Abstract
We hypothesized that the AAV2 vector is targeted for destruction in the cytoplasm by the host cellular kinase/ubiquitination/proteasomal machinery and that modification of their targets on AAV2 capsid may improve its transduction efficiency. In vitro analysis with pharmacological inhibitors of cellular serine/threonine kinases (protein kinase A, protein kinase C, casein kinase II) showed an increase (20–90%) on AAV2-mediated gene expression. The three-dimensional structure of AAV2 capsid was then analyzed to predict the sites of ubiquitination and phosphorylation. Three phosphodegrons, which are the phosphorylation sites recognized as degradation signals by ubiquitin ligases, were identified. Mutation targets comprising eight serine (S) or seven threonine (T) or nine lysine (K) residues were selected in and around phosphodegrons on the basis of their solvent accessibility, overlap with the receptor binding regions, overlap with interaction interfaces of capsid proteins, and their evolutionary conservation across AAV serotypes. AAV2-EGFP vectors with the wild-type (WT) capsid or mutant capsids (15 S/T→alanine [A] or 9 K→arginine [R] single mutant or 2 double K→R mutants) were then evaluated in vitro. The transduction efficiencies of 11 S/T→A and 7 K→R vectors were significantly higher (∼63–90%) than the AAV2-WT vectors (∼30–40%). Further, hepatic gene transfer of these mutant vectors in vivo resulted in higher vector copy numbers (up to 4.9-fold) and transgene expression (up to 14-fold) than observed from the AAV2-WT vector. One of the mutant vectors, S489A, generated ∼8-fold fewer antibodies that could be cross-neutralized by AAV2-WT. This study thus demonstrates the feasibility of the use of these novel AAV2 capsid mutant vectors in hepatic gene therapy.
Gabriel and colleagues examine the in vitro and in vivo efficacy of novel AAV2 vectors, which are modified at critical serine/threonine/lysine residues of the vector capsid. In vitro, they find that the transduction efficiencies of 11 S/T → A and 7 K → R vectors are significantly higher than the AAV2-wild type (WT) vectors. In vivo, they find that hepatic gene transfer of these mutant vectors results in higher vector copy numbers (up to 4.9-fold) and transgene expression (up to 14-fold) than observed from the AAV2-WT vector.
Introduction
Recombinant adeno-associated viral (AAV) vectors based on serotype 2 have been used successfully for in vivo gene transfer in numerous preclinical animal models (Mingozzi and High, 2011). AAV2 vectors have shown sustained clinical benefit when targeted to immune-privileged sites such as for Leber's congenital amaurosis (Simonelli et al., 2010). However, their therapeutic efficiency when targeted to other organ systems, such as during hepatic gene transfer in patients with hemophilia B, is suboptimal because of the CD8+ T cell response directed against the AAV capsid particularly at higher administered vector doses (≥2×1012 viral genomes [VG]/kg) (Manno et al., 2006). A similar theme of vector dose-dependent immunotoxicity has emerged from the use of alternative AAV serotypes in other clinical trials as well (Stroes et al., 2008). More recently, in the recombinant AAV8-mediated gene transfer for hemophilia B (Nathwani et al., 2011), two patients who received the highest dose (2×1012 VG/kg) of vector required glucocorticoid therapy to attenuate a capsid-specific T cell response developed against capsid. Therefore, irrespective of whether an alternative AAV serotype (other than AAV2) or an immune suppression protocol is used, it is important to develop novel AAV vectors that provide enhanced gene expression at significantly lower vector doses to achieve successful gene transfer in humans.
Although conventional wild-type AAV2 (AAV2-WT) vectors can transduce a variety of cell types and tissues, the onset of gene expression is slow and they typically require several weeks to achieve sustained, steady state levels of transgene expression (Buning et al., 2008). The AAV capsid has been reported to influence transduction efficiency at many steps, including vector binding to cell surface receptors, internalization, cytoplasmic trafficking to the nuclear membrane, and viral uncoating (Nonnenmacher and Weber, 2012). It has been shown that epidermal growth factor receptor protein tyrosine kinase (EGFR-PTK)-mediated tyrosine phosphorylation of capsid surface-exposed AAV2 residues leads to ubiquitination and proteasomal degradation of viral particles (Jayandharan et al., 2008; Zhong et al., 2008b). The use of proteasomal inhibitors is known to result in an ∼2-fold increase in gene expression from AAV vectors (Monahan et al., 2010). However, systemic administration of these proteasomal inhibitors leads to severe side effects (Rajkumar et al., 2005). Alternatively, altering the enzymatic (kinase/ubiquitin ligase) targets on AAV capsid may be a rational approach to circumvent capsid ubiquitination and increase the transduction efficiency of these vectors.
AAV capsid is composed of three proteins—VP1, VP2, and VP3—generated from a single cap gene by alternative splicing (Becerra et al., 1985; Trempe and Carter, 1988). Specific residues/motifs on AAV capsid are known to interact with viral receptors on the cell membrane, help in the endosomal escape of the vector (Girod et al., 2002), and, importantly, determine the serotype of the vector. Hence it is but logical to assume that capsid mutagenesis of AAV vectors can introduce functional changes in the vector. To this end, the generation of hybrid serotypes by capsid fusion of multiple serotypes and capsid mutations has been reported (Choi et al., 2005; Koerber et al., 2008). Earlier studies, wherein random capsid mutations of AAV2 were introduced, have demonstrated that such modifications could alter the efficiency of vector packaging, receptor binding, intracellular trafficking, or transgene expression (Kern et al., 2003; Opie et al., 2003; Lochrie et al., 2006). More recently, site-specific mutagenesis of AAV2 capsid to generate tyrosine-mutant AAV2 vectors has demonstrated increased gene expression in vitro and in vivo (Zhong et al., 2008b; Li et al., 2010). However, because serine (8%), threonine (7.2%), and lysine (4%) residues are more abundant on AAV2 capsid than are tyrosine residues (3.5%), we hypothesized that mutating amino acids other than tyrosines on AAV2 capsid may provide further opportunities to augment AAV-mediated gene expression. This hypothesis is supported by several studies. Targeted inhibition of the serine/threonine kinase phosphorylation of a cellular protein, FK506-binding protein 52 (FKBP52), improves AAV-mediated gene transfer by 30-fold compared with the ∼5-fold increase seen by inhibition of tyrosine kinases alone (Zhao et al., 2006). It is also known that lysine residues are direct targets for host cell ubiquitination (Hatakeyama et al., 2005) and therefore modifying them is likely to reduce vector ubiquitination and subsequent proteasome-mediated degradation. On the basis of these data, the present study was designed to test the in vitro and in vivo efficacy of novel AAV2 vectors that are modified at critical serine/threonine/lysine residues of the vector capsid.
Materials and Methods
Cell lines and reagents
Human cervical carcinoma cell line HeLa and human embryonic kidney cell line HEK-293 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The packaging cell line for the vectors, AAV-293, was obtained from Stratagene/Agilent Technologies (Palo Alto, CA). Cells were maintained as monolayer cultures in Iscove's modified Dulbecco's medium (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 1% by volume of a 100× stock solution of antibiotics (penicillin–streptomycin), and sodium bicarbonate (1.2 g/liter; Sigma-Aldrich). Small-molecule inhibitors of protein kinase A (PKA) (PKA inhibitor fragment 6–22 amide), PKC (inhibitor Gö 6983), and casein kinase II (CKII) (inhibitor TBB) were purchased from Sigma-Aldrich. Fragment 6–22 amide is derived from the active portion of the heat-stable PKA inhibitor protein PKI. Go6983 is a direct inhibitor of L type Ca2+ channel and can selectively inhibit several PKC isozymes. TBB (4,5,6,7-tetrabromobenzotriazole) is a highly selective, ATP/GTP-competitive inhibitor of casein kinase II.
