Abstract
Background
The normal RPE sheet in the C57BL/6J mouse is subclassified into two major tiling patterns: a regular generally hexagonal array covering most of the surface and a “soft network” near the ciliary body made of irregularly shaped cells. Physics models predict these two patterns based on contractility and elasticity of the RPE cell, and strength of cellular adhesion between cells.
Hypothesis
We hypothesized and identified major changes in RPE regular hexagonal tiling pattern in rd10 compared to C57BL/6J mice.
Results
In rd10 mice, RPE sheet damage was extensive but occurred later than expected, after most retinal degeneration was complete. RPE sheet changes occur in zones with a bullseye pattern. In the posterior zone, around the optic nerve, RPE cells take on larger irregular and varied shapes to maintain an intact monolayer. In mid periphery, RPE cells have a compressed or convoluted morphology that progress into ingrown layers of RPE under the retina. Cells in the periphery maintain their shape and size until the late stages of the RPE reorganization. The number of neighboring cells varies widely depending on zone and progression. RPE morphology continues to deteriorate after the photoreceptors have degenerated.
Conclusions
The RPE cells are bystanders to photoreceptor degeneration in the rd10 model, and the collateral damage to the RPE results in changes in morphology as early as 30 days old. Quantitative measures of the tiling patterns and histopathology detected here were scripted in a pipeline written in Perl and Cell Profiler (an open source MatLab plugin) and are directly applicable to RPE sheet images from noninvasive fundus autofluorescence (FAF), adaptive optics confocal scanning laser ophthalmoscope (AO-cSLO), and spectral domain optical coherence tomography (SD-OCT) of patients with early stage AMD or RP.
XX.1 Introduction
We are interested in measuring changes in RPE cell morphology that occur in disease states. We hypothesized that changes in the local environment of the RPE cells as a result of retinal degeneration would result in changes in the morphology of RPE cells. To test this hypothesis, we examined the morphology of RPE cells in rd10 mice.
Rd10 mice have a missense mutation in phosphodiesterase 6B. PDE6B is the beta subunit of the phosphodiesterase that hydrolyzes cGMP in the phototransduction cascade. These mice are a model of autosomal recessive retinitis pigmentosa (RP) and have a retinal degeneration that begins at postnatal day 16 and is complete at 60 days old (Farber et al. 1988; Bowes et al. 1990; Chang et al. 2007).
The forces that organize RPE cell-cell contacts include adhesion, tension, and contraction. Tight junctions hold adjacent RPE cells together (Rizzolo 2007). A subcortical actin-myosin cytoskeleton contributes contractile forces that lead to regular polygonal (mostly hexagonal) shapes. One of the tight junction adhesion molecules that contribute to the characteristic RPE cell shape is zona occludens 1 (ZO-1), which also serves as a high quality marker of RPE cell borders. Trafficking and movement of proteins that are part of the adhesion, tension, or contraction mechanism will lead to changes in force balance, which result in remodeling of cell shape and cell packing, and consequently measurable alteration or rearrangement of the patterns and tiling of the RPE sheet.
The physical description of patterning and tiling of endothelial cells in two dimensions is clinically useful in assessing pathology of corneal diseases. Here, we apply similar analyses to RPE sheets that may help predict disease stage, time course, and progression for retinal degeneration and AMD. Patterns and cell shape of the RPE sheet are evident in images from FAF and AO-cSLO, which are obtained noninvasively (de Bruin et al. 2008; Morgan et al. 2008; Geng et al. 2009; Schmitz-Valckenberg et al. 2010). Thus, analyses of these RPE patterns may become diagnostic and prognostic.
Here we begin to build a dynamic physical description of RPE sheet morphology and compare it to disease stage in the rd10 mouse, a genetic model of RP caused by a lesion in the PDE6B gene.
