Abstract
Adeno-associated virus (AAV)-based vectors are promising tools for gene therapeutic applications, in part because AAVs are nonpathogenic viruses, and vectors derived from them can drive long-term transgene expression without integration of the vector DNA into the host genome. AAVs are not strongly immunogenic, but they can, nonetheless, give rise to both a cellular and humoral immune response. As a result, a significant fraction of potential patients for AAV-based gene therapy harbors pre-existing antibodies against AAV. Because even very low levels of antibodies can prevent successful transduction, antecedent anti-AAV antibodies pose a serious obstacle to the universal application of AAV gene therapy. In this review, we discuss the current knowledge of the role of anti-AAV antibodies in AAV-based gene therapy with a particular emphasis on approaches to overcome the hurdle that they pose.
Introduction
Adeno-associated virus (AAV) has originally been identified as a contamination in a simian adenovirus preparation (Atchison et al., 1965). It has later been demonstrated that AAV is a replication-deficient virus that can only efficiently replicate in the presence of a helper virus such as adenovirus (Atchison et al., 1965), herpes virus (Atchison, 1970), or papilloma virus (Ogston et al., 2000). Taxonomically, AAV has been classified as a member of the family parvoviridae, the subfamily parvovirinae, and is the founding member of the genus dependovirus. Like all members of the parvoviridae family, AAV is a small (∼25 nm), non-enveloped virus with an icosahedral capsid and a single-stranded DNA genome, approximately 5 kb in size (Kerr, 2006), that is flanked by two inverted terminal repeats (ITRs) (Lusby et al., 1980, 1981). Two genes are located between the two ITRs. The first gene encodes for the four replication (Rep) proteins (Mendelson et al., 1986), which are involved in DNA replication and packaging and other viral function. The second gene encodes the three capsid proteins (Cassinotti et al., 1988) and, on an alternative reading frame, the assembly-activating protein (AAP), which is required for capsid assembly (Sonntag et al., 2010, 2011).
A unique property of AAV is that, in the absence of a helper virus, the wild-type virus—but not AAV vectors—can integrate its genome site-specifically into the long-arm of human chromosome 19 (Kotin et al., 1990, 1992; Samulski et al., 1991; Linden et al., 1996). When integrated into the human genome, AAV enters a latent phase, from which it can be rescued by a superinfection with one of its helper viruses. An important step toward the development of AAV vectors was the isolation of an infectious clone (Samulski et al., 1982). Shortly after the isolation of an infectious clone, the generation of recombinant AAV vectors, in which part or—with the exception of the ITRs—all of the viral genome has been replaced by transgene expression cassettes, was achieved (Hermonat and Muzyczka, 1984; Senapathy et al., 1984; Tratschin et al., 1984), thus setting the stage for using recombinant AAVs as gene transfer vectors.
AAV as a Gene Therapy Vector
There are several reasons why, in recent years, AAV vectors have gained increasing attention as gene therapy vectors. Among them are the nonpathogenic nature of AAV; the ability of AAV vectors to drive long-term transgene expression, at least in nondividing cells (Clark et al., 1997); and the broad tropism of the AAV serotypes and more than 100 AAV variants described in the literature (Gao et al., 2002, 2004; Asokan et al., 2012; Nonnenmacher et al., 2012). Nonetheless, AAV gene therapy still faces significant hurdles. One of the obstacles is that, despite the significant number of serotypes and variants with diverse capsids, certain tissues and cell types remain refractory to transduction. Conversely, for some applications it is preferable to transduce only a specific tissue, and all current AAV vectors transduce more than one tissue, albeit to varying degrees (Rapti et al., 2012). Another hurdle that has gained increasing attention is an immune response against the AAV capsid, both cellular and humoral. In this review, we summarize our knowledge regarding the humoral response against the capsid, with a particular emphasis on the role of pre-existing neutralizing antibodies (NAbs) in AAV gene therapy and approaches on how to overcome the challenge posed by these antecedent antibodies. A detailed discussion of the cellular immune response against the AAV capsid is unfortunately beyond the scope of this review.
Cellular Immune Response
Recombinant AAV vectors do not express any viral genes. Consequently, it is not surprising that AAV vectors do not give rise to a strong cellular immune response against the vector (Chirmule et al., 1999). Nonetheless, a fairly weak immune response can be generated against the AAV capsids, and even this modest cellular immune response can result in the ablation of transduced cells, and in some instances results in a failure to establish long-term transgene expression (Mingozzi et al., 2009). This is, however, not uniformly the case. While all patients in a phase II clinical trial to treat α1-antitrypsin deficiency by intramuscular injection of AAV1.hAAT (human α1-antitrypsin) developed interferon-γ enzyme-linked immunospot responses, hAAT levels were nonetheless elevated in all dose groups up to at least 90 days post injection. This was true even in patients who displayed a transient increase in serum creatinine kinase and showed evidence of inflammatory cells in muscle biopsies (Flotte et al., 2011). These data suggest that cellular immune responses against the AAV capsid might vary not only with the vector dose but possibly also with the AAV serotype used and/or the route of vector administration. A cellular immune response against the AAV capsid can clearly be triggered by the administration of recombinant AAVs, but it has also to be pointed out that a transient immunosuppression might be sufficient to circumvent potential problems posed by such a response (Nathwani et al., 2011). On the other hand, immunosuppression has its own set of drawbacks, such as an increased risk for infection, and the precise immunosuppression regimen used can be critical. Thus, it has been reported that, depending on the protocol used, immunosuppression can give rise to antibodies against the transgene, presumably as a result of a depletion of CD4+CD25+FoxP3+ regulatory T cells (Tregs) (Mingozzi et al., 2007). Taken together, these and other results obtained with immunosupression in AAV gene therapy suggest that while immunosuppression will likely be an important tool, it is unlikely to be a panacea against capsid-specific cellular immune responses.
