Abstract
Cell shape is critical for cell function. However, despite the importance of cell morphology, little is known about how individual cells generate specific shapes. Drosophila tracheal terminal cells have become a powerful genetic model to identify and elucidate the roles of genes required for generating cellular morphologies. Terminal cells are a component of a branched tubular network, the tracheal system that functions to supply oxygen to internal tissues. Terminal cells are an excellent model for investigating questions of cell shape as they possess two distinct cellular architectures. First, terminal cells have an elaborate branched morphology, similar to complex neurons; second, terminal cell branches are formed as thin tubes and contain a membrane-bound intracellular lumen. Quantitative analysis of terminal cell branch number, branch organization and individual branch shape, can be used to provide information about the role of specific genetic mechanisms in the making of a branched cell. Analysis of tube formation in these cells can reveal conserved mechanisms of tubulogenesis common to other tubular networks, such as the vertebrate vasculature. Here we describe techniques that can be used to rapidly fix, image, and analyze both branching patterns and tube formation in terminal cells within Drosophila larvae. These techniques can be used to analyze terminal cells in wild-type and mutant animals, or genetic mosaics. Because of the high efficiency of this protocol, it is also well suited for genetic, RNAi-based, or drug screens in the Drosophila tracheal system.
Keywords: Developmental Biology, Issue 77, Genetics, Molecular Biology, Cellular Biology, Biochemistry, Biophysics, Bioengineering, Cellular Structures, Epithelial Cells, Drosophila melanogaster, Microscopy, Phase-Contrast Microscopy, Fluorescence Microscopy, genetics (animal and plant), animal biology, animal models, Respiratory System, trachea, terminal cell, intact animal, larvae, cell morphology, Drosophila, fluorescence, branching, lumen, fruit fly, animal model
Introduction
Cell shape is critical for function of individual cells within an organism, as well as cells that function as part of a tissue or organ. We use Drosophila tracheal terminal cells, a component of the insect respiratory system, to investigate the molecular mechanisms that participate in controlling two conserved types of cellular morphology: branching and tube formation (lumenogenesis). Terminal cells are located at the tips of a network of branched tubes that functions to deliver oxygen to internal tissues1 and have an elaborate branched morphology which depends on an FGF signaling pathway that is controlled by local oxygen levels within target tissues2. Terminal cell branches are thin tubes, with a gas-filled subcellular lumen running through each branch. The distinct cellular architectures of terminal cells, along with the ease by which genetic analysis can be performed in Drosophila, make these cells an excellent model for investigating mechanisms of cellular outgrowth, branching, and intracellular tube formation. Terminal cells have proved a useful model for understanding some of the signaling pathways leading to branched cell differentiation, outgrowth, and maturation2-4. Using this system unbiased, forward genetic screens for cell morphogenesis mutants have been performed, yielding insights into mechanisms controlling cell shape5,6. For instance, these screens have revealed that a specific RabGAP is required for cytoskeletal polarity and vesicle trafficking in lumen formation and positioning7; that integrin-mediated adhesion is required for branch stability8; and that epithelial PAR-polarity proteins regulate polarized membrane trafficking required for both branching and lumen formation9. Other studies in terminal cells have shown that asymmetric actin accumulation and microtubule organization is required for cell elongation and lumenogenesis10. Thus, diverse, conserved cell biological mechanisms contribute to terminal cell morphogenesis.
Here, we describe a method to rapidly fix intact third-instar Drosophila larvae for analysis of terminal cell branching and lumen formation. This protocol can also be carried out on both first and second instar animals. Key to this technique is the ability to visualize terminal cells that are genetically labeled by fluorescent protein expression directly through the larval cuticle of intact animals. Since this procedure does not require any post-fixation manipulations, such as antibody staining, to observe the cells, it is well suited to high throughput analysis, including genetic or drug screening. Fluorescent protein expression reveals the structure of the cytoplasmically-filled branches. Tube formation can be monitored in parallel using brightfield microscopy to identify the gas-filled lumen, which contrasts with the surrounding fluid-filled tissues.
