Abstract
We are at an inflection point in our study of the human genome as it relates to neurodegenerative disease. The sequencing of the human genome, cataloguing of human genetic variation and rapid technological as well as methodological development, introduced a period of rapid gene discovery over the past decade. These efforts have yielded many new insights and will continue to uncover the genetic architecture of syndromically-defined neurodegenerative diseases in the coming decades. More recently, these successful study designs have been applied to the investigation of intermediate traits that relate to and inform our understanding of clinical syndromes, and the architecture of chromatin (the epigenome), the higher order structure of DNA that dictates the expression of a given genetic risk factor. While still nascent - given the challenges of accumulating large numbers of subjects with detailed phenotypes and technological hurdles in characterizing the state of chromatin – these efforts represent key investments that will enable the characterization of the functional consequences of a genetic risk factor and, eventually, its contribution to the clinical manifestations of a given disease. As a community of investigators, we are therefore at an exciting inflection point at which gene discovery efforts are transitioning towards the functional characterization of implicated genetic variation crucial for understanding the molecular, cellular, and systemic events that lead to a syndromic diagnosis for neurodegenerative diseases.
Over the past decade, efforts to catalogue human genetic variation, such as the HapMap1 and, more recently, the 1000 Genomes project2, have synergized with rapidly evolving technologies to yield a wealth of insights into the genetic architecture of many human diseases. We are now at an inflection point where we are shifting from gene discovery for common disease-associated variants to the exploration of the higher-order architecture of the genome, which is captured by the term “epigenome.” Neurologic disease research provides a good illustration of this trend. Researchers have successfully exploited the genome-wide association study design, and are now initiating epigenome-wide scans for common diseases.
Here, we review our current understanding of the genetic architecture of four common neurodegenerative diseases – Alzheimer's Disease (AD), Amyotrophic Lateral Sclerosis (ALS), Multiple Sclerosis (MS), and Parkinson's Disease (PD) – and use them to highlight successful strategies for gene discovery as well as emerging strategies being deployed to address the next series of challenges that we face in investigating the pathophysiological basis of these diseases. For all four diseases, which have long prodromal phases, intermediate traits that relate to the ultimate clinical manifestation and syndromic classification of these diseases will play an increasingly important role as we continue to dissect the sequence of events that lead to these neurodegenerative processes and their ultimate expression as clinical disease.
Allelic spectrum of syndromically defined subject classes
The allelic spectrum of neurodegenerative disease ranges from rare variation with profound effects, such as those found in families with fully penetrant Mendelian versions of these diseases, to common variants (variants having a frequency > 0.05) with modest effects. Most gene discoveries have occurred at the extremes of this distribution because genotyping technologies, analytic methods, and study designs converged to maximize our power to discover variants in those aspects of the allelic spectrum. Coarse genome-wide genotyping, a linkage strategy, and highly selected collections of families with a Mendelian pattern of inheritance led to the identification early onset AD genes (such as amyloid precursor protein and, and the presenilin 1 and 2 genes)3, early onset PD genes (such as alpha-synuclein, parkin, PINK1, DJ-1 and ATP13A2)4, and familial ALS genes (such as SOD1, SETX, VAPB, ALSIN and DCTN1)5. With the advent of next-generation sequencing, this study design has undergone a renaissance, with new susceptibility genes affected by rare variation of profound effect being discovered in MS (CYP27B1)6 and PD (VPS35)7,8, for example. Thus, linkage study as a strategy for gene discovery has been clearly validated and continues to yield new insights as technology advances.