Humoral Immune Response and Pre-existing Anti-AAV Antibodies
For the universal application of AAV gene therapy, the presence of (neutralizing) anti-AAV antibodies, both pre-existing and generated as a result of the therapeutic use of recombinant AAVs, is probably more problematic than the cellular immune responses against the AAV capsid. Whereas pre-existing anti-AAV antibodies might prevent the use of AAV gene therapy in a particular individual altogether, administration of recombinant AAV will give rise to NAbs even in immunologically naive patients, thus precluding repeat vector administration, which would be beneficial in certain instances.
It is not surprising then that there is a large body of literature describing the prevalence of (neutralizing) antibodies against the various AAV serotypes, especially against AAV2. Indeed, the first report of antibodies against AAV dates back to the 1960s when Rowe and colleagues published the prevalence of NAbs against AAV2 to be approximately 40% (Blacklow et al., 1968). In broad strokes, all later studies confirmed this seroprevalence of NAbs against AAV2. However, it is worth noting that the assay measuring NAbs has not been standardized. Indeed, many assays use conditions under which AAV transduction is enhanced by addition of either adenovirus, chemical drugs, or by the use of genetically manipulated cells (Supplementary Table 1; Supplementary Data available online at www.liebertonline.com/hgtb). In addition, the amount of virus used in a particular assay as well as the reporter gene employed is not uniform across studies. Even the definition of what neutralizing titer qualifies an individual as being considered seropositive varies between studies, although most studies used a cutoff of 1/20 (Table 1). Notably, only two studies used a definition of seropositivity as being a neutralizing titer of greater than or equal to 3.1 (Murphy et al., 2009; Mingozzi et al., 2012a). From a clinical point of view, neutralizing antibody titers are very significant because the presence of NAbs against the AAV serotype used is a critical exclusion criterion in the clinics.
Table 1.
Prevalence of Neutralizing Antibodies Against AAV Serotypes
| Study | Dilution | AAV1 | AAV2 | AAV5 | AAV6 | AAV7 | AAV8 | AAV9 |
|---|---|---|---|---|---|---|---|---|
| Boutin et al., 2010 | 1/20 | 50 | 59 | 3 | 37 | 19 | 33 | |
| Chirmule et al., 1999 | 1/20 (?) | 32 | ||||||
| Murphy et al.,2009 | 1/3.1 | 38 | ||||||
| Calcedo et al.,2009; Australia | 1/20 | 30 | 35 | 29 | 27 | |||
| Calcedo et al.,2009; Europe | 1/20 | 27 | 35 | 25 | 22 | |||
| Calcedo et al.,2009; Africa | 1/20 | 43 | 56 | 31 | 31 | |||
| Calcedo et al.,2009; United States* | 1/20 | 20 | 28 | 12 | 14 | |||
| Halbert et al., 2006* | 30 | 18 | 30 | 14 | 30 | |||
| Parks et al., 1970 | 1/10 | 40 | ||||||
| Blacklow et al.,1968 | 1/10 | 40 | ||||||
| Ito et al., 2009 | 1/20 | 40 | ||||||
| Moss et al., 2004 | ? | 32 | ||||||
| Wagner et al., 2002 | 1/20 | 22 | ||||||
| Erles et al.,1999* | 50 | 50 | ||||||
| Veron et al.,2012 | 1/2 | 59 | ||||||
| Mingozzi et al.,2012a | 1/10 | 82 | 27 | 64 | 50 | |||
| 1/3.1 | 100 | 36 | 91 | 90 |
The numbers in the columns of specific AAV serotypes indicate the percentage of subjects whose serum inhibited transduction by ≥50% at the indicated serum dilution.
Approximate values.
Prevalence of antibodies against the commonly used AAV serotypes
The most commonly used AAV serotypes in basic, preclinical, and clinical research are serotypes 1 to 9 and, so far, our knowledge of the prevalence of pre-existing NAbs is mostly limited to these serotypes, and studies describing the prevalence of antibodies against AAV2 are by far the most common. To date, Wilson and colleagues provided the most comprehensive study of anti-AAV NAbs in several countries and continents and against several serotypes (Calcedo et al., 2009). In this study, in all populations tested, the prevalence of NAbs against AAV2 was higher than the prevalence of NAbs against other AAV serotypes. Nonetheless, it is fair to say that the prevalence of NAbs against AAV1, AAV2, AAV7, and AAV8 did not vary drastically, although the NAb prevalence against AAV7 and AAV8 was somewhat lower than the prevalence of NAbs against both AAV1 and AAV2 (Calcedo et al., 2009).
The majority of studies investigating the prevalence of antibodies against AAV used neutralization as a method of choice to detect anti-AAV antibodies. It needs to be stressed, however, that even non-neutralizing antibodies can trigger vector clearance by the immune system. Consistent with this, it has been demonstrated that in nonhuman primates with pre-existing immunity against AAV8, vector particles fail to reach the liver; instead, the AAV particles preferentially accumulate in the spleen, which is not the case in animals without pre-existing immunity (Wang et al., 2011).