Included in this protocol is the method for generating genetic mosaics based on the MARCM system11, to produce homozygous mutant terminal cells labeled with fluorescent proteins in otherwise unlabeled animals. This is necessary, since terminal cells only elaborate their complex structures relatively late in development; genetic mosaics allow for bypass of gene requirements in other tissues earlier in development. To generate MARCM clones, trachea are labeled using the tracheal-specific driver breathless (btl)12. Described here is the protocol for the Drosophila X chromosome; for other chromosomes, a similar procedure can be used, with genetic reagents appropriate to the chromosome being examined. Here, trachea are labeled by expression of a cytoplasmically localized GFP, but the procedure works equally well with expression of other fluorescent proteins, such as DsRed.
Additionally, we have included a method to quantify branching patterns and lumen formation in terminal cells, based on methods developed for characterizing neuronal branch patterns13. This kind of quantitative data can be critical in discerning the precise role of genes in the branching or lumenogenesis process, as well as allowing for direct comparisons between different mutants9.
Protocol
1. Mosaic Generation
Genetic Crossw FRT19A tub:GAL80 FLP122/Y; Btl-GAL4, UAS-GFP (males) X * FRT19A/Bal (females), where * represents the mutant to be examined. Note: Set up cross at least one day prior to experiment, as this allows animals to mate appropriately.
Pre-lay: Transfer cross to fly-food vials with a small smear of fresh yeast paste placed on the media. Allow to lay for 1 hr at 25 °C.
Lay: Transfer animals from pre-lay vial to a fresh vial with a small smear of fresh yeast paste. Allow to lay for 6 hr at 25 °C.
Transfer adults back to pre-lay vial for storage and use in subsequent experiments. Immediately heat shock the lay vial containing 0-6 hr old embryos for 45 min at 38 °C, in a circulating water bath. Note: The surface of the food should be below the water level to verify all embryos are appropriately heat shocked.
Incubate heat shocked vials at 25 °C for 4-5 days until third instar larvae start to wander.
2. Screen for Mosaic Animals
Using fine forceps gently pick wandering third-instar larvae from the sides of the vial and place them into chilled 100% glycerol in a small plastic plate.
Examine larvae using a high magnification-dissecting microscope equipped with fluorescent optics. Identify larvae with mosaic expression of fluorescently labeled tracheal cells. Typically, this can be done at 10-20X magnification.
3. Heat Fixation
Using fine forceps carefully pick mosaic animals out of the dish and place in a drop of fresh 100% glycerol on a 75x25x1 mm white glass microscope slide. Note: multiple animals can be placed in the drop (Figure 1A). Animals can also be placed in a drop of glycerol directly onto a coverslip, but care must be taken during fixation (step 3.2), as the animals will heat very quickly and can become over-fixed.
Place glass slide onto 70 °C heat block until the animals just stop moving, no more than 20 sec for a slide, and 10 sec for a cover slip (Figure 1B). Animals will wriggle initially but stop and become rigid and elongated once dead. Note: overlong exposure to heat will damage terminal branches and lumens as well as make the GFP signal dim and diffuse. For optimal efficiency it is recommended to have the heat block in the same room as the microscopes.
Using fine forceps carefully orient all larvae in the drop in a single direction, such that all larvae on the slide are oriented parallel to one another (Figure 1C).
Gently place an 18x18mm micro cover glass on top of larvae (or invert the coverslip onto a glass slide) and avoid forming bubbles (Figure 1D). Note: coverslip may not lie exactly flat over larvae, this is not a problem.
Image larvae within 30 min. Heat fixed terminal cells will degrade and GFP signal will become very diffuse.
4. In vivo Imaging of Tracheal Terminal Cells
Place slide on the stage of a compound stereomicroscope. Using a 5X objective locate the two parallel dorsal trunks running from anterior to posterior on the dorsal surface of the animal (Figure 1F). Locate the two dorsal terminal cells, directly between the two dorsal trunks. This helps identify the appropriate segment for analysis.
Once oriented, rotate the animals to image desired terminal cells. To do this, carefully push the coverslip on the edge with the forceps perpendicular to the long axis of the larvae, slowly rolling the larvae underneath the coverslip (Figures 1E and 1G).