At the other extreme, genotyping arrays, enhanced by linkage disequilibrium (LD)-based imputation methods that capture a large fraction of the common genetic variation in the human genome, were deployed in a simple association study design and have yielded common variants with modest effects on susceptibility for one of these syndromic, neurodegenerative phenotypes. The number of validated susceptibility alleles is rapidly increasing, as expected with the increasing coverage of the genome coupled with the increasing sample sizes included in studies led by consortia of investigators. As suggested by analyses in MS9, it is likely that there are hundreds of variants that influence susceptibility to neurodegenerative diseases, and, as study sizes increase, we will continue to discover more variants with increasingly small effects. It is not yet clear at what point such associations are no longer meaningful. Critics suggest that even the current batch of validated susceptibility alleles is not very useful given their minuscule effects on syndromically-defined disease susceptibility for an individual subject. However, as will be discussed below, these variants of modest effects have already made important contributions by providing a robust platform with which to explore the earliest events in a disease. Further, the use of syndromic phenotypes in gene discovery, while expeditious to accumulate the required large sample sizes by pooling phenotypes common across many studies which contribute to consortia, may not be the optimal strategy to deconstruct neurodegenerative diseases as it obscures the clinical reality of phenotypic heterogeneity. Recognizing this phenotypic architecture of the patient population may well enhance our efforts and clarify the effect size of known variants that may have an apparently weak effect in the syndromically-defined population but a much stronger effect on a particular endophenotype or in a subset of subjects with a shared pathophysiologic mechanism. Thus, identifying common variants will continue to yield new, useful insights into neurodegenerative disease susceptibility. However, we will eventually become limited by the availability of samples with a given diagnosis and by the diminishing returns of increasing sample sizes, that are predicted by the flattening of the curves for statistical power in the association study design.10
The center of the allelic spectrum – less common variants (<0.05 frequency) with moderate effect sizes - has been the most difficult to explore to date; it contains the bulk of single nucleotide polymorphisms (SNPs), insertion/deletion (indel) polymorphisms and other variants that have been catalogued in human populations but are typically poorly captured by current imputation methods. They essentially require direct genotyping, which is not yet available at a practical cost with available genotyping technologies. Whole genome sequencing will eventually enable the comprehensive genotyping of study subjects but remains impractical for large-scale studies today. Nonetheless, an excellent assessment of the success of this possible strategy is underway, facilitated by the creation of “exome chips” that genotype the subset of less common variants found in or near exonic features of genes in ∼12,000 individuals that have undergone whole exome sequencing (http://genome.sph.umich.edu/wiki/Exome_Chip_Design). While not comprehensive in assessing all variants in this allelic spectrum, the exome chip content, when coupled with large sample sizes, will powerfully interrogate an interesting subset of these variants that, a priori, are more likely to have a strong effect on gene function and hence human susceptibility to disease. Such assessments of loci influencing susceptibility to AD, MS and other diseases are under way and will provide a useful, systematic assessment of the role of this class of alleles in neurodegenerative diseases. In terms of study design, large studies of these less common variants are necessary to capture enough subjects with each variant to enable robust analyses: the variance in a trait explained by a less common variant of moderate effect will probably be similar to that of a common variant of modest effect. We can calibrate the design of these exome chip studies using known disease-associated variants that have been reported such as TNFRSF1A R92Q in MS11, GBA N370S and L444P in PD12, as well as TREM2 R47H in AD13,14. The fact that some disease-associated variants are found in this allelic spectrum suggests that these “exome chip” studies will successfully identify additional novel susceptibility genes.
Shared genetic architecture in susceptibility to neurodegenerative diseases
As the repertoire of susceptibility loci becomes more comprehensive, it is likely that neurodegenerative diseases will have both disease-specific risk alleles, and alleles that influence risk via less specific processes such as neuronal loss, regardless of the pathologic process that challenges neuronal survival. This is now well documented for inflammatory diseases.15 In neurodegenerative disease, we are beginning to see evidence for such a shared architecture as the complement of susceptibility loci becomes better established. Examples of this shared architecture include TARDBP with associations to frontotemporal lobar degeneration and amyotrophic lateral sclerosis, as well as the MAPT locus where different variants are associated with Parkinson's disease, frontotemporal dementia with parksinonism linked to chromosome 17, and progressive supranuclear palsy (OMIM entries 138945 and 605078). The extent of the shared nature of genetic architecture will also require the careful selection of the traits to be compared. For example, MS susceptibility alleles - which overlap extensively with those of other, episodic inflammatory diseases – may not be pertinent to the genetic architecture of the other neurodegenerative conditions that present with a more insidious, progressive course. However, variants that relate to features of MS such as its accelerated brain atrophy and the progressive course displayed by a subset of subjects may have effects that are shared across neurodegenerative diseases. This point illustrates the need for the detailed characterization of pertinent, intermediate traits in all of these diseases; this strategy, while challenging because of the effort and cost entailed by detailed phenotyping of subjects, is nonetheless a clear goal for the near future. Current efforts are already illustrating the utility of these strategies.