To detect such non-neutralizing antibodies, several studies determined the prevalence of anti-AAV antibodies by ELISA. In most of these studies, there was a significant—but not perfect—correlation between the anti-AAV immunoglobulin titers determined by ELISA and the neutralizing titers (Chirmule et al., 1999; Erles et al., 1999; Murphy et al., 2009; Boutin et al., 2010; Veron et al., 2012). Interestingly, however, there appear to be differences between the serotypes. Whereas there was a strong correlation between ELISA and neutralizing titers for AAV1, AAV2, AAV6, AAV8, and AAV9, this was not the case for AAV5 (Boutin et al., 2010).
Age-dependence of prevalence of anti-AAV antibodies
Especially for genetic diseases it is likely often desirable to administer gene therapy shortly after birth or in early childhood. Consequently, the age at which individuals become seropositive for AAV is important. Interestingly, antibodies against AAVs can already be detected at birth, suggesting vertical transmission of maternal antibodies (Calcedo et al., 2011). After birth, antibody levels appear to decrease over the first year of life (Calcedo et al., 2011). The immunoglobulin G (IgG) levels then increase again and reach a plateau in adolescence (Erles et al., 1999). Unexpectedly, at least in individuals younger than 60 years, a substantial fraction of individuals that harbor anti-AAV IgGs was also seropositive for IgM, suggesting a reinfection, possibly in the presence of a helper-virus (Erles et al., 1999; Murphy et al., 2009). Of the immunoglobulin subclasses, IgG1 levels were usually the highest, although in some individuals IgG2/3 levels exceed those of IgG1 (Murphy et al., 2009). Maybe most interestingly, high levels of IgG1, IgG2, and IgGM correlate well with neutralizing antibody titers, whereas this is not the case for IgG3 and IgG4 levels (Murphy et al., 2009).
Overall, the data available paint a complex picture of the humoral immune response against AAVs, and future research will be needed to determine if antibodies of different IgG subclasses affect transduction efficiencies similarly or not (Murphy et al., 2009).
Approaches to Overcome Inhibition of Transduction by (Neutralizing) Anti-AAV Antibodies
Use of alternative serotypes
The conceptually simplest approach to overcome NAbs against a specific AAV serotype is the use of a different serotype or naturally occurring AAV variant. It has indeed been shown in rodent (Halbert et al., 2000; Gao et al., 2002) and nonhuman primate models (Davidoff et al., 2005) that successful transduction can be achieved in animals that harbor antibodies against AAV serotypes other than the serotype used to deliver the transgene. The successful use of serotype-switching to overcome pre-existing NAbs, however, relies on two conditions: (1) that the alternative AAV serotype/variant has a similar tissue tropism and (2) that there is no crossreactivity between the two serotypes/variants. In this context, it must be pointed out that it is not unusual that AAV serotypes have overlapping tissue tropism. For instance, in mice, AAV1, 6, 8, and 9 show robust transduction of the myocardium, albeit to different degrees. On the other hand, the co-prevalence of anti-AAV antibodies between the serotypes can be substantial. Boutin et al. (2010) reported that of patients who were seropositive for AAV2, 93%, 52%, 59% 57%, and 58% were also seropositive for AAV1, 5, 6, 8, and 9, respectively. Interestingly, despite the fact that the capsid sequences of AAV1 and AAV6 differ by only six amino acids, only 73% of individuals seropositive for AAV1 also harbored immunoglobulins against AAV6. Intriguingly, if patients were seropositive for any of the serotypes 1, 5, 6, 8, or 9, they were also seropositive for AAV2. For all the serotype combinations, crossreactivity always exceeded 50% (Boutin et al., 2010).
In addition to the most commonly used AAV serotypes, more than 100 AAV variants have been isolated, and the crossreactivity among these variants and the AAV serotypes, and the variants themselves, has so far only been incompletely explored. Theoretically, it might be possible to identify AAV serotypes/variants for which a specific patient is seronegative. However, even if it were possible to identify for each individual an AAV serotype/variant with the desired tropism and for which the particular individual is seronegative, it is highly unlikely that such an individualized approach is practical. Clearly, while potentially useful, the use of alternative serotypes and variants is insufficient to overcome universally the challenges posed by pre-existing anti-AAV antibodies.
Development of NAb-Resistant AAV Variants by Mutations in the AAV Capsid
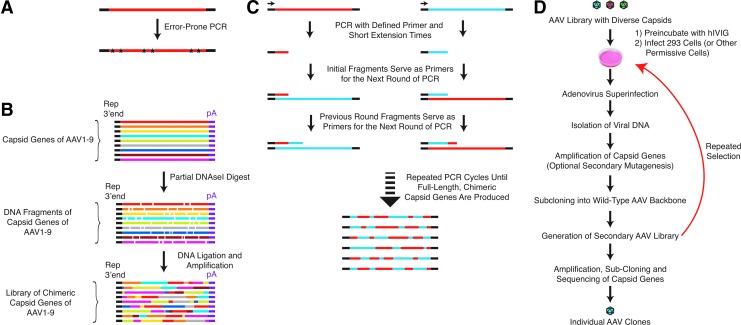
Over the past years, significant effort has been put into the development of “designer” AAV variants that show increased resistance to NAbs. Some of these approaches are based on the targeted alteration of known antigenic regions of a particular serotype (Lochrie et al., 2006), others are based on the selection of AAV variants with increased resistance to NAbs from a large library of AAV variants (Maheshri et al., 2006) or a combination of both approaches (Maersch et al., 2010) in a process called directed evolution (Fig. 1D). The first step toward the creation of libraries of AAV variants is the creation of a plasmid library that encodes variants of AAV capsid proteins. Several methods exist that allow the generation of such plasmid libraries, the most important of which are discussed below.
