such as neu-specific Ab [17], Fas-Fas ligand interactions, or Granzyme B-mannose-6-phosphate receptor interactions may also contribute to the neu antigen loss. These possibilities remain to be investigated. It is also likely that MMC clones with low expression of neu antigen may not survive for 3–4 wk after removal of IFN-γ in vitro, while neu-positive MMC may survive and become predominant. Although downregulation of the neu protein was detected in MMC at the time of tumor rejection, treatment of animals with GR20 Ab to block IFN-γ receptor in vivo did not prevent the rejection of MMC or downregulation of neu expression in wild-type FVB mice (data not shown). This indicates that IFN-γ partially, not solely, contributes in the tumor rejection or induction of epigenetic changes in MMC resulting in downregulation of the neu antigen. Interestingly, IFN-γ receptor-positive MMC clones (MMCr+) relapsed in vivo in the presence of CD8+ T cells and absence of CD4+ T cells or anti-neu Ab. Relapsed tumors were ANV. On the other hand, IFN-γ receptor-negative MMCr− tumors were rejected, and animals remained tumor-free during the trial. These in vivo studies suggest that the status of the IFN-γ receptor in tumors will determine rejection or relapse of MMC in FVB mice (manuscript in preparation). It has been shown that loss/downregulation of neu in FVBN202-transgenic mice is controlled by hypermethylation of the MMTV promoter in CpG sites within region 1 [13]. Zhou and coworkers [13] identified ten potential sites of methylation within CpG islands. However, we identified one additional potential site of methylation (position 213) in this region. We detected hypermethylation of the MMTV promoter during antigen loss induced by IFN-γ treatment in vitro. ANV cells isolated from animals with intact T cells also showed hypermethylation of MMTV. This further confirmed that IFN-γ-mediated epigenetic changes in MMC result in neu antigen loss and tumor escape. Treatment of ANV with 3 μM of a demethylating agent (5-aza-deoxy-cytidine) was able to reverse neu expression in ANV.
Our findings are consistent with other reports showing that IFN-γ can promote immune-mediated tumor escape by downregulation of gp70 in CT26 tumors [10]. The downstream signaling pathways leading to IFN-γ-mediated antigen loss remain to be determined. Interestingly, expression of HER-2/neu in human breast carcinomas is regulated by the AP-2 transcription factor, which binds a CpG-rich promoter region of the HER-2/neu gene [18]. Therefore, it is likely that methylation of this promoter region induced by IFN-γ may inhibit the expression of HER-2/neu by AP-2. It was reported that treatment of HER-2/neu-positive human ovarian carcinoma cells with IFN-γ reduced the expression of HER-2/neu at both mRNA and protein levels [19]. We also detected IFN-γ-mediated downregulation of HER-2/neu in human breast carcinoma cell lines (manuscript in preparation).
Secondly, we showed that residual neu-negative clones in MMC (MMCneg) and residual neu-positive clones in ANV (ANVpos) differ from ANV and MMC, respectively. ANVpos showed intermediate levels of neu expression as compared to MMC cells, which indicates that ANVpos cells were not MMC left behind following neu-targeted immune responses. The differences between ANV and MMCneg also do not support the hypothesis that ANV might be derived from MMCneg following eradication of neu-positive clones under immune pressure. The heterogenic nature of MMC and ANV allowed us to isolate MMCneg and ANVpos cells and perform comparative studies. The Darwinian selection hypothesis also predicts the presence of such residual clones. These residual neu-negative and neu-positive clones were not artifacts of in vitro cell culture, because the status of neu expression in these tumors was validated in several tumor clones, and similar observations were made in fractions of freshly isolated MMC and ANV [16, 17, 20]. In addition, MMCneg and ANVpos were highly stable for the lack of or intermediate expression of neu over 45 passages in vitro. Thus, no residual neu-positive clones or neu-negative clones were detected in MMCneg or ANVpos, respectively. Patterns of methylation of the MMTV promoter in four cell lines corresponded to the patterns of surface neu expression in these cell lines. There was one clone in ANV with complete demethylation and two clones in MMC with partial methylation. These clones may represent residual neu-positive and neu-negative clones of ANV and MMC, respectively. The four tumor lines appeared to have distinct gene expression profiles as well as different morphology and proliferation rates. Hundreds of genes were upregulated or downregulated when comparing MMC with ANV, MMC with ANVpos, or ANV with MMCneg. Gene array analyses showed that epithelial markers such as claudin 3 (CLDN3), CLDN4, and occluding (OCLN) [21] were markedly increased in MMC versus ANV or MMC versus ANVpos, while these genes were not expressed in MMCneg. In addition, mesenchymal markers such as procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide (P4HA1), and snail homolog 1 (SNAI1) as well as actin, alpha 2, smooth muscle, and aorta (ACTA2) [21] were markedly higher in ANV versus MMC or ANVpos versus MMC. Expression of vimentin (VIM) was higher in ANV than in MMC. These findings are consistent with previous reports [20].