Leveraging intermediate traits
Most successful gene discovery studies to date have focused on syndromic phenotypes given the availability of large numbers of subjects that fit the clinical definitions of AD, ALS, MS and PD that can be merged from multiple sources. However, this approach, while convenient and reasonable as a first effort, ignores the fact that large fractions of the control populations used in these studies have subclinical features of the disease. This is particularly true for AD and PD, and probably to a lesser extent for ALS. This includes the accumulation of neuritic amyloid plaques, neuronal loss in the substantia nigra and anterior horn, and other pathologies, or symptoms such as subtle cognitive impairment, bradykinesia, and muscle atrophy and weakness, that do not fulfill a syndromic definition.16,17 This has most likely reduced the statistical power of studies of AD and perhaps PD; ALS and MS, because of their low incidence rate in the general population, have been less affected by this problem. Intermediate traits (also referred to as endophenotypes) that capture pertinent features of a neurodegenerative disease have been suggested to have greater statistical power for gene discovery efforts than syndromic phenotypes: for example, the known APOE AD-associated alleles have much larger effects on AD neuropathology and trajectories of cognitive decline than on a syndromic diagnosis of AD when investigated in the same set of deeply phenotyped subjects.18 The endophenotype strategy has been implemented in several studies, but its success is clearly dependent on the quality and statistical properties of the trait being considered. Further, for the more distal phenotypes that capture pre-diagnosis features of the disease, such studies have been hampered by (1) the lack of consistency in the manner and frequency in which intermediate traits are measured across subject collections and (2) the nature of the subject collections which range from population-based samples to subjects selected in specialized clinics of tertiary care centers or samples of convenience collected for other purposes. Estimates of the needed sample size for a study of cognitive decline, for example, appear not to be too different from those required for syndromic traits19, and the recent successful GWAS for loci influencing hippocampal and intracranial volume required in one case a discovery study of more than 9,000 subjects and, in the other, more than 7,000 subjects.20,21 The latter examples speak eloquently to the challenge of combining a trait that is measured in different ways on many different platforms, limiting the power of meta-analysis of different subject collections.
While these clinical, imaging, and pathologic endophenotypes that are relatively distal on the causal chain linking genetic risk factors to a syndromic phenotype (Figure 1) have proven to be challenging to dissect genetically, they have been critical in beginning to elucidate the functional consequences of the validated disease-associated variants. APOEε4 with its very large effect size highlights this strategy well18,22, but it has already born fruit with common variants such as the AD-associated variants in CR1 and PICALM that have been implicated in the amyloid pathology that plays an important and early role in AD.23-26 These and many other studies will gradually identify the pathophysiologic consequences of disease-associated variants and will help to assemble them into molecular pathways whose alterations leads to disease. Further, they will play an important role in the detailed dissection of associated loci: helping to (1) identify what may be the causal variant if there are several equivalent candidates at the end of the discovery genome-wide association study and/or (2) to map the location of variants, within a susceptibility locus, with independent effects on the neurodegenerative process being studied.27 The early examples cited above and many other studies illustrate that this approach will be fruitful, particularly as larger, well-phenotyped sample collections are assembled; however, the more proximal intermediate phenotypes – molecular events such as transcription of genes found in the associated loci – has proven even more tractable.
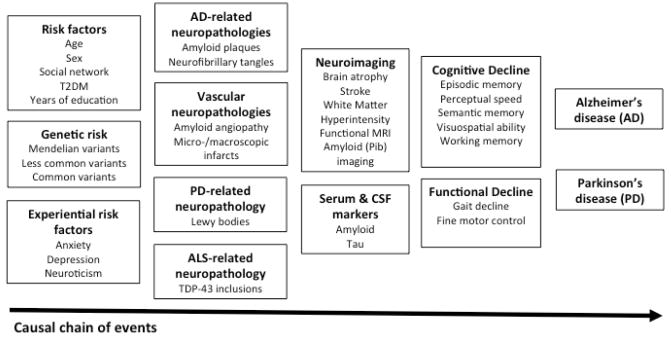
Figure 1. A causal chain linking risk factors to the syndromic phenotypes of AD and PD.