XX.2 Methods
XX.2.1 RPE flatmount technique
Mice were euthanized with CO2 in accordance with Emory IACUC guidelines and The Association for Research in Vision and Ophthalmology guidelines for treatment of animals. Eyes were marked on the superior side with a blue sharpie and then enucleated, fixed for 10 min in 10% neutral buffered formalin, and then washed 3 times with PBS. Extra tissue was removed from the outside of the globe. Flat mounting was done by making 4 radial cuts from the center of the cornea back towards the optic nerve. A drop of PBS was placed on the eye to keep it moist. The flaps were pealed away from the lens and the lens removed. The iris and retina were removed using forceps. Tension from the sclera was relieved by making cuts halfway through each flap at the ciliary body/cornea margin and small cuts through the ciliary body.
XX.2.2 ZO-1 staining
The RPE flatmounts were blocked with HBSS + 0.01% Tween-20 and 1% BSA (antibody buffer) for 30 min. Immunostaining with a 1:100 dilution of rabbit anti-ZO-1 antibody (Invitrogen 61-7300) in antibody buffer was done for approximately 16 hours at room temperature. The flatmounts were washed 5 times with HBSS + 0.01% Tween-20 (wash buffer) for 2 min and then stained for 1 hour with Oregon Green conjugated goat anti-rabbit IgG secondary antibody (Invitrogen O11038) in antibody buffer and then washed 5 times with wash buffer. The flatmounts were mounted with Vectashield hardset (Vector Laboratories H-1400) and allowed to harden overnight.
XX.2.3 Imaging
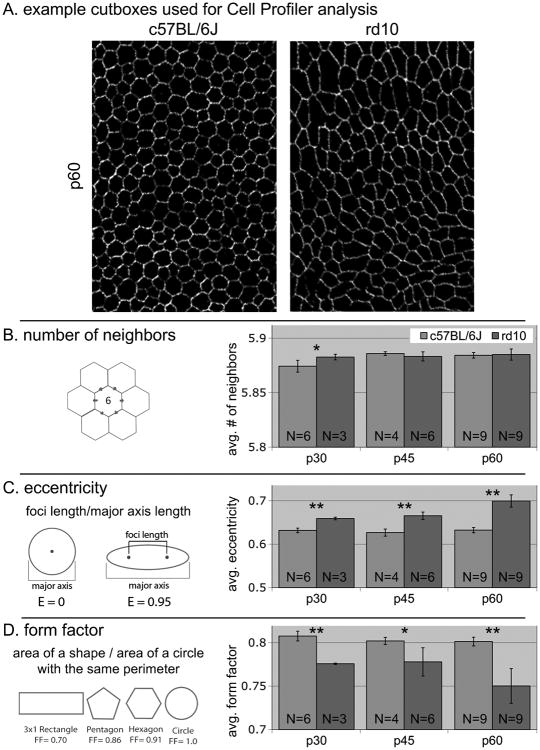
Imaging of the flatmounts was performing using a Nikon C1 confocal imaging system with Argon laser excitation at 488 nm. Confocal images were stitched together using Adobe Photoshop CS2. Cut boxes of equal size (181 × 266 pixels; 225 × 331 μm) were cropped from the merged flatmount image from areas devoid of dissection artifact. As many cut boxes as possible were taken from each image (45-60 cut boxes per image). Morphometric measurements including cell density, cell area, solidity, eccentricity, form factor, and number of neighbors were calculated using Cell Profiler (Lamprecht et al. 2007).
XX.2.4 Statistics
Mean data from each flatmount were used for comparisons between genetic groups and time points. Experimental repeats were averaged and standard deviations of the averages between individual animals are represented as error bars on figure 2. Cochran's t-test for two sample sets with unequal variances was used to determine significance.
Fig.XX.2.