Since microarray analysis showed differentially expressed H-2q, STAT-1, and Fas in MMC, ANV, MMCneg, and ANVpos, we used semi-quantitative RT-PCR to further validate the microarray findings. These findings in biological replicates of the cells further confirmed differential expression of these molecules in MMC versus ANVpos and ANV versus MMCneg. These results are consistent with our previous findings showing that MMC and ANV have distinct proteomic profiles [16], and these tumor lines express comparable levels of IFN-γ receptors, while expression of STAT-1 is downregulated in ANV [14]. This also suggests that ANV should be more resistant than MMC to IFN-γ-induced apoptosis in vivo. Others have also reported that relapsed tumors are refractory to IFN-γ-mediated T cell responses [22].
It has been reported that IFN-γ downregulates the NKG2D ligand H60 on tumors, rendering them resistant to NK-mediated killing [23]. Therefore, the epigenetic effect of IFN-γ is not restricted to particular tumor antigens. On the other hand, depending on the antigenic system and mechanism of antigen expression, IFN-γ may or may not modulate particular tumor antigens. Regulatory functions of IFN-γ on the expression of other tumor antigens that are regulated by hypomethylation of the gene promoter remain to be investigated. Our findings are consistent with other reports [10] and further support the hypothesis that IFN-γ simultaneously induces apoptosis and antigen loss in tumors. Therefore, the outcome of anti-tumor immune responses will depend on the balance between these tumor inhibitory and tumor immunoediting effects. IFN-γ is one of the components of anti-tumor immune responses that might be actively involved in neu antigen loss due to the induction of epigenetic changes in primary tumors. Whether other components of the immune response such as Fas ligand, granzyme, or TRAIL contribute to tumor rejection or neu antigen loss also remains to be determined. It has been reported that anti-neu Ab can induce neu antigen loss in mammary carcinomas of FVBN202-transgenic mice [16]. Identification of antigenic epitopes in HER-2/neu that may induce either immune-mediated tumor rejection or tumor relapse would improve peptide-based vaccination approaches to overcome tumor relapse [24, 25].
Materials and methods
Mice
Wild-type FVB (Jackson Laboratories) and FVBN202-transgenic female mice (Charles River Laboratories) were used throughout these studies. FVBN202 is the rat neu-transgenic mouse model in which 100% of females develop spontaneous mammary tumors by 8–10 months of age, with many features similar to human breast cancer. These mice overexpress an unactivated rat neu transgene under the regulation of the MMTV promoter [26]. The studies have been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Commonwealth University.
Tumor cell lines
The MMC cell line was established from a spontaneous tumor harvested from FVBN202-transgenic mice as previously described, with minor modifications [16, 20]. Tumors were sliced into pieces and treated with 0.25% trypsin at 4°C for 12–16 h. Cells were then incubated at 37°C for 30 min, washed, and cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) [27]. The ANV cell line was generated by s.c. inoculation of MMC (5 × 106 to 6 × 106) into the right mid-dorsum of a non-transgenic wild-type FVB mouse. In this mouse, anti-neu immune responses can be elicited, resulting in initial tumor rejection and relapse of ANV after a long latency. MMCneg and ANVpos cell lines were derived from MMC and ANV, respectively, using the Beckman Coulter EPICS Elite sorter. These cells were then cultured in RMPI 1640 supplemented with 10% FBS and were analyzed for the expression of rat neu protein before use. In some experiments, MMC (4 × 106 cells) were cultured in the presence or absence of IFN-γ (50 ng/mL; Serotec, NC).