Selecting the appropriate terminal cells is critical for consistent results and comparisons between mutants. The fat body branch (FB) terminal cells are easily found adjacent to the dorsal trunk, lateral to the midline. The lateral group G terminal cells (LG) are easily found and are located just lateral of the ventral midline (Figure 1H). Note: in the most anterior (Tr1) and posterior (Tr10) segments the arrangement of terminal cell is divergent from other segments. We only analyze terminal cells in segments Tr2-Tr9. Dorsal terminal cells have a more stereotyped branching pattern than other tracheal terminal cells. This may depend on additional, non-cell autonomous signals, and this should be considered when deciding whether to include them in analysis.
Once a terminal cell of interest has been identified, capture an image using the desired magnification; typically 10X or 20X is best (Figure 2A). Note: due to variable branching patterns, try to capture images that include as many branches and branch tips as possible.
Without moving the stage, switch off the fluorescence and turn on the transmitted light to capture a brightfield image of the same cell and focal plane (Figure 2B). The lumen will be visualized as darkly contrasting within the terminal cell space. If planning on quantify terminal cell branches and lumens, collect multiple images in multiple focal planes of each terminal cell.
5. Analysis and Quantification of Terminal Cell Morphology Using ImageJ
Open the fluorescent and brightfield images using the ImageJ (http://rsb.info.nih.gov/ij/) plug-in NeuronJ (http://www.imagescience.org/meijering/software/neuronj/).
Using the "Add tracings" tool, manually trace the branches and lumen of the entire terminal cell.
Using the "Label tracings" tool, rename and recolor to assign each line within the trace a specific designation.
Using the "Set parameters" tool, adjust the line width from 6-10 pixels (for 512x512 images obtained from the digital camera connected to the compound microscope. For higher or lower resolution cameras, the pixel width should be scaled appropriately), using all other default settings, and choose "OK". Then use the "Make snapshot" tool to take an image. Choose "Draw trace" to get a snapshot of the trace that can be shown side-by-side with the original.
Select the "Measure tracings" tool and select the tracing type (i.e. which branches to measure based on the specific designations assigned in step 5.3). Next, select "Display tracing measurements" and "Clear previous measurements" then click "RUN". This will produce a spreadsheet that contains the name, label, length (in pixels), and other data for the image. This data can then be saved as a Microsoft Excel spreadsheet for further statistical analysis.
Representative Results
Results are shown in Figure 2. A single lateral group (LG) terminal cell shows extensive subcellular branching (visualized by GFP; A) and a gas-filled subcellular lumen running through each of the branches (visualized by brightfield microscopy; B). These images were collected from a mosaic L3 larva, generated using the MARCM system described in sections 1 & 2, and heat fixed and imaged, as described in sections 3 & 4. Panels C & D show a NeuronJ generated trace, as described in section 5, of the branches and the lumen respectively of the images shown in A & B. Panels E & F show the location of the fat body (FB) branch in a larva prepared for imaging as described in sections 3 & 4. Note that in this example, the animal is not a mosaic, and GFP is expressed throughout the entire tracheal system. Panels G and H show an example of a GFP-labeled terminal cell that was heat fixed for too long a time. GFP is diffuse (G), branches have broken down and portions of lumens are no longer gas filled, thus appearing as breaks in the brightfield image.
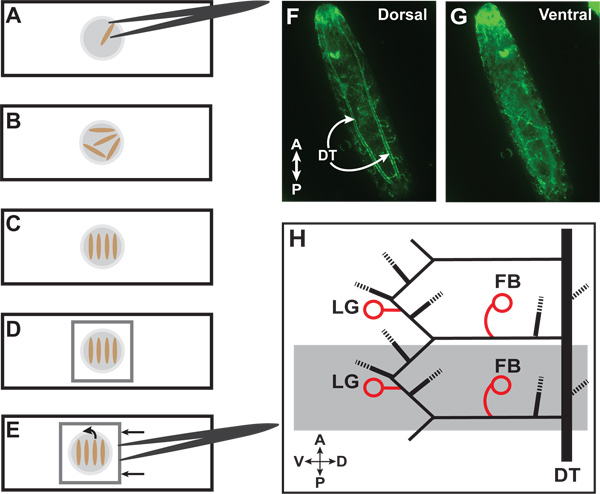 Figure 1. Larvae preparation and identification of terminal cells.(A) Place larva in a drop of 100% glycerol on a glass slide. (B) Multiple larvae can be placed for heat fixation. (C) After heat fixation, organize larvae parallel to each other and perpendicular to the long axis of the slide. (D) Place cover glass over larvae. (E) To reorient larvae, carefully push cover glass with forceps to roll the animals. (F) Wild-type third instar larva with GFP expressed throughout the tracheal system. The paired dorsal trunks (DT) are visible on the dorsal side. (G) The same larva after the rolling technique with the ventral side now facing upwards. (H) Diagram of lateral view of two third-instar tracheal hemisegments (one hemisegment is highlighted in grey). Circles indicate lateral group (LG) and fat body (FB) terminal cells which we use for quantitation. Dashed lines represent other branches of the tracheal system which we do not routinely quantitate. For a full description of the tracheal branches in a larval segment, refer to Ref 1.