Structural analysis of AAV2 capsid
The three-dimensional structure of the AAV2 capsid from the Protein Data Bank (Berman et al., 2000) (PDB accession number 1LP3) (Xie et al., 2002) was analyzed extensively. Protein–protein interaction interface residues on the capsid proteins were determined by a distance-based method using computer programs described elsewhere (De et al., 2005). Briefly, a residue pair from the adjacent subunits is said to be in the interaction interface if the distance between the two interacting atoms is greater than the sum of their van der Waals radii plus 0.5 Å. Solvent accessibility values of the residues were determined with the NACCESS program (Hubbard and Thornton, 1993). Phosphorylation sites in capsid proteins were predicted with NetPhosK (http://www.cbs.dtu.dk/services/NetPhosK/), Phosida (Phosphorylation Site Database; http://www.phosida.com/), KinasePhos (http://kinasephos.mbc.nctu.edu.tw/), and Scansite (http://scansite.mit.edu/) prediction servers whereas ubiquitination sites were predicted with UbiPred (http://iclab.life.nctu.edu.tw/ubipred/), Composition of k-Spaced Amino Acid Pairs (CKSAAP_UbSite; http://protein.cau.edu.cn/cksaap_ubsite/), and Prediction of Ubiquitination Sites with Bayesian Discriminant Method (BDM-PUB; http://bdmpub.biocuckoo.org/index.php) prediction servers.
Structures were visualized with the PyMOL software package (DeLano, 2002). To assess the conservation of the predicted phosphorylation as well as ubiquitination sites, multiple sequence alignment of the VP1 sequence across the 10 serotypes was generated with ClustalW (Chenna et al., 2003). The nomenclature of the target amino acids is based on the AAV2 VP1 reference sequence in the NCBI database (accession number NC_001401).
Site-directed mutagenesis
Serine (S)→alanine (A), threonine (T)→A (alanine), and lysine (K)→arginine (R) mutations were introduced into the AAV2 rep/cap plasmid (p.ACG2; a kind gift from A. Srivastava, University of Florida, Gainesville, FL) with a QuikChange II XL site-directed mutagenesis kit (Stratagene/Agilent Technologies) in accordance with the manufacturer's protocol. Briefly, a one-step PCR amplification of the target sites was performed for 18 cycles with the primers (Supplementary Table S1; supplementary data are available online at http://www.liebertpub.com/hgtb) followed by DpnI digestion for 1 hr. Two microliters of this digested PCR product was then transformed into XL10-Gold ultracompetent cells (Stratagene/Agilent Technologies). After plasmid isolation, the presence of the desired point mutation was verified by restriction digestion analysis and DNA sequencing (Applied Biosystems 3130 genetic analyzer; Life Technologies, Warrington, UK).
Generation of recombinant vectors
Highly purified stocks of self-complementary (sc) AAV2-WT or 26 capsid mutants of AAV2 vectors or AAV8-WT vector carrying the enhanced green fluorescent protein (EGFP) gene driven by the chicken β-actin promoter were generated by polyethyleneimine-based triple transfection of AAV-293 cells (Ling et al., 2011). Briefly, forty 150-mm2 dishes 80% confluent with AAV-293 cells were transfected with AAV2 rep/cap (p.ACG2), transgene (dsAAV2-EGFP), and AAV-helper free (p.helper) plasmids. Cells were collected 72 hr post-transfection, lysed, and treated with Benzonase nuclease (25 units/ml; Sigma-Aldrich). Subsequently, the vectors were purified by iodixanol gradient ultracentrifugation (OptiPrep; Sigma-Aldrich) (Zolotukhin et al., 1999) followed by column chromatography (HiTrap SP column; GE Healthcare Life Sciences, Pittsburgh, PA). The vectors were finally concentrated to a final volume of 0.5 ml in phosphate-buffered saline (PBS), using Amicon Ultra 10K centrifugal filters (Millipore, Bedford, MA). The physical particle titers of the vectors were quantified by slot-blot analysis and expressed as vector genomes per milliliter (Kube and Srivastava, 1997).
Recombinant AAV2 vector transduction assays in vitro
To assess the effect of pharmacological inhibition of cellular serine/threonine kinases on AAV2 transduction, approximately 1.6×105 HeLa cells were mock (PBS)-treated or pretreated with optimal concentrations of PKA inhibitor (25 nM), PKC inhibitor (70 nM), or CKII inhibitor (1 μM), or with a combination of each of these inhibitors overnight and transduced with AAV2-WT vector at 2×103 VG/cell. The safe and effective concentration of kinase inhibitors used was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, performed with three 10-fold dilutions around the median inhibition constant (IC50) values for these small-molecule inhibitors. Twenty-four hours later, transgene expression was measured by flow cytometry (FACS Calibur; BD Biosciences, San Jose, CA). A total of 1×104 events were analyzed for each sample. Mean values of percent EGFP positivity from three replicate samples were used for comparison between treatment groups.
To assess the efficacy of the novel mutant vectors generated, HeLa or HEK-293 cells were mock-infected or infected with either AAV2-WT or AAV2 S/T/K mutant vector (2×103 VG/cell). Forty-eight hours post-transduction, transgene expression was quantitated by flow cytometry (FACSCalibur; BD Biosciences) or captured by EGFP imaging. For flow cytometric analysis, HeLa or HEK-293 cells were trypsinized (0.05% trypsin; Sigma-Aldrich) and rinsed twice with PBS (pH 7.4). A total of 1×104 events were analyzed for each sample. In total, three independent experiments were performed including three intraassay replicates in each of the experiment. Mean values of percent GFP positivity from these nine replicate samples were used for comparison between AAV2-WT- and AAV2 S/T/K-infected cells.
Recombinant AAV2 vector transduction studies in vivo
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved and carried out according to the institutional guidelines for animal care (Christian Medical College, Vellore, India). Groups (n=4 per group) of 8- to 12-week-old C57BL/6 mice were mock-injected or injected with 5×1010 VG each of scAAV2-WT or scAAV2 S/T/K mutant vector carrying the EGFP transgene, via the tail vein. Mice were killed 4 weeks after vector administration. Cross-sections from three hepatic lobes of the mock-injected and vector-injected groups were assessed for EGFP expression by fluorescence microscopy.
Estimation of AAV2 vector genome copies and EGFP expression in murine hepatocytes by quantitative PCR analysis
To quantitate the transduction efficiency of AAV2 vectors in vivo, liver tissue samples were collected from each of the mice injected with either AAV2-WT or AAV2 S/T/K mutant vector, 4 weeks after vector administration. Genomic DNA was isolated with a QIAamp DNA mini kit (Qiagen, Valencia, CA). Vector genome copy numbers per diploid genome were quantified with TaqMan probes and primers designed against the AAV2 inverted terminal repeat (ITR) sequence and estimated as described previously (Aurnhammer et al., 2011), using a low-ROX quantitative PCR MasterMix according to the protocol of the manufacturer (Eurogentec, Seraing, Belgium).
To measure EGFP transcript levels, total RNA was isolated from murine hepatocytes 4 weeks after vector administration, using TRIzol reagent (Sigma-Aldrich). Approximately 1 μg of RNA was reverse transcribed, using Verso reverse transcriptase according to the manufacturer's protocol (Thermo Scientific, Surrey, UK). TaqMan PCR was done with primers and probe against the EGFP gene (forward primer, CTTCAAGATCCGCCACAACATC; reverse primer, ACCATGTGATCGCGCTTCTC; probe, FAM-CGCCGACCACTACCAGCAGAACACC-TAMRA), according to the manufacturer's protocol (Eurogentec). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the housekeeping control gene. Data were captured and analyzed with ABI Prism 7500 sequence detection system version 1.1 software (Life Technologies).
Estimation of neutralizing antibodies against S/T/K AAV2 vectors
Heat-inactivated serum samples from AAV2-WT-injected or S→A and K→R mutant AAV2-injected C57BL/6 mice were assayed for neutralizing antibody (NAb) titers as described previously (Calcedo et al., 2009). Briefly, groups of mice (n=4) were administered 5×1010 VG of AAV2-WT and AAV2 S/T/K mutant vectors via tail vein injections. Four weeks after vector delivery, animals were killed and serum was collected. The pooled serum was snap frozen to −80°C. Samples were analyzed by the Immunology Core (Gene Therapy Program, Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA). The NAb titer was reported as the highest serum dilution that inhibited AAV transduction of Huh7 cells by 50% or more compared with that for the naive serum control.