Flow cytometry
A single staining flow cytometry analysis of the mammary tumor cells (106 cells/tube) was carried out using mouse anti-neu (Ab-4) Ab (Calbiochem, San Diego, CA), mouse anti-H-2q Ab, isotype control Ig, and FITC-conjugated anti-mouse Ig (BD Pharmingen, San Diego, CA) at the concentrations recommended by the manufacturer. Cells were finally washed, fixed with 1% ultra-pure formaldehyde, and analyzed with the Beckman Coulter EPICS XL within 24 h of fixing. Double staining of viable cells was also performed using Annexin V and propidium iodide (PI) as previously described [28].
ELISPOT assay
Splenocytes of FVB and FVBN202-transgenic mice (inoculated with MMC) were subjected to ELISPOT assay as previously described by our group [12]. Briefly, 96-well filtration plates (Millipore, Bedford, MA) were coated with 10 lg/mL rat anti-mouse IFN-γ Ab (BD PharMingen, San Diego, CA) and subsequently blocked with RPMI 1640 medium containing 10% FBS. RBC were lysed with Tris-NH4Cl, and 50 μL of the splenocytes (5 × 105 cells/well) were added to each well and incubated with 50 lL Con A (5 lg/mL) or irradiated MMC (15 000 rad; 6:1 E:T ratios) in complete medium (10% FBS, 50 U/mL penicillin/streptomycin, 2 mM L-glutamine, 1 mM 2-ME) at 37°C in an atmosphere of 5% CO2 for 20–24 h. The plates were then washed extensively and incubated with 5 lg/mL biotinylated anti-mouse IFN-γ Ab (BD PharMingen), followed by a pulse with 0.2 U/mL alkaline phosphatase avidin D (Vector Laboratories, Burlingame, CA). Positive spots were developed by adding 50 lL/well 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium solution (Roche, Indianapolis, IN).
Microarray analysis
The Affymetrix® protocol has been described elsewhere [29, 30]. The GeneChip® Mouse Genome 430A 2.0 array provides comprehensive coverage of the transcribed murine genome by including over 22 600 probe sets that analyze the expression level of over 14 000 murine transcripts. Each chip was scanned at a high resolution, with pixelations ranging from 2.5 μM down to 0.51 μM, with the Affymetrix GeneChip® Scanner 3000 according to the GeneChip® Expression Analysis Technical Manual procedures (Affymetrix, Santa Clara, CA). After scanning, the raw intensities for each probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip® Operating Software (GCOSv1.1) (Affymetrix). Normalization, background subtraction, and expression values for each probe set were calculated using a method developed by others [31], which is an effective expression summary motivated by a log scale linear additive model. This summary statistics, referred to as the log scale robust multi-array analysis (RMA), uses .CEL files to calculate probe set expression summaries.
Statistical analysis
The microarray analysis was performed with the BRB-ArrayTools v3.1.0 [32], an Excel Add-in that collates microarray data with sample data. Moreover, the “significance-score” algorithm (S-score) was used to produce a score for all the comparisons of the expression summaries between MMC and ANV, MMC and ANVpos, and ANV and MMCneg [33]. The S-score produces a robust measure of expression changes by weighting oligonucleotide pairs according to their signal strength above empirically determined noise levels. The procedure produces scores centered around “0” (no change) with a standard deviation of 1. Thus, scores >2 or <−2 from a single comparison have, on average, a 95% chance of being significant hybridization changes, corresponding to p<0.05.