FIG. 1.
Creation of libraries of AAV variants and directed evolution for the isolation of AAV variants with increased resistance to neutralizing antibodies. (A) Error-prone PCR: Creation of libraries of AAV variants through the introduction of mutations into the AAV capsid by error-prone PCR. The fidelity of the PCR reaction is either reduced by the use of a DNA polymerase with decreased accuracy or by modification of the PCR conditions. Examples include altered nucleotide composition, the addition of high amounts of magnesium or the substitution of magnesium by manganese. (B) DNA shuffling: Libraries of AAV variants with chimeric capid proteins composed of stretches of multiple AAV serotypes are created by limited DNAse digestion of the capsid DNA of several AAV serotypes followed by PCR amplification. (C) Staggered extension PCR: AAV variants with chimeric capsid proteins are created by repetitive PCR reaction with very short extension times. For simplicity, only one strand of two serotypes are depicted. However, staggered extension PCR can, at least in principle, be used to assemble AAV capsids with capsid proteins composed of stretches of multiple AAV serotypes. (D) Directed evolution to isolate AAV variants with increase resistance to neutralizing antibodies: A library of AAV variants is incubated with high amounts of neutralizing antibodies, for example, human IVIG, and used to infect permissive cells. Adenovirus superinfection triggers the amplification of AAV genomes that successfully reached the nucleus. The capsid genes from these successful AAV variants can then be isolated by PCR and subcloned to generate a secondary AAV library. Optionally, the diversity of this secondary library can be increased by additional mutagenesis. After several rounds of selection, individual variants are isolated by PCR amplification and subcloning.
Error-prone polymerase chain reaction
Likely due to its relative simplicity, error-prone polymerase chain reaction (PCR) is a popular method to create DNA libraries for directed evolution. Error-prone PCR is based on the introduction of random mutations during DNA polymerization in a polymerase chain reaction (Fig. 1A). While point mutations are the most common alteration of the DNA sequence, deletions and frameshift mutations can also occur. Common ways to reduce the fidelity of DNA duplication in error-prone PCR (Labrou, 2010) are the substitution of Mg2+ by Mn2+, which reduces base-pairing specificity (Dube et al., 1975; Beckman et al., 1985), or the use of deoxynucleotide triphosphate (dNTP) mixtures that are not composed of equal amounts of the four bases (Chaput et al., 2008). Alternative methods include the use of increased Mg2+ concentrations, which results in the stabilization of mismatched bases (Eckert et al., 1991), or the use of high polymerase concentration (Labrou, 2010), which facilitates the extension of misprimed termini. In principle, the easiest approach for error-prone PCR would appear to be the use of a polymerase with very low fidelity. Unfortunately, however, currently available low-fidelity polymerases show considerable mutational bias, i.e., certain base substitutions are significantly more probable than others, resulting in libraries with reduced diversity (Tindall et al., 1988; Cline et al., 1996; Labrou, 2010).
DNA shuffling and staggered extension PCR
With the exception of AAV4 and AAV5, the capsid sequences of AAV1 to AAV9 are ≥80% homologous on the protein level and are highly homologous on the DNA level as well. For instance, the DNA homology for AAV1 and AAV9 in the capsid region is 79%. This high degree of homology makes it possible to create viral libraries with “shuffled capsids” (i.e., libraries with AAV variants whose capsid proteins are composed of stretches of several AAV serotypes), which results in different biochemical properties, including altered tropism and sensitivity to NAbs. Two principal methods to create libraries with shuffled capsids exist (Fig. 1B and C). In the first method, the capsid DNA of several AAV serotypes is subjected to a limited DNAseI digestion. Because of the comparatively high homology of the AAV serotypes, it is possible to “stitch together” a full-length AAV capsid sequence from these fragments by PCR. In the first step of this PCR reaction, the DNA fragments of the serotypes serve simultaneously as template and internal primers, yielding full-length chimeric capsid coding sequences, which are then further amplified with primers lying just outside the capsid region (Fig. 1B) (Stemmer, 1994).
An alternative method is so-called staggered-extension PCR. As illustrated in Figure 1C, this method uses capsid DNA of two or more serotypes to assemble full-length chimeric capsids, that is, capsid composed of capsid proteins that have stretches of multiple serotypes within any given capsid protein. In this method, fixed primers just outside the capsid coding sequence are used to perform PCR cycles with very short extension times. In the first round, this results in very short fragments—both from the 5’- and 3’-primers. In the following rounds, these initial short fragments can then serve as primers themselves. Because of the high homology between most AAV serotypes, these fragments cannot only anneal with the parent serotype DNA but also cross-anneal to capsid DNA of a different serotype. Over multiple rounds, this will eventually generate full-length, chimeric capsid proteins. For simplicity, Figure 1C depicts staggered extension PCR between only two serotypes, and only one DNA strand is shown. To our knowledge, this method has yet to be used to generate chimeric capsids composed of multiple serotypes. However, staggered extension has been used to increase successfully the diversity of an AAV-capsid library created by error-prone PCR (Maheshri et al., 2006).
