Abstract
Recombinant adeno-associated virus vectors based on serotype 8 (AAV8) have shown significant promise for liver-directed gene therapy. However, to overcome the vector dose dependent immunotoxicity seen with AAV8 vectors, it is important to develop better AAV8 vectors that provide enhanced gene expression at significantly low vector doses. Since it is known that AAV vectors during intracellular trafficking are targeted for destruction in the cytoplasm by the host–cellular kinase/ubiquitination/proteasomal machinery, we modified specific serine/threonine kinase or ubiquitination targets on the AAV8 capsid to augment its transduction efficiency. Point mutations at specific serine (S)/threonine (T)/lysine (K) residues were introduced in the AAV8 capsid at the positions equivalent to that of the effective AAV2 mutants, generated successfully earlier. Extensive structure analysis was carried out subsequently to evaluate the structural equivalence between the two serotypes. scAAV8 vectors with the wild-type (WT) and each one of the S/T→Alanine (A) or K-Arginine (R) mutant capsids were evaluated for their liver transduction efficiency in C57BL/6 mice in vivo. Two of the AAV8-S→A mutants (S279A and S671A), and a K137R mutant vector, demonstrated significantly higher enhanced green fluorescent protein (EGFP) transcript levels (∼9- to 46-fold) in the liver compared to animals that received WT-AAV8 vectors alone. The best performing AAV8 mutant (K137R) vector also had significantly reduced ubiquitination of the viral capsid, reduced activation of markers of innate immune response, and a concomitant two-fold reduction in the levels of neutralizing antibody formation in comparison to WT-AAV8 vectors. Vector biodistribution studies revealed that the K137R mutant had a significantly higher and preferential transduction of the liver (106 vs. 7.7 vector copies/mouse diploid genome) when compared to WT-AAV8 vectors. To further study the utility of the K137R-AAV8 mutant in therapeutic gene transfer, we delivered human coagulation factor IX (h.FIX) under the control of liver-specific promoters (LP1 or hAAT) into C57BL/6 mice. The circulating levels of h.FIX:Ag were higher in all the K137R-AAV8 treated groups up to 8 weeks post-hepatic gene transfer. These studies demonstrate the feasibility of the use of this novel AAV8 vectors for potential gene therapy of hemophilia B.
Sen and colleagues generated AAV8 capsid point mutants by replacing specific serine/threonine kinase or ubiquitination target residues. Two of the mutants yielded significantly higher transgene expression over AAV8 when injected into mice, and the best performing vector also exhibited significantly reduced capsid ubiquitination, innate immune response activation, and neutralizing antibody formation.
Introduction
Recombinant adeno-associated virus (AAV) vectors have gained significant attention as a gene therapy vector due to their lack of pathogenicity and their ability to infect different tissues (Flotte and Carter, 1995; Choi et al., 2005; Wu et al., 2006; Mueller and Flotte, 2008). So far, 12 serotypes of AAV (AAV1 to AAV12) vectors have been studied extensively as gene therapy vectors (Schultz and Chamberlain, 2008). These AAV serotypes share a common genome, but their unique capsid structure helps them recognize different cell-surface receptors (Grimm and Kay, 2003). AAV serotype 2 is the most extensively studied but has demonstrated only limited preclinical success (Snyder et al., 1997, 1999; Wang et al., 1999; Mount et al., 2002; Nathwani et al., 2002). The capsid antigens of AAV2 are known to elicit a cytotoxic T-cell response as evidenced during liver-directed gene therapy in patients with hemophilia B (Nakai et al., 2002; Manno et al., 2006). To overcome such limitations imposed by AAV2 vectors, alternate AAV serotypes are being rigorously evaluated for hepatic gene transfer (Nakai et al., 2002). AAV serotype 8, in particular, has consistently demonstrated remarkable transduction efficiency (10- to 100-fold) and lower immunogenicity in comparison to AAV2 vectors (Gao et al., 2002; Davidoff et al., 2005; Wang et al., 2005; Nathwani et al., 2006, 2007; Vandenberghe et al., 2006). The rapid uncoating of the AAV8 vector post-internalization overcomes many of the limitations of AAV2 (Thomas et al., 2004). Importantly, in the context of hepatic gene transfer, peripheral venous administration of AAV8 vectors is very effective, suggesting the high liver-targeting capacity of these vectors (Davidoff et al., 2005). In addition, the long-term safety and efficacy of AAV8 vectors during hepatic gene therapy has been established in not only murine but also canine and primate models (Davidoff et al., 2005; Wang et al., 2005; Nathwani et al., 2006, 2007). These studies formed the basis for the successful AAV8-mediated gene transfer in patients with hemophilia B (Nathwani et al., 2011). In this trial, two patients who received the highest dose (2×1012 vg/kg) of the vector developed capsid-specific T cells that required glucocorticoid therapy to attenuate this response. Therefore, irrespective of the serotype employed, the concept of vector dose-dependent immune response persists. It is thus important to develop novel AAV8 vectors that provide enhanced gene expression at significantly less vector doses to achieve successful gene transfer in humans.
A major barrier that negatively affects AAV-mediated gene expression is the degradation of the viral particles during their intracellular trafficking via the ubiquitination-proteasomal degradation machinery (Zhong et al., 2008). As a measure to evade phosphorylation and subsequent ubiquitination leading to vector loss, next generation of AAV2 vectors were designed with point mutations on surface exposed tyrosine residues on the capsid protein creating vectors that had higher transduction in both in vitro and in vivo settings (Zhong et al., 2008). The superimposition of similar tyrosine→phenylalanine mutations in the AAV8 capsid did not alter the gene expression from these vectors in muscle and heart tissue (Qiao et al., 2012). However, like in the case of AAV2, pretreatment of cells with a proteasomal inhibitor (MG132) with AAV8 has been shown to increase their nuclear translocation and their gene expression (Liu et al., 2012), although its systemic administration is likely to have severe side effects (Rajkumar et al., 2005). These data suggest that amino acids other than tyrosines may be recognized on the AAV8 capsid by the host–cellular kinase machinery. Our recent studies in AAV2 vectors have implicated serine (S) and threonine (T) or lysine (K) amino acids as potential cellular kinase or ubiquitination targets, which, when substituted with compatible amino acids, improved AAV2 transduction in vitro and in vivo (Gabriel et al., 2013). Based on these data, the present study was designed to test the efficacy of AAV8 vectors, which are modified at critical S/T/K residues on the vector capsid.
Material and Methods
Animals
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved and carried out according to the Institutional Guidelines for Animal Care (Christian Medical College, Vellore, India).
Site-directed mutagenesis
S→A, T→A, and K→R mutations were introduced on the AAV8 rep/cap plasmid by QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, Agilent Technologies, La Jolla, CA) following the manufacturer's protocol (Table 1). The sites on the AAV8 capsid were selected for site-directed mutagenesis on the basis of their equivalence to the mutated residues of effective AAV2 mutants generated earlier (Gabriel et al., 2013). Mutations were carried out at S279A, S501A, S671A, T252A, and K137R. The presence of the desired point mutation was verified by restriction enzyme analysis and DNA sequencing (Applied Biosystems 3130 Genetic Analyzer; Life Technologies, Warrington, United Kingdom). The amino acids on AAV8 capsid are numbered according to the National Center for Biotechnology Information (NCBI) database (accession ID: NC_006261.1).
Table 1.