Semi-quantitative RT-PCR
According to the differences detected by microarray analyses, three immunologically relevant genes (neu, STAT-1, and Fas) were selected to validate the differences in gene expression between primary MMC versus ANVpos or relapsed ANV versus MMCneg. Total RNA (1.5 lg) from four tumor lines was used as templates in a reverse transcription reaction system (total volume of 20 lL). The cDNA were then transferred to a PCR master mixture containing 1× PCR buffer, 1.5 mM MgCl2, 2.5 U Taq polymerase, and 1 μM gene-specific primers: Neu [5′-ATGATCATCATGGAGCTGGCG-3′ (sense) and 5′-CTAG-GATCTCAGGGTTCTCTGCA-3′ (anti-sense)]; STAT-1 and Fas [34]; and β-actin [5′-GTGGGCCGCTCTAGGCACCAA-3′ (sense) and 5′-CTCTTTGATGTCACGCACGATTTC-3′ (anti-sense)]. PCR conditions were as follows: Neu: 94°C 5 min, 94°C 1 min, 66°C 1 min, 72°C 3 min (40 cycles) followed by 10 min extension at 72°C; STAT-1 and Fas: 94°C 5 min, 94°C 30 s, 60°C 30 s, 72°C 1 min (35 cycles) followed by 5 min extension at 72°C; β-actin: 94°C 5 min, 94°C 30 s, 52°C 30 s, 72°C 1 min (30 cycles) followed by 5 min extension at 72°C. Amplified fragments were visualized by ethidium bromide staining of the agarose gel and photography under UV light in Gel Doc 2000™ (BioRad). Quantity One 1-D analysis Software was used to quantitate each PCR product. Data were normalized using β-actin as an internal control. The highest value in the MMC-positive control was adjusted to 100%, and values for samples were calculated proportionally.
In vivo tumor challenge studies
Female FVB mice (n=4) were inoculated s.c. with MMC or ANV (5 × 106 cells/mouse). Animals were inspected twice every week for the development of tumors. Masses were measured with calipers along the two perpendicular diameters. Tumor volume was calculated by: V(volume) = L(length) × W(width)2 ÷ 2. Mice were killed before the tumor mass exceeded 2000 mm3. FVBN202-transgenic mice (n=3) were also inoculated with MMC or ANV.
Bisulfite genomic sequencing
Genomic DNA was isolated from the tumor cells (6 × 106) using the ZR Genomic DNA Kit (Zymo Research, Orange, CA). For DNA methylation analysis, 0.5 lg DNA was treated with bisulfate using the EZ DNA Methylation Kit according to the manufacturer’s protocol (Zymo Research). The reaction was performed using the FastStart High Fidelity PCR System (Roche) and the following primers: forward 5′-GAGAAGTAGT-TAAGGGGTTGTTTTTTAT-3′; reverse 5′-AAATTAACTA-TAATCCTTACCCCAAAAA-3′. PCR reactions were carried out as described previously [13]. The resulting PCR fragments were ligated into the pGEM-T Easy vector (Promega, Madison, WI) and were sequenced.
Demethylation studies
ANV cells were treated with 1 or 3 μM 5-aza-deoxycytidine (EMD Biosciences, San Diego, CA) for 3 days. MMC and ANV were used as positive and negative controls for neu expression. RNA was then isolated and used in a two-step RT-PCR reaction using neu-specific primers that amplified a 474-bp fragment of neu mRNA: sense 5′-AACAGCTCAGAGACCTGCTTTGGA-3′ and anti-sense 5′-TGATCCAAGCACCTTCACCTTCCT-3′. β-ac-was amplified as an internal control.
Acknowledgements
This work was supported by the NIH (R01 CA104757), the Susan G. Komen Foundation for Breast Cancer Research (BCTR0504184), and the flow cytometry shared resources facility (supported in part by NIH grant P30CA16059). We thank Hooman Nikizad, Julie S. Farnsworth, and Teri L. Russell for their assistance with flow cytometry and production of GK1.5, 2.43, and GR20 Ab using hybridomas. We also thank Dr. Ian MacDonald for critical reading of the manuscript. We gratefully acknowledge helpful discussions from Dr. Shirley Taylor and the support of the VCU Massey Cancer Center and the Commonwealth Foundation for Cancer Research.