Mutagenesis of known epitopes
At present, the antigenic regions in the AAV capsid have only been characterized in some detail for AAV2 (Moskalenko et al., 2000), although specific epitopes in AAV8 have recently been reported as well (Gurda et al., 2012). Modification of known epitopes has obvious advantages over random mutations across the entire capsid region, because it limits alteration of the capsid sequence to the precise regions where the changes are required to increase resistance to NAbs. Consequently, because mutations are restricted to antigenic regions of the capsid, an AAV library with mutated epitopes has a much higher “functional” complexity when compared to a library with an equivalent number of AAV variants that has the mutations spread across the entire capsid. This method was used successfully by Perabo and colleagues to identify AAV variants with decreased sensitivity to NAbs (Perabo et al., 2006). They selected five positions in the AAV capsid that are known to lie in antigenic regions and submitted these positions to saturation mutagenesis. Put simply, mutants with each of the 20 possible amino acids at all five positions are represented in the library. The theoretical complexity of such a library is 205 (3.2×106) mutants, and the complexity of the library that Maersch et al. (2010) obtained was 6×106 clones. This means that each possible mutant should be represented approximately twice. Interestingly, the two mutants that showed the highest increase in resistance to NAbs both contained amino acids at positions 459, 493, and 551, which differed from “NAb-resistant” AAV variants with mutations at the same positions that were reported previously (Maheshri et al., 2006; Perabo et al., 2006). The simplest explanation for this observation is that certain regions/amino acids exert a dominant effect on the Nab-sensitivity of AAV vectors, and that the nature of the mutation introduced at these positions might matter less than the fact that a particular amino acid is mutated.
Directed evolution for the isolation of NAb-resistant AAV variants
A common feature of the methods using AAV libraries to isolate AAV variants with decreased sensitivity to NAbs is that they take advantage of directed evolution, which is schematically depicted in Figure 1D. This method consists of an iterative selection of AAV variants that display increasing resistance to NAbs. In short, cells that are permissive to the starting serotype used, for example, HEK293 or HeLa cells in the case of AAV2, are infected with a library of AAV variants in the presence of high amounts of neutralizing antibodies (e.g., human intravenous immunoglobulin [IVIG]). After superinfection with adenovirus to allow AAV replication, the capsid DNA is amplified by PCR and subcloned in a wild-type AAV backbone. In one approach, a new AAV library is created directly from the plasmids carrying the capsid sequences that were isolated in the first round of selection. Preferably, in order to increase further the complexity, the plasmid library is further diversified by either error-prone PCR or staggered extension PCR (Maheshri et al., 2006). This process is then repeated several times until the variants that are most resistant to NAbs emerge. If coupled with secondary rounds of mutagenesis during each step, this directed evolution approach has the advantage that it increases diversity at each step, and that it mimics the way in which naturally occurring viruses acquire mutations when under evolutionary pressure to increase their resistance to NAbs—namely beneficial mutations accumulate in the capsid or envelope proteins over time.
While directed evolution and other methods of capsid engineering have been quite successful in isolating AAV variants with improved neutralization profiles (Bartel et al., 2011), none of the variants identified are completely resistant to NAbs and, as mutations accumulate, both the infectivity of the mutants as well as their tissue tropism can be affected. Clearly, additional approaches to identify AAV variants with an improved resistance to NAbs are still needed.
Shielding of AAV by Chemical Modification
The AAV capsid cannot only be modified by the creation of novel, genetically diverse AAV mutants, but also by the chemical modification of the AAV capsid. The basic idea in this chemical modification approach is that the chemical attachment of ligands to the AAV capsid will shield the capsid from binding of NAbs while retaining receptor binding and other necessary functions for transduction. The chemical modification of proteins has long been used to alter the pharmacokinetic properties and toxicological profile of protein therapeutics (Harris and Chess, 2003) and, because of its flexibility, relative hydrophilicity and the ability to easily modulate the shielding by varying the chain-length of the ligand, polyethylene glycol (PEG) has emerged as the preferred ligand. Importantly, PEGylation—that is, the process of attaching PEG—has been used previously to prevent neutralization of adenoviral vectors upon repeated vector administration (O'Riordan et al., 1999; Croyle et al., 2001, 2002). It is not surprising then that several groups tested whether PEGylation or modification with other ligands of AAV could render AAV vectors resistant to NAbs (Le et al., 2005; Lee et al., 2005; Carlisle et al., 2008). The picture that emerged from these studies is that chemical surface modification is challenging and, overall, only resulted in moderate increase to neutralization by pre-existing antibodies (Le et al., 2005; Carlisle et al., 2008). This can likely be attributed to the fact that, because of the small size and compactness of the AAV capsid, antigenic regions and regions essential for transduction are in close proximity. Hence, surface modification of the capsid that is sufficient to shield the virion from NAbs might not be compatible with the retention of infectivity of the vector.