Nucleotide Sequence of the Mutation Primers Used for Site-Directed Mutagenesis in Adeno-Associated Virus Serotype 8 Capsid, and the Change in Nucleotide and Restriction Patterns of Silent Mutation Obtained with the Primers
| Residue | Sequence (5’ to 3’) | Nucleotide change | Change in restriction enzyme due tosilent mutation |
|---|---|---|---|
| S279A | Wild-type primer sequence: | AGC→GCC | SfcI disappears |
| CTACTTCGGCTACAGCACCCCCTGGGGG | |||
| Mutant Primer Sequence: | |||
| CTACTTCGGCTACGCCACCCCCTGGGGG | |||
| S501A | Wild-type primer sequence: | AGC→GCC | G→T AgeI appears |
| GACAACCGGGCAAAACAACAATAGCAACTTTGCCTGGACT | |||
| Mutant primer sequence: | |||
| GACAACCGGTCAAAACAACAATGCCAACTTTGCCTGGACT | |||
| S671A | Wild-type primer sequence: | TCT→GCT | T→A AluI disappears |
| CCTTCAACCAGTCAAAGCTGAACTCTTTCATCACGCAATACA | |||
| Mutant primer sequence: | |||
| CCTTCAACCAGTCAAAGCAGAACGCTTTCATCACGCAATACA | |||
| T252A | Wild-type primer sequence: | ACC→GCC | C→G NaeI appears |
| ACCCGAACCTGGGCCCTGCCCACCTACAACAACCACCTCTAC | |||
| Mutant primer sequence: | |||
| ACCCGAACCTGGGCCCTGCCGGCCTACAACAACCACCTCTAC | |||
| K137R | Wild-type primer sequence: GGTTGAGGAAGGCGCTAAGACGGCTCCTGG | AAG→AGG | C→G HhaI disappears |
| Mutant primer sequence: | |||
| GGTTGAGGAAGGGGCTAGGACGGCTCCTGG |
In silico analysis of AAV8 mutants: Prediction of phospho and ubiquitination sites
Sites of phosphorylation and ubiquitination on the AAV8 capsid were predicted using online prediction tools, namely NetphosK (www.cbs.dtu.dk/services/NetPhosK/), Phosida (www.phosida.com/), Kinasephos (http://kinasephos.mbc.nctu.edu.tw/), and Scansite (http://scansite.mit.edu/) for phosphosites, and UbPred (www.ubpred.org), Composition of K Spaced Amino Acid Pairs (CKSAAP_ubsite; http://protein.cau.edu.cn/cksaap_ubsite/), and Prediction of Ubiquitination site with Bayesian Discriminant Method (BDM_PUB; http://bdmpub.biocuckoo.org/index.php) for ubiquitination site prediction.
Analysis of three-dimensional structure
The available three-dimensional (3-D) structure of the AAV8 capsid (PDB code:2qa0) (Nam et al., 2007) was analyzed extensively to determine interaction interfaces of capsid protein chains. Accessibility-based method was employed to determine the residues participating in the protein–protein interactions between capsid proteins. Briefly, solvent accessibility of every residue was computed using NACCESS tool (Hubbard, 1993), and the residues were grouped as solvent-exposed if the solvent accessibility values were more than 7%, while those with lesser accessibility were called buried residues. The residues were called interface residues if they were buried (accessibility <7%) in protein complex while being solvent-exposed (accessibility >10%) in isolated chains. The structure of the viral capsid was visualized using PYMOL software (DeLlano, 2002) and was compared to the structure of AAV2 capsid using DALIlite tool (Holm and Park, 2000) for structure-based comparison.
Prediction of antigenicity
Antigenicity of the capsid proteins was predicted using online tools, namely BCPREDS (Chen et al., 2007), ABCpred (Saha and Raghava, 2006), and BepiPred (Larsen et al., 2006), which employ machine-learning algorithms.
Generation of recombinant vectors
Highly purified stocks of self-complementary wild-type (WT) AAV8, or the mutant AAV8 vectors encoding the enhanced green fluorescent protein (EGFP) gene driven by the chicken β-actin promoter containing the cytomegalovirus (CMV) enhancer and SV40 poly A signal or the human coagulation factor IX (h.FIX) under the control of liver-specific promoters, human alpha-1-antitrypsin (hAAT) (Manno et al., 2006), or LP1 promoter (consisting of core liver-specific elements from human apolipoprotein hepatic control region) (Nathwani et al., 2011) were generated by polyethyleneimine-based triple transfection of AAV-293 cells (Stratagene). Briefly, forty numbers of 150-mm2 dishes 80% confluent with AAV 293 cells were transfected with AAV8 rep-cap, transgene-containing and AAV-helper free (p.helper) plasmids. Cells were collected 72 hr post-transfection, lysed, and treated with 25 units/ml of benzonase nuclease (Sigma Aldrich, St Louis, MO). Subsequently, the vectors were purified by iodixanol gradient ultracentrifugation (Optiprep, Sigma Aldrich) (Zolotukhin et al., 1999) followed by column chromatography (HiTrap Q column, GE Healthcare, Pittsburgh, PA). The vectors were finally concentrated to a final volume of 0.5 ml in phosphate buffered saline (PBS) using Amicon Ultra 10K centrifugal filters (Millipore, Bedford, MA). The physical particle titers of the vectors were quantified by slot blot analysis and expressed as viral genomes (vgs)/ml (Kube and Srivastava, 1997). The p.AAV-CB-EGFP, p.AAV-LP1-h.FIX, and p.AAV-hAAT-h.FIX constructs were kind gifts from Dr. Arun Srivastava, University of Florida, Gainesville; Dr. Amit Nathwani, University College London; and Dr. Katherine High, Children's Hospital of Philadelphia, Philadelphia, PA, respectively.
Recombinant AAV8 vector transduction studies in vivo
Groups of 8- to 12-week-old C57BL/6 mice (n=4–8 per group) were mock-injected or injected with 5×1010 vgs each of WT-AAV8 or AAV8 mutant vectors, containing EGFP as the transgene, via the tail vein. Mice were euthanized 2 or 4 weeks after vector administration. Cross-sections from three hepatic lobes of the mock-injected and vector-injected groups were assessed for EGFP expression by a fluorescence microscope (Leica CTR6000; GmbH, Stuttgart, Germany). Images from five visual fields of mock-infected and vector-infected cells were analyzed by ImageJ analysis software (NIH, Bethesda, MD). Transgene expression (mean value) was assessed as total area of green fluorescence and expressed as mean pixels per visual field (mean±SD). The best-performing AAV8 capsid mutant, along with the WT-AAV8 vector containing h.FIX as the transgene (under LP1 and hAAT promoter), were administered into 8- to 12-week-old male C57BL/6 mice (n=3–4 per group) intravenously at different doses (2.5×1010 and 1×1011 vgs per mouse). Blood was collected retro-orbitally 2, 4, and 8 weeks post–vector administration. The antigenic activity of hF.IX (FIX:Ag) was measured using a commercial kit (Asserachrom, Diagnostica Stago, Asniers, France).
Biodistribution studies
Liver, spleen, lung, heart, kidney, and muscle tissue were collected from each of the mice administered with either WT-AAV8 or the mutant vectors, 2 or 4 weeks after gene transfer. Genomic DNA was isolated using the QIAamp® DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Quantitative polymerase chain reaction (PCR) was used to estimate the vector copy numbers in 100 ng of template genomic DNA by amplifying the viral inverted terminal repeats (ITRs) with specific probes/primers as described (Aurnhammer et al., 2012) using a low ROX qPCR mastermix according to manufacturer's protocol (Eurogentec, Seraing, Belgium). Data was captured and normalized to mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping control gene and analyzed in an ABI Prism 7500 Sequence Detection System Version 1.1 Software (Life Technologies, Applied Biosystems).
Real-time PCR assay
Groups (n=4 per group) of 8- to 12-week-old C57BL/6 mice were mock-injected or injected with 1×1011 vgs each of WT-AAV8 or K137R-AAV8 mutant vectors intravenously. Two hours later, mice were euthanized. Total RNA was isolated from liver sections of each mouse using the NucleoSpin® RNA isolation kit (Machery-Nagel, Düren, Germany). Approximately 2 μg of RNA was reverse transcribed using the first-strand RT kit (Qiagen, SABiosciences). The transcript levels of interleukin (IL) 1; IL6; tumor necrosis factor (TNF) α; Kupffer cells (KC); regulated on activation, normal T cell expressed and secreted (RANTES); IL12; toll-like receptor (TLR) 2; and TLR9 was measured using primers described earlier (Hussain et al., 2008; Jayandharan et al., 2011) with a PCR master mix (MESA GREEN Mastermix plus, Eurogentec).
To measure EGFP transcript levels, total RNA was isolated from the murine liver samples 2 or 4 weeks post–vector administration (5×1010 vgs per mouse) using TRIZOL® reagent (Sigma Aldrich). Approximately 1μg of RNA was reverse-transcribed using Verso™ Reverse Transcriptase according to the manufacturer's protocol (Thermo Scientific, Surrey, United Kingdom). TAQMAN® PCR was done using primers/probe against EGFP gene (Forward Primer: CTTCAAGATCCGCCACAACATC; Reverse Primer: ACCATGTGATCGCGCTTCTC; Probe: FAM-CGCCGACCACTACCAGCAGAACACC-TAMRA) and according to the manufacturer's protocol (Eurogentec). GAPDH was used as the housekeeping control gene. Data was captured and analyzed using the ABI Prism 7500 Sequence Detection (Life Technologies, Applied Biosystems).