Abbreviations
- ANV
neu antigen-negative variant
- MMC
neu overexpressing mouse mammary carcinoma
- MMTV
mouse mammary tumor virus
References
- 1.Maeurer MJ, Gollin SM, Storkus WJ, Swaney W, Karbach J, Martin D, Castelli C, et al. Tumor escape from immune recognition: lethal recurrent melanoma in a patient associated with downregulation of the peptide transporter protein TAP-1 and loss of expression of the immunodominant MART-1/Melan-A antigen. J. Clin. Invest. 1996;98:1633–1641. doi: 10.1172/JCI118958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Marincola FM, Rivoltini L, Parmiani G, Ferrone S. Selective histocompatibility leukocyte antigen (HLA)-A2 loss caused by aberrant pre-mRNA splicing in 624MEL28 melanoma cells. J. Exp. Med. 1999;190:205–215. doi: 10.1084/jem.190.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamshchikov GV, Mullins DW, Chang CC, Ogino T, Thompson L, Presley J, Galavotti H, et al. Sequential immune escape and shifting of Tcell responses in a long-term survivor of melanoma. J. Immunol. 2005;174:6863–6871. doi: 10.4049/jimmunol.174.11.6863. [DOI] [PubMed] [Google Scholar]
- 4.Facoetti A, Nano R, Zelini P, Morbini P, Benericetti E, Ceroni M, Campoli M, Ferrone S. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clin. Cancer Res. 2005;11:8304–8311. doi: 10.1158/1078-0432.CCR-04-2588. [DOI] [PubMed] [Google Scholar]
- 5.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFN-γ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 6.Engel AM, Svane IM, Rygaard J, Werdelin O. MCA sarcomas induced in scid mice are more immunogenic than MCA sarcomas induced in congenic, immunocompetent mice. Scand. J. Immunol. 1997;45:463–470. doi: 10.1046/j.1365-3083.1997.d01-419.x. [DOI] [PubMed] [Google Scholar]
- 7.Svane IM, Engel AM, Nielsen MB, Ljunggren HG, Rygaard J, Werdelin O. Chemically induced sarcomas from nude mice are more immunogenic than similar sarcomas from congenic normal mice. Eur. J. Immunol. 1996;26:1844–1850. doi: 10.1002/eji.1830260827. [DOI] [PubMed] [Google Scholar]
- 8.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 9.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 10.Beatty GL, Paterson Y. IFN-γ can promote tumor evasion of the immune system in vivo by down-regulating cellular levels of an endogenous tumor antigen. J. Immunol. 2000;165:5502–5508. doi: 10.4049/jimmunol.165.10.5502. [DOI] [PubMed] [Google Scholar]
- 11.Beatty GL, Paterson Y. Regulation of tumor growth by IFN-γ in cancer immunotherapy. Immunol. Res. 2001;24:201–210. doi: 10.1385/IR:24:2:201. [DOI] [PubMed] [Google Scholar]
- 12.Manjili MH, Wang X-Y, Chen X, Martin T, Repasky EA, Henderson R, Subjeck JR. HSP110-HER2/neu chaperone complex vaccine induces protective immunity against spontaneous mammary tumors in HER-2/neu transgenic mice. J. Immunol. 2003;171:4054–4061. doi: 10.4049/jimmunol.171.8.4054. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Chen WD, Qin X, Lee K, Liu L, Markowitz SD, Gerson SL. MMTV promoter hypomethylation is linked to spontaneous and MNU associated c-neu expression and mammary carcinogenesis in MMTV c-neu transgenic mice. Oncogene. 2001;20:6009–6017. doi: 10.1038/sj.onc.1204830. [DOI] [PubMed] [Google Scholar]
- 14.Manjili MH, Kmieciak M, Keeler J. Comment on “tumor progression can occur despite the induction of very high levels of self/tumor antigen-specific CD8+ T cells in patients with melanoma”. J. Immunol. 2006;176:4511. doi: 10.4049/jimmunol.176.8.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 16.Manjili MH, Arnouk H, Knutson KL, Kmieciak M, Disis ML, Subjeck JR, Kazim AL. Emergence of immune escape variant of mammary tumors that has distinct proteomic profile and a reduced ability to induce “danger signals”. Breast Cancer Res. Treat. 2006;96:233–241. doi: 10.1007/s10549-005-9044-4. [DOI] [PubMed] [Google Scholar]