Risk factors are presented in the left of the figure, and their functional consequences progress along the horizontal axis, culminating in the clinical expression of symptoms that meet syndromic definitions for these two neurodegenerative diseases. (Adapted from an original model devised by D. Bennett of Rush University, Chicago IL)
Indeed, there are now many examples in which the effect size of a disease-associated variant on transcription of a nearby gene (referred to as a “cis-expression quantitative trait locus” (cis-eQTL) effect is many orders of magnitude greater than its effect on disease susceptibility.28 Further, its effect on biological functions such as the cytotoxic function of natural killer cells29 can be studied in much smaller sample sizes, particularly when accessible cell types such as those in peripheral blood are pertinent to the biology of the disease or contain a molecular pathway shared with CNS cells that are not readily accessible. As recently reviewed elsewhere30, such data, when captured transcriptome-wide in large numbers of subjects provide an excellent substrate for the systematic examination of the variants associated with a given condition. Large-scale characterizations of RNA expression in the brain have already begun31,32 and will doubtlessly play an important role as we move forward in exploring the cascade of molecular events that lead from a susceptibility allele to a syndromic condition. While more challenging, both broad33 and targeted34 characterizations of the proteome as well as other traits of potential interest such as drug response35 are underway and will further inform our investigations of the functional consequences of genetic variation.
These various insights will be critical to understanding the biology of neurodegenerative diseases because, by their nature of being present since the conception of an individual, disease-associated genetic variation provide a view on the earliest events involved in disease susceptibility. This has proven particularly informative in MS: the large collection of validated susceptibility alleles36,37 clearly identifies it as an inflammatory disease that emerges from altered immunological function and then triggers a neurodegenerative process. Interestingly, while we have a less complete picture of the function of susceptibility alleles involved in AD, ALS, and PD, several of them also appear to have a clear role of the immune process. Overall, genetics is providing a glimpse into the very early elements of the pathophysiologic cascade that eventually leads to symptoms of disease. In addition to a better understanding of these diseases, they offer new targets for treatment and suggest new approaches with which to intervene in the disease process instead of managing the symptoms of these diseases.
The asymptomatic and minimally symptomatic phase of disease
From the treatment of MS, we have empiric evidence from several phase III trials that intervening at the earliest possible time after the appearance of the first symptoms decreases the likelihood of accumulation of disability.38,39 The challenge in this and the other neurodegenerative diseases is identifying subjects in their earliest, or even presymptomatic phase, during which it is hypothesized that treatments will be most efficacious. This is illustrated most clearly in AD, in which treatment of symptomatic subjects has proven to be very challenging and new study designs are emerging to develop approaches to treating subjects with subclinical disease.40,41 The use of key biomarker data such as radiolabelled positron emission tomography agents for amyloid imaging has opened up the design of trials in this vein. Genetic data, when combined with pertinent biomarker data (some of which may generated through genetic insights), may provide an efficient manner to stratify subpopulations of subjects in terms of their risk for a given disease. For example, APOE is sufficiently common and the associated risk sufficiently high that if knowledge of allele status lead to a clinical therapeutic decision, the field would consider recommending population screening. Other methodologies for predicting risk are well known and were developed by leveraging non-genetic risk factors such as those involved in calculating the Framingham Risk Score for cardiovascular disease.42 This methodology is flexible and can readily integrate genetic and non-genetic variables to provide aggregate estimates of risk that are informative. Further, the initial REVEAL study and its successors have demonstrated that we can safely communicate this type of information to asymptomatic individuals.43
Genetic data will eventually emerge in clinical practice in some form. It is unlikely (aside from the highly penetrant Mendelian variants) that genetic data will be sufficient, by itself, to be informative in a clinical setting. However, given the ease, precision and cost of their measurement, genetic variants provide robust, if modest, information that can be integrated with other forms of information, such as cerebrospinal fluid biomarkers and imaging, for integrated risk assessments. Further, they are excellent candidates as a first line of diagnostic tools for paradigms that involve successive steps of profiling, in which only the higher risk individuals from a given stratum are interrogated with the more costly or invasive profiling (lumbar puncture, imaging) of the later steps in the evaluation process. This strategy is currently deployed for in utero profiling for birth defects and provides a roadmap for what could be done in neurodegenerative diseases.