Ubiquitin conjugation assay and immunoblotting
A ubiquitination assay of viral capsids was done with a ubiquitin–protein conjugation kit according to the protocol of the manufacturer (Boston Biochem, Cambridge, MA). Briefly, 10× energy solution, conjugation fraction A, conjugation fraction B, and ubiquitin were mixed to a final reaction volume of 100 μl. The conjugation reaction was then initiated by adding 3×108 heat-denatured AAV2-WT, AAV2 mutant vector, or AAV5 viral particles and incubated at 37°C for 4 hr. Equal volumes of sodium dodecyl sulfate (SDS)-denatured ubiquitinated samples were then resolved on a 4–20% gradient gel. The ubiquitination pattern for the various viral particles was detected by immunoblotting of the samples with mouse anti-ubiquitin monoclonal antibody (P4D1) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG1 secondary (Cell Signaling Technology, Boston, MA). VP1, VP2, and VP3 capsid proteins were detected with AAV clone B1 antibody (Fitzgerald, North Acton, MA) and HRP-conjugated anti-mouse IgG1 secondary antibody (Cell Signaling Technology).
Histological examination
Liver tissues from mock-injected mice or those injected with either AAV2-WT or AAV2 mutant vector were collected 4 weeks postinjection, fixed in 10% buffered formalin, and processed for microscopy. Three-micron-thick liver sections were cut and stained with hematoxylin and eosin. The degree of lobular and portal inflammation was scored (inflammation score, IS) by a pathologist, who was blinded to the experimental conditions. Inflammation scores were based on the degree of lobular and portal inflammation and expressed as follows: 0, no inflammation; 1, minimal inflammation; 2, mild inflammation; 3, moderate inflammation. The median score for each group (n=3) was calculated.
Results
Structural analysis of AAV2 capsid to select amino acids for mutagenesis
To select mutation positions in the AAV2 capsid, the three-dimensional structure available for the capsid (Protein Data Bank accession number 1LP3) (Xie et al., 2002), was analyzed extensively. Sites for phosphorylation and the kinases involved in this process as well as ubiquitination sites were predicted with various software tools, as mentioned in Materials and Methods. Most commonly, the sites predicted were probable targets of the kinases PKA, PKC, and CKII. The consensus residues, predicted by most of the prediction tools, were given higher preference and selected as mutation targets.
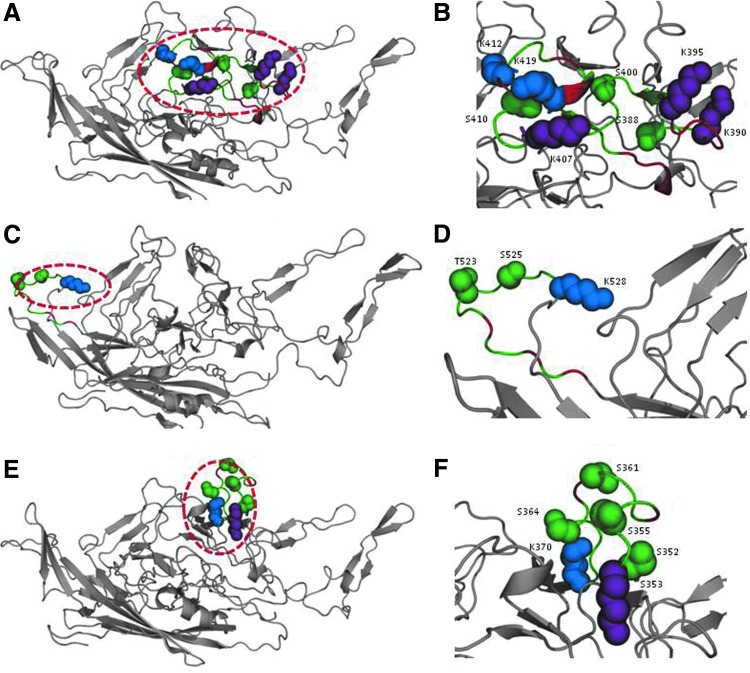
The phosphorylation and ubiquitination sites forming phosphodegrons were then identified in the AAV2 capsid. It is known that the serine/threonine residues in phosphodegrons reside in the vicinity of lysine residues (within 9–13 residues in the sequence), allowing them to be identified as a degradation signal by the ubiquitin ligase enzyme (Wu et al., 2003). Also, a negative charge often accumulates near the phosphosite and there are multiple phosphosites in one phosphodegron (Wang et al., 2012). The region separating phosphosite and ubiquitination site is largely unstructured and solvent exposed (Inobe et al., 2011). With this information, three phosphodegrons were identified in the AAV2 capsid as shown in Fig. 1. Interactions between the capsid proteins need to be critically maintained to preserve the capsid geometry. Hence, the interaction interfaces were determined from the capsid structure, using both the distance criterion and the accessibility criterion (De et al., 2005), as mentioned in Materials and Methods. Thus, in selecting mutation targets, care was taken that the residues did not belong to these interaction interfaces. A group of positively charged residues on the AAV2 capsid, distributed in three clusters, mediates binding of AAV2 to heparin sulfate receptors (Kern et al., 2003; Opie et al., 2003). Hence, lysines in the receptor-binding regions, if lying in/around phosphodegrons, were still selected and mutated to arginine residues but the serines and threonines were left unaltered. Conservation of a residue across AAV serotypes was considered an added advantage in selection for mutation (Fig. 2). Table 1 summarizes the features of the three phosphodegrons identified and highlights the selected mutation targets within the phosphodegron sequences.
FIG. 1.
Structural analysis of phosphodegrons 1–3 in the AAV2 capsid. (A), (C), and (E) show phosphodegrons 1, 2, and 3 colored in green, respectively, and corresponding zoomed-in regions of the three phosphodegrons are shown in (B), (D), and (F), respectively. Phosphodegrons in the AAV2 capsid are largely present in the loop regions and are solvent exposed as shown. The phosphorylation and ubiquitination sites in the phosphodegrons are shown as green and blue spheres, respectively. Receptor-binding residues that have also been predicted as ubiquitination sites are shown as purple spheres. The acidic residues in phosphodegrons 1 and 3 and prolines in phosphodegron 2 are colored red whereas the rest of the protein structure is shown in gray. The images were generated with PyMOL software (DeLano, 2002). Color images available online at www.liebertpub.com/hgtb
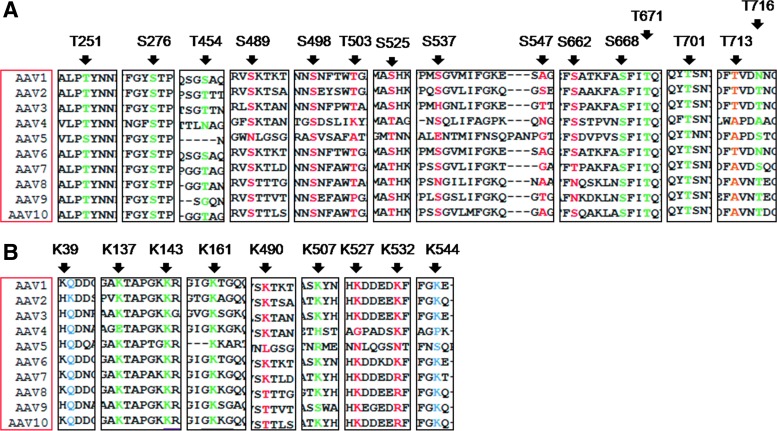
FIG. 2.
Schematic representation and conservation status of the various serine (S), threonine (T), and lysine (K) residues mutated in the AAV2 capsid. VP1 protein sequences from AAV serotypes 1 through 10 were aligned with ClustalW and the conservation status of each of the mutated sites is given. S/T residues are shown in (A) and lysine residues are shown in (B). S/T/K residues within phosphodegrons 1, 2, and 3 are shown in red whereas those chosen on the basis of evolutionary conservation are shown in green. Those residues that were chosen on the basis of either in silico prediction to be a part of a phosphosite or high ubiquitination score with the UbiPred tool are shown in blue. A control threonine mutation shown in brown was chosen as a negative control for the mutation experiments. Color images available online at www.liebertpub.com/hgtb
Table 1.
