Abstract
We review recent examples of the burgeoning literature on three gases that have major impacts in biology and microbiology. NO, CO and H2S are now co-classified as endogenous gasotransmitters with profound effects on mammalian physiology and, potentially, major implications in therapeutic applications. All are well known to be toxic yet, at tiny concentrations in human and cell biology, play key signalling and regulatory functions. All may also be endogenously generated in microbes. NO and H2S share the property of being biochemically detoxified, yet are beneficial in resisting the bactericidal properties of antibiotics. The mechanism underlying this protection is currently under debate. CO, in contrast, is not readily removed; mounting evidence shows that CO, and especially organic donor compounds that release the gas in biological environments, are themselves effective, novel antimicrobial agents.
Introduction and chronology
Gases play critical roles in life on earth. In microbiology, the best-known gases are nutrients or metabolites: dioxygen, carbon dioxide, dinitrogen, dihydrogen, methane and others [1]. This article highlights the extraordinary advances that have been made in the biology of three other gases – nitric oxide (nitrogen monoxide, NO), carbon monoxide (CO) and hydrogen sulfide (H2S).
Great strides forward in understanding how and why these gases are important in mammalian physiology have kick-started a new area of “gasotransmitters”, i.e. small gaseous molecules that play key roles in biology, illustrated by a large number of reviews published in the last few years (see below). All these gases penetrate membranes, are poisons in excess, are endogenously generated and have important biological targets, especially metalloproteins [2-4]. The discovery in the 1980s that NO produced by mammalian NO synthases is important in the cardiovascular, immune and nervous systems [5-8] was the first impetus to this new area of biology [9]. A decade later, CO emerged as a neurotransmitter and regulator of the cardiovascular and immune systems [10-13]. Most recently, H2S has emerged as a third gasotransmitter, with key roles in the nervous and cardiovascular systems and regulation of cellular and whole body metabolism (reviewed in [14,15]). The literature on the biology of these three gases is now so vast that these and other reviews must act as a surrogate for the relevant papers: [16-20]. In the microbial world too, all three gases have important long-recognised roles: NO is an intermediate in denitrification [21] and is detoxified by pathogens [22,23], CO is an unusual carbon and energy source [24], and H2S is well known to all microbiologists as a product of anoxic sulfate respiration [25]. Each gas is suggested to have valuable therapeutic applications [26,27].
Here, we present recent advances in the field that cement the view that each of these gases is not only toxic but also plays essential roles in metabolism. Toxicity has been most clearly harnessed by the innate immune response in the use of NO as a chemical weapon against pathogens, but may also find future applications via the therapeutic delivery of CO to pathogens. We look at the surprising evidence that NO and H2S protect bacteria from the lethal effects of antibiotics. As a corollary, we report on two new papers that challenge the premise that these gases act as antioxidants.
Lessons from mammalian physiology
The era of gas biology started with the recognition that NO was the elusive “endothelium-derived relaxing factor” (EDRF), a messenger (or gasotransmitter in current nomenclature) produced by endothelial cells that leads to vascular muscle relaxation. In 1998, this work led to the award of the Nobel Prize for Physiology or Medicine to Murad [28], Furchgott [8] and Ignarro [7]. A fourth major proponent of this idea, Salvador Moncada, was not so honoured. The story of these discoveries and the ensuing escalation of research on NO are now well known. NO is generated in man by a family of NO synthases (NOS, i.e. inducible NO synthase, endothelial NO synthase etc.) from arginine and has many critical roles in the cardiovascular, immune and central nervous systems. NO is the most chemically reactive of the gasotransmitters considered here and is a toxic radical: it is employed in the immune response of macrophages, constituting a key component of the arsenal of damaging species directed at engulfed pathogens [23]. Although Sjostrand provided experimental evidence for the endogenous production of CO in man in 1950, it was only in 1991 that Marks and others [10] hypothesised that CO might be another gasotransmitter, produced by haem oxygenases. Indeed, many of its physiological effects are similar to those of NO: roles in neurotransmission and platelet aggregation and anti-apoptotic activity are well documented. Nevertheless, CO is more notorious as the “silent killer”, while H2S is more toxic to humans than HCN [3]. Less is known about H2S, but it is synthesised endogenously and opens vascular smooth muscle channels [29].
All these compounds are gases at atmospheric pressure, all are more (H2S) or less soluble in water and, being small and uncharged, move freely through membranes. No dedicated transporters are expected for the gases, but the nitrosating agent S-nitrosoglutathione (which is widely used experimentally as a source of “reactive nitrogen species”) is modified on transport through the periplasm and cytoplasmic membrane (Fig. 1). The jury is out on whether CO-releasing molecules must be transported inwards or can be expelled from cells. The sulfide anion that accumulates in the bacterial cytoplasm can be moved out by a specific transporter. Some of the salient physicochemical features of these gases and a comparison with oxygen are summarised in Table 1 and Fig. 1 shows their origins and effects.
Figure 1. Simplified schematic diagram of the origins of NO, CO and H2S in bacteria and their major physiological effects.
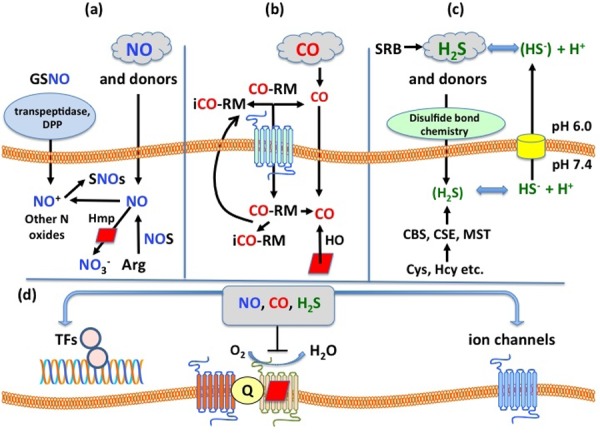
(a) NO accesses the cell interior via free passage through the membrane. The commonly used nitrosating agent, S-nitrosoglutathione (GSNO), a tripeptide, is hydrolysed in the periplasm and the nitrosated dipeptide transported inwards via a dipeptide permease (DPP) [32]. Intracellular NO is also generated by bacterial NO synthases (bNOS) [59] from arginine or anaerobically by nitrite reduction (not shown). NO or GSNO exhibit complex biological chemistry leading to the formation of various N oxides, especially S-nitrosothiols (SNOs) [22,32]. The best understood route for NO detoxification is a dioxygenase or denitrosylase reaction with oxygen catalysed by the flavohaemoglobin Hmp [22].
(b) CO accesses the cell interior via free passage through the membrane. CO-releasing molecules (CO-RMs) may or may not require transport systems. Release of CO from CO-RMs yields an “inactive” form (iCO-RM) either inside or outside the cell, the biology of which needs careful consideration [81] The fate of CO-RM or iCO-RM is unknown but efflux systems might operate. CO is also generated endogenously in certain bacteria by haem oxygenases (HO) from the breakdown of haem (red diamonds).