Figure 1. Larvae preparation and identification of terminal cells.(A) Place larva in a drop of 100% glycerol on a glass slide. (B) Multiple larvae can be placed for heat fixation. (C) After heat fixation, organize larvae parallel to each other and perpendicular to the long axis of the slide. (D) Place cover glass over larvae. (E) To reorient larvae, carefully push cover glass with forceps to roll the animals. (F) Wild-type third instar larva with GFP expressed throughout the tracheal system. The paired dorsal trunks (DT) are visible on the dorsal side. (G) The same larva after the rolling technique with the ventral side now facing upwards. (H) Diagram of lateral view of two third-instar tracheal hemisegments (one hemisegment is highlighted in grey). Circles indicate lateral group (LG) and fat body (FB) terminal cells which we use for quantitation. Dashed lines represent other branches of the tracheal system which we do not routinely quantitate. For a full description of the tracheal branches in a larval segment, refer to Ref 1.
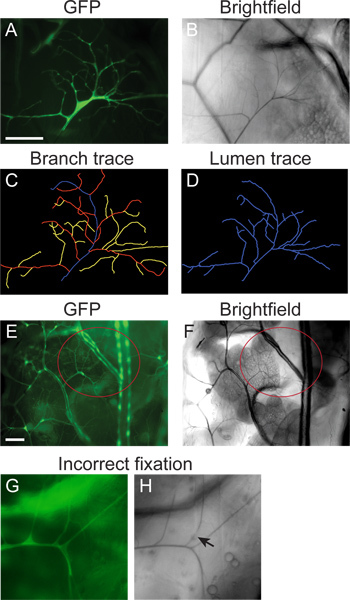 Figure 2. Representative images.(A-D) Mosaic L3 larvae were generated using the MARCM technique (section 1) and fixed and imaged using the protocol in sections 2-4. The branching pattern of a single LG terminal cell was visualized by mosaic expression of GFP (A); the gas-filled lumen visualized with brightfield microscopy (B). (C) Tracing of the branching pattern of the cell in A, generated using NeuronJ (section 5). (D) Tracing of the gas-filled lumen of the cell in B, generated using NeuronJ (protocol section 5). (E, F) A fat body (FB) terminal cell (highlighted by the red circle) visualized by GFP expression throughout the tracheal system (E) and brightfield microscopy (F) in a larva prepared by the protocol in sections 2-4. (G, H) Example of fixation artifacts obtained when the sample is heated for too long a period. GFP is diffuse and small branches have degraded (G). Areas of lumen also no longer appear air-filled (arrow in H). Scale bar: 100 μm.
Figure 2. Representative images.(A-D) Mosaic L3 larvae were generated using the MARCM technique (section 1) and fixed and imaged using the protocol in sections 2-4. The branching pattern of a single LG terminal cell was visualized by mosaic expression of GFP (A); the gas-filled lumen visualized with brightfield microscopy (B). (C) Tracing of the branching pattern of the cell in A, generated using NeuronJ (section 5). (D) Tracing of the gas-filled lumen of the cell in B, generated using NeuronJ (protocol section 5). (E, F) A fat body (FB) terminal cell (highlighted by the red circle) visualized by GFP expression throughout the tracheal system (E) and brightfield microscopy (F) in a larva prepared by the protocol in sections 2-4. (G, H) Example of fixation artifacts obtained when the sample is heated for too long a period. GFP is diffuse and small branches have degraded (G). Areas of lumen also no longer appear air-filled (arrow in H). Scale bar: 100 μm.