Location and Amino Acid Sequence of the Three Phosphodegrons in the AAV2 Capsida
| Phosphodegron | Amino acid position (NCBI numbering) | Amino acid sequence (N-C terminus) | Average solvent accessibility (%) |
|---|---|---|---|
| 1 | 525–564 | ShKddeekffpqSgvlifgKqgsektnvdiekvmitdeee | 23.6 |
| 2 | 652–665 | pvpanpstTfSaaK | 35.0 |
| 3 | 489–507 | SKtsadnnnSeyswTgatK | 24.5 |
The predicted phosphorylation and ubiquitination sites (shown in boldface) that are highly conserved among all the serotypes of AAV within the phosphodegron region (shown enlarged) are listed. All three phosphodegrons are solvent accessible as shown by their high average solvent accessibility.
Pharmacological inhibition of cellular serine/threonine kinases improves AAV2-mediated gene expression in vitro
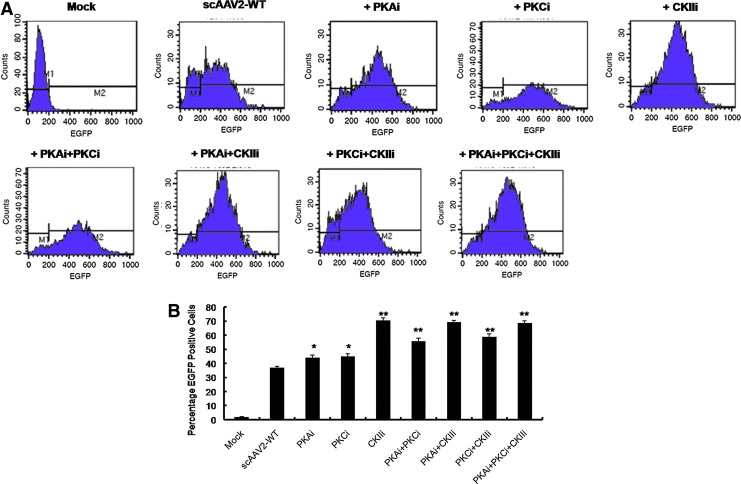
Our in silico analysis of the AAV2 capsid structure, using various phosphorylation prediction tools, identified PKA, PKC, and CKII as major binding partners of phosphodegrons of the AAV2 capsid. Because these enzymes are primarily serine/threonine kinases with an ability to phosphorylate S/T residues, we hypothesized that the inhibition of these viral capsid phosphorylating kinases could augment AAV2 transduction. To test whether the host cellular PKA, PKC, and CKII serine/threonine kinases play a rate-limiting role in AAV2 transduction, we inhibited the kinase activity by specific small-molecule inhibitors and then infected HeLa cells with scAAV2-EGFP vector. As can be seen in Fig. 3A and B, significantly higher gene expression from the AAV2-WT vector was observed when HeLa cells were pretreated with these kinase inhibitors, with a maximal 90% increase seen in cells treated with the CKII inhibitor. This demonstrates that one or more surface-exposed serine and/or threonine amino acids in the AAV2 capsid is phosphorylated within the host cell by PKA, PKC, and CKII serine/threonine kinases and that specific inhibition of this process improves gene expression from the AAV vectors. Because systemic administration of serine/threonine kinase inhibitors in an in vivo setting is likely to be toxic (Force and Kolaja, 2011), we instead chose to modify the kinase target substrates in the AAV2 capsid to further improve the transduction efficiency of AAV2 vectors.
FIG. 3.
Effect of pharmacological inhibition of host cellular serine/threonine kinases on AAV2-mediated gene expression. (A) HeLa cells were mock (PBS)-treated or pretreated with protein kinase A (PKA), protein kinase C (PKC), and casein kinase II (CKII) inhibitors (PKAi, PKCi, and CKIIi, respectively) either alone or in the combinations shown, 24 hr before transduction with AAV2-EGFP vectors. Twenty-four hours post-transduction, cell suspensions were analyzed for EGFP expression by flow cytometry. (B) Quantitative representation of the data from (A). One-way analysis of variance (ANOVA) was used for statistical analysis. *p<0.05; **p<0.01 versus AAV2-WT-infected cells. Color images available online at www.liebertpub.com/hgtb
AAV2 serine/threonine/lysine mutant vectors demonstrate significantly improved transduction efficiency in vitro
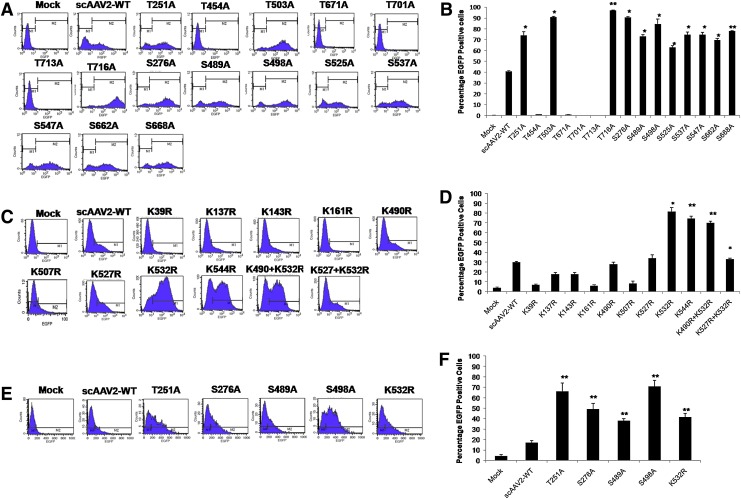
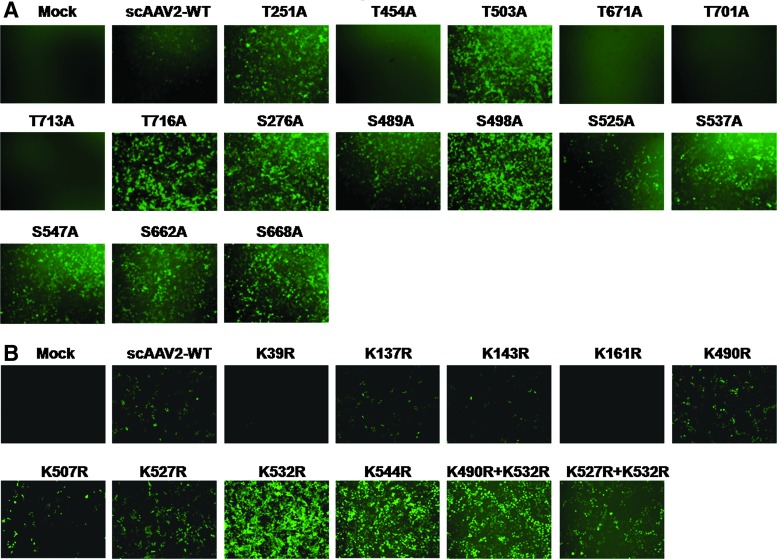
Each of the S/T/K residues identified in the vicinity of phosphodegrons (Figs. 1 and 2) was mutated either as a single mutant (n=24) or as a double mutant (n=2). The vast majority of these S/T/K mutant capsids did not affect the vector packaging efficiency (Table 2), suggesting that modification of these specific amino acids had negligible effect on the capsid structure. Only four of the mutants generated, S276A (1.65×1010 VG/ml), T454A (2.5×1010 VG/ml), T503A (5.25×1010 VG/ml), and T716A (5.25×1010 VG/ml), had consistently 8- to 24-fold lower average packaging titers compared with the AAV2-WT vector and were used only for the in vitro transduction studies. Among the 15 S/T→A mutant AAV2 vectors tested for their transduction efficiency at a multiplicity of infection (MOI) of 2000 in HeLa cells, 11 had a significantly higher increase in EGFP-positive cells (63–97%) compared with AAV2-WT vector-infected cells (41%) by FACS analysis (Fig. 4).