(c) H2S accesses the cell interior via free passage through the membrane but the hydrosulfide anion, which is prominent intracellularly by virtue of the pH gradient, may be exported by a specific transporter (yellow) [91]. Three endogenous mechanisms for H2S generation from Hcy and Cys have been identified [99].
(d) Global consequences of the three gases include activation of gas-specific transcription factors (TFs), inhibition of respiratory oxygen reduction by binding to the haem(s) of terminal oxidases, and modulation of bacterial ion transport [77]. Q, quinones involved in respiration.
Table 1. Physicochemical properties of oxygen and three gasotransmitters.
| Oxygen, O2 | Carbon monoxide, CO | Nitric oxide, NO | Hydrogen sulfide, H2S* | |
|---|---|---|---|---|
| Molar mass, g/mol | 32 | 28 | 30 | 34 |
| Solubility in water, 20 oC | 0.0043 | 0.0028 | 0.0062 | 0.40 |
| Molecular size (pm) | 121 | 113 | 115 | 134 |
| Importance of redox metabolism in biology | +++ | + | ++ (radical) | ++ |
| Haem ligand? | yes | yes | yes | yes |
| Metal binding | +++ | +++ | +++ | +++ |
| Major mechanisms of endogenous generation | Catalase, photosynthesis | Haem oxygenases | NO synthases, nitrate/nitrite respiration, from SNOs (minor) | CBS, CSE, MST, sulfate reduction |
| Important roles in microbes | Major biological oxidant and co-substrate | Carbon and energy source | Intermediate in denitrification, antibiotic resistance | Metabolic intermediate, antibiotic resistance |
* pKa is 6.8, so HS− is dominant species in biology
Nitric oxide – the old hand
Nitric oxide and reactive nitrogen species now occupy central positions in contemporary medicine, physiology, biochemistry and microbiology. All these species are generated in biological systems from initial formation of NO (from nitrite, NO synthases, or other sources). The major targets of NO and reactive nitrogen species are metal centres (Table 1) and thiols, so that numerous critical biomolecules are at risk. Not surprisingly, microbes have evolved mechanisms for resisting or reversing such damage and many of these are critical for pathogenicity; such measures include the activities of hemoglobins that enzymically detoxify NO (to nitrate) [30], respiratory and NADH-linked NO reductases [31] and repair mechanisms (for example, those that reverse S-nitrosothiol formation) [32]. Microbial resistance to these stresses is generally inducible via the action of NO-sensing transcriptional regulators (such as Fnr, NorR, NsrR, NssR) [33,34]. However, a phenomenal growth in the literature on NO has often not gone hand in hand with an understanding of the intricacies and complexities of nitrogen chemistry. The distinctiveness of NO and its chemical cousins, nitrosonium (NO+), nitroxyl (NO−, HNO), peroxynitrite (ONOO−), nitrite (NO2 −) and nitrogen dioxide (NO2) has been critically reviewed recently [22]. Unfortunately, some authors write “NO” when it is actually meant “in a generic sense”, thus confusing the biological literature. It should not be necessary to write “NO radical” to eliminate the possibility that one is actually meaning NO+ or some other congener. There is only one NO.
There is a growing literature on signalling functions for NO in microbes. An interesting example is the influence of NO on biofilm formation. Recently it has been reported that NO derived from periplasmic reductase positively influences biofilm formation in Azospirillum brasilensae [35]. Conversely, NO donors aid in dispersing biofilms of Pseudomonas aeruginosa [36]. Recently, an NO sensor has been identified in Vibrio fischeri: H-NOX appears to be part of a signal transduction pathway to detect host-produced NO and influence iron utilisation genes [37,38].
NO is most efficiently detoxified in bacteria by the activity of haemoglobins that catalyse the conversion of NO and dioxygen to nitrate anion (Fig. 1). The best characterised such proteins are flavohaemoglobins but, even now, 15 years after this reaction was first proposed for the Escherichia coli protein [39], and 17 years since the finding that flavohaemoglobin gene expression was dramatically up-regulated by NO [40], there is still controversy about the reaction mechanism. Two recent reviews [41,42] assume the reaction to proceed via an NO dioxygenation mechanism in which oxygen binds to the ferrous haem, followed by NO. However, since 2001, Stamler's group have favoured an alternative denitrosylase mechanism in which the protein binds NO during turnover in situ, and rapidly catalyses nitrate formation across a wide range of oxygen concentrations [43]. These alternative views are debated in letters [44,45] that promptly followed the Forrester review [42]; however, it is not disputed that flavohaemoglobins do catalyse the rapid detoxification of NO to nitrate and play a critical role in the response of pathogenic microorganisms to the innate immune system. An interesting computational approach to the complexity of NO biochemical networks has just been published [46] and concluded that Hmp is the dominant NO-consuming pathway at oxygen concentrations down to about 35 μM but, interestingly, loses effectiveness as NO delivery rates increase.
Even though (almost) all haemoglobins (flavohaemoglobins, single-domain and truncated globins) seem to possess NO-consuming activity, this physiological function in vivo has been demonstrated for only a few globins. Indeed, of the very large number of globin sequences identified in prokaryotic genomes (c. 1161 globin-containing genomes), only a handful has been subjected to rigorous genetic and physiological characterisation and the vast majority of the evidence pertains to flavohaemoglobins [47]. The most notable exception is the single-domain globin of Campylobacter jejuni, a myoglobin-like protein that is expressed in bacteria on challenge with NO, and mutation of which elicits an NO-sensitive phenotype [47,48]. The true function of most microbial globins is unknown [47].
Although many globin crystal structures from all classes are available (at least 15 bacterial and archaeal globins), understanding globin function(s) has not been greatly facilitated by these structures or by the numerous elegant kinetic experiments on ligand binding to the purified proteins. For example, the structure of the second (truncated) haemoglobin from C. jejuni has been solved [49-51] and its oxygen affinity is remarkably high [52], but we still do not understand its role in vivo.
The expression of globins in a convenient heterologous host is often exploited to test functional properties when genetic manipulation of the native host is difficult. An especially useful model is the flavohaemoglobin-lacking E. coli (hmp mutant), which is hypersensitive to NO. When complementation of the phenotype is observed, it is generally interpreted to demonstrate detoxification properties for the heterologously expressed globin (for a recent example, see [53]). However, the discovery of a function in a heterologous organism does not necessarily tell us that the protein will have the same function in the native host, especially when the bacteria are phylogenetically distant. For example, the requirement for interaction with a cognate reductase by single-domain or truncated globins (as in [53]) and/or for oxygen for the NO → NO3 − reaction may be accomplished in E. coli but not in the native host (reviewed in [47]).