Ubiquitin conjugation assay and immunoblotting
Ubiquitination assay of viral capsids were done using the Ubiquitin-Protein Conjugation kit according to the manufacturer's protocol (Boston Biochem, Cambridge, MA). Briefly, energy solution, conjugation fraction A, conjugation fraction B, and ubiquitin were mixed to a final reaction volume of 100 μL. The conjugation reaction was then initiated by adding 3×108 heat-denatured WT-AAV8, mutant AAV8, or WT-AAV5 viral particles and incubated at 37°C for 4 hr. Equal volumes of sodium dodecyl sulfate (SDS) denatured samples were then resolved on a 4–20% gradient gel. The ubiquitination pattern for the different viral particles was detected by immunoblotting of the samples with mouse anti-ubiquitin monoclonal antibody (P4D1) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG1 secondary antibody (Cell Signaling Technology, Boston, MA). To check for equal loading of the samples, 3×108 particles of WT-AAV8 and K137R-AAV8 vectors were heat denatured with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibiter cocktail (Cell Signaling Technology). The samples were then resolved on a 4–20% gradient gel. The VP1, VP2, and VP3 capsid proteins were detected with AAV clone B1 antibody (Fitzgerald, North Acton, MA) and HRP-conjugated anti-mouse IgG1 secondary antibody (Cell Signaling Technology).
Estimation of neutralizing antibodies against AAV8 vectors
Heat inactivated serum samples from WT-AAV8 or S→A and K→R mutant AAV8-injected animals were assayed for the neutralizing antibody (NAb) titers as described previously (Calcedo et al., 2009) by the Immunology Core at the University of Pennsylvania. The NAb titer is reported as the highest plasma dilution that inhibited AAV transduction of Huh7 cells by 50% or more compared with that of the naive serum control.
Results
Criteria for choosing the specific serine/threonine/lysine residues in the AAV8 capsid for mutagenesis
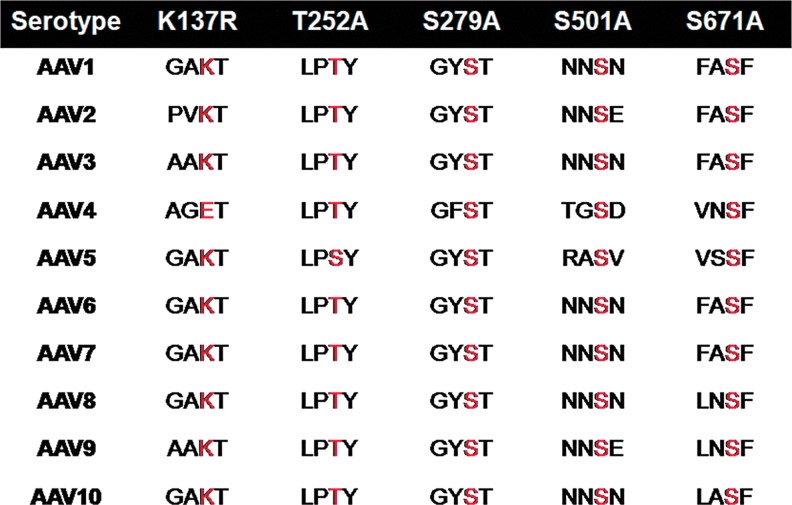
Our recent studies on bioengineering of AAV2 capsid at specific S/T/K sites have demonstrated an increase in the transduction efficiency of the mutant vectors both in vitro and in vivo (Gabriel et al., 2013). However, since the potential of AAV2 as a vector for hepatic gene transfer is limited due to higher prevalence of neutralizing antibodies against AAV2 than AAV8 vectors (43.5 vs. 22.6%) (Li et al., 2012), and the recent success demonstrated with AAV8 vectors for gene therapy of hemophilia B (Nathwani et al., 2011), we reasoned that modified AAV8 vectors will be a better alternative for hepatic gene therapy. Thus, in the present studies we selected specific AAV8 serine (S279, S501, and S671) and threonine (T252) residues whose mutagenesis on equivalent residues in AAV2 sequence showed the highest increase in transduction efficiency. Incidentally, the residues chosen for mutagenesis in AAV8 capsid are also conserved in at least 9 out of the 10 AAV capsid sequences analyzed (Fig. 1). The lysine K137 residue was chosen based on the highest probability of being ubiquitinated as predicted by the UbPred software (www.ubpred.org). Except for the T252A mutant, which had a 50-fold lower titer, all the other S/K mutant vectors had comparable packaging efficiency to WT-AAV8 vectors suggesting that S→A, K→R mutations were compatible modifications on the AAV8 capsid (Table 2).
FIG. 1.
Schematic representation of the serine (S), threonine (T), and lysine (K) residues mutated in adeno-associated virus serotype 8 (AAV8) and their conservation status across other AAV serotypes. VP1 protein sequences from AAV serotypes 1 through 10 were aligned by ClustalW, and the conservation status of each of the target sites for mutagenesis is shown in red. Color images available online at www.liebertpub.com/hgtb
Table 2.
Physical Particle Packaging Titers (Viral Genomes/ml) of Various Serine/Threonine/Lysine Mutant AAV8 Vectors
| Serine (S)>alanine (A) | Threonine (T)>alanine (A) | Lysine (K)>arginine (R) |
|---|---|---|
| S279A (8×1012) | T252A (8×1011) | K137R (4×1013) |
| S501A (4×1013) | ||
| S671A (4×1013) |
Average packaging titers from at least two packaging experiments are shown. The titer of wild-type self-complementary AAV8 vectors was 4×1013 viral genomes/ml in the laboratory.
Structure–function analysis of the proposed sites of mutagenesis
The mutants generated were subjected to extensive computational analysis to evaluate structural and functional equivalence between the two serotypes, AAV2 and AAV8, as opposed to the sequence equivalence, which was the criteria behind the selection of mutation positions. These mutation positions were scrutinized with respect to their inclusion in phosphodegrons (phosphorylation sites recognized as degradation initiation signals by ubiquitin ligases), they being either phosphosites or ubiquitination sites, or their participation in the protein–protein interactions of the capsid proteins. Also, inclusion of these positions in the antigenic segments on capsid proteins and/or in the receptor-binding regions were investigated further.
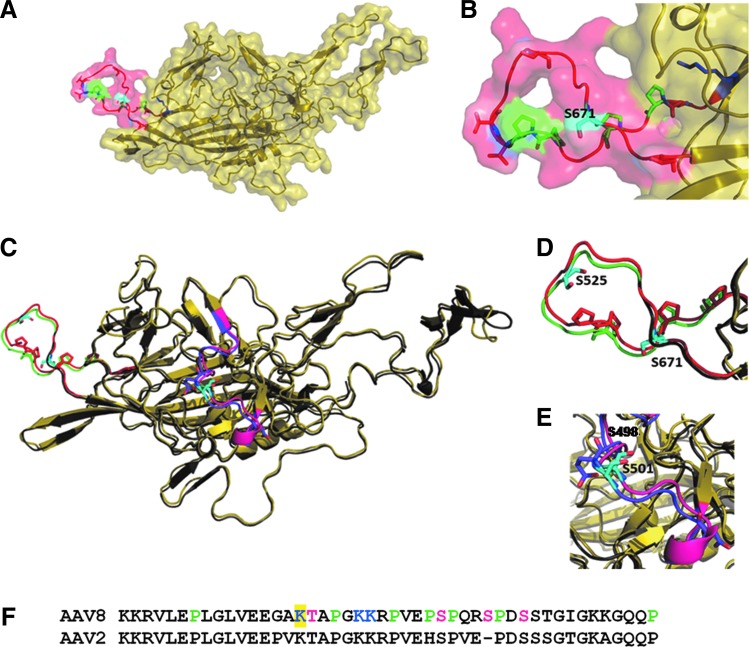
As summarized in Table 3, S671 lies in the phosphodegron region (652–671) rich in prolines. The presence of phosphosites (S667, S671, T654, T662, T663, and T674) and a Ub-site (K652) were predicted to lie within the phosphodegron region, as highlighted in Figure 2A and B. This serine residue neither participates in the interaction interface nor in receptor binding. This phosphodegron region containing S671 has also been predicted to be highly antigenic (presence of B-cell epitopes) by all the three prediction tools independently. Thus, mutation in the AAV8 capsid protein at S671 can be expected to be able to evade host degradation machinery effectively. Structure comparison between AAV2 and AAV8 serotypes, as shown in Figure 2C and D, revealed that the particular phosphodegron is well conserved across the two serotypes.