Table 2.
Physical Particle Packaging Titers (Viral Genomes/ml) of AAV2 Serine/Threonine/Lysine Mutant Vectors
| Serine (S)→Alanine (A)a | Threonine (T)→Alanine (A)a | Lysine (K)→Arginine (R)a |
|---|---|---|
| S276A (1.65×1010) | T251A (1.8×1012) | K39R (2.4×1011) |
| S489A (3.2×1012) | T454A (2.5×1010) | K137R (3×1012) |
| S498A (1×1012) | T503A (5.25×1010) | K143R (2.3×1012) |
| S525A (3.2×1012) | T671A (1.6×1012) | K161R (9×1011) |
| S537A (8×1011) | T701A (3.2×1012) | K490R (2.3×1011) |
| S547A (1.6×1012) | T713A (3.2×1012) | K507R (2×1011) |
| S662A (3.2×1012) | T716A (5.25×1010) | K527R (3.2×1011) |
| S668A (4×1011) | K532R (2.4×1012) | |
| K544R (3×1011) | ||
| K527R+K532R (6×1011) | ||
| K490R+K532R (2×1011) |
Average packaging titers from at least two packaging experiments. Vectors were generated by polyethyleneimine-based triple transfection of AAV-293 cells. The vectors were purified by iodixanol gradient ultracentrifugation and column chromatography (mentioned in Materials and Methods) and resuspended in a final volume of 0.5 ml of phosphate-buffered saline. The titers of wild-type self-complementary AAV2 vectors ranged between 4×1011 and 1×1012 VG/ml in the laboratory. VG, viral genomes.
FIG. 4.
AAV2 serine/threonine/lysine mutant vectors demonstrate increased transduction efficiency in vitro. HeLa cells were either mock-infected or infected at 2×103 viral genome (VG)/cell with AAV2-WT or AAV2 S/T→A (A) or AAV2 K→R (C) mutant vectors and cells were analyzed for EGFP expression 48 hr later by flow cytometry. The percentage of EGFP-positive cells posttransduction with either serine/threonine mutants (B) or lysine mutants (D) is shown. Similar experiments were carried out in HEK-293 cells with AAV2-WT or AAV2 S/T/K mutant vectors at an MOI of 2×103 VG/cell (E). Quantitative analysis of these data by flow cytometric analysis is shown in (F). The data depicted in (A), (C), and (E) are representative histograms whereas the data in (B), (D), and (F) are means of triplicate analyses. One-way analysis of variance (ANOVA) was used for statistical analysis.*p<0.05, **p<0.01 versus AAV2-WT-infected cells. Color images available online at www.liebertpub.com/hgtb
We then assessed the transduction potential of the nine single-mutant and two double-mutant AAV2 K→R vectors in HeLa cells at an MOI of 2000. The K532R and K544R single mutants and one double mutant (K490+532R) showed significantly higher transduction compared with the AAV2-WT vector (82–70 vs. 30%) by flow cytometric analysis (Fig. 4C and D). To further rule out the possibility that this increase in transgene expression was cell line specific, the best-performing AAV2 mutant vectors—T251A, S276A, S489A, S498A, and K532R—were further tested in HEK-293 cells, which showed a similar increase in EGFP expression by FACS analysis (38–71 vs. 17%) (Fig. 4E). These data were further corroborated by fluorescence imaging of AAV vector-infected cells, which demonstrated higher EGFP expression with most of the AAV S/T/K mutant vectors (Fig. 5A and B).
FIG. 5.
Fluorescence imaging of HeLa cells infected with AAV2 wild-type or S/T/K mutant vectors. HeLa cells were either mock-infected or infected with AAV2-WT or AAV2 S/T/K mutant vectors at 2×103 VG/cell. Forty-eight hours later, the cells were analyzed by fluorescence microscopy. (A) Visual comparison of AAV2 S/T→A mutants compared with AAV2-WT vectors. (B) Visual comparison of AAV2 K→R mutants compared with AAV2-WT vectors. Color images available online at www.liebertpub.com/hgtb
AAV2 S/T/K mutant vectors improve hepatic gene transfer in C57BL/6 mice in vivo
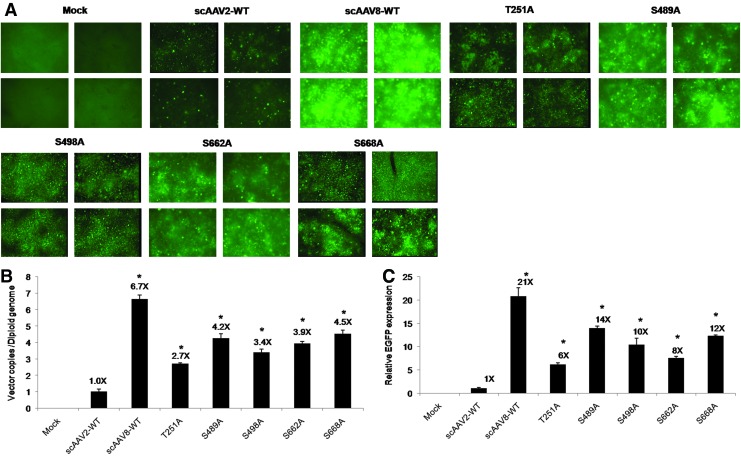
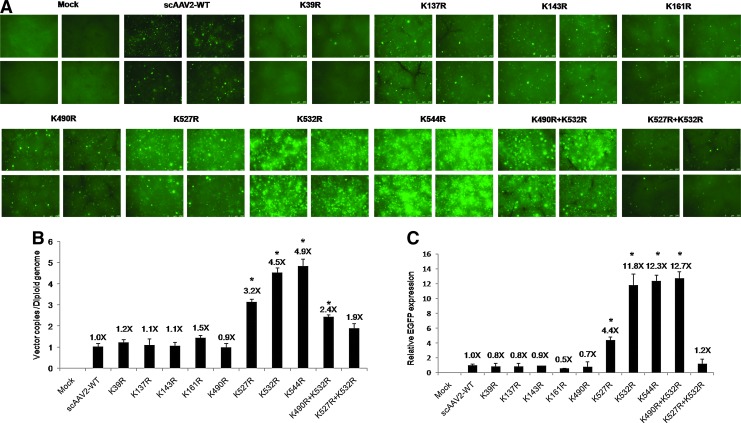
To analyze the liver-directed gene expression of the various AAV2 mutant vectors, we next examined their potential efficacy in vivo. AAV2 S/T/K mutant vectors that package as efficiently as the AAV2-WT vector and those that showed enhanced transgene expression in vitro were administered at a dose of 5×1010 VG/animal. Consistent with our in vitro studies, liver tissues of mice administered the four S→A mutants (S489A, S498A, S662A, and S668A) and the T251A mutant showed higher levels of EGFP reporter when compared with animals injected with AAV2-WT vector and analyzed by fluorescence microscopy (Fig. 6A). A similar increase in EGFP levels was noted after hepatic gene transfer with the AAV2 lysine mutants K532R, K544R, and K490R+K532R (Fig. 7A). To confirm this phenomenon, we then measured AAV vector genome copy numbers in the liver tissue of vector- or mock-injected mice. As shown in Figs. 6B and 7B, a significant increase in vector copies per diploid genome (up to 4.9-fold) was observed in animals injected with S/T/K mutant vectors in comparison with animals that received the AAV2-WT vector alone. To further corroborate these data, we then measured the transcript levels of EGFP in hepatic RNA isolated from these mice. Our studies demonstrate higher levels of transgene transcript expression (up to 14-fold) after hepatic gene transfer, in AAV2 S/T/K mutant-administered mice in comparison with AAV2-WT vector-injected animals (Figs. 6C and 7C). In all these studies, AAV8-injected animals were used as a control group for hepatic gene transfer. Taken together, our data clearly suggest that select S/T→A and K→R mutations can augment the transduction efficiency of AAV2 vectors in vivo.
FIG. 6.