Although generally harmful, NO may have beneficial effects, as in the case of the squid-Vibrio light-organ symbiosis, where NO serves as a signal, antioxidant and specificity determinant [54-56]. A recent high-profile case concerns the NO generated endogenously by certain bacteria from arginine by the activity of bacterial NO synthase (Figs. 1,2). Two papers reported that this NO protects bacteria against oxidative stress [57,58]. Later, many antibacterial agents were shown to suppress growth of a nos mutant lacking bacterial NO synthase [59]. The authors suggest that NO-mediated antibiotic resistance is explained in two ways: first, by chemical modification of the toxic compounds (for example, the nitrosation by NO+ of aromatic amino groups on acriflavine) and, second, by NO-protecting bacterial cells against the oxidative stress that antibiotics like quinolones generate (Fig. 2). However, two very recent papers in Science question the premise that killing by bactericidal antibiotics depends on reactive oxygen species (see below).
Figure 2. Proposed interactions between NO, CO and H2S with antibiotics and oxidative stress in bacteria.
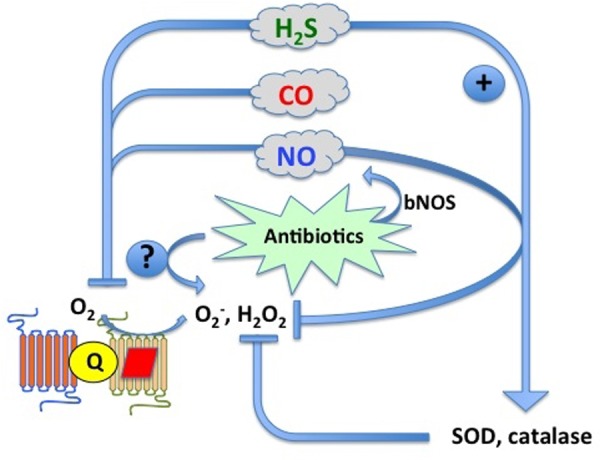
All three gases are potent inhibitors of terminal oxidase activity in bacteria, leading to the accumulation of ROS (reactive oxygen species), initially superoxide anion (O2 −) and hydrogen peroxide (H2O2). ROS are detoxified by superoxide dismutases (SOD) and catalases. ROS may also be generated by chemically unrelated antibiotics [102], although this is now disputed [104,105]. Antibiotics may also increase NO formation from bNOS [59] while H2S induces activity of SOD and catalase.
Carbon monoxide – the new NO?
CO is among the most abundant air pollutants in developed countries and its dangers are compounded by difficulties in detecting it. Tragic cases of CO fatalities are frequent in the media, including one recently involving many street children in China (http://www.bbc.co.uk/news/world-asia-china-20389004). Nevertheless, the ability of animals, plants and pathogenic microbes to make CO via haem oxygenase enzymes is widespread and the CO generated has potent beneficial biological consequences [60]. CO is now identified as a “gasotransmitter” with wide-ranging effects on vasodilation and inflammation [10-12]. The emerging view is that CO and CO-releasing molecules have biological effects far-removed from the classical view that CO is a toxic gas, useful to haem biochemists, but merely a by-product of haem oxygenase-catalysed haem degradation.
In certain bacteria, CO is an oxidisable substrate and must be sensed. In Rhodospirillum rubrum and Carboxydothermus hydrogenoformus [61], the coo operon encodes a CO dehydrogenase, which allows the anaerobic metabolism of CO. The expression of this operon is regulated by the CO sensor CooA [62], a member of the CRP/FNR family. In response to a reducing cellular environment, CO binds to ferrous haem of CooA, causing a conformational change that permits DNA binding and therefore transcription of the coo operon [63,64]. Recently, an ingenious use of CooA has been described in which the natural sensitivity of CooA to CO has been exploited in a sensitive fluorescent protein reporter for CO imaging in living cells [65]. However, homologues of this canonical CO oxidation system, including CooA, CO dehydrogenase, and a CO-dependent Coo hydrogenase, are also present in the sulfate-reducing bacterium Desulfovibrio vulgaris, although it grows only poorly on CO. Recent global transcriptional analyses suggests that CooA and CO dehydrogenase are used during normal metabolism of this bacterium [66]. Another haem-containing CO sensor, RcoM from Burkholderia xenovorans, which is also able to bind NO, regulates the expression of genes in response to the redox state of the cell [67,68].
CO also has useful antibacterial features. It enhances phagocytosis of E. coli [69] by macrophages and shows promising effects on bacteraemia in sepsis [12,70]. Because of the difficulties of studying CO in the laboratory and using it therapeutically (see, however, [71]), CO-releasing molecules, mostly metal carbonyl compounds, developed decades ago by chemists, have more recently been used in biological studies [72]. These release CO in biological milieu with defined kinetics and stoichiometry, promoted by reaction with other species [73] or enzymic or photochemical activation [74]. CORM-3 (Ru(CO)3Cl(glycinate)) is a water-soluble CO-releasing molecule that has numerous beneficial effects in mammalian systems and is an effective antibacterial agent [75]. CORM-3 decreased bacterial counts in the spleen and increased survival of mice following experimental bacteraemia [76]; injection into mice increased phagocytosis of Enterococcus faecalis and rescued haem oxygenase-deficient mice from sepsis-induced lethality [70]. Thus, therapies involving haem oxygenases, CO or CO-releasing molecules have great potential for treating infections [12], but the microbiological literature on these compounds is very limited and only a handful of the hundreds of CO-releasing molecules made over the past few years have been tested against microbes. At the time of writing, for example, no papers on the effects on microbes of any photo-CORM, which releases CO on demand after photolysis, have appeared. Our rudimentary knowledge of the modes of actions of CO and CO-releasing molecules, and the difficulty of drawing conclusions from so few CO-releasing molecules limits progress.
In many cases, the antimicrobial effects of CO-releasing molecules are clearly caused by the released CO, as control molecules that do not release CO do not induce these effects [75,77]. However, there is mounting evidence that CO-releasing molecules are much more powerful antibacterial agents than CO [75,78] and elicit biological effects beyond those of CO gas. For example, CO-releasing molecules exhibit antimicrobial effects even under highly aerobic conditions where CO gas is ineffective (because the gas is a competitive inhibitor with oxygen) [75]. Furthermore, a recent study of the effects of CORM-3 on the respiration of a range of bacteria found that this CO-RM elicited in E. coli transient stimulation of respiration, prior to respiratory inhibition; these effects were not mimicked by CO gas [77]. Uncoupling of respiration by promotion of proton flux, as proposed in mitochondria [79,80], was ruled out. In E. coli, the working hypothesis is that CO-releasing molecules cause the opening of other ion channels (K+, Na+), which transiently collapses the protonmotive force and disturbs ion transport. The reason that CO-releasing molecules potentiate the effects of CO may be that they deliver CO directly into bacterial cells, thereby allowing a high concentration of this gas to accumulate at the sites of action [75] – the “Trojan Horse” mechanism [77].