Averaged RPE cell morphometrics. A – examples of cutboxes used for analysis, these were taken from all areas of the flatmount where there were no dissection artifacts. B – number of neighbors is consistent suggesting that there is no cell death at these time points. C and D – measures of cell shape to used to determine if we can detect RPE cell differences early in this degeneration. * P<0.05, ** P<0.01
XX.3 Results
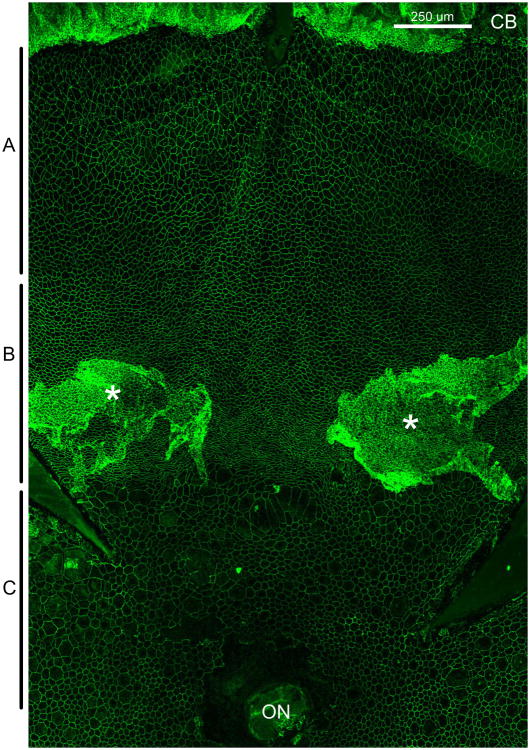
Initially, we made RPE flatmounts from c57BL/6J (wildtype) and rd10 mice from 100, 180 and 330 day old mice and determined that there were massive changes in RPE cell morphology that occurred following retinal degeneration in the rd10 mice (data from 100 day old shown in figure 1). Figure 1 shows a superior portion of a 100 day old rd10 RPE flatmount with ciliary body at the top and the optic nerve at the bottom of the image. This image shows dramatic changes in RPE morphology from wildtype (see figure 2 for an example of wildtype morphology). Wildtype RPE cell morphology is mostly homogenous with an array of hexagonal cells (as shown in the top panel of figure 2) throughout the RPE sheet with a looser network of cells in the periphery at the margins of the ciliary body.
Fig.XX.1.
Superior flap of an RPE flatmount stained with ZO-1 to outline RPE cells from a 100 day old rd10 mouse. A indicates the peripheral region, B indicates the mid-periphery and C indicates the posterior region of the RPE sheet. CB – ciliary body, ON – optic nerve, * - areas of RPE ingrowth away from Bruch's membrane. Size bar is 250 μm.
We decided to look at earlier timepoints to see if we could find changes in RPE morphology that precede the obvious changes that we see in figure 1. The top panel of figure 2 provides examples of the cut boxes that are used in Cell Profiler for analysis. Of the many analyses that can be run with Cell Profiler, we find that number of neighbors, eccentricity, and form factor are the most useful measures for RPE cell morphology.
The number of neighbors is very similar in all comparisons between wildtype and rd10. This is expected as at these timepoints there does not appear to be very much death of RPE cells. This indicates that from 30-60 days old, the changes in cell shape are not due to cells moving to fill in empty spots after cells have died.
Typically RPE cells have a regular hexagonal shape. Stretching or compression of the cells results in a distorted cell shape which can be measured as eccentricity. A typical circle has one focal point in the middle of the circle. As a circle is changed to an ellipse, the focal point splits into 2 foci that spread as the shape becomes more elliptical. The eccentricity measurement in Cell Profiler assigns 2 foci to each cell and provides the measurement between those foci. A larger number indicates more compressed/stretched cell morphology. We find that there is greater eccentricity in RPE cells from rd10 versus wildtype as early as 30 days old.
Form factor is another measure of distortion from a circular shape: it measures the overall shape of the cell in terms of perimeter versus cell area. This is calibrated to a circle of the same perimeter. As such, cell shape distortion from a normal hexagonal pattern reduces their similarity to a circle and generates a lower number in this measure. RPE cells from rd10 mice have a lower form factor than wildtype as early as 30 days old.
XX.4 Discussion
There are major changes to the morphology of RPE cells following retinal degeneration in rd10 mice from 100-330 days old. In figure 1 we showed data from 100 day old rd10 mice to demonstrate some of these disruptions. At later time points the morphological changes spread to the whole retina and the patterning becomes more disorganized than what we see at 100 days old (data not shown).