Table 3.
Structure Analysis of Mutation Positions
| Serial no. | Mutation | Phosphodegron prediction | Phosphosite prediction | Ub-site prediction | Interface residue | Receptor-binding region | Antigenic nature (B-cell epitope prediction) |
|---|---|---|---|---|---|---|---|
| 1 | S671A | Yes (652–671) | Yes | — | No | No | Yes |
| 2 | K137R | Yes (122–167) | — | Yes | — | No | Yes |
| 3 | S501A | No | No | — | Yes | Yes | Weak |
| 4 | S279A | No | No | — | Yes | No | Weak |
| 5 | T252A | No | Yes | — | No | No | No |
Mutation positions were analyzed extensively based on various criteria as listed.
FIG. 2.
The phosphodegron in AAV8 capsid structure. (A) The figure shows the capsid protein from AAV8 (PDB id: 2qa0), which is colored yellow. S671 (colored cyan) lies in phosphodegron region (652–674), which is colored red. The phosphodegron is rich in prolines, which are colored green. The predicted phosphosites (serines and threonine) are shown in red while the predicted ubiquitination sites are in blue. (B) The zoomed-in view of phosphodegron region (652–674) containing S671. (C) Comparison of phosphodegrons in AAV8 and AAV2. The figure shows structural superimposition of AAV8 (PDB id: 2qa0) and AAV2 (PDB id: 1lp3) in yellow and gray, respectively. Phosphodegron in AAV8, colored red, is equivalent to phosphodegron2 in AAV2 (515–528), which is colored green. Phosphodegrons in both AAV2 and 8 are rich in proline residues. The residue S525 in AAV2 (colored cyan) lies in the phosphodegron and has been shown to increase the transduction efficiency (Gabriel et al., 2013). (D) The zoomed-in view of the phosphodegron2 in AAV2 and AAV8. (E) The zoomed-in view of phosphodegron3 of AAV2 in comparison with that in AAV8. The presence of another phosphodegron region in AAV2 (phosphodegron 3: residues 489-507), which is rich in acidic residues is colored purple. S489 and S498 in this region of AAV2 have shown to increase the transduction efficiency in vivo. Whereas, the equivalent region in AAV8, colored pink, lacks the acidic residues. (F) The figure shows the predicted phosphodegron stretch (122–137) in AAV8, which contains K137, highlighted in yellow. The phosphodegron is rich in proline and is colored green. The predicted phosphosites (T138, S149, S153, and S156) are colored magenta while the predicted ubiquitination sites (K137, K142, and K143) are colored blue. Color images available online at www.liebertpub.com/hgtb
However, same is not the case with the other two phosphodegrons seen in AAV2. As indicated in the Figure 2C, the crucial acidic residues of the phosphodegron 3 of AAV2 have been replaced in AAV8 capsid protein, along with some of the phosphosites in the region. Hence, the equivalent of this phosphodegron is absent in AAV8. Replacing residue S501 in this region may not be very effective in increasing the transduction efficiency of the vector, although the equivalent residue S498 in AAV2 was found to be very effective in improving the transduction of AAV2 (Gabriel et al., 2013). Also, S501 residue lies in the receptor-binding region of AAV8, which marks one of the few differences between the two serotypes.
The residue K137 is not part of the available AAV8 capsid structure. However, thorough sequence analysis of the region indicated presence of phosphodegron-like region nearby, namely the residues 122–167 that are rich in prolines. Interestingly, the prolines in this region seem to be well conserved in AAV2, as can be seen in Figure 2F. Besides K137, the phosphosites (T138, S149, S153, S156) and Ub-sites (K142, K143) have been predicted to lie within this phosphodegron region in AAV8. Also, the K137 residue does not belong to the receptor-binding region. The phosphodegron-like region has been predicted to be a B-cell epitope, which suggests that it could be solvent exposed and highly antigenic. Thus, K137 can be an excellent target to be mutated to improve transduction efficiency of the vector by increasing its half-life in the host cell.
The positions T252 and S279 appear to be weak targets for mutagenesis in AAV8, although their sequence counterparts in AAV2 were effective in improving transduction efficiency of the vector upon mutagenesis (Gabriel et al., 2013). As can be seen in Table 3, S279 lies in the interaction interface of the capsid proteins. As the two residues have neither been predicted as phosphosites nor been predicted to be antigenic, mutating these residues may not be very effective in improving the transduction efficiency of AAV8 vector.
Despite the strong indications from the structural studies mentioned above, as some of the proposed mutations may be only marginally effective in improving the transduction efficiency, all the mutants were generated subsequently. Thorough experimental analysis of these mutants helped in verifying the apparent structural differences between the two serotypes despite high sequence identity and their implications in functioning of the viruses.
AAV8-EGFP serine and lysine mutant vectors demonstrate higher hepatic gene transfer efficiency in vivo
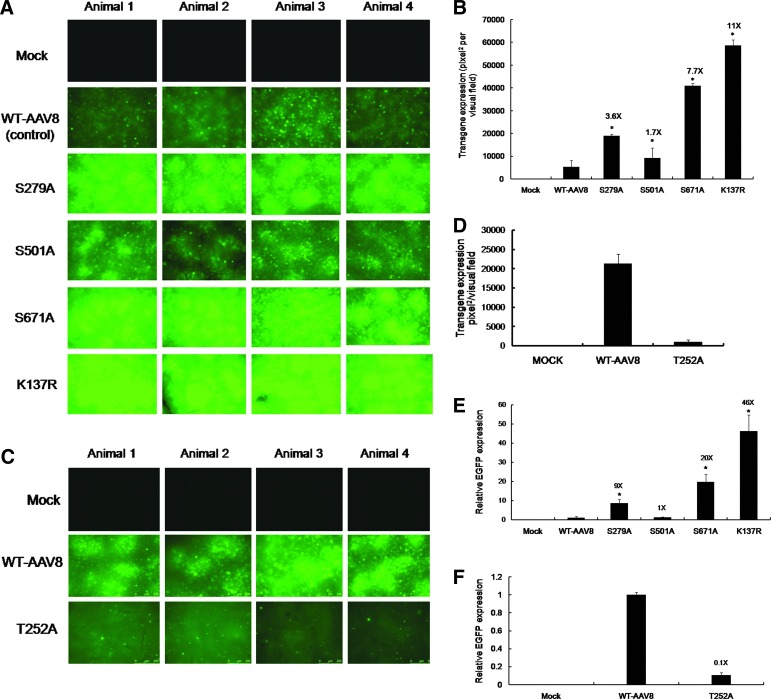
In anticipation of achieving a robust liver-directed expression of the various AAV8 mutant vectors, as mentioned above, their potential efficacy was examined in vivo. AAV8 S/T/K mutant vectors were administered at a dose of 5×1010 vgs/animal. Two of the S→A mutants (S279A and S671A) and the K137R mutant tested had a 3.6- to 11-fold higher EGFP expression by fluorescence imaging (Fig. 3A and B) and a 9- to 46-fold higher EGFP transcript level as analyzed by quantitative PCR (Fig. 3E). The T252A mutant had lower levels of EGFP expression when compared to the WT-AAV8 vector, 2 weeks postadministration (Fig. 3C, D, and F). When the transduction efficiency of the K137R mutant was assessed in a more immunogenic BALB/c strain of mice (Michou et al., 1997; Breous et al., 2010), a similar increase in transduction was noted for the K137R mutant (14-fold by fluorescence imaging, data not shown) as compared to the WT-AAV8 vector. To further corroborate this data, the biodistribution of AAV8 vectors in recipient mice was analyzed by quantitative PCR measurement of vector genomes in various tissues. Table 4 demonstrates that the K137R mutant had the best tropism for liver as compared to the WT-AAV8 (106 vs. 7.7 vector copies/mouse diploid genome). Interestingly, a preferential transduction of the lungs (0.13 vs. 0.03 vector copies/mouse diploid genome), and muscle tissue (1.38 vs. 0.45 vector copies/mouse diploid genome) was also noted for the K137R mutant suggesting that this vector has a better systemic transduction profile when compared to the other vectors tested. Similarly, the S→A mutant vectors (S279A, S501A, and S671A) generally showed increased transduction of the liver and muscle compared to WT-AAV8 vectors (Table 4). However, the T252A mutant was considerably retargeted to other tissues such as the lungs (Table 4), which may explain the low levels of EGFP expression seen in the liver (Fig. 3C, D, and F). These results clearly indicate that the S→A and K→R mutant capsids augment the hepatic transduction of AAV8 vectors, with K137R being the most effective. Hence, this mutant in particular was subjected to further analysis to gain insights into mechanisms by which it exhibits its effects on transduction of the vector.