Pharmacological Inhibition of Neutralization
A potentially attractive approach to overcome the hurdle of antecedent NAbs is the elimination of antibody-producing cells by pharmacological means. While this approach remains to be tested rigorously, two studies suggest that it might be a promising method (Mingozzi et al., 2012a, 2012b). In the first study, High and colleagues injected rhesus macaques with AAV8 encoding for human factor IX and analyzed the effect of B-cell depletion with the monoclonal anti-CD20 antibody rituximab, combined with cyclosporine A, on the development of antibodies against human factor IX (Mingozzi et al., 2012b). In line with clinical observations of factor IX protein replacement therapy, both monkeys injected with AAV8-F.IX developed antibodies against human (but not rhesus) factor IX. Importantly, Mingozzi and colleagues (2012b) could demonstrate that following immunosuppression with rituximab and cyclosporine A, the level of anti-factor IX antibodies returned to baseline. At the same time, the authors also analyzed the levels of NAbs against AAV8 and AAV6. As expected, AAV8-F.IX administration resulted in a drastic increase in NAbs against AAV8, from undetectable to a titer of >1/1000. Immune suppression was unable to lower the titer of anti-AAV8 NAbs below the upper level of detection, that is, >1/1000. Interestingly, however, in at least one animal, the neutralizing titer against AAV6 dropped from 1/31.6 to undetectable. Furthermore, injection of AAV6-F.IX resulted in a strong increase in F.IX expression in the same animals that were previously injected with AAV8-F.IX (Mingozzi et al., 2012b). In the second study, High and colleagues analyzed the effect of B-cell depletion with rituximab on NAb titers against AAV2 in patients with rheumatoid arthritis (Mingozzi et al., 2012a), for whom rituximab is a standard treatment. Significantly, in two patients, the NAb titers fell below 3.16 after a single dose of rituximab (Mingozzi et al., 2012a). Clearly, immunosuppression to eliminate NAbs is an area of research that deserves further investigation.
Plasmapheresis and Saline Flushing
During plasmapheresis, blood is withdrawn from a patient and the plasma and blood cells are separated by either centrifugation or hollow fiber filtration. The blood cells are then returned to the patient together with either treated plasma or replacement fluids, such as a 4.5% human albumin in saline. A common use of therapeutic apheresis is the removal of undesired immunoglobulins. For instance, in the case of thrombocytopenia purpura, antibodies against a metalloproteinase are removed, and in Guillain-Barré syndrome, anti-ganglioside antibodies are depleted. Hence, at least in principle, plasmapheresis represents an attractive approach to deplete anti-AAV antibodies.
A recent publication (Monteilhet et al., 2011) supports the possible utility of plasmapheresis to deplete NAbs prior to vector administration. In their study, Masurier and colleagues analyzed the presence of neutralizing factors against AAV1, 2, 6, and 8 in 10 patients who were undergoing repeated plasmapheresis for a variety of clinical indications (Monteilhet et al., 2011). Importantly, they were able to demonstrate that frequent plasmapheresis could lead to a substantial reduction in neutralizing factors. In several patients who had low neutralizing titers (≤1/20) before initiation of the plasmapheresis protocol, they were able to deplete neutralizing factors to undetectable levels after several rounds of plasmapheresis (Monteilhet et al., 2011). Similarly, Hurlbut and colleagues demonstrated that a single round of plasmapheresis can result in an almost three-fold drop in neutralizing antibody titers (Hurlbut et al., 2010). These authors conclude that, if combined with high vector doses, plasmapheresis would allow the treatment of up to 90% of potential patients with AAV8-based vectors (Hurlbut et al., 2010)—although this might be an overly optimistic estimate. These results are promising, but it seems more realistic that, in its present form, plasmapheresis will likely only be valuable in patients that have relatively low neutralizing titers but that would, nonetheless, disqualify from treatment in the absence of antibody depletion.
The underlying principle of plasmapheresis is that it prevents the vector from coming in contact with neutralizing antibodies. Most recently, promising results of a study based on the same principle, but using a different approach, have been published (Raper et al., 2013). In their investigation, Sakata and colleagues investigated the value of flushing the liver with saline prior to administration of AAV8 encoding factor IX in macaques with moderate titers of neutralizing antibodies against AAV8. Interestingly, they could demonstrate that saline flushing before vector administration led to the expression of therapeutic levels of factor IX in animals with neutralizing titers as high as 1/56 (Raper et al., 2013), suggesting that this might be a promising approach to circumvent challenges posed by pre-existing neutralizing antibodies, at least for liver-directed gene therapy in patients with moderate neutralizing titers.
Conclusions
With the recent approval of AAV gene therapy for the treatment of lipoprotein lipase deficiency in Europe (Gruber, 2012), the first such approval in the Western World, AAV gene therapy has at last entered the clinic. There is little doubt that AAV gene therapy for other diseases, such as hemophilias and diseases of the eye, are only a short step away from being used clinically. Given this success, it is all the more important that AAV gene therapy can be used in the largest segment of the patient population possible. At present, pre-existing NAbs against the various AAV serotypes pose a significant hurdle to the universal application of AAV gene therapy. However, recent advances in the development of new AAV variants with different immunological profiles, chemical vector modification, immunosuppression, and plasmapheresis indicate that it is likely that AAV gene therapy can be extended to a significant number of patients that currently would have to be excluded because of antecedent NAbs, especially if the different modalities can be combined.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants HL100396 (to R.J.H.), HL088434 (to R.J.H.), and HL007824 (to V.L.J.)
Author Disclosure Statement
Dr. Hajjar is the scientific cofounder of Celladon, which plans to commercialize AAV1.SERCA2a for the treatment of heart failure. All other authors declare no competing financial interests.