The Epigenome: exploring the role of cell state
Aside from a small proportion of families with Mendelian inheritance and APOE, genetic variation associated with risk of neurodegenerative disease is not deterministic. Other factors, loosely captured by the terms of “environmental” or “experiential” factors, have validated effects on risk of neurodegenerative disease, and, for most, the molecular mechanism linking these non-genetic factors to clinical disease remains unclear. One promising area of investigation in this respect is the study of chromatin conformation because this structure - consisting of the DNA strand and its associated histones and other proteins - has recently been shown to be much more plastic and responsive to the environment than was previously appreciated.44 The study of chromatin and of the transcriptional potential of a given cell is referred to as epigenomics, a rapidly evolving field that is currently driving technological advances for measuring epigenomic marks and the development of novel analytic tools to leverage increasingly complex datasets.
Epigenomics represents the natural progression of the study of the human genome: as we complete our catalogue of genetic variation and their associated human traits, it is clear that we must explore the three-dimensional structure of chromosomes to understand whether the potential impact of an allele is realized in a given cell. It is the local architecture of chromatin that dictates whether a segment of DNA is actively transcribed, is repressed or is in another state, such as a “poised promoter” that has a certain probability of becoming transcribed given the correct stimulus. This architecture is determined in part by a range of epigenomic marks on the DNA strand itself or on the histones and other proteins on which the DNA is strand is bound. We are currently still deciphering the manner by which this information is encoded, and coordinated efforts such as those of the National Institute of Health's Epigenomic Roadmap Initiative45, the ENCODE project46, and other international efforts are enabling the identification of markers that capture chromatin information. They are creating reference profiles that can be used to understand the correlation structure of these markers and their relevance to transcription. These efforts, coupled with new analytic methods to reduce the complexity of the data and identify major “chromatin states”47, have led to major advances in the field that are already beginning to help the interpretation of genetic associations to disease.48 However, major challenges remain in the study of the epigenome: (1) unlike DNA, chromatin is plastic, responding to its environment over the life course of an individual, (2) unlike genomic DNA which has one sequence per person, there are numerous chromatin marks across the DNA and attendant histones and other proteins, each of which requires unique profiling, (3) while there are shared patterns, each cell type (and possibly every cell) of an individual organism has a unique epigenome, and (4) the technology to produce reliable results in large numbers of subjects epigenome-wide does not yet exist.
Today, a first generation of disease-related epigenomic studies are being performed and are beginning to be reported. They have focused on two approaches that are feasible today: (1) the generation of reference chromatin maps pertinent to the study of neurodegenerative diseases and (2) a first generation of epigenome-wide screens leveraging technologies that measure DNA methylation. For example, through the NIH Roadmap and the Broad Institute's Reference Epigenomic Mapping Center, data with which to produce chromatin maps of seven brain regions have been generated and distributed publicly (http://www.genboree.org/epigenomeatlas/index.rhtml). Specifically, chromatin immunoprecipitation of six histone marks that capture different aspects of chromatin conformation has been used to identify DNA segments that are bound to each mark in each of the seven profiled brain regions (Figure 2): angular gyrus, anterior caudate, cingulate gyrus, hippocampus, inferior temporal lobe, midfrontal lobe, and susbstantia nigra. Using next-generation sequencing, the DNA segments associated with a given epigenomic mark are identified; they are aligned to the reference human genome; and DNA segments enriched for each chromatin mark are annotated. The six marks can then be considered together to generate a chromatin map using existing computational approaches.47 An important limitation of this and many other efforts is that they represent profiles of human tissues rather than purified cell populations. Thus, while they are informative and provide a first look at the differences between seven distinct brain regions, these profiles must be seen as an important first step towards more granular profiles of important CNS regions that target specific cell populations and subpopulations. However, such efforts await the miniaturization of these assays, and their deployment in disease association studies requires that they become more robust, high-throughput, and cost-effective.