The changes to cell morphology occur in 3 zones in a bullseye pattern around the optic nerve. In the posterior section of the RPE sheet around the optic nerve there are many cells that are larger than normal with much variability in cell size. In the mid peripheral region, cells have a compressed appearance and more tortuosity. In many samples from the 100 day rd10 mice there is a ring of RPE in growth in this region. Parts of the RPE appear to be detached from Bruch's membrane and growing in multiple layers under the retina. In later time points there are patches of this RPE in growth throughout the RPE sheet (data not shown). In the periphery near the ciliary body at 100 days old, RPE morphology looks quite similar to wildtype controls with a soft network of cells of uniform size. This area becomes disrupted later.
Having identified these qualitative differences in the RPE between wildtype and rd10 mice, we decided to use morphometric analysis software to quantify changes to RPE morphology at timepoints earlier than 100 days old. We analyzed RPE flatmounts from 30, 45 and 60 days old. Thirty days old is a point in the rd10 retinal degeneration when there are approximately 10% of photoreceptors remaining, and qualitatively there does not appear to be much effect on the RPE at this point. We theorized and have demonstrated that minute changes in the RPE morphology that are undetectable qualitatively by visual inspection can be detected quantitatively using computational analysis.
There are distinct changes that occur in the morphology of RPE cells in response to the retinal degeneration that occurs in rd10 mice (figure 2). These changes are detectable as early as 30 days old, well before the retina is completely degenerated. We have demonstrated that the use of morphometric analysis software can detect these early changes. With the improvements that are occuring in in vivo imaging technology, application of these morphometrics measurements to human disease will be possible.
XX.5 Conclusions
As rd10 is a model for retinitis pigmentosa, we predict that similar changes to RPE are occurring in RP patients, and there may be therapeutic complications with changes in RPE function. Understanding RPE cell morphology and how it pertains to normal RPE cell function is also important for early AMD diagnosis and therapy. It may be possible to predict the progression of retinal disease by noninvasive imaging of RPE sheets using AO-cSLO, SD-OCT, or FAF imaging to determine the health of RPE cells.
Acknowledgments
The authors would like to acknowledge the support from the following organizations: NIH R01EY016470, NIH R24EY017045, NIH P30EY06360, Research to Prevent Blindness, and Foundation Fighting Blindness.
References
- Bowes C, Li T, Danciger M, et al. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- Chang B, Hawes NL, Pardue MT, et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 2007;47:624–633. doi: 10.1016/j.visres.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin DM, Burnes DL, Loewenstein J, et al. In vivo three-dimensional imaging of neovascular age-related macular degeneration using optical frequency domain imaging at 1050 nm. Invest Ophthalmol Vis Sci. 2008;49:4545–4552. doi: 10.1167/iovs.07-1553. [DOI] [PubMed] [Google Scholar]
- Farber DB, Park S, Yamashita C. Cyclic GMP-phosphodiesterase of rd retina: biosynthesis and content. Exp Eye Res. 1988;46:363–374. doi: 10.1016/s0014-4835(88)80026-5. [DOI] [PubMed] [Google Scholar]
- Geng Y, Greenberg KP, Wolfe R, et al. In vivo imaging of microscopic structures in the rat retina. Invest Ophthalmol Vis Sci. 2009;50:5872–5879. doi: 10.1167/iovs.09-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Hunter JJ, Masella B, et al. Light-induced retinal changes observed with high-resolution autofluorescence imaging of the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2008;49:3715–3729. doi: 10.1167/iovs.07-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolo LJ. Development and role of tight junctions in the retinal pigment epithelium. Int Rev Cytol. 2007;258:195–234. doi: 10.1016/S0074-7696(07)58004-6. [DOI] [PubMed] [Google Scholar]
- Schmitz-Valckenberg S, Steinberg JS, Fleckenstein M, et al. Combined confocal scanning laser ophthalmoscopy and spectral-domain optical coherence tomography imaging of reticular drusen associated with age-related macular degeneration. Ophthalmology. 2010;117:1169–1176. doi: 10.1016/j.ophtha.2009.10.044. [DOI] [PubMed] [Google Scholar]