As described above, CO is a classical inhibitor of respiration in both eukaryotes and prokaryotes, and CO from CO-releasing molecules shares this property. CO is delivered from CO-releasing molecules directly to the haems of cytochromes bd and bo' of the aerobic respiratory chain of E. coli and transcriptomic profiling has shown that exposure of E. coli to CORM-3 results in down-regulation of the cyo genes encoding cytochrome bo' and slight up-regulation of the cydAB genes encoding cytochrome bd-I [75], suggesting a role for cytochrome bd-I in resisting CORM-3. Subsequent time-resolved analysis revealed that the transcription of appBC, which encodes cytochrome bd-II is not affected in response to CORM-3 [81]. In support, a strain expressing cytochrome bd-I as the sole oxidase is less susceptible to both respiratory and growth inhibition by this compound [82], consistent with the established role of this oxidase in resisting a variety of environmental stresses including NO [83]. In the case of NO, it has been proposed that cytochrome bd resists inhibition because of an exceptionally high off rate (koff 0.163 s−1 for cytochrome bd, compared to 0.03 s−1 for cytochrome bo' [83]), although it is difficult to draw such conclusions for CO due to discrepancies in the literature as to the off rate of this gas from cytochrome bd-I (see references in [82]).
CO is implicated in many recent studies as an antioxidant, for example in ischemia-reperfusion injury in vitro [84]. On the other hand, because CO is a potent inhibitor of respiration, it may increase generation of reactive oxygen species in mitochondria [85,86] or bacteria [87] (Fig. 2). One recent study [88] suggests that CORM-2 promotes the formation of reactive radical species, even in the absence of cells, and that such species are largely responsible for CORM-mediated bacterial killing. This assertion appears to be supported by the oft reported, but poorly understood, observation that N-acetylcysteine protects against CORM-2 and CORM-3 toxicity (e.g. [76,89]). Other data, however, contradict this view [81]; some antioxidants do not share this property and N-acetylcysteine is effective at inhibiting CO-RM uptake by E. coli, presenting us with a new testable hypothesis for the protective effects of N-acetylcysteine [82]. The field is in a state of flux and further insights are anticipated but, unlike the case for NO (above) and H2S (below), there appears to be no clear evidence yet that CO or CO-releasing molecules either protect against antibiotics or potentiate their activities.
Hydrogen sulfide – an (un)welcome gate crasher?
Hydrogen sulfide is a toxic, freely permeable, malodorous gas. It is very soluble in water (Table 1) and the equilibrium between its three states (H2S, HS− and S2−) is pH-dependent. Here, we write “H2S” or “sulfide” (HS−) while recognising the importance of pH in controlling speciation [90]. At 37 oC the pKa is 6.76; since the pH of the human large intestine is in the range 5.5-7.0, H2S and HS− are the predominant sulfide species, and the hydrosulfide anion (HS−) will be the dominant species inside gut bacteria [91]. An exporter for this anion has been identified in Clostridium difficile [91]. Recently, H2S has been recognized as an important gasotransmitter in higher organisms (reviewed by [15,92,93]). This gas is abundant in the anaerobic lumen of the gut (1-2 mM total sulfide) with ~60 μM being present in a free, unbound form where it is generated by the reduction of sulfate and the decomposition of sulfur-containing organic compounds, such as cysteine, by resident microflora. Sulfate-reducing bacteria generate H2S by the reduction of various sulfur compounds, e.g. SO42− or S0 under anaerobic conditions, often using H2 or simple organic molecules as an electron donor. H2S and SO42− are also generated by bacterial disproportionation of thiosulfate. Furthermore, human faecal slurries ferment cysteine, (via the action of cysteine desulfurases, which yield H2S, ammonia and pyruvate) and other organic sulfur compounds [94]. Dietary polysulfides from “healthy” sources (garlic, onions etc.) further increase formation of H2S. Thus, H2S is abundant in the anaerobic environment of the mammalian lower intestine and consequently the gut epithelium is exposed to this potent signalling molecule and toxin.
Exogenous H2S modulates the behaviour of intestinal epithelium cells [95]; however, the effects of exposure to exogenous H2S are considered less significant than those elicited by endogenous H2S production by the enzymes cystathionine-β-synthase and cystathionine-γ-lyase [92,96]. Endogenous H2S levels are uncertain because of extraction and binding issues, but tissue homogenates and other material produce 1-10 μM of free H2S in the presence of cysteine and low O2. These significant concentrations control the relaxation of blood vessels, inhibit inflammation, and modulate neuronal activity [92]. The precise molecular mechanisms by which H2S elicits these positive effects are not well established. However, the thiol groups of protein cysteine residues are modified by H2S yielding hydropersulfide moieties (S-sulfhydration), which can result in altered activity. For example, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity is enhanced seven-fold by S-sulfhydration of Cys-150 [97]. Thus, S-sulfhydration is an important post-translational modification in higher organisms, with implications for protein function and transduction of the H2S signal into altered behaviour.
As well as acting as an important signalling molecule, H2S damages cell components and mediates its toxic effects, at least in part, by inhibiting respiration, reacting with other metals and by reducing protein disulfide bonds. Terminal respiratory oxidases have been known to be inhibited by H2S for many decades [2]. Although the significance of H2S signalling in higher organisms is now established, essentially nothing is known about the effect of H2S on gut bacteria that are exposed to fluctuating concentrations of exogenous H2S in their preferred niche and/or that generate endogenous H2S [98]. The major source of endogenous H2S production by E. coli, identified by Shatalin et al. [99], was the combined action of the L-cysteine transaminase (AspC) and the 3-mercaptopyruvate sulfurtransferase SseA (MST). However, there are several other potential H2S-generating enzymes in E. coli, including CysJI, MetC, TnaA and H2S-consuming enzymes, e.g. CysEK and CysM. In other bacteria like Bacillus anthracis, Staphylococcus aureus and P. aeruginosa, the main mechanisms are via cystathionine-β-synthase/cystathionine-γ-lyase [99]. We know much less about potential resistance mechanisms for H2S in bacteria, but plant pathogenic bacteria use the bigR operon for H2S detoxification via the action of a sulfur dioxygenase (Blh) and a sulfite exporter [100].
Emerging information on the biological roles of H2S is likely to have significant impact in the foreseeable future, following the demonstration [99] that H2S protects bacteria from a diversity of antibiotics by mitigating the effects of oxidative stress. Note, however, that this contrasts with a previous report that H2S potentiates hydrogen peroxide-induced killing of E. coli [101]. The suggestion that H2S can act as “a universal defence against antibiotics in bacteria” [99] is predicated on the assumption, first, that H2S is an antioxidant and, second, that numerous antibiotics exert their bactericidal activities by mitigating oxidative stress. Let us now examine the second of these.
Gas interactions with microbes, antibiotics and other antimicrobial agents - do we really know how antibiotics work?