FIG. 3.
AAV8 serine and lysine mutant vectors exhibit enhanced transduction upon hepatic gene transfer in vivo. (A) Representative images from each animal (n=4 for mock, S279A, S501A, and S671A; n=8 for WT and K137R) showing the transgene expression as detected by fluorescence microscopy 4 weeks postinjection with 5×1010 particles/animal of wild-type or S→A and K→R mutant AAV8 via the tail vein. (B) Quantitative analyses of the data from (A). (C) Representative images from each animal (n=4) showing the transgene expression as detected by fluorescence microscopy 2 weeks postinjection with 5×1010 particles of wild-type or T→A mutant AAV8 vectors. (D) Quantitative analyses of the data from (C). All the images were taken at the same exposure within each group (A or C) tested [207 milliseconds for (A) and 1.2 sec for (C), gain (n=2) and intensity (n=4) in a fluorescence microscope (Leica CTR6000). (E) Enhanced green fluorescent protein (EGFP) transcript levels normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in murine hepatocytes 4 weeks or (F) 2 weeks post–vector administration as measured by quantitative polymerase chain reaction (PCR). Analysis of variance (ANOVA) was used to compare EGFP expression between wild-type (WT) or mutant AAV8 treated groups. *p<0.05. Color images available online at www.liebertpub.com/hgtb
Table 4.
Vector Biodistribution in Various Organs in C57BL/6 Mice 2 or 4 Weeks Post–Gene Transfer with the WT-AAV8 and S/T/K Mutant AAV8 Vectors
| Vector type | Muscle | Liver | Kidney | Lung | Spleen | Heart |
|---|---|---|---|---|---|---|
| WT-AAV8 (4 weeks) | 0.45±0.2 | 7.77±0.8 | 0.0016±0.00045 | 0.03±0.07 | 0.004±0.0009 | 0.0003±0.00006 |
| K137R-AAV8 (4 weeks) | 1.38±0.4 | 106.8±11 | 0.0011±0.0003 | 0.13±0.09 | 0.004±0.0006 | 0.0006±0.00004 |
| S279A-AAV8 (4 weeks) | 2.84±0.5 | 30.3±3.2 | 0.0029±0.0007 | 0.01±0.003 | 0.002±0.0005 | 0.0007±0.00001 |
| S501A-AAV8 (4 weeks) | 3.07±0.6 | 15±1.9 | 0.003±0.0008 | 0.11±0.08 | 0.0009±0.0008 | 0.003±0.0002 |
| S671A-AAV8 (4 weeks) | 1.89±0.3 | 65.89±5.6 | 0.0006±0.00012 | 0.051±0.01 | 0.001±0.0006 | 0.001±0.0008 |
| WT-AAV8 (2 weeks) | 0.008±0.08 | 0.51±0.1 | 0.0006±0.0002 | 0.0008±0.0002 | 0.022±0.009 | 0.0001±0.00005 |
| T252A (2 weeks) | 0.01±0.005 | 0.01±0.002 | 0.0001±0.00006 | 0.01±0.004 | 0.0005±0.00009 | 0.0006±0.00009 |
Values are shown as mean (±SD) vector copy numbers per mouse diploid genome.
K137R mutant vector demonstrates higher hF.IX expression in vivo
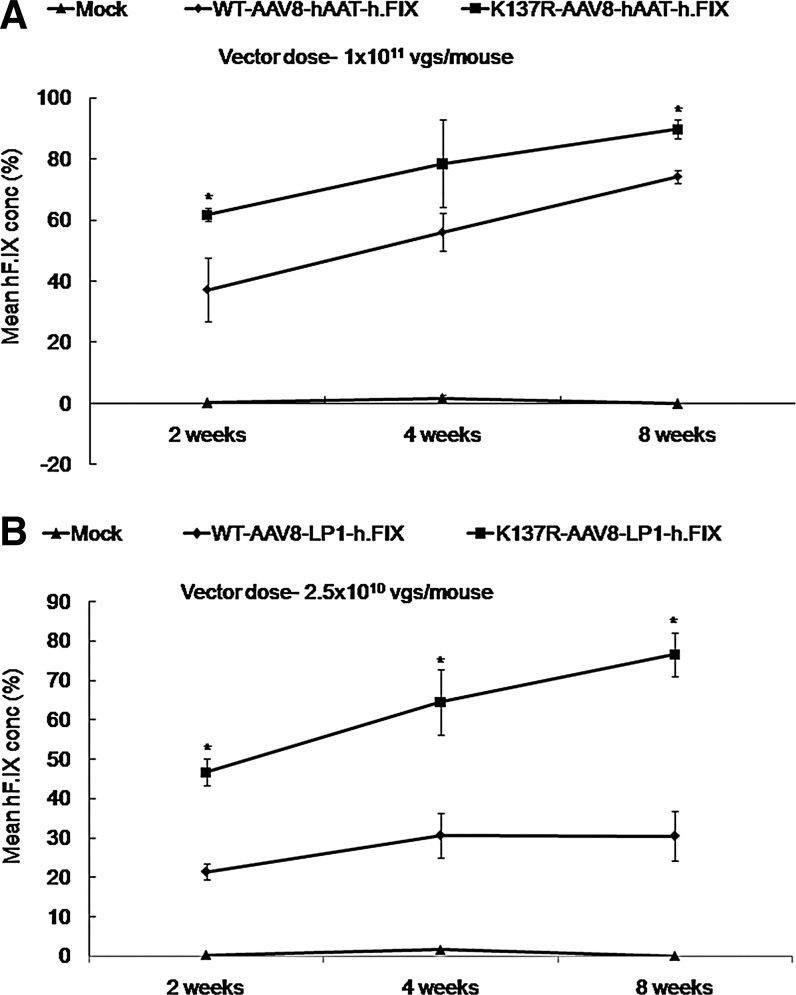
To further study the utility of the K137R-AAV8 mutant in therapeutic gene transfer, we delivered h.FIX under the control of liver-specific promoters at two different doses (2.5×1010 vgs per mouse for LP1-F.IX and 1×1011 vgs per mouse for hAAT-F.IX) in 8- to 12-week-old male C57BL/6 mice. As can be seen in Figure 4, the circulating levels of h.FIX were higher in all the K137R-AAV8-treated groups as compared to the WT-AAV8-treated groups either at 2 weeks (62% vs. 37% for hAAT constructs and 47% vs. 21% for LP1 constructs), 4 weeks (78% vs. 56% for hAAT constructs and 64% vs. 30% for LP1 constructs), or 8 weeks (90% vs. 74% for hAAT constructs and 77% vs. 31% for LP1 constructs) post-hepatic gene transfer. These results further corroborate the potential of the K137R mutant for hepatic gene therapy of hemophilia B.
FIG. 4.
K137R-AAV8 vector demonstrates superior hF.IX expression in comparison to WT-AAV8 vectors in C57BL/6 mice. Increased h.FIX expression from transgene constructs driven by either hAAT (A) or LP1 (B) promoters from animals injected with K137R-AAV8 vector and compared to the WT-AAV8 vector up to 8 weeks after hepatic gene transfer. *p<0.05 vs. WT-AAV8 injected mice.