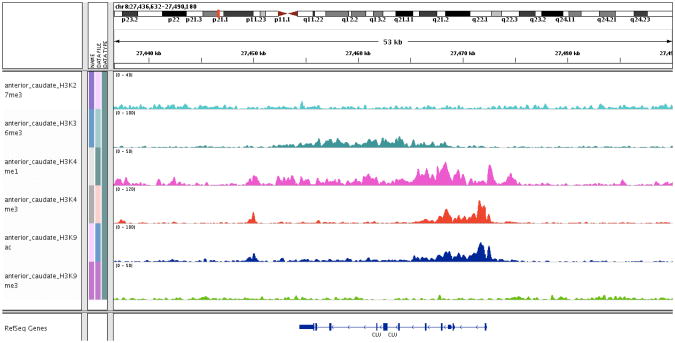
Figure 2.
Representative chromatin data generated from frozen, post-mortem anterior caudate using chromatin immunoprecipitation with high-throughput sequencing for 6 chromatin marks (left column). The x-axis is the physical position along chromosome 8, which is detailed at the top of the figure; the clusterin(CLU) locus was selected for visualization. The distribution of the CLU exons is shown at the bottom of the figure. Each chromatin mark is shown in a horizontal track and has its own distribution over the region selected for viewing; some marks, such as H3K9Ac (acetylation of the 9th Lysine of histone H3) and H3K4Me3 (trimethylation of the 4th Lysine of histone H3), have very similar but not identical distributions. Peaks in each track identify positions where the chromatin mark of interest is enriched. Each mark is associated with different functional features, such as a peak of H3K9Ac at the promoter of actively transcribed genes.
The construction of reference maps that help to interpret results of other studies will also enhance the design of future studies that implement genome-wide scans of chromatin marks such as DNA methylation in large subject collections. Currently, the advent of technologies that can measure a large number of epigenomic marks, such as the methylation of a CG dinucleotide, in a high-throughput manner while minimizing technical variation has enabled the execution of disease-association studies. These are beginning to be reported and will doubtlessly lead to rapid improvement of the platforms for the typing of DNA methylation. Today, platforms such as the Illumina HumanMethylation450 beadset interrogate over 450,000 CG dinucleotides and offer a reasonable first assessment of DNA methylation epigenome-wide. However, the CG being typed represent a small, highly selected, gene-centric subset of the CG that can be potentially methylated throughout the epigenome. We will learn a lot from this and other platforms, which will guide the development of the next generation of platforms. It is already clear that there is an extensive correlation structure among DNA methylation data, suggesting that, as with SNPs, we will not have to interrogate every single CG dinucleotide: we will be able to select tag-CG that capture the state of methylation of a group of CG. It is important to consider that, in many cases, the state of methylation of an individual CG is not likely to have a strong biological effect. Rather, it is the state of methylation of a small chromosomal segment containing a genomic feature that is likely to be meaningful.
Overall, the study of the epigenome in neurodegenerative disease has accelerated significantly as new tools and methods have emerged in recent years, and this rapid evolution is likely to continue and to open up the possibility to develop and execute new study designs. The epigenome is of particular interest not just because it will refine our interpretation of the genetic sequence but also because there are many suggestions that it may be one way in which life experience can shape brain function. It thus has an intriguing potential to be a critical node in the interaction of genetic and environmental risk factors for disease.
Future
We are now at an exciting point in our investigation of the human genome for loci that influence the onset and course of neurodegenerative diseases. As illustrated by studies exploring the genetic architecture of the four more common neurodegenerative diseases (AD, ALS, MS, and PD), the focus on maximizing sample size for subjects meeting a syndromic diagnosis has been very fruitful and validates the association study design for gene discovery in the CNS. This experimental strategy - coupled with the linkage study design that continues to yield new genes associated with rare, highly penetrant genetic variation – has provided us with a rich picture of the allelic spectrum of neurodegenerative diseases that ranges from Mendelian variants with strong effects on disease susceptibility to common polymorphisms with modest effects. These study designs work well when applied to the right population and the right number of subjects; they will continue to yield insights on susceptibility loci for syndromic phenotypes, particularly as new technologies enable a more comprehensive assessment of the allelic spectrum and the characterization of regions that are difficult to genotype. These studies can and should be performed; however, it is clear that the richer areas of investigations are elsewhere.