The papers cited above from Shatalin [58,99], Gusarov [57,59] and others suggest that, separately, NO and H2S production may constitute a natural bacterial defence method against antimicrobial compounds. The specific proposal is that these gases (but not CO, yet) partially neutralise the effectiveness of antibiotics by suppressing the oxidative stress that antibiotics induce. This model of antibiotic action [102,103] appears to be a unified explanation of the effectiveness of numerous bactericidal compounds that all induce the formation of reactive oxygen species by inhibiting the electron transport chain. However, this model fails to explain many aspects of antibiotic action, such as the effectiveness of bactericidal antibiotics against Streptococcus pneumoniae, which lacks any electron transport chain. Recently, Keren et al. [104] have produced persuasive evidence that a cell's probability of survival in the presence of an antibiotic does not correlate with the level of reactive oxygen species. Furthermore, thiourea, a reagent that quenches reactive oxygen species, is equally effective at protecting cells from antibiotics under anaerobic conditions, when reactive oxygen species cannot be formed. Likewise, Liu and Imlay [105] show that antibiotic treatment does not promote hydrogen peroxide generation or elevate intracellular free iron, a precursor to reactive oxygen species-induced damage via Fenton chemistry.
Perhaps curiously, neither of the most recent Science papers [104,105] mention the implications of their findings for the protection by NO and H2S against bactericidal antibiotics. We are therefore left with puzzles: what is the mechanism of gas-mediated antibiotic resistance, does CO share this remarkable property and can the clinical effectiveness of antibiotics be modulated by endogenous microbial gas production? Only time will tell.
Questions, controversies, and conflicts
There is no doubt that NO, CO and H2S play key roles in bacterial life, survival strategies and metabolism, undreamt of a few decades ago. Despite the rapid progress on all three gases in the mammalian field, only NO has achieved a degree of prominence in microbiology as a gasotransmitter or signalling molecule. CO and H2S deserve more attention and the possible link between the latter and antibiotic tolerance should kick-start such activity. In the case of CO, all the evidence points at present to its potential as an antimicrobial agent, rather than a protective molecule for bacteria. In fact, we are unaware that antibiotic tolerance elicited by CO has ever been reported. CO and other products of haem degradation are considered as antioxidants in mammalian systems, but it remains to be seen whether CO can exert such a role in bacteria, or even whether the concept that antibiotics kill bacteria by stimulating reactive oxygen species formation withstands further careful scrutiny.
Since NO and H2S appear to act as bacterial defences against antibiotics, future therapeutic strategies might involve targeting the gas-generating enzymes – bacterial NO synthase [58,59,106] and the three H2S-synthesising enzymes identified thus far [99]. In the case of CO, a more productive strategy may be to develop clinically effective and permissible CO-releasing molecules. The most commonly used, CORM-2 and CORM-3, are both ruthenium metal carbonyls and there is likely to be resistance to using a “non-biological” delivery molecule; on the other hand, witness the widespread use of cis-diamminedichloroplatinum(II) (Cisplatin), a chemotherapy drug that is given to treat testicular, bladder, and other cancers.
Despite an accelerating interest from microbiologists in these three remarkable gases, we urgently need more fundamental research into their synthesis, functions, and potential roles in therapy against pathogenic microbes.
Acknowledgments
Work in the laboratory of the authors was supported by the Biotechnology and Biological Sciences Research Council (BBSRC, UK) and Consejo Nacional de Ciencia y Tecnologia (Mexico) through grant number 99171 and Consejo Estatal de Ciencia, Tecnología e Innovación de Michoacán through grant number 007 (Mariana Tinajero-Trejo).
Abbreviations
- CORM-2
tricarbonyldichloroRu(II) dimer
- CORM-3
Ru(CO)3Cl(glycinate)
- DPP
dipeptide permease
- EDRF
endothelial-derived relaxing factor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GSNO
S-nitrosoglutathione
- Hcy
homocysteine
- iCO-RM
“inactive” form of CO-RM after CO release
- MST
3-meracptopyruvate sulfurtransferase
- Q
quinones involved in respiration
- SNOs
S-nitrosothiols
- SOD
superoxide dismutases
- TFs
transcription factors
Disclosure
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/5/28
Contributor Information
Mariana Tinajero-Trejo, Email: mbp09mt@sheffield.ac.uk.
Helen E. Jesse, Email: mbp09hej@sheffield.ac.uk.
Robert K. Poole, Email: r.poole@sheffield.ac.uk.
References
- 1.Poole RK, Dow CS, editors. Microbial gas metabolism. Mechanistic, metabolic and biotechnological aspects. London: Academic Press; 1985. p. 304. [Google Scholar]
- 2.Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–39. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Moore PK. An overview of the biological significance of endogenous gases: new roles for old molecules. Biochem Soc Trans. 2007;35:1138–41. doi: 10.1042/BST0351138. [DOI] [PubMed] [Google Scholar]
- 4.Fukuto JM, Carrington SJ, Tantillo DJ, Harrison JG, Ignarro LJ, Freeman BA, Chen A, Wink DA. Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem Res Toxicol. 2012;25:769–93. doi: 10.1021/tx2005234. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030788
- 5.Ignarro LJ. NO more heart disease. New York: St. Martin's Griffin; 2005. p. 248. [Google Scholar]
- 6.Murad F. Discovery of some of the biological effects of nitric oxide and its role in cell signaling. Biosci Rep. 1999;19:133–54. doi: 10.1023/A:1020265417394. [DOI] [PubMed] [Google Scholar]
- 7.Ignarro LJ. Nitric oxide: A unique endogenous signaling molecule in vascular biology. Biosci Rep. 1999;2:51–71. doi: 10.1023/A:1020150124721. [DOI] [PubMed] [Google Scholar]
- 8.Furchgott RF. Endothelium-derived relaxing factor: Discovery, early studies, and identifcation as nitric oxide. Biosci Rep. 1999;4:235–51. doi: 10.1023/A:1020537506008. [DOI] [PubMed] [Google Scholar]
- 9.Wang R. The evolution of gasotransmitter biology and medicine. From atmospheric toxic gases to endogenous gaseous signaling molecules. In: Wang R, editor. Signal Transduction and the Gasotransmitters. NO, CO and H2S in Biology and Medicine. Totowa, New Jersey: Humana Press; 2004. pp. 3–31. [DOI] [Google Scholar]
- 10.Marks GS, Brien JF, Nakatsu K, McLaughlin BE. Does Carbon-Monoxide Have a Physiological-Function. Trends Pharmacol Sci. 1991;12:185–88. doi: 10.1016/0165-6147(91)90544-3. [DOI] [PubMed] [Google Scholar]
- 11.Mann BE. Carbon Monoxide: An Essential Signalling Molecule. Med Organomet Chem. 2010;32:247–85. doi: 10.1007/978-3-642-13185-1_10. [DOI] [Google Scholar]