K137R mutation decreases ubiquitination of AAV8 viral capsid
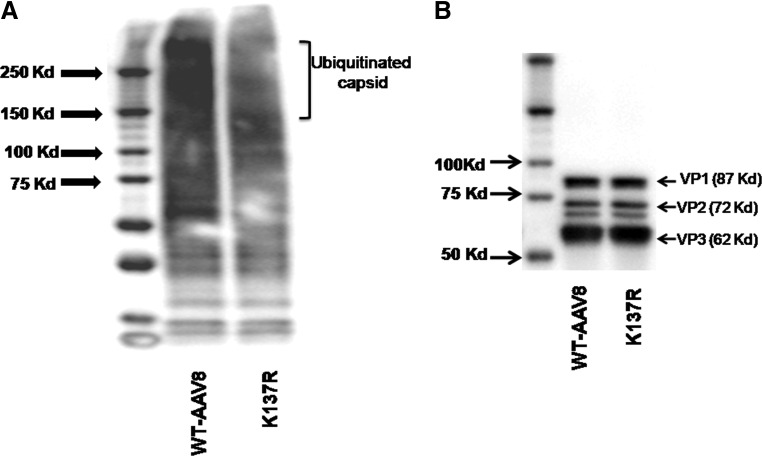
To understand if the improved transduction achieved with the lysine mutant vector (K137R) is due to reduced ubiquitination of viral capsid, an in vitro ubiquitination assay was performed, followed by western blotting to detect the levels of mono- and poly-ubiquitin moieties on the AAV capsid (Fig. 5A). The AAV8 K137R mutant vector had significantly reduced ubiquitination pattern compared to WT-AAV8 vector. AAV8 capsid proteins VP1 (87kDa), VP2 (72 kDa), and VP3 (62 kDa) were probed as gel-loading controls, which showed similar levels of these proteins across the samples tested (Fig. 5B). These data provide direct evidence that the superior transduction achieved with the K137R mutant vector is due to the reduced ubiquitination of the viral capsid, which possibly results in rapid intracellular trafficking of the virus and improved gene expression.
FIG. 5.
K137R-AAV8 lysine mutant vector demonstrates reduced ubiquitination in comparison to WT-AAV8 vector. (A) Approximately 3×108 viral particles of WT-AAV8 and K137R-AAV8 vectors were denatured at 95°C for 5 minutes. The denatured viral particles were then used to perform the ubiquitin conjugation assay according to the manufacturer's protocol. The processed samples were electrophoresed on a 4–20% denaturing polyacrylamide gel and the ubiquitination pattern detected by immunoblotting using an anti-ubiquitin antibody. The mono-to-polyubiquitin conjugates are detected as a smear at molecular weight >150Kda. (B) Approximately 3×108 viral particles of WT-AAV8 and K137R-AAV8 vectors were denatured with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail at 95°C for 10 minutes. The samples were resolved in a 4–20% denaturing polyacrylamide gel and the VP1 (87Kda), VP2 (72 KDa), and VP3 (62 Kda) capsid proteins were detected with AAV clone B1 antibody as described in the Materials and Methods section.
The K137R mutant vector demonstrates reduced inflammatory cytokine and cross-neutralizing antibody response
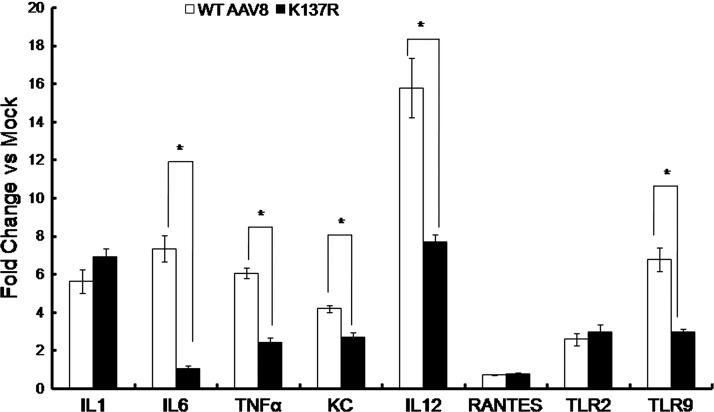
As can be seen in Figure 6, the levels of inflammatory cytokines such as IL1, IL6, TNFα, IL12, KC, RANTES, and innate immune responsive TLRs 2 and 9 were upregulated in the hepatocytes within 2hr of the WT-AAV8 administration indicating the activation of innate immune response toward the virus as reported earlier (Jayandharan et al., 2011; Martino et al., 2011). Interestingly, the K137R vector had a significantly reduced activation of IL6 (6.9-fold), TNFα (2.5-fold), KC (1.5-fold), IL12 (2-fold), and TLR9 (2.2-fold) compared to the WT-AAV8 vectors (Fig. 6). Further measurement of neutralizing antibodies against the various mutants demonstrated a 2-fold reduction in the neutralizing antibody titer for the K137R-AAV8 vector (Table 5). These results imply that the mutant K137R vector is significantly less immunogenic when compared to WT-AAV8 vectors.
FIG. 6.
K137R-AAV8 vector elicits reduced inflammatory cytokine response compared to WT-AAV8 vectors in vivo. Quantitative PCR was used to profile the hepatic expression of key proinflammatory cytokines and other markers of innate immune response as described in the Materials and Methods section. The relative fold change in the target gene expression from mice injected with WT-AAV8 and K137R-AAV8 vector in comparison to mock-injected animals are shown. *p<0.05 denotes statistical significance as compared to the WT-AAV8-injected mice.
Table 5.
Neutralization Antibody Formation Against Wild-Type or Mutant AAV8 Vectors
| S. No. | Groups | Reciprocal NAb titer |
|---|---|---|
| 1 | Mock | 0 |
| 2 | WT-AAV8 | 320 |
| 3 | S279A | 320 |
| 4 | S501A | 640 |
| 5 | S671A | 320 |
| 6 | K137R | 160 |
| 7 | Anti-AAV8 rabbit control serum | 20480 |
Pooled serum samples from WT-AAV8 or S/K-AAV8 mutant injected mice (n=4 per group) 4 weeks after vector administration was studied. Heat-inactivated serum samples were assayed for the neutralizing antibody (NAb) titers as described in Materials and Methods. The NAb titer is reported as the highest plasma dilution that inhibited AAV transduction of Huh7 cells by 50% or more compared with that of the naive serum control. Limit of detection of the assay was 1/5 dilution.
Discussion
Among the various alternate AAV serotypes (Schultz and Chamberlain, 2008) and a new generation of hybrid vectors (Choi et al., 2005) available for gene delivery, there is tremendous interest with AAV8 vectors. In particular, its ability to uncoat and release its genome rapidly after cellular entry may enable therapeutic transgene expression using a lower dose of vector (Thomas et al., 2004; Nakai et al., 2005). Also, the relatively shorter persistence of AAV8 capsid proteins in hepatocytes compared to AAV2 decreases the risk of an anti-capsid immune response (Thomas et al., 2004). The ability of AAV8 vector to transduce liver efficiently by peripheral vein delivery is another factor that augurs well for widespread use of AAV8 vectors (Nathwani et al., 2001, 2011; Davidoff et al., 2005). AAV8 vectors also have an ability to bypass natural neutralizing antibodies to AAV2, and combined with its low seroprevalence in humans, make it an ideal vector for gene therapy (Erles et al., 1999; Li et al., 2012). It is these unique biological characteristics that contributed to the success of AAV8 vectors in the conversion of six patients with severe hemophilia B into a moderate disease (Nathwani et al., 2011). For this approach to be widely applicable and therapeutically successful, several issues have to be resolved (High, 2012), including the need for improvements in AAV8 vectors that can elicit lesser immune response or have the capability to express high levels of transgene at lower doses.