The application of these study designs to more deeply characterized subjects has begun and will be a major focus of the coming decade in neurodegenerative disease genetics. Given the appropriate number of subjects for a given intermediate trait – a few dozen to a few hundred for transcriptional and other molecular traits but thousands for imaging and clinical traits – association studies will be successful in discovering novel loci that influence neurodegenerative disease. However, their more profound impact will probably come from the use of endophenotypes to map the cascade of molecular events that lead from a known susceptibility allele to a syndromic phenotype. As has been well illustrated, this process has already begun and will be key in assembling susceptibility loci that converge on shared pathophysiologic feature of disease. Beyond this activity, the consideration of endophenotypes and their relation to non-disease associated variants in susceptibility loci will also yield key insights into the biology of susceptibility genes that will enrich our understanding of molecular pathways and targets that are pertinent to disease. Endophenotypes also hold great potential in understanding the shared elements of the genetic architecture for susceptibility to neurodenerative diseases in human populations. For example, it is interesting that the general theme of immune system dysfunction is appearing to a greater or lesser degree in susceptibility to each of the four diseases that we have reviewed here. A critical feature of the study of endophenotypes is that they will require the collaboration of large groups of investigators that coordinate the manner in which they collect their phenotypes to maximize sample size. The consortium model that has worked well for susceptibility gene discovery is one model for what may be a new generation of consortia that assemble the relevant expertise and resources to execute these studies. Industry may play an important role in endophenotype studies given their accumulation of longitudinal data on relevant biomarkers and intermediate traits on large numbers of subjects.
A different set of consortia is emerging in the field of epigenomics: the current collaborative groups have appropriately focused on the development of novel technologies, analytic methodologies, and reference maps of chromatin. However, the maturation of experimental platforms has now reached a stage where the assembly of disease-focused investigators into consortia to enable the large-scale study of the epigenome is propitious. The first generation of DNA methylation studies are now being completed, but it is clear that the integration of data across studies and their analysis in the context of an increasingly complex picture of the three-dimensional structure of chromatin in CNS tissue and cell types will require an integrated approach by a field of committed, interdisciplinary investigators. The potential for new insights into the molecular mechanism of human disease from epigenomics is as vast as the challenges that investigators confront today in deciphering a molecular architecture of chromatin, an architecture that is much more complex and dynamic than that of DNA sequence. Thus, it is a field that will demand our attention in the coming decade.
We are at an inflection point in our study of the human genome as it relates to neurodegenerative disease. The great successes in gene discovery over the past two decades promise continued novel findings that relate to syndromic diagnoses in the short term, but these efforts are now mature and will run their course. Deploying these successful approaches in the realm of intermediate phenotypes and adapting them to the more complex tasks of exploring the epignomic architecture of disease is where the larger insights will emerge in the coming decade. Leveraging the spirit and model of the consortia brought together for the study of syndromic phenotypes, collaborative groups that include the appropriate involvement of industry will generate the novel insights that inform our study of neurodegenerative diseases. It will also inform the development of algorithms that are clinically meaningful and are used to safely inform patients as they make decisions on their management from the presymptomatic phase of disease with their physicians.