AAV2 serine/threonine mutant vectors exhibit enhanced transduction on hepatic gene transfer in vivo. (A) Transgene expression was detected by fluorescence microscopy 4 weeks post-injection of scAAV2-EGFP, scAAV8-EGFP, or AAV2 mutant S/T vectors at 5×1010 vector particles per animal. Representative images of hepatic tissues from four different animals in each group are shown. (B) Estimation of vector genome copies in liver after AAV-mediated gene transfer. Genomic DNA was isolated from the liver tissue of C57BL/6 mice 4 weeks after vector administration and viral copy numbers were estimated by quantitative PCR as described in Materials and Methods. (C) Analysis of EGFP transcript levels by real-time quantitative PCR. Hepatic RNA isolated from animals injected with AAV2-WT, AAV8-WT, or AAV2 S/T vector was analyzed for EGFP expression; the data are normalized to the GAPDH reference gene. One-way analysis of variance (ANOVA) was used for the statistical comparisons. *p<0.05 versus AAV2-WT-injected animals. Color images available online at www.liebertpub.com/hgtb
FIG. 7.
Analysis of AAV2 lysine mutant vector-mediated EGFP expression in hepatocytes of normal C57BL/6 mice in comparison with wild-type AAV2 vector-mediated EGFP expression. (A) Transgene expression was detected by fluorescence microscopy 4 weeks post-injection of scAAV2-EGFP or AAV2 K→R mutant vector at 5×1010 vector particles per animal. Representative images of hepatic tissues from four different animals in each group are shown. (B) Estimation of vector genome copies in liver after AAV-mediated gene transfer. Genomic DNA was isolated from the liver tissue of C57BL/6 mice 4 weeks after vector administration and the viral copy numbers were estimated by quantitative PCR as described in Materials and Methods. (C) Analysis of EGFP transcript levels by real-time quantitative PCR. Hepatic RNA isolated from animals injected with AAV2-WT or K→R mutant vector was analyzed for EGFP expression; the data are normalized to the GAPDH reference gene. One-way analysis of variance (ANOVA) was used for the statistical comparisons. *p<0.05 versus AAV2-WT-injected animals. Color images available online at www.liebertpub.com/hgtb
AAV2 S489A mutant vector demonstrates significantly lower neutralizing antibody formation in vivo
Serially diluted serum samples from animals injected with AAV2-WT or with AAV2 S489A, S525A, S537A, S547A, or S662A vector were assayed for neutralizing antibody formation against these vectors (Table 3). The S489A vector-injected group had an 8-fold lower neutralization antibody titer compared with animals injected with AAV2-WT vector. These results imply that the S→A mutation at amino acid position 489 in AAV capsid generated fewer antibodies that could be cross-neutralized by AAV2-WT vectors. Interestingly, the S489A vector also demonstrated 14-fold higher EGFP transcript levels over AAV2-WT vectors in transduced liver (Fig. 6C).
Table 3.
Neutralizing Antibody Titers: AAV2 S→A Vectors Compared with AAV2-WTa
| Serum no. | Group | Reciprocal NAb Titer |
|---|---|---|
| 1 | scAAV2-WT | 5,120 |
| 2 | S489A | 640 |
| 3 | S525A | 5,120 |
| 4 | S537A | 5,120 |
| 5 | S547A | 5,120 |
| 6 | S662A | 5,120 |
| 7 | Anti-AAV2 rabbit control serum | 81,920 |
AAV2 S489A vector demonstrates lower neutralization antibody titers compared with the WT-AAV2 vector. Pooled serum samples from WT-AAV2- or AAV2 mutant-injected mice (n=4 per group) were analyzed for neutralizing antibodies 4 weeks after vector administration. Values are the reciprocal of the serum dilution at which relative luminescence units (RLUs) were reduced 50% compared with virus control wells (no test samples).
Targeted mutagenesis of lysine residue on AAV2 reduces ubiquitination of AAV vectors
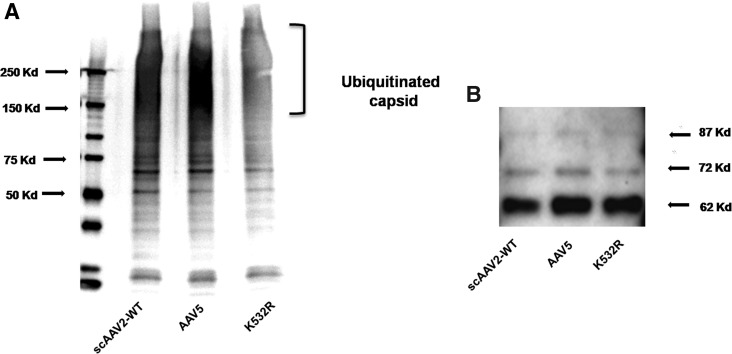
To understand whether the improved transduction achieved with the lysine mutant vectors is due to decreased ubiquitination of viral capsid, we performed an in vitro ubiquitination assay followed by Western blotting to detect the levels of mono- and polyubiquitin moieties in the AAV2 capsid. As can be seen in Fig. 8, the AAV2 K532R mutant vector demonstrated significantly reduced ubiquitination compared with either the AAV2-WT or AAV5-WT vector. Interestingly, AAV5 capsid had higher ubiquitination than did AAV2-WT capsid, a phenomenon that has been reported previously (Yan et al., 2002). These data provide direct evidence that the superior transduction achieved with the AAV2 K532R mutant vector is due to reduced ubiquitination of the viral capsid, which possibly results in rapid intracellular trafficking of the virus and improved gene expression, as has been suggested previously for the AAV2 tyrosine mutant vectors (Zhong et al., 2008a).
FIG. 8.
AAV2 lysine mutant K532R demonstrates reduced ubiquitination compared with the AAV2-WT and AAV5-WT vectors. (A) Approximately 3×108 viral particles of AAV2-WT, AAV5-WT, and AAV2 K532R vectors were denatured at 95°C for 5 min. The denatured viral particles were then used to perform the ubiquitin conjugation assay according to the manufacturer's protocol. The processed samples were electrophoresed on a 5–20% denaturing polyacrylamide gel and the ubiquitination pattern was detected by immunoblotting with an anti-ubiquitin antibody. The mono- to-polyubiquitin conjugates were detected as a smear at molecular mass >150 kDa. (B) Capsid VP1, VP2, and VP3 proteins were used as loading control.
AAV2 S/T/K mutant vectors do not cause any adverse event in C57BL/6 mice
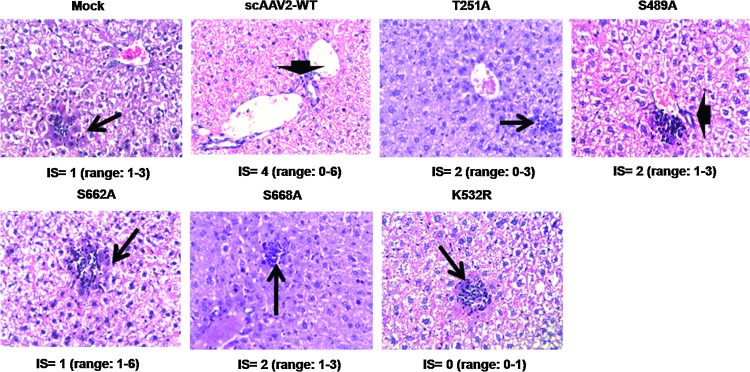
The in vivo administration of AAV2 S/T/K mutant vectors did not lead to any significant histological abnormalities in the livers of C57BL/6 mice 4 weeks after vector administration. Livers of mice injected with either AAV2-WT or AAV2 S/T/K mutant vectors were grossly normal with comparable inflammation scores. A set of representative data, shown in Fig. 9, corroborate that AAV2 S/T/K mutant vectors were generally nontoxic and that no adverse events were evident at the 4-week post-injection time point.
FIG. 9.