- 17.Knutson KL, Almand B, Dang Y, Disis ML. Neu antigen-negative variants can be generated after neu-specific Ab therapy in neu transgenic mice. Cancer Res. 2004;64:1146–1151. doi: 10.1158/0008-5472.can-03-0173. [DOI] [PubMed] [Google Scholar]
- 18.Vernimmen D, Begon D, Salvador C, Gofflot S, Grooteclaes M, Winkler R. Identification of HTF (HER2 transcription factor) as an AP-2 (activator protein-2) transcription factor and contribution of the HTF binding site to ERBB2 gene overexpression. Biochem. J. 2003;370:323–329. doi: 10.1042/BJ20021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marth C, Muller-Holzner E, Greiter E, Cronauer MV, Zeimet AG, Doppler W, Eibl B, et al. Gamma-interferon reduces expression of the protooncogene c-erbB-2 in human ovarian carcinoma cells. Cancer Res. 1990;50:7037–7041. [PubMed] [Google Scholar]
- 20.Knutson KL, Lu H, Stone B, Reiman JM, Behrens MD, Prosperi CM, Gad EA, et al. Immunoediting of cancers may lead to epithelial to mesenchymal transition. J. Immunol. 2006;177:1526–1533. doi: 10.4049/jimmunol.177.3.1526. [DOI] [PubMed] [Google Scholar]
- 21.Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, Raaka BM. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science. 2004;306:2261–2264. doi: 10.1126/science.1101968. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg SA, Sherry RM, Morton KE, Scharfman WJ, Yang JC, Topalian SL, Royal RE, et al. Tumor progression can occur despite the induction of very high levels of self/tumor antigen-specific CD8+ T cells in patients with melanoma. J. Immunol. 2005;175:6169–6176. doi: 10.4049/jimmunol.175.9.6169. [DOI] [PubMed] [Google Scholar]
- 23.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J. Immunol. 2006;176:905–913. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- 24.Hueman MT, Dehqanzada ZA, Novak TE, Gurney JM, Woll MM, Ryan GB, Storrer CE, et al. Phase I clinical trial of a HER-2/neu peptide (E75) vaccine for the prevention of prostate-specific antigen recurrence in high-risk prostate cancer patients. Clin. Cancer Res. 2005;11:7470–7479. doi: 10.1158/1078-0432.CCR-05-0235. [DOI] [PubMed] [Google Scholar]
- 25.Peoples GE, Gurney JM, Hueman MT, Woll MM, Ryan GB, Storrer CE, Fisher C, et al. Clinical trial results of a HER2/neu (E75) vaccine to prevent recurrence in high-risk breast cancer patients. J. Clin. Oncol. 2005;23:7536–7545. doi: 10.1200/JCO.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 26.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell MJ, Wollish WS, Lobo M, Esserman LJ. Epithelial and fibroblast cell lines derived from a spontaneous mammary carcinoma in a MMTV/neu transgenic mouse. In vitro Cell. Dev. Biol. Anim. 2002;38:326–333. doi: 10.1290/1071-2690(2002)038<0326:EAFCLD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 28.Eray M, Matto M, Kaartinen M, Andersson L, Pelkonen J. Flow cytometric analysis of apoptotic subpopulations with a combination of annexin V-FITC, propidium iodide, and SYTO 17. Cytometry. 2001;43:134–142. doi: 10.1002/1097-0320(20010201)43:2<134::aid-cyto1028>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 29.Affymetrix . Affymetrix Microarray Suite User’s Guide, version 5.0. Affymetrix; Santa Clara, CA: 2001. [Google Scholar]
- 30.Sugita M, Geraci M, Gao B, Powell RL, Hirsch FR, Johnson G, Lapadat R, et al. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- 31.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Wang L, Ravindranathan A, Miles MF. A new algorithm for analysis of oligonucleotide arrays: application to expression profiling in mouse brain regions. J. Mol. Biol. 2002;317:225–235. doi: 10.1006/jmbi.2001.5350. [DOI] [PubMed] [Google Scholar]
- 34.Liu K, Caldwell SA, Abrams SI. Immune selection and emergence of aggressive tumor variants as negative consequences of Fas-mediated cytotoxicity and altered IFN-γ-regulated gene expression. Cancer Res. 2005;65:4376–4388. doi: 10.1158/0008-5472.CAN-04-4269. [DOI] [PubMed] [Google Scholar]