- 12.Motterlini R, Otterbein LE. The therapeutic potential of carbon monoxide. Nat Rev Drug Discov. 2010;9:728–43. doi: 10.1038/nrd3228. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030789
- 13.Davidge KS, Motterlini R, Mann BE, Wilson JL, Poole RK. Carbon monoxide in biology and microbiology: surprising roles for the “Detroit perfume”. Adv Microb Physiol. 2009;56:85–167. doi: 10.1016/S0065-2911(09)05603-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang R. The gasotransmitter role of hydrogen sulfide. Antiox Redox Signal. 2003;5:493–501. doi: 10.1089/152308603768295249. [DOI] [PubMed] [Google Scholar]
- 15.Kashfi K, Olson KR. Biology and therapeutic potential of hydrogen sulfide and hydrogen sulfide-releasing chimeras. Biochem Pharmacol. 2013;85:689–703. doi: 10.1016/j.bcp.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kajimura M, Fukuda R, Bateman RM, Yamamoto T, Suematsu M. Interactions of Multiple Gas-Transducing Systems: Hallmarks and Uncertainties of CO, NO, and H(2)S Gas Biology. Antiox Redox Signal. 2010;13:157–92. doi: 10.1089/ars.2009.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajimura M, Nakanishi T, Takenouchi T, Morikawa T, Hishiki T, Yukutake Y, Suematsu M. Gas biology: Tiny molecules controlling metabolic systems. Respir Physiol Neurobiol. 2012;184:139–48. doi: 10.1016/j.resp.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Hsu A, Moore PK. Actions and interactions of nitric oxide, carbon monoxide and hydrogen sulphide in the cardiovascular system and in inflammation--a tale of three gases! Pharmacol Ther. 2009;123:386–400. doi: 10.1016/j.pharmthera.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Prabhakar NR, Semenza GL. Gaseous messengers in oxygen sensing. J Mol Med. 2012;90:265–72. doi: 10.1007/s00109-012-0876-1. [DOI] [PubMed] [Google Scholar]
- 20.Semenza GL, Prabhakar NR. Gas biology: small molecular medicine. J Mol Med. 2012;90:213–15. doi: 10.1007/s00109-012-0877-0. [DOI] [PubMed] [Google Scholar]
- 21.Lundberg JO, Weitzberg E, Cole JA, Benjamin N. Nitrate, bacteria and human health. Nat Rev Microbiol. 2004;2:593–602. doi: 10.1038/nrmicro929. [DOI] [PubMed] [Google Scholar]
- 22.Bowman LAH, McLean S, Poole RK, Fukuto J. The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins. Adv Microb Physiol. 2011;59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 23.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2:820–32. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 24.King GM, Weber CF. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol. 2007;5:107–18. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- 25.Slonczewski JL, Foster JW. Microbiology, An Evolving Science. New York, London: W. W. Norton; 2009. p. 1096. [Google Scholar]
- 26.Bannenberg GL, Vieira HL. Therapeutic applications of the gaseous mediators carbon monoxide and hydrogen sulfide. Expert Opin Ther Pat. 2009;19:663–82. doi: 10.1517/13543770902858824. [DOI] [PubMed] [Google Scholar]
- 27.Szabo C. Gaseotransmitters: New Frontiers for Translational Science. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murad F. Discovery of some of the biological effects of nitric oxide and its role in cell signaling. Biosci Rep. 1999;3:133–54. doi: 10.1023/A:1020265417394. [DOI] [PubMed] [Google Scholar]
- 29.Liu YH, Yan CD, Bian JS. Hydrogen Sulfide: A Novel Signaling Molecule in the Vascular System. J Cardiovasc Pharmacol. 2011;58:560–69. doi: 10.1097/FJC.0b013e31820eb7a1. [DOI] [PubMed] [Google Scholar]
- 30.Poole RK, Hughes MN. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–83. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- 31.Fang H, Caranto JD, Mendoza R, Taylor AB, Hart PJ, Kurtz DM., Jr Histidine ligand variants of a flavo-diiron protein: effects on structure and activities. J Biol Inorg Chem. 2012;17:1231–9. doi: 10.1007/s00775-012-0938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laver JR, McLean S, Bowman LAH, Harrison LJ, Read RC, Poole RK. Nitrosothiols in Bacterial Pathogens and Pathogenesis. Antiox Redox Signal. 2013;18:309–22. doi: 10.1089/ars.2012.4767. [DOI] [PubMed] [Google Scholar]
- 33.Spiro S. Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors. Phil Trans Roy Soc B - Biol Sci. 2012;367:1213–25. doi: 10.1098/rstb.2011.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiro S, D'Autreaux B. Non-Heme Iron Sensors of Reactive Oxygen and Nitrogen Species. Antiox Redox Signal. 2012;17:1264–76. doi: 10.1089/ars.2012.4533. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030790
- 35.Arruebarrena Di Palma A, Pereyra CM, Moreno Ramirez L, Xiqui Vazquez ML, Baca BE, Pereyra MA, Lamattina L, Creus CM. Denitrification-derived nitric oxide modulates biofilm formation in Azospirillum brasilense. FEMS Microbiol Lett. 2013;338:77–85. doi: 10.1111/1574-6968.12030. [DOI] [PubMed] [Google Scholar]
- 36.Barnes RJ, Bandi RR, Wong WS, Barraud N, McDougald D, Fane A, Kjelleberg S, Rice SA. Optimal dosing regimen of nitric oxide donor compounds for the reduction of Pseudomonas aeruginosa biofilm and isolates from wastewater membranes. Biofouling. 2013;29:203–12. doi: 10.1080/08927014.2012.760069. [DOI] [PubMed] [Google Scholar]
- 37.Wang YL, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A. 107:8375–80. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030774
- 38.Pietra F. Understanding How H-NOX (Heme Nitric Oxide/Oxygen) Domain Works Needs First Clarifying How Diatomic Gases Are Relocated Inside This Sensing Protein. A Molecular-Mechanics Approach. Chem Biodivers. 2012;9:606–14. doi: 10.1002/cbdv.201100382. [DOI] [PubMed] [Google Scholar]
- 39.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:10378–83. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030775
- 40.Poole RK, Anjum MF, Membrillo-Hernández J, Kim SO, Hughes MN, Stewart V. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol. 1996;178:5487–92. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner PR. Hemoglobin: a nitric-oxide dioxygenase. Scientifica. 2012;34 doi: 10.6064/2012/683729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forrester MT, Foster MW. Protection from nitrosative stress: A central role for microbial flavohemoglobin. Free Radic Biol Med. 2012;52:1620–33. doi: 10.1016/j.freeradbiomed.2012.01.028. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030791
- 43.Hausladen A, Gow A, Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci U S A. 2001;98:10108–12. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030776
- 44.Hausladen A, Stamler JS. Is the flavohemoglobin a nitric oxide dioxygenase? Free Rad Biol Med. 2012;53:1209–10. doi: 10.1016/j.freeradbiomed.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 45.Forrester MT, Foster MW. Response to “Is flavohemoglobin a nitric oxide dioxygenase?”. Free Rad Biol Med. 2012;53:1211–12. doi: 10.1016/j.freeradbiomed.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 46.Robinson JL, Brynildsen MP. A Kinetic Platform to Determine the Fate of Nitric Oxide in Escherichia coli. PLoS Comput Biol. 2013;9:e1003049. doi: 10.1371/journal.pcbi.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718009419