References
- Asokan A. Schaffer D.V. Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison R.W. The role of herpesviruses in adenovirus-associated virus replication in vitro. Virology. 1970;42:155–62. doi: 10.1016/0042-6822(70)90248-5. [DOI] [PubMed] [Google Scholar]
- Atchison R.W. Casto B.C. Hammon W.M. Adenovirus-associated defective virus particles. Science. 1965;149:754–6. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- Bartel M. Schaffer D. Buning H. Enhancing the clinical potential of AAV vectors by capsid engineering to evade pre-existing immunity. Front Microbiol. 2011;2:204. doi: 10.3389/fmicb.2011.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman R.A. Mildvan A.S. Loeb L.A. On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry. 1985;24:5810–7. doi: 10.1021/bi00342a019. [DOI] [PubMed] [Google Scholar]
- Blacklow N.R. Hoggan M.D. Kapikian A.Z., et al. Epidemiology of adenovirus-associated virus infection in a nursery population. Am. J. Epidemiol. 1968;88:368–78. doi: 10.1093/oxfordjournals.aje.a120897. [DOI] [PubMed] [Google Scholar]
- Boutin S. Monteilhet V. Veron P., et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–12. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- Calcedo R. Vandenberghe L.H. Gao G., et al. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–90. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcedo R. Morizono H. Wang L., et al. Adeno-associated virus antibody profiles in newborns, children, and adolescents. Clin. Vaccine Immunol. 2011;18:1586–8. doi: 10.1128/CVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle R.C. Benjamin R. Briggs S.S., et al. Coating of adeno-associated virus with reactive polymers can ablate virus tropism, enable retargeting and provide resistance to neutralising antisera. J. Gene Med. 2008;10:400–11. doi: 10.1002/jgm.1161. [DOI] [PubMed] [Google Scholar]
- Cassinotti P. Weitz M. Tratschin J.D. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology. 1988;167:176–84. [PubMed] [Google Scholar]
- Chaput J.C. Woodbury N.W. Stearns L.A. Williams B.A. Creating protein biocatalysts as tools for future industrial applications. Expert Opin. Biol. Ther. 2008;8:1087–98. doi: 10.1517/14712598.8.8.1087. [DOI] [PubMed] [Google Scholar]
- Chirmule N. Propert K. Magosin S., et al. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6:1574–83. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- Clark K.R. Sferra T.J. Johnson P.R. Recombinant adeno-associated viral vectors mediate long-term transgene expression in muscle. Hum. Gene Ther. 1997;8:659–69. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- Cline J. Braman J.C. Hogrefe H.H. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–51. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croyle M.A. Chirmule N. Zhang Y. Wilson J.M. “Stealth” adenoviruses blunt cell-mediated and humoral immune responses against the virus and allow for significant gene expression upon readministration in the lung. J. Virol. 2001;75:4792–801. doi: 10.1128/JVI.75.10.4792-4801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croyle M.A. Chirmule N. Zhang Y. Wilson J.M. PEGylation of E1-deleted adenovirus vectors allows significant gene expression on readministration to liver. Hum. Gene Ther. 2002;13:1887–900. doi: 10.1089/104303402760372972. [DOI] [PubMed] [Google Scholar]
- Davidoff A.M. Gray J.T. Ng C.Y., et al. Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol. Ther. 2005;11:875–88. doi: 10.1016/j.ymthe.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Dube D.K. Loeb L.A. Manganese as a mutagenic agent during in vitro DNA synthesis. Biochem. Biophys. Res. Commun. 1975;67:1041–6. doi: 10.1016/0006-291x(75)90779-2. [DOI] [PubMed] [Google Scholar]
- Eckert K.A. Kunkel T.A. DNA polymerase fidelity and the polymerase chain reaction. PCR Methods Appl. 1991;1:17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- Erles K. Sebökovà P. Schlehofer J.R. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV) J. Med. Virol. 1999;59:406–11. doi: 10.1002/(sici)1096-9071(199911)59:3<406::aid-jmv22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Flotte T.R. Trapnell B.C. Humphries M., et al. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing alpha1-antitrypsin: interim results. Hum. Gene Ther. 2011;22:1239–47. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G. Vandenberghe L.H. Alvira M.R., et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004;78:6381–8. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.P. Alvira M.R. Wang L., et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U S A. 2002;99:11854–9. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber K. Europe gives gene therapy the green light. Lancet. 2012;380:e10. doi: 10.1016/s0140-6736(12)61992-8. [DOI] [PubMed] [Google Scholar]
- Gurda B.L. Raupp C. Popa-Wagner R., et al. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J. Virol. 2012;86:7739–51. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert C.L. Rutledge E.A. Allen J.M., et al. Repeat transduction in the mouse lung by Using adeno-associated virus vectors with different serotypes. J. Virol. 2000;74:1524–32. doi: 10.1128/jvi.74.3.1524-1532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.M. Chess R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003;2:214–21. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- Hermonat P.L. Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA. 1984;81:6466–70. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut G.D. Ziegler R.J. Nietupski J.B., et al. Pre-existing immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol. Ther. 2010;18:1983–94. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J.R. Parvoviruses. Hodder Arnold; distributed in the United States by Oxford University Press; London/New York: 2006. [Google Scholar]
- Kotin R.M. Linden R.M. Berns K.I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–8. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin R.M. Siniscalco M. Samulski R.J., et al. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2211–5. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrou N.E. Random mutagenesis methods for in vitro directed enzyme evolution. Curr. Protein Pept. Sci. 2010;11:91–100. doi: 10.2174/138920310790274617. [DOI] [PubMed] [Google Scholar]