Discussion
The heat fixation technique described here is a rapid and convenient tool for imaging Drosophila larval tracheal terminal cells. Here, we use this technique to examine the branching and lumen pattern of wild-type cells. Tracheal cells expressing GFP, driven by the tracheal specific promoter breathless, can be easily visualized through the larval cuticle after heat fixation. The specific branching patterns of individual terminal cells, as well as those of the air-filled lumen, can be quickly visualized and measured using this method. This technique can also be performed on both first and second instar larvae. However, care must be taken in fixing smaller animals, as fluorescent protein expression can easily be disrupted.
In the method shown here, we describe an analysis of mosaic animals with GFP expressed in single tracheal cells. However, the same techniques are applicable to analysis of tracheal cells under a number of experimental manipulations. For instance, animals in which gene expression has been altered by molecular approaches, throughout the tracheal system or in individual cells by expression of RNAi transgenes or modified proteins, can also be examined with this approach. Non-genetic treatments which affect terminal cell development, such as drugs or hypoxia2,14, can also be characterized using these methods. In these latter cases, it is necessary to start with a Drosophila stock constitutively expressing GFP or another fluorescent protein throughout its tracheal system in order to visualize branching patterns. Gas-filling can be examined regardless of fluorescent protein expression.
Here, we only show examples of untagged (cytoplasmically localized) GFP being used to label the tracheal system. However, the methods described here are also suitable for detection of other native fluorescent proteins, such as DsRed, as well as fluorescent proteins which have been modified to be localized to specific subcellular structures. For instance, actin:GFP or tubulin:GFP fusions can be used to visualize the tracheal cytoskeleton10.
Disclosures
The authors declare that they have no competing financial interest.
Acknowledgments
We thank Gillian Stanfield for comments on the manuscript. T.A.J. is supported by the University of Utah Genetics Training Grant T32-GM007464 from NIH NIGMS.
References
- Manning G, Krasnow MA. Development of the Drosophila Tracheal System. The Development of Drosophila melanogaster. 1993. pp. 609–685.
- Jarecki J, Johnson E, Krasnow MA. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell. 1999;99(2):211–220. doi: 10.1016/s0092-8674(00)81652-9. [DOI] [PubMed] [Google Scholar]
- Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441(7094):746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Ruiz OE, Nikolova LS, Metzstein MM. Drosophila Zpr1 (Zinc Finger Protein 1) Is Required Downstream of Both EGFR And FGFR Signaling in Tracheal Subcellular Lumen Formation. PLoS ONE. 2012;7(9):e45649. doi: 10.1371/journal.pone.0045649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial AS, Levi BP, Krasnow MA. A systematic screen for tube morphogenesis and branching genes in the Drosophila tracheal system. PLoS Genetics. 2011;7(7):e1002087. doi: 10.1371/journal.pgen.1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer MM, Bilstein A, Leptin M. A clonal genetic screen for mutants causing defects in larval tracheal morphogenesis in Drosophila. Genetics. 2007;176(4):2279–2291. doi: 10.1534/genetics.107.074088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottenfeld-Roames J, Ghabrial AS. Whacked and Rab35 polarize dynein-motor-complex-dependent seamless tube growth. Nat. Cell Biol. 2012;14(4):386–393. doi: 10.1038/ncb2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi BP, Ghabrial AS, Krasnow MA. Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development. 2006;133(12):2383–2393. doi: 10.1242/dev.02404. [DOI] [PubMed] [Google Scholar]
- Jones TA, Metzstein MM. A novel function for the PAR complex in subcellular morphogenesis of tracheal terminal cells in Drosophila melanogaster. Genetics. 2011;189(1):153–164. doi: 10.1534/genetics.111.130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais L, Casanova J. In vivo coupling of cell elongation and lumen formation in a single cell. Curr. Biol. 2010;20(4):359–366. doi: 10.1016/j.cub.2009.12.043. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Shiga Y, Tanaka-Matakatsu M, Hayashi S. A nuclear GFP/ß-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Differ. 1996;38:99–106. [Google Scholar]
- Meijering E, Jacob M, Sarria J-CF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry. Part A: The Journal of the International Society for Analytical Cytology. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Centanin L, Dekanty A, Romero N, Irisarri M, Gorr TA, Wappner P. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev. Cell. 2008;14(4):547–558. doi: 10.1016/j.devcel.2008.01.020. [DOI] [PubMed] [Google Scholar]