- 47.Vinogradov SN, Tinajero-Trejo M, Poole RK, Hoogewijs D. Bacterial and archaeal globins - A revised perspective. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 48.Elvers KT, Wu G, Gilberthorpe NJ, Poole RK, Park SF. Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J Bacteriol. 2004;186:5332–41. doi: 10.1128/JB.186.16.5332-5341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nardini M, Pesce A, Labarre M, Richard C, Bolli A, Ascenzi P, Guertin M, Bolognesi M. Structural determinants in the group III truncated hemoglobin from Campylobacter jejuni. J Biol Chem. 2006;281:37803–12. doi: 10.1074/jbc.M607254200. [DOI] [PubMed] [Google Scholar]
- 50.Nardini M, Pesce A, Milani M, Bolognesi M. Protein fold and structure in the truncated (2/2) globin family. Gene. 2007;398:2–11. doi: 10.1016/j.gene.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 51.Arroyo Manez P, Lu C, Boechi L, Marti MA, Shepherd M, Wilson JL, Poole RK, Luque FJ, Yeh SR, Estrin DA. Role of the distal hydrogen-bonding network in regulating oxygen affinity in the truncated hemoglobin III from Campylobacter jejuni. Biochemistry. 2011;50:3946–56. doi: 10.1021/bi101137n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wainwright LM, Wang YH, Park SF, Yeh SR, Poole RK. Purification and spectroscopic characterization of ctb, a group III truncated hemoglobin implicated in oxygen metabolism in the food-borne pathogen Campylobacter jejuni. Biochemistry. 2006;45:6003–11. doi: 10.1021/bi052247k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coppola D, Giordano D, Tinajero-Trejo M, di Prisco G, Ascenzi P, Poole RK, Verde C. Antarctic bacterial haemoglobin and its role in the protection against nitrogen reactive species. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Dunn AK, Karr EA, Wang YL, Batton AR, Ruby EG, Stabb EV. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol Microbiol. 2010;77:44–55. doi: 10.1111/j.1365-2958.2010.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030777
- 55.Wang YL, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol. 2010;78:903–15. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030778
- 56.Wang YL, Ruby EG. The roles of NO in microbial symbioses. Cell Microbiol. 2011;13:518–26. doi: 10.1111/j.1462-5822.2011.01576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030792
- 57.Gusarov I, Nudler E. NO-mediated cytoprotection: Instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci U S A. 2005;102:13855–60. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci U S A. 2008;105:1009–13. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/1099101
- 59.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–4. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/1166017
- 60.Boczkowski J, Poderoso JJ, Motterlini R. CO-metal interaction: vital signaling from a lethal gas. Trends Biochem Sci. 2006;31:614–21. doi: 10.1016/j.tibs.2006.09.001. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030793
- 61.Clark RW, Lanz ND, Lee AJ, Kerby RL, Roberts GP, Burstyn JN. Unexpected NO-dependent DNA binding by the CooA homolog from Carboxydothermus hydrogenoformans. Proc Natl Acad Sci U S A. 2006;103:891–96. doi: 10.1073/pnas.0505919103. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030779
- 62.Roberts GP, Kerby RL, Youn H, Conrad M. CooA, a paradigm for gas sensing regulatory proteins. J Inorg Biochem. 2005;99:280–92. doi: 10.1016/j.jinorgbio.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 63.Clark RW, Youn H, Lee AJ, Roberts GP, Burstyn JN. DNA binding by an imidazole-sensing CooA variant is dependent on the heme redox state. J Biol Inorg Chem. 2007;12:139–46. doi: 10.1007/s00775-006-0168-8. [DOI] [PubMed] [Google Scholar]
- 64.Ibrahim M, Kuchinskas M, Youn H, Kerby RL, Roberts GP, Poulos TL, Spiro TG. Mechanism of the CO-sensing heme protein CooA: New insights from the truncated heme domain and UVRR spectroscopy. J Inorg Biochem. 2007;101:1776–85. doi: 10.1016/j.jinorgbio.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Karpus J, Zhao BS, Luo Z, Chen PR, He C. A selective fluorescent probe for carbon monoxide imaging in living cells. Angew Chem Int Ed Engl. 2012;51:9652–6. doi: 10.1002/anie.201203684. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/717963533
- 66.Rajeev L, Hillesland KL, Zane GM, Zhou AF, Joachimiak MP, He ZL, Zhou JZ, Arkin AP, Wall JD, Stahl DA. Deletion of the Desulfovibrio vulgaris Carbon Monoxide Sensor Invokes Global Changes in Transcription. J Bacteriol. 2012;194:5783–93. doi: 10.1128/JB.00749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kerby RL, Youn H, Roberts GP. RcoM: A new single-component transcriptional regulator of CO metabolism in bacteria. J Bacteriol. 2008;190:3336–43. doi: 10.1128/JB.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030780
- 68.Marvin KA, Kerby RL, Youn H, Roberts GP, Burstyn JN. The transcription regulator RcoM-2 from Burkholderia xenovorans is a cysteine-ligated hemoprotein that undergoes a redox-mediated ligand switch. Biochemistry. 2008;47:9016–28. doi: 10.1021/bi800486x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otterbein LE, May A, Chin BY. Carbon monoxide increases macrophage bacterial clearance through toll-like receptor (TLR)4 expression. Cell Mol Biol. 2005;51:433–40. [PubMed] [Google Scholar]
- 70.Chung SW, Liu X, Macias AA, Baron RM, Perrella MA. Heme oxygenase-1-derived carbon monoxide enhances the host defense response to microbial sepsis in mice. J Clin Invest. 2008;118:239–47. doi: 10.1172/JCI32730. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030781
- 71.Mitchell LA, Channell MM, Royer CM, Ryter SW, Choi AM, McDonald JD. Evaluation of inhaled carbon monoxide as an anti-inflammatory therapy in a nonhuman primate model of lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2010;299:L891–7. doi: 10.1152/ajplung.00366.2009. [DOI] [PubMed] [Google Scholar]
- 72.Motterlini R, Mann BE, Foresti R. Therapeutic applications of carbon monoxide-releasing molecules. Expert Opin Investig Drugs. 2005;14:1305–18. doi: 10.1517/13543784.14.11.1305. [DOI] [PubMed] [Google Scholar]
- 73.McLean S, Mann BE, Poole RK. Sulfite species enhance carbon monoxide release from CO-releasing molecules: Implications for the deoxymyoglobin assay of activity. Anal Biochem. 2012;427:36–40. doi: 10.1016/j.ab.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 74.Schatzschneider U. PhotoCORMs: Light-triggered release of carbon monoxide from the coordination sphere of transition metal complexes for biological applications. Inorg Chim Acta. 2011;374:19–23. doi: 10.1016/j.ica.2011.02.068. [DOI] [Google Scholar]
- 75.Davidge KS, Sanguinetti G, Yee CH, Cox AG, McLeod CW, Monk CE, Mann BE, Motterlini R, Poole RK. Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. J Biol Chem. 2009;284:4516–24. doi: 10.1074/jbc.M808210200. [DOI] [PubMed] [Google Scholar]