The targeted modification of the AAV8 vector capsid, such as tyrosine to phenylalanine mutations, has been successful during gene transfer to the retina (Petrs-Silva et al., 2011; Deng et al., 2012) but their efficacy has been modest when targeted to other tissues such as muscle, heart (Qiao et al., 2012). This suggests that cell-specific barriers affect their transduction potential. It is known that serine/threonine kinases are abundant in murine liver compared to tyrosine kinases (Villen et al., 2007), and since S/T/K are abundant (18.3%) on the AAV8 capsid over tyrosine residues (4.4%), we reasoned that mutating amino acids other than tyrosines on AAV8 capsid may provide further opportunities to augment AAV8-mediated gene expression. Toward this, S/T/K residue positions were selected for mutagenesis by sequence comparison between the effective AAV2 capsid mutants generated earlier (Gabriel et al., 2013) and the AAV8 capsid. Systematic computational analysis carried out subsequently helped us point out the similarities and differences between the capsid structures of the two serotypes with respect to the conservation of phosphodegron regions along with phosphosites and ubiquitination sites, receptor-binding regions, and interaction interfaces between the capsid proteins. This analysis thus enabled us to envisage and understand the effects of the capsid mutations on transduction efficiency of the vectors. Indeed, our studies demonstrate that these selective modifications at S/K residues enhanced the liver-directed EGFP gene expression of AAV8 vectors. Certain mutant vectors such as S501A and S671A showed altered, but higher, transduction of the muscle tissue, which might make them useful for gene therapy of muscular dystrophy (Bowles et al., 2012) or cardiovascular diseases (Zincarelli et al., 2010; Pacak and Byrne, 2011). In particular, the K137R mutant had significantly higher systemic transduction efficiency, possibly due to decreased ubiquitination of the viral capsid resulting in rapid intracellular trafficking of the virus and improved gene expression. In addition, the potential therapeutic benefit of K137R mutant has been demonstrated with increased levels of h.FIX expression up to 2 months post-hepatic gene transfer. Further ongoing studies in hemophilia B mice are likely to shed more light on the potential of this vector for gene therapy of hemophilia B in humans.
The K137R mutant generated in this study was also less immunogenic when compared to WT-AAV8 vectors. Previous studies have demonstrated that ubiquitinated and proteasomally processed AAV peptides are transported to the endoplasmic reticulum and are restricted by MHC Class I molecules (Yan et al., 2002; Vandenberghe et al., 2006; Finn et al., 2010). This presentation flags hepatocytes for recognition and destruction by capsid-specific CD8+ T cells. In addition, the processed vector can also be taken up the professional antigen-presenting cells, which after MHC class II restriction can activate a CD4+ T-cell response (Chen et al., 2006; High, 2012). In line with these observations, the use of proteasomal inhibitors prior to AAV8 administration has shown reduced immune response and increased transduction efficiency in vivo (Karman et al., 2012; Liu et al., 2012). More importantly, K137 is known to be within a previously described MHC class II T-cell recognition epitope (L126-P140) of AAV8 in both humans and mice (Sabatino et al., 2005; Chen et al., 2006). This residue is also in the vicinity of a previously characterized AAV8 neutralizing antibody epitope (N113-R132) in humans (Wobus et al., 2000; Gurda et al., 2012). Based on these data and the reduced ubiquitination seen on the K137R capsid, it is possible that the K137R mutant has reduced antigen recognition/presentation of vector capsid in comparison to WT-AAV8 vectors. However, further detailed studies are needed to confirm this phenomenon.
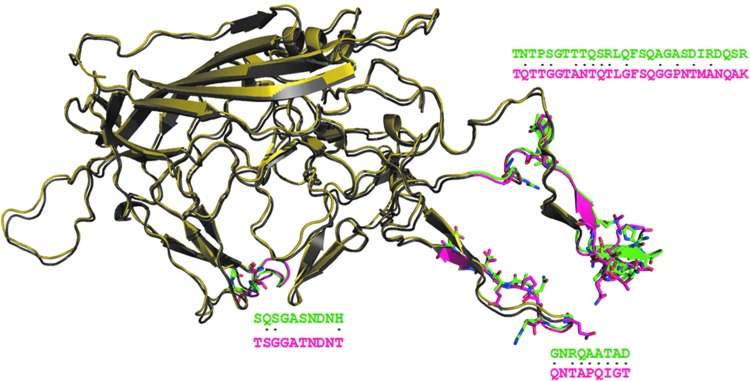
Interestingly, when the same mutation, K137R, was carried out in AAV2 (Gabriel et al., 2013) that is at the equivalent and conserved position in the sequence of AAV2 capsid, it did not improve its transduction efficiency. Sequence comparison of the two serotypes revealed conservation of the lysine residue as well as the phosphodegron-like neighborhood of it. In absence of the crystal structure for the region of about 200 residues encompassing K137 in both AAV2 and AAV8, it is very difficult to pinpoint the differences, if any. Nevertheless, a theoretical possibility can be envisaged that may provide a plausible explanation for the differential effects of this K137R mutation in the two serotypes. The 200-residue stretch, that is absent in the crystal structures of the two capsids, equivalent to a separate domain, can be imagined to interact with the surface of a domain from the crystal structure. The possible surfaces of the AAV2 and AAV8 capsid crystal structures that could interact with the conserved region encompassing K137 have been compared, as shown in the Figure 7. When such a comparison was carried out, three distinct regions were identified, which showed drastic residue differences between AAV2 and AAV8. Thus, the differential effects of the mutation may be arising from the interactions of the conserved regions with the unconserved regions in the two serotypes. Potential differences in the interactions involving the region with K137 could result in a different chemical and structural environment around K137 that may manifest into different effects in AAV2 and AAV8 for the mutation K137R. These possibilities, however, need an experimental validation in terms of determination of crystal structure of the full-length capsid protein.
FIG. 7.
Comparison of AAV8 and AAV2 capsid structures. Superimposition of AAV2 (colored yellow) and AAV8 (colored gray) capsid structures are shown. The green-colored region in AAV2 and the magenta-colored region in AAV8 show drastic residue substitutions. The residues that are drastically substituted are represented as sticks and are also marked as dots (.) in the sequence alignment between AAV2 and AAV8. These regions with drastic substitutions are away from the phosphodegron-containing and receptor-binding regions. It can be speculated that these regions could interact with the N-terminal region of the capsid structure for which the crystal structure is currently not available. Drastic differences in the interface could have an influence on the interaction with the N-terminal region of capsid structure flanking the K137. This could affect the conformation of region spanning K137 in AAV2 and AAV8 and thus contribute to their varied transduction efficiency as seen experimentally with the K137R mutation in AAV2 and AAV8 serotypes (Gabriel et al., 2013). Color images available online at www.liebertpub.com/hgtb
These optimized AAV8 capsid–mutant AAV vectors in general, and K137R mutant in particular, have the potential to become important tools for therapeutic hepatic gene transfer, such as in hemophilia or alpha 1–antitrypsin deficiency. Although this study has shown proof-of-concept—the efficacy of these single-mutant AAV8 vectors with a limited number of serine or lysine modifications—our ongoing studies with multiple combinations of these mutant vectors is likely to further improve the efficiency of these second-generation AAV8 vectors.
Acknowledgments
We thank Mr. Sathish Y., Laboratory Animal Core Facility, Centre for Stem Cell Research, Vellore, for animal care. G.R.J. is supported by research grants from Department of Science of Technology, Government of India (Swarnajayanti Fellowship 2011), Department of Biotechnology (DBT), Government of India (Innovative Young Biotechnologist award 2010: BT/03/IYBA/2010; Grant: BT/PR14748/MED/12/491/2010; Grant: BT/01/COE/08/03) and an early career investigator award, 2010, from Bayer Hemophilia Awards program, Bayer Inc, Montville, New Jersey. R.A.G. and G.S. are supported by DST women scientist program and DBT research fellowships, respectively. N.S. acknowledges the support of DBT, government of India. We thank Immunology Core, Gene Therapy Program, Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, for estimation of neutralizing antibodies against AAV8.
Author Disclosure Statement
No competing financial interests exist for all the authors.