- Le H.T. Yu Q.C. Wilson J.M. Croyle M.A. Utility of PEGylated recombinant adeno-associated viruses for gene transfer. J. Control Release. 2005;108:161–77. doi: 10.1016/j.jconrel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Lee G.K. Maheshri N. Kaspar B. Schaffer D.V. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol. Bioeng. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- Linden R.M. Ward P. Giraud C., et al. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11288–94. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie M.A. Tatsuno G.P. Christie B., et al. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006;80:821–34. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusby E. Fife K.H. Berns K.I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J. Virol. 1980;34:402–9. doi: 10.1128/jvi.34.2.402-409.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusby E. Bohenzky R. Berns K.I. Inverted terminal repetition in adeno-associated virus DNA: independence of the orientation at either end of the genome. J. Virol. 1981;37:1083–6. doi: 10.1128/jvi.37.3.1083-1086.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maersch S. Huber A. Büning H., et al. Optimization of stealth adeno-associated virus vectors by randomization of immunogenic epitopes. Virology. 2010;397:167–75. doi: 10.1016/j.virol.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Maheshri N. Koerber J.T. Kaspar B.K. Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Mendelson E. Trempe J.P. Carter B.J. Identification of the trans-acting Rep proteins of adeno-associated virus by antibodies to a synthetic oligopeptide. J. Virol. 1986;60:823–32. doi: 10.1128/jvi.60.3.823-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. Hasbrouck N.C. Basner-Tschakarjan E., et al. Modulation of tolerance to the transgene product in a nonhuman primate model of AAV-mediated gene transfer to liver. Blood. 2007;110:2334–41. doi: 10.1182/blood-2007-03-080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. Meulenberg J.J. Hui D.J., et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood. 2009;114:2077–86. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. Chen Y. Edmonson S.C., et al. Prevalence and pharmacological modulation of humoral immunity to AAV vectors in gene transfer to synovial tissue. Gene Ther. 2012a doi: 10.1038/gt.2012.55. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. Chen Y. Murphy S.L., et al. Pharmacological Modulation of Humoral Immunity in a Nonhuman Primate Model of AAV Gene Transfer for Hemophilia B. Mol. Ther. 2012b;20:1410–6. doi: 10.1038/mt.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteilhet V. Saheb S. Boutin S., et al. A 10 patient case report on the impact of plasmapheresis upon neutralizing factors against adeno-associated virus (AAV) types 1, 2, 6, and 8. Mol Ther. 2011;19:2084–91. doi: 10.1038/mt.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko M. Chen L. Van Roey M., et al. Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: implications for gene therapy and virus structure. J. Virol. 2000;74:1761–6. doi: 10.1128/jvi.74.4.1761-1766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.L. Li H. Mingozzi F., et al. Diverse IgG subclass responses to adeno-associated virus infection and vector administration. J. Med. Virol. 2009;81:65–74. doi: 10.1002/jmv.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C. Tuddenham E.G. Rangarajan S., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–65. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M. Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther. 2012;19:649–58. doi: 10.1038/gt.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogston P. Raj K. Beard P. Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J. Virol. 2000;74:3494–504. doi: 10.1128/jvi.74.8.3494-3504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan C.R. Lachapelle A. Delgado C., et al. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum Gene Ther. 1999;10:1349–58. doi: 10.1089/10430349950018021. [DOI] [PubMed] [Google Scholar]
- Perabo L. Endell J. King S., et al. Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J. Gene Med. 2006;8:155–62. doi: 10.1002/jgm.849. [DOI] [PubMed] [Google Scholar]
- Raper S.E. Wilson J.M. Nunes F.A. Flushing out antibodies to make aav gene therapy available to more patients. Mol Ther. 2013;21:269–271. doi: 10.1038/mt.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti K. Hajjar R.J. Weber T. Novel approaches to deliver molecular therapeutics in cardiac disease using adeno-associated virus vectors. Translational Cardiology. 2012:391–458. [Google Scholar]
- Samulski R.J. Berns K.I. Tan M. Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl. Acad. Sci. U.S.A. 1982;79:2077–81. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R.J. Zhu X. Xiao X., et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–50. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P. Tratschin J.D. Carter B.J. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep- mutants by a wild-type genome or an ori- mutant and correction of terminal palindrome deletions. J. Mol. Biol. 1984;179:1–20. doi: 10.1016/0022-2836(84)90303-6. [DOI] [PubMed] [Google Scholar]
- Sonntag F. Schmidt K. Kleinschmidt J.A. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. USA. 2010;107:10220–5. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F. Kother K. Schmidt K., et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011;85:12686–97. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–91. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- Tindall K.R. Kunkel T.A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–13. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Tratschin J.D. West M.H. Sandbank T. Carter B.J. A human parvovirus, adeno-associated virus, as a eucaryotic vector: transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol. Cell Biol. 1984;4:2072–81. doi: 10.1128/mcb.4.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron P. Leborgne C. Monteilhet V., et al. Humoral and cellular capsid-specific immune responses to adeno-associated virus type 1 in randomized healthy donors. J Immunol. 2012;188:6418–24. doi: 10.4049/jimmunol.1200620. [DOI] [PubMed] [Google Scholar]
- Wang L. Calcedo R. Bell P., et al. Impact of pre-existing immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum. Gene Ther. 2011;22:1389–401. doi: 10.1089/hum.2011.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.