- 76.Desmard M, Davidge KS, Bouvet O, Morin D, Roux D, Foresti R, Ricard JD, Denamur E, Poole RK, Montravers P, Motterlini R, Boczkowski J. A carbon monoxide-releasing molecule (CORM-3) exerts bactericidal activity against Pseudomonas aeruginosa and improves survival in an animal model of bacteraemia. FASEB J. 2009;23:1023–31. doi: 10.1096/fj.08-122804. [DOI] [PubMed] [Google Scholar]
- 77.Wilson JL, Jesse HE, Hughes BM, Lund V, Naylor K, Davidge KS, Cook GM, Mann BE, Poole RK. Ru(CO)3Cl(glycinate) (CORM-3): a CO-releasing molecule with broad-spectrum antimicrobial and photosensitive activities against respiration and cation transport in Escherichia coli. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nobre LS, Seixas JD, Romao CC, Saraiva LM. Antimicrobial action of carbon monoxide-releasing compounds. Antimicrob Agents Chemother. 2007;51:4303–07. doi: 10.1128/AAC.00802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030782
- 79.Lo Iacono L, Boczkowski J, Zini R, Salouage I, Berdaux A, Motterlini R, Morin D. A carbon monoxide-releasing molecule (CORM-3) uncouples mitochondrial respiration and modulates the production of reactive oxygen species. Free Rad Biol Med. 2011;50:1556–64. doi: 10.1016/j.freeradbiomed.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 80.Sandouka A, Balogun E, Foresti R, Mann BE, Johnson TR, Tayem Y, Green CJ, Fuller B, Motterlini R. Carbon monoxide-releasing molecules (CO-RMs) modulate respiration in isolated mitochondria. Cell Mol Biol. 2005;51:425–32. [PubMed] [Google Scholar]
- 81.McLean S, Begg R, Jesse HE, Mann BE, Sanguinetti G, Poole RK. Analysis of the Bacterial Response to Ru(CO)Cl(Glycinate) (CORM-3) and the Inactivated Compound Identifies the Role Played by the Ruthenium Compound and Reveals Sulfur-Containing Species as a Major Target of CORM-3 Action. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2012.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jesse HE, Nye TL, McLean S, Green J, Mann BE, Poole RK. The terminal oxidase cytochrome bd-I in Escherichia coli has lower susceptibility than cytochromes bd-II or bo' to inhibition by the carbon monoxide-releasing molecule, CORM-3: N-acetylcysteine reduces CO-RM uptake and inhibition of respiration. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- 84.Berne JP, Lauzier B, Rochette L, Vergely C. Carbon Monoxide Protects Against Ischemia-reperfusion Injury in Vitro via Antioxidant Properties. Cell Physiol Biochem. 2012;29:475–84. doi: 10.1159/000338501. [DOI] [PubMed] [Google Scholar]
- 85.D'Amico G, Lam F, Hagen T, Moncada S. Inhibition of cellular respiration by endogenously produced carbon monoxide. J Cell Sci. 2006;119:2291–98. doi: 10.1242/jcs.02914. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030783
- 86.Lee SJ, Ryter SW, Xu JF, Nakahira K, Kim HP, Choi AMK, Kim YS. Carbon Monoxide Activates Autophagy via Mitochondrial Reactive Oxygen Species Formation. Am J Respir Cell Mol Biol. 2011;45:867–73. doi: 10.1165/rcmb.2010-0352OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith H, Mann BE, Motterlini R, Poole RK. The carbon monoxide-releasing molecule, CORM-3 (Ru(CO)3Cl(Glycinate)), targets respiration and oxidases in Campylobacter jejuni, generating hydrogen peroxide. IUBMB Life. 2011;63:363–71. doi: 10.1002/iub.476. [DOI] [PubMed] [Google Scholar]
- 88.Tavares AFN, Teixeira M, Romao CC, Seixas JD, Nobre LS, Saraiva LM. Reactive oxygen species mediate bactericidal killing elicited by carbon monoxide-releasing molecules. J Biol Chem. 2011;286:26708–17. doi: 10.1074/jbc.M111.255752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Desmard M, Foresti R, Morin D, Dagoussat M, Berdeaux A, Denamur E, Crook SH, Mann BE, Scapens D, Montravers P, Boczkowski J, Motterlini R. Differential Antibacterial Activity Against Pseudomonas aeruginosa by Carbon Monoxide-Releasing Molecules. Antiox Redox Signal. 2012;16:153–63. doi: 10.1089/ars.2011.3959. [DOI] [PubMed] [Google Scholar]
- 90.Hughes MN, Centelles MN, Moore KP. Making and working with hydrogen sulfide The chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: A review. Free Rad Biol Med. 2009;47:1346–53. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/718030784
- 91.Czyzewski BK, Wang DN. Identification and characterization of a bacterial hydrosulphide ion channel. Nature. 2012;483:494–U155. doi: 10.1038/nature10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gadalla MM, Snyder SH. Hydrogen sulfide as a gasotransmitter. J Neurochem. 113:14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030785
- 93.King SB. Potential biological chemistry of hydrogen sulfide (H2S) with the nitrogen oxides. Free Rad Biol Med. 2013;55:1–7. doi: 10.1016/j.freeradbiomed.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith EA, Macfarlane GT. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe. 1997;3:327–37. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- 95.Bouillaud F, Blachier F. Mitochondria and Sulfide: A Very Old Story of Poisoning, Feeding, and Signaling? Antiox Redox Signal. 2011;15:379–91. doi: 10.1089/ars.2010.3678. [DOI] [PubMed] [Google Scholar]
- 96.Kimura H. Hydrogen Sulfide: From Brain to Gut. Antiox Redox Signal. 12:1111–23. doi: 10.1089/ars.2009.2919. [DOI] [PubMed] [Google Scholar]
- 97.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu WT, Gazi SK, Barrow RK, Yang GD, Wang R, Snyder SH. H2S Signals Through Protein S-Sulfhydration. Science Signaling. 2009;2 doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030786
- 98.Lloyd D. Hydrogen sulfide: clandestine microbial messenger? Trends Microbiol. 2006;14:456–62. doi: 10.1016/j.tim.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: A universal defense against antibiotics in bacteria. Science. 2011;334:986–90. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/13376990
- 100.Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CCC, Leme AFP, Murakami MT, Benedetti CE. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286:26148–57. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/11951957
- 101.Berglin EH, Carlsson J. Potentiation by sulfide of hydrogen peroxide-induced killing of Escherichia coli. Infect Immun. 1985;49:538–43. doi: 10.1128/iai.49.3.538-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/718030787
- 102.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/1092245
- 103.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–9. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keren I, Wu YX, Inocencio J, Mulcahy LR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–16. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]; f1000.com/prime/717988030
- 105.Liu YY, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339:1210–13. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]; f1000.com/prime/717988031
- 106.Chung MC, Narayanan A, Popova TG, Kashanchi F, Bailey CL, Popov SG. Bacillus anthracis-derived nitric oxide induces protein S-nitrosylation contributing to macrophage death. Biochem Biophys Res Commun. 2013;430:125–30. doi: 10.1016/j.bbrc.2012.11.042. [DOI] [PubMed] [Google Scholar]