References
- Aurnhammer C. Haase M. Muether N., et al. Universal Real-Time PCR for the Detection and Quantification of Adeno-Associated Virus Serotype 2-Derived Inverted Terminal Repeat Sequences. Hum. Gene Ther. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- Bowles D.E. Mcphee S.W. Li C., et al. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breous E. Somanathan S. Wilson J.M. BALB/c mice show impaired hepatic tolerogenic response following AAV gene transfer to the liver. Mol. Ther. 2010;18:766–774. doi: 10.1038/mt.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcedo R. Vandenberghe L.H. Gao G., et al. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Wu Q. Yang P., et al. Determination of specific CD4 and CD8 T cell epitopes after AAV2- and AAV8-hF.IX gene therapy. Mol. Ther. 2006;13:260–269. doi: 10.1016/j.ymthe.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Chen J. Liu H. Yang J. Chou K.C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33:423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- Choi V.W. Mccarty D.M. Samulski R.J. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 2005;5:299–310. doi: 10.2174/1566523054064968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff A.M. Gray J.T. Ng C.Y., et al. Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol. Ther. 2005;11:875–888. doi: 10.1016/j.ymthe.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Delano W. The PyMOL molecular graphics system. 2002. www.pymol.org/ [May 8;2012 ]. www.pymol.org/
- Deng W.T. Dinculescu A. Li Q., et al. Tyrosine-mutant AAV8 delivery of human MERTK provides long-term retinal preservation in RCS rats. Invest. Ophthalmol. Vis. Sci. 2012;53:1895–1904. doi: 10.1167/iovs.11-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K. Sebokova P. Schlehofer J.R. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV) J. Med. Virol. 1999;59:406–411. doi: 10.1002/(sici)1096-9071(199911)59:3<406::aid-jmv22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Finn J.D. Hui D. Downey H.D., et al. Proteasome inhibitors decrease AAV2 capsid derived peptide epitope presentation on MHC class I following transduction. Mol. Ther. 2010;18:135–142. doi: 10.1038/mt.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotte T.R. Carter B.J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- Gabriel N. Hareendran S. Sen D., et al. Bioengineering of AAV-2 capsid at specific serine, threonine or lysine residues improves its transduction efficiency in vitro and in vivo. Hum. Gene Ther. Methods. 2013 doi: 10.1089/hgtb.2012.194. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.P. Alvira M.R. Wang L., et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. Kay M.A. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- Gurda B.L. Raupp C. Popa-Wagner R., et al. Mapping a Neutralizing Epitope onto the Capsid of Adeno-Associated Virus Serotype 8. J. Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High K.A. The gene therapy journey for hemophilia: are we there yet? Blood. 2012;120:4482–4487. doi: 10.1182/blood-2012-05-423210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J. Thornton J.M. Department of Biochemistry and Molecular Biology, University College; London: 1993. NACCESS [computer program] [Google Scholar]
- HUSSAIN T. NASREEN N. LAI Y., et al. Innate immune responses in murine pleural mesothelial cells: Toll-like receptor-2 dependent induction of beta-defensin-2 by staphylococcal peptidoglycan. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295:L461–470. doi: 10.1152/ajplung.00276.2007. [DOI] [PubMed] [Google Scholar]
- Jayandharan G.R. Aslanidi G. Martino A.T., et al. Activation of the NF-kappaB pathway by adeno-associated virus (AAV) vectors and its implications in immune response and gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3743–3748. doi: 10.1073/pnas.1012753108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Karman J. Gumlaw N.K. Zhang J., et al. Proteasome inhibition is partially effective in attenuating pre-existing immunity against recombinant adeno-associated viral vectors. PLoS One. 2012;7:e34684. doi: 10.1371/journal.pone.0034684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube D.M. Srivastava A. Quantitative DNA slot blot analysis: inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997;25:3375–3376. doi: 10.1093/nar/25.16.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J.E. Lund O. Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Narkbunnam N. Samulski R.J., et al. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther. 2012;19:288–294. doi: 10.1038/gt.2011.90. [DOI] [PubMed] [Google Scholar]
- Liu Y. Joo K.I. Wang P. Endocytic processing of adeno-associated virus type 8 vectors for transduction of target cells. Gene Ther. gt. 20122012:41. doi: 10.1038/gt.2012.41. [DOI] [PubMed] [Google Scholar]
- Manno C.S. Pierce G.F. Arruda V.R., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- Martino A.T. Suzuki M. Markusic D.M., et al. The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver. Blood. 2011;117:6459–6468. doi: 10.1182/blood-2010-10-314518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michou A.I. Santoro L. Christ M., et al. Adenovirus-mediated gene transfer: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther. 1997;4:473–482. doi: 10.1038/sj.gt.3300412. [DOI] [PubMed] [Google Scholar]
- Mount J.D. Herzog R.W. Tillson D.M., et al. Sustained phenotypic correction of hemophilia B dogs with a factor IX null mutation by liver-directed gene therapy. Blood. 2002;99:2670–2676. doi: 10.1182/blood.v99.8.2670. [DOI] [PubMed] [Google Scholar]
- Mueller C. Flotte T.R. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- Nakai H. Thomas C.E. Storm T.A., et al. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in recombinant adeno-associated virus-mediated liver transduction. J. Virol. 2002;76:11343–11349. doi: 10.1128/JVI.76.22.11343-11349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H. Fuess S. Storm T.A., et al. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H.J. Lane M.D. Padron E., et al. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C. Davidoff A. Hanawa H., et al. Factors influencing in vivo transduction by recombinant adeno-associated viral vectors expressing the human factor IX cDNA. Blood. 2001;97:1258–1265. doi: 10.1182/blood.v97.5.1258. [DOI] [PubMed] [Google Scholar]
- Nathwani A.C. Davidoff A.M. Hanawa H., et al. Sustained high-level expression of human factor IX (hFIX) after liver-targeted delivery of recombinant adeno-associated virus encoding the hFIX gene in rhesus macaques. Blood. 2002;100:1662–1669. doi: 10.1182/blood-2002-02-0589. [DOI] [PubMed] [Google Scholar]
- Nathwani A.C. Gray J.T. Ng C.Y., et al. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood. 2006;107:2653–2661. doi: 10.1182/blood-2005-10-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C. Gray J.T. Mcintosh J., et al. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109:1414–1421. doi: 10.1182/blood-2006-03-010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C. Tuddenham E.G. Rangarajan S., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak C.A. Byrne B.J. AAV vectors for cardiac gene transfer: experimental tools and clinical opportunities. Mol. Ther. 2011;19:1582–1590. doi: 10.1038/mt.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H. Dinculescu A. Li Q., et al. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011;19:293–301. doi: 10.1038/mt.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C. Yuan Z. Li J., et al. Single tyrosine mutation in AAV8 and AAV9 capsids is insufficient to enhance gene delivery to skeletal muscle and heart. Hum. Gene Ther. Methods. 2012;23:29–37. doi: 10.1089/hgtb.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar S.V. Richardson P.G. Hideshima T. Anderson K.C. Proteasome inhibition as a novel therapeutic target in human cancer. J. Clin. Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Sabatino D.E. Mingozzi F. Hui D.J., et al. Identification of mouse AAV capsid-specific CD8+ T cell epitopes. Mol. Ther. 2005;12:1023–1033. doi: 10.1016/j.ymthe.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Saha S. Raghava G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Schultz B.R. Chamberlain J.S. Recombinant adeno-associated virus transduction and integration. Mol. Ther. 2008;16:1189–1199. doi: 10.1038/mt.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R.O. Miao C.H. Patijn G.A., et al. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat. Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- Snyder R.O. Miao C. Meuse L., et al. Correction of hemophilia B in canine and murine models using recombinant adeno-associated viral vectors. Nat. Med. 1999;5:64–70. doi: 10.1038/4751. [DOI] [PubMed] [Google Scholar]
- Thomas C.E. Storm T.A. Huang Z. Kay M.A. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe L.H. Wang L. Somanathan S., et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat. Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- Villen J. Beausoleil S.A. Gerber S.A. Gygi S.P. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Takabe K. Bidlingmaier S.M., et al. Sustained correction of bleeding disorder in hemophilia B mice by gene therapy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3906–3910. doi: 10.1073/pnas.96.7.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Zhu T. Qiao C., et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- Wobus C.E. Hugle-Dorr B. Girod A., et al. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 2000;74:9281–9293. doi: 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. Asokan A. Samulski R.J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Yan Z. Zak R. Luxton G.W., et al. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L. Li B. Jayandharan G., et al. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C. Soltys S. Rengo G., et al. Comparative cardiac gene delivery of adeno-associated virus serotypes 1-9 reveals that AAV6 mediates the most efficient transduction in mouse heart. Clin. Transl. Sci. 2010;3:81–89. doi: 10.1111/j.1752-8062.2010.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S. Byrne B.J. Mason E., et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]