Histological examination of C57BL/6 liver samples 4 weeks postinjection of AAV2-WT or mutant vector. Hepatic sections were fixed in 10% buffered formalin and stained with hematoxylin–eosin. The median inflammation score (IS) for each group is indicated below the images (original magnification, ×40) with the range of values given within parentheses. Arrowheads and arrows denote portal and focal lobular inflammation, respectively. A representative image of one animal liver from each group (n=3) is shown. Color images available online at www.liebertpub.com/hgtb
Discussion
The collective experience from various AAV2-mediated clinical trials suggests that strategies to improve the transduction efficiencies of these vectors are needed to circumvent the dose-dependent immune response directed against them and to achieve successful long-term gene transfer (Jiang et al., 2006; Jayandharan et al., 2008). Consequently, there has been tremendous interest in evaluating other naturally occurring isolates of AAV (AAV1 through AAV12) or bioengineered AAV strains (Choi et al., 2005; Zincarelli et al., 2008) for gene transfer, each validated for their own desirable properties such as tissue tropism or other clinically relevant challenges. Despite this, AAV2 remains the predominant serotype vector currently in use in human gene therapy applications (High, 2011) as it is the best characterized in terms of vector toxicology. However, its optimal use is contingent on a thorough understanding of the fundamental steps in virus–host cell interactions, which include viral binding and entry (Summerford and Samulski, 1998), intracellular trafficking (Duan et al., 1999), nuclear transport, uncoating (Shi et al., 2006), and viral second-strand DNA synthesis. As previously noted, viral intracellular trafficking is an important rate-limiting step that directly influences the efficiency of transgene expression (Sanlioglu et al., 2001). Because it is known that this process is regulated largely by host cellular phosphorylation of the viral capsid, strategies aimed at reversing this block by concurrent administration of pharmacological inhibitors may possibly work, as demonstrated in our present and previous studies (Monahan et al., 2010). However, their applicability in human gene therapy is likely to be limited because of toxicity concerns (Ding et al., 2006). Alternatively, to scale up this approach for possible use in liver-directed human gene therapy, modification of specific phosphorylation targets is likely to be a viable strategy.
The concept of mutagenesis of the AAV capsid sequence has been previously employed to generate novel AAV vectors either by targeted evolution or by targeted design. Directed evolution of AAV vectors to generate chimeric AAVs with enhanced gene transfer to the airway epithelia, CNS tissue, or retina has been reported. Similarly, rationally designed AAV strains with robust abilities to transfer genes into muscle (AAV2.5) or with enhanced immunogenic profiles to act as vaccine candidates (Lin et al., 2009) are also available. Mutagenesis of the surface-exposed tyrosines (Y) to phenylalanine (F) has been shown to substantially improve gene expression by up to severalfold in a variety of tissues such as the liver, retina, and musculoskeletal targets (Zhong et al., 2008b; Petrs-Silva et al., 2009, 2011). However, the transduction efficiency of these tyrosine mutants varies according to the target cell type. For example, the AAV2 Y730F mutant shows enhanced gene transfer into hepatocytes but when directed to stem cells or the retina, its efficiency is modest (Kauss et al., 2010; Ryals et al., 2011). This may possibly be due to varying levels of tyrosine kinase activity in these tissues or to the differential accessibility of the cellular tyrosine kinase to the tyrosines on the AAV2 capsid, the surface exposure of which may be determined in part by specific receptor and coreceptor binding as well as by the varied endosomal processing in these tissues (Qing et al., 1999; Kaludov et al., 2001; Kashiwakura et al., 2005; Seiler et al., 2006). However, we reasoned that apart from targeting tyrosine kinase targets on the AAV capsid, modifying other kinase targets such as S/T residues or ubiquitination targets such as K residues on the AAV2 capsid is likely to further improve its gene delivery. It is important to note that phosphorylation of the viral capsid serves as a trigger for uncoating and release of viral nuclear material inside the host cell. Hence, phosphorylation sites must be mutated more strategically and cannot be replaced at random. Keeping this in mind, a thorough analysis of viral capsid structure was carried out. Three phosphodegron sequences were identified in the AAV2 capsid. The phosphorylation sites within the phosphodegrons were thought to be effective and safer targets to mutate as they are the ones used by the host as a signal for clearance of the virus. These residues are thus expected to have minimal influence on the capsid-uncoating processes, essential for the virus inside the host cell. Also, to preserve capsid geometry, only those residues that lie outside the interaction interfaces in the phosphodegron were selected for mutagenesis.
Our hypothesis was further supported by our preliminary studies, in which specific inhibition of CKII serine/threonine kinase increased the transduction profile of AAV2-WT vectors. Subsequently, 24 single S/T/K residues in and around phosphodegrons were selected as targets for site-directed mutagenesis, and our data show that selective modification of these targets on the AAV2 capsid substantially improved gene expression from AAV2 vectors both in vitro (up to 97%) and in vivo (up to 14-fold). The enhanced transduction seen with the S→A mutants in our study is similar to that with S→V (valine) mutations, which have been shown to be efficacious in gene delivery into dendritic cells in vitro. (Aslanidi et al., 2012). As highlighted in Table 2 and Fig. 2, residues S489 and S498 are located in phosphodegron 3, residues S662 and S668 are in/near phosphodegron 2, and residue K532 is part of phosphodegron 1. The effect of these mutations thus corroborates our selection process for the mutagenesis targets. Further ongoing studies with the optimal S/T/K-mutant AAV2 vectors expressing human coagulation factor IX in preclinical models of hemophilia B will demonstrate the feasibility of the use of these novel vectors for potential gene therapy of hemophilia B.
Interestingly, previous mutations at the K532 residue have shown disparate effects on vector infectivity and heparin binding. Opie and colleagues (2003) demonstrated that substitution of K532/K527 with alanine had a modest effect on heparin binding but that the mutant was ∼5 logs less infectious than AAV2-WT. Kern and colleagues (2003) have shown that the K532A mutant had similar infectivity but reduced heparin binding. In the present study, the packaging titer of the K532R mutant was 10 times higher and ∼6-fold higher infectivity was seen when compared with the AAV2-WT vector (Kern et al., 2003). Taken together, these data suggest that AAV2 K532 might not be as important as other basic residues (R585 and R588) for effective heparin binding (Opie et al., 2003). This can be further substantiated by the fact that both AAV1 (which binds poorly to heparin) and AAV3 (which binds to heparin effectively) have conserved K532. However, it is possible that our choice to replace the lysine amino acid with a structurally compatible arginine instead of alanine perhaps contributed to the observed increase in packaging titers and also its infectivity by minimizing the charge switch on the AAV2 capsid surface. It has been demonstrated that AAV2 capsid mutants generated with various amino acid substitutions can have varied transduction efficiencies (Aslanidi et al., 2012). Hence, the choice of amino acid for mutagenesis has a significant effect on AAV2 vector packaging and transduction efficiency.
The availability of superior AAV2 S/T/K mutant vectors presents several possibilities. First, about ∼30% of the S/T/K residues that we mutated are conserved in AAV serotypes 1–10. It is therefore tempting to speculate that S/T/K mutations on other AAV serotypes (1–12) are likely to increase the transduction capabilities of these vectors as well. Second, multiple combinations of these AAV S/T/K mutants are also possible and this is likely to further minimize the overall phosphorylation and ubiquitinated amino acid content of the AAV capsid. Further ongoing studies on the above-mentioned strategies are likely to offer a vast repertoire of these S/T/K mutants and a tool kit of superior AAV vectors.
Supplementary Material
Acknowledgments
The authors thank Dr. R. Sumathy and Mr. Y. Sathish (Laboratory Animal Core Facility, Centre for Stem Cell Research, Vellore) for animal care. G.R.J. is supported by research grants from the Department of Science and Technology, Government of India (Swarnajayanti Fellowship 2011); the Department of Biotechnology (DBT), Government of India (Innovative Young Biotechnologist award 2010: BT/03/IYBA/2010; grant BT/PR14748/MED/12/491/2010; grant BT/01/COE/08/03); and an early career investigator award (2010) from the Bayer Hemophilia Awards program (Bayer). R.A.G. is supported by a grant under the Women Scientists Programme from the Department of Science and Technology (New Delhi, India). G.S. is supported by a Ph.D. student fellowship from the DBT (New Delhi, India). N.S. acknowledges the support of the DBT, Government of India.