Bibliography
- 1.International HapMap, C. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 2.Genomes Project, C et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Human molecular genetics. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 5.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nature reviews Neuroscience. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 6.Ramagopalan SV, et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Annals of neurology. 2011;70:881–886. doi: 10.1002/ana.22678. [DOI] [PubMed] [Google Scholar]
- 7.Vilarino-Guell C, et al. VPS35 mutations in Parkinson disease. American journal of human genetics. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimprich A, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. American journal of human genetics. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Multiple Sclerosis Genetics, C et al. Evidence for polygenic susceptibility to multiple sclerosis--the shape of things to come. Am J Hum Genet. 2010;86:621–625. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 11.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shulman JM, De Jager PL, Feany MB. Parkinson's disease: genetics and pathogenesis. Annual review of pathology. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson T, et al. Variant of TREM2 Associated with the Risk of Alzheimer's Disease. N Engl J Med. 2012 doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerreiro R, et al. TREM2 Variants in Alzheimer's Disease. N Engl J Med. 2012 doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett DA, Wilson RS, Boyle PA, Buchman AS, Schneider JA. Relation of neuropathology to cognition in persons without cognitive impairment. Ann Neurol. 2012;72:599–609. doi: 10.1002/ana.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchman AS, et al. Nigral pathology and parkinsonian signs in elders without Parkinson disease. Ann Neurol. 2012;71:258–266. doi: 10.1002/ana.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009;72:1495–1503. doi: 10.1212/WNL.0b013e3181a2e87d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Jager PL, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiology of aging. 2012;33:1017 e1011–1015. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein JL, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bis JC, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiman EM, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chibnik LB, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias-Sonnenschein LS, Bertram L, Visser PJ. Relationship between genetic risk factors and markers for Alzheimer's disease pathology. Biomarkers in medicine. 2012;6:477–495. doi: 10.2217/bmm.12.56. [DOI] [PubMed] [Google Scholar]
- 25.Brouwers N, et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Molecular psychiatry. 2012;17:223–233. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schjeide BM, et al. The role of clusterin, complement receptor 1, and phosphatidylinositol binding clathrin assembly protein in Alzheimer disease risk and cerebrospinal fluid biomarker levels. Archives of general psychiatry. 2011;68:207–213. doi: 10.1001/archgenpsychiatry.2010.196. [DOI] [PubMed] [Google Scholar]
- 27.Keenan BT, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Hum Mol Genet. 2012;21:2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers AJ, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiology of disease. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Xia Z, et al. A 17q12 allele is associated with altered NK cell subsets and function. J Immunol. 2012;188:3315–3322. doi: 10.4049/jimmunol.1102775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranger BE, De Jager PL. Coordinating GWAS results with gene expression in a systems immunologic paradigm in autoimmunity. Current opinion in immunology. 2012;24:544–551. doi: 10.1016/j.coi.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou F, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster JA, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lourdusamy A, et al. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 2012;21:3719–3726. doi: 10.1093/hmg/dds186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauwe JS, et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc Natl Acad Sci U S A. 2008;105:8050–8054. doi: 10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choy E, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patsopoulos NA, et al. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International Multiple Sclerosis Genetics, C et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kappos L, et al. Effect of early versus delayed interferon beta-1b treatment on disability after a first clinical event suggestive of multiple sclerosis: a 3-year follow-up analysis of the BENEFIT study. Lancet. 2007;370:389–397. doi: 10.1016/S0140-6736(07)61194-5. [DOI] [PubMed] [Google Scholar]
- 39.Comi G, et al. Effect of glatiramer acetate on conversion to clinically definite multiple sclerosis in patients with clinically isolated syndrome (PreCISe study): a randomised, double-blind, placebo-controlled trial. Lancet. 2009;374:1503–1511. doi: 10.1016/S0140-6736(09)61259-9. [DOI] [PubMed] [Google Scholar]
- 40.Morris JC, et al. Developing an international network for Alzheimer research: The Dominantly Inherited Alzheimer Network. Clinical investigation. 2012;2:975–984. doi: 10.4155/cli.12.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reiman EM, et al. Alzheimer's Prevention Initiative: a plan to accelerate the evaluation of presymptomatic treatments. Journal of Alzheimer's disease : JAD. 2011;26(3):321–329. doi: 10.3233/JAD-2011-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson PW, et al. Cumulative effects of high cholesterol levels, high blood pressure, and cigarette smoking on carotid stenosis. The New England journal of medicine. 1997;337:516–522. doi: 10.1056/NEJM199708213370802. [DOI] [PubMed] [Google Scholar]
- 43.Green RC, et al. Disclosure of APOE genotype for risk of Alzheimer's disease. N Engl J Med. 2009;361:245–254. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]