Author Disclosure Statement
No competing financial interests exist.
References
- Aslanidi G.V. Rivers A.E. Ortiz L., et al. High-efficiency transduction of human monocyte-derived dendritic cells by capsid-modified recombinant AAV2 vectors. Vaccine. 2012;30:3908–3917. doi: 10.1016/j.vaccine.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurnhammer C. Haase M. Muether N., et al. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2011;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- Becerra S.P. Rose J.A. Hardy M., et al. Direct mapping of adeno-associated virus capsid proteins B and C: A possible ACG initiation codon. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7919–7923. doi: 10.1073/pnas.82.23.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M. Westbrook J. Feng Z., et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buning H. Perabo L. Coutelle O., et al. Recent developments in adeno-associated virus vector technology. J. Gene Med. 2008;10:717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- Calcedo R. Vandenberghe L.H. Gao G., et al. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R. Sugawara H. Koike T., et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi V.W. McCarty D.M. Samulski R.J. AAV hybrid serotypes: Improved vectors for gene delivery. Curr. Gene Ther. 2005;5:299–310. doi: 10.2174/1566523054064968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S. Krishnadev O. Srinivasan N. Rekha N. Interaction preferences across protein–protein interfaces of obligatory and non-obligatory components are different. BMC Struct. Biol. 2005;5:15. doi: 10.1186/1472-6807-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. The PyMOL molecular graphics system. 2002. http://www.pymol.org/ http://www.pymol.org/
- Ding Q. Dimayuga E. Markesbery W.R. Keller J.N. Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 2006;20:1055–1063. doi: 10.1096/fj.05-5495com. [DOI] [PubMed] [Google Scholar]
- Duan D. Sharma P. Dudus L., et al. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 1999;73:161–169. doi: 10.1128/jvi.73.1.161-169.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force T. Kolaja K.L. Cardiotoxicity of kinase inhibitors: The prediction and translation of preclinical models to clinical outcomes. Nat. Rev. Drug Discov. 2011;10:111–126. doi: 10.1038/nrd3252. [DOI] [PubMed] [Google Scholar]
- Girod A. Wobus C.E. Zadori Z., et al. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. Matsumoto M. Nakayama K.I. Mapping of ubiquitination sites on target proteins. Methods Enzymol. 2005;399:277–286. doi: 10.1016/S0076-6879(05)99019-8. [DOI] [PubMed] [Google Scholar]
- High K.A. Gene therapy for haemophilia: A long and winding road. J. Thromb. Haemost. 2011;9(Suppl. 1):2–11. doi: 10.1111/j.1538-7836.2011.04369.x. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J. Thornton J.M. Department of Biochemistry and Molecular Biology, University College London; London, UK: 1993. NACCESS. [Google Scholar]
- Inobe T. Fishbain S. Prakash S. Matouschek A. Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayandharan G.R. Zhong L. Li B., et al. Strategies for improving the transduction efficiency of single-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2008;15:1287–1293. doi: 10.1038/gt.2008.89. [DOI] [PubMed] [Google Scholar]
- Jiang H. Lillicrap D. Patarroyo-White S., et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood. 2006;108:107–115. doi: 10.1182/blood-2005-12-5115. [DOI] [PubMed] [Google Scholar]
- Kaludov N. Brown K.E. Walters R.W., et al. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwakura Y. Tamayose K. Iwabuchi K., et al. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J. Virol. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss M.A. Smith L.J. Zhong L., et al. Enhanced long-term transduction and multilineage engraftment of human hematopoietic stem cells transduced with tyrosine-modified recombinant adeno-associated virus serotype 2. Hum. Gene Ther. 2010;21:1129–1136. doi: 10.1089/hum.2010.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A. Schmidt K. Leder C., et al. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber J.T. Jang J.H. Schaffer D.V. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube D.M. Srivastava A. Quantitative DNA slot blot analysis: Inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997;25:3375–3376. doi: 10.1093/nar/25.16.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Jayandharan G.R. Li B., et al. High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum. Gene Ther. 2010;21:1527–1543. doi: 10.1089/hum.2010.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Calcedo R. Vandenberghe L.H., et al. A new genetic vaccine platform based on an adeno-associated virus isolated from a rhesus macaque. J. Virol. 2009;83:12738–12750. doi: 10.1128/JVI.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C. Lu Y. Cheng B., et al. High-efficiency transduction of liver cancer cells by recombinant adeno-associated virus serotype 3 vectors. J. Vis. Exp. 2011;49:2538. doi: 10.3791/2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie M.A. Tatsuno G.P. Christie B., et al. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno C.S. Pierce G.F. Arruda V.R., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- Mingozzi F. High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Monahan P.E. Lothrop C.D. Sun J., et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: A strategy for broad clinical application. Mol. Ther. 2010;18:1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C. Tuddenham E.G. Rangarajan S., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M. Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther. 2012;19:649–658. doi: 10.1038/gt.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opie S.R. Warrington K.H., Jr. Agbandje-McKenna M., et al. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H. Dinculescu A. Li Q., et al. High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol. Ther. 2009;17:463–471. doi: 10.1038/mt.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H. Dinculescu A. Li Q., et al. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011;19:293–301. doi: 10.1038/mt.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing K. Mah C. Hansen J., et al. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- Rajkumar S.V. Richardson P.G. Hideshima T. Anderson K.C. Proteasome inhibition as a novel therapeutic target in human cancer. J. Clin. Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Ryals R.C. Boye S.L. Dinculescu A., et al. Quantifying transduction efficiencies of unmodified and tyrosine capsid mutant AAV vectors in vitro using two ocular cell lines. Mol. Vis. 2011;17:1090–1102. [PMC free article] [PubMed] [Google Scholar]
- Sanlioglu S. Monick M.M. Luleci G., et al. Rate limiting steps of AAV transduction and implications for human gene therapy. Curr. Gene Ther. 2001;1:137–147. doi: 10.2174/1566523013348788. [DOI] [PubMed] [Google Scholar]
- Seiler M.P. Miller A.D. Zabner J. Halbert C.L. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum. Gene Ther. 2006;17:10–19. doi: 10.1089/hum.2006.17.10. [DOI] [PubMed] [Google Scholar]
- Shi X. Fang G. Shi W. Bartlett J.S. Insertional mutagenesis at positions 520 and 584 of adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors with eliminated heparin-binding ability and introduced novel tropism. Hum. Gene Ther. 2006;17:353–361. doi: 10.1089/hum.2006.17.353. [DOI] [PubMed] [Google Scholar]
- Simonelli F. Maguire A.M. Testa F., et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 2010;18:643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroes E.S. Nierman M.C. Meulenberg J.J., et al. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler. Thromb. Vasc. Biol. 2008;28:2303–2304. doi: 10.1161/ATVBAHA.108.175620. [DOI] [PubMed] [Google Scholar]
- Summerford C. Samulski R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J.P. Carter B.J. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 1988;62:3356–3363. doi: 10.1128/jvi.62.9.3356-3363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Guan S. Acharya P., et al. Multisite phosphorylation of human liver cytochrome P450 3A4 enhances Its gp78-, CHIP-mediated ubiquitination: A pivotal role of its Ser-478 residue in the gp78-catalyzed reaction. Mol. Cell. Proteomics. 11:M111.010132. doi: 10.1074/mcp.M111.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. Xu G. Schulman B.A., et al. Structure of a β-TrCP1-Skp1-β-catenin complex: Destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Xie Q. Bu W. Bhatia S., et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Zak R. Luxton G.W., et al. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. Zhong L. Wu J., et al. Role of cellular FKBP52 protein in intracellular trafficking of recombinant adeno-associated virus 2 vectors. Virology. 2006;353:283–293. doi: 10.1016/j.virol.2006.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L. Li B. Jayandharan G., et al. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008a;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L. Li B. Mah C.S., et al. Next generation of adeno-associated virus 2 vectors: Point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. U.S.A. 2008b;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C. Soltys S. Rengo G. Rabinowitz J.E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S. Byrne B.J. Mason